Transcriptional dynamics during parasitism in the parasitic weed Cuscuta pentagona reveals increased expression of genes encoding transporters and stimulus response regulators, and a decrease in the expression of genes encoding photosynthetic proteins.
Abstract
Parasitic flowering plants are one of the most destructive agricultural pests and have major impact on crop yields throughout the world. Being dependent on finding a host plant for growth, parasitic plants penetrate their host using specialized organs called haustoria. Haustoria establish vascular connections with the host, which enable the parasite to steal nutrients and water. The underlying molecular and developmental basis of parasitism by plants is largely unknown. In order to investigate the process of parasitism, RNAs from different stages (i.e. seed, seedling, vegetative strand, prehaustoria, haustoria, and flower) were used to de novo assemble and annotate the transcriptome of the obligate plant stem parasite dodder (Cuscuta pentagona). The assembled transcriptome was used to dissect transcriptional dynamics during dodder development and parasitism and identified key gene categories involved in the process of plant parasitism. Host plant infection is accompanied by increased expression of parasite genes underlying transport and transporter categories, response to stress and stimuli, as well as genes encoding enzymes involved in cell wall modifications. By contrast, expression of photosynthetic genes is decreased in the dodder infective stages compared with normal stem. In addition, genes relating to biosynthesis, transport, and response of phytohormones, such as auxin, gibberellins, and strigolactone, were differentially expressed in the dodder infective stages compared with stems and seedlings. This analysis sheds light on the transcriptional changes that accompany plant parasitism and will aid in identifying potential gene targets for use in controlling the infestation of crops by parasitic weeds.
Most land plants are sessile, obtaining water and mineral nutrients from the soil, and are autotrophic, relying on photosynthesis for carbohydrates. However, numerous heterotrophic plants, commonly called parasitic plants, acquire water and nutrients by utilizing specialized structures, called haustoria, to attach to, penetrate, and establish vascular connections with a host plant. Approximately 4,000 plant species, distributed in at least 13 families, are parasitic to some extent and are classified as stem or root parasites, ranging from facultative to hemiparasitic to holoparasitic, based on the degree of host dependence (Yoder and Scholes, 2010). Parasitic weeds severely compromise growth and development of the host and cause annual yield losses of more than $7 billion in sub-Saharan Africa alone (Ejeta, 2007). Despite the economic importance of parasitic weeds, little is known about the basic molecular, genetic, and biochemical mechanisms regulating host plant infection by parasitic weeds.
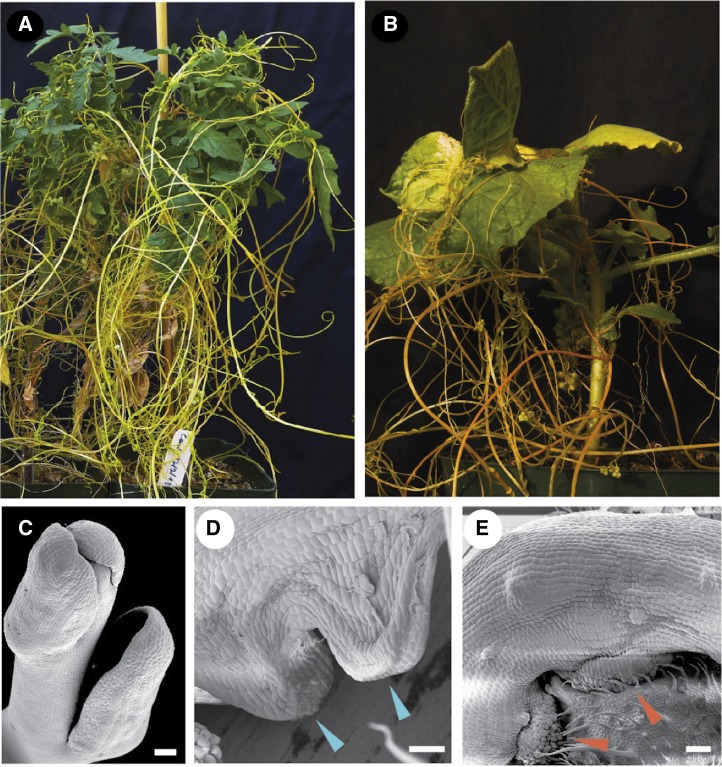
The genus Cuscuta, a member of the family Convolvulaceae, consists of approximately 170 species and includes successful and devastating obligate stem holoparasitic plants distributed throughout the world (Dawson et al., 1994). Cuscuta pentagona, commonly known as dodder, is one of the most widespread and agriculturally destructive species, with a broad host range (Fig. 1, A and B; Malik and Singh, 1979; Dawson et al., 1994; Furuhashi et al., 2011). Dodder seedlings are unable to survive more than a few days after germination without finding a host plant. Mature dodder plants lack roots and have highly reduced leaves at the shoot apex and nodes (Fig. 1C). Coiling of the thread-like dodder stem around the host stem induces the formation of prehaustoria and the subsequent development of haustoria by dedifferentiation and reprogramming of dodder cells (Fig. 1, D and E; Kuijt, 1983; Vaughn, 2003; Yoshida and Shirasu, 2012).
Figure 1.
Parasitism of host plants by dodder. A and B, Tomato (A) and tobacco (B) plants infected with dodder. C, Scanning electron microscopy (SEM) image of the dodder shoot apex with reduced leaves. D, SEM image of prehaustoria (arrowheads). E, SEM image of mature haustoria (arrowheads) penetrating host tissue in the dodder-tomato interaction. Bars = 100 μm.
Upon parasitization, dodder is intimately associated with the host plant and can benefit from the herbicide resistance mechanisms of the host, making it difficult to achieve effective control without detriment to the host (Cook et al., 2009; Nadler-Hassar et al., 2009). Agronomic and traditional breeding practices have limited efficacy in the control of parasitic plant infection (Musselman et al., 2001; Gurney et al., 2006; Mishra, 2009; Sandler, 2010; Westwood et al., 2010). Recently, cross-species transfer of small RNAs has been suggested as a promising agrobiotechnological tool for controlling parasitic plant infections, as haustorial connections have been shown to facilitate RNA and protein movement between the parasite and host (Roney et al., 2007; David-Schwartz et al., 2008; Yoder et al., 2009; Runo et al., 2011). Consistent with the increased expression of SHOOT MERISTEMLESS-like (STM) at the time of haustoria induction, the ability to make haustoria was attenuated in dodder grown on host transgenics expressing dodder STM RNA interference using a phloem-specific promoter (Alakonya et al., 2012). Similar transspecific silencing of parasite genes encoding key enzymes caused reduced viability of the root parasites Orobanche aegyptiaca and Triphysaria versicolor (Aly et al., 2009; Bandaranayake and Yoder, 2013). However, manipulation of the expression levels of none of these genes provides complete control of the parasite, suggesting that suitable target genes for cross-species RNA silencing that will confer complete parasitic weed control remain to be identified. The development of comprehensive genomic and molecular resources for these species may lead to the identification of such targets.
Recently, de novo assembly and analysis of transcriptomes of root parasites, belonging to family Orobanchaceae, provided important insight into the process of parasitism, showing the association of photosynthesis-related genes and cell wall-modifying β-expansin in the process of parasitism (Wickett et al., 2011; Honaas et al., 2013). To understand the molecular mechanisms underlying dynamic morphological and functional changes in haustoria development, however, detailed transcriptional profiling along developmental stages of parasitism is necessary. In this study, we use diverse tissue and developmental stages to assemble and annotate a reference transcriptome for the rootless stem parasite dodder (Supplemental Fig. S1). This transcriptome was then used to investigate the dynamic gene expression patterns across different developmental stages of dodder and identified statistically robust Gene Ontology (GO) categories and underlying genes associated with the prehaustorial and haustorial stages of dodder development.
RESULTS
De Novo Assembly, Refinement, and Quality Assessment of the Dodder Transcriptome
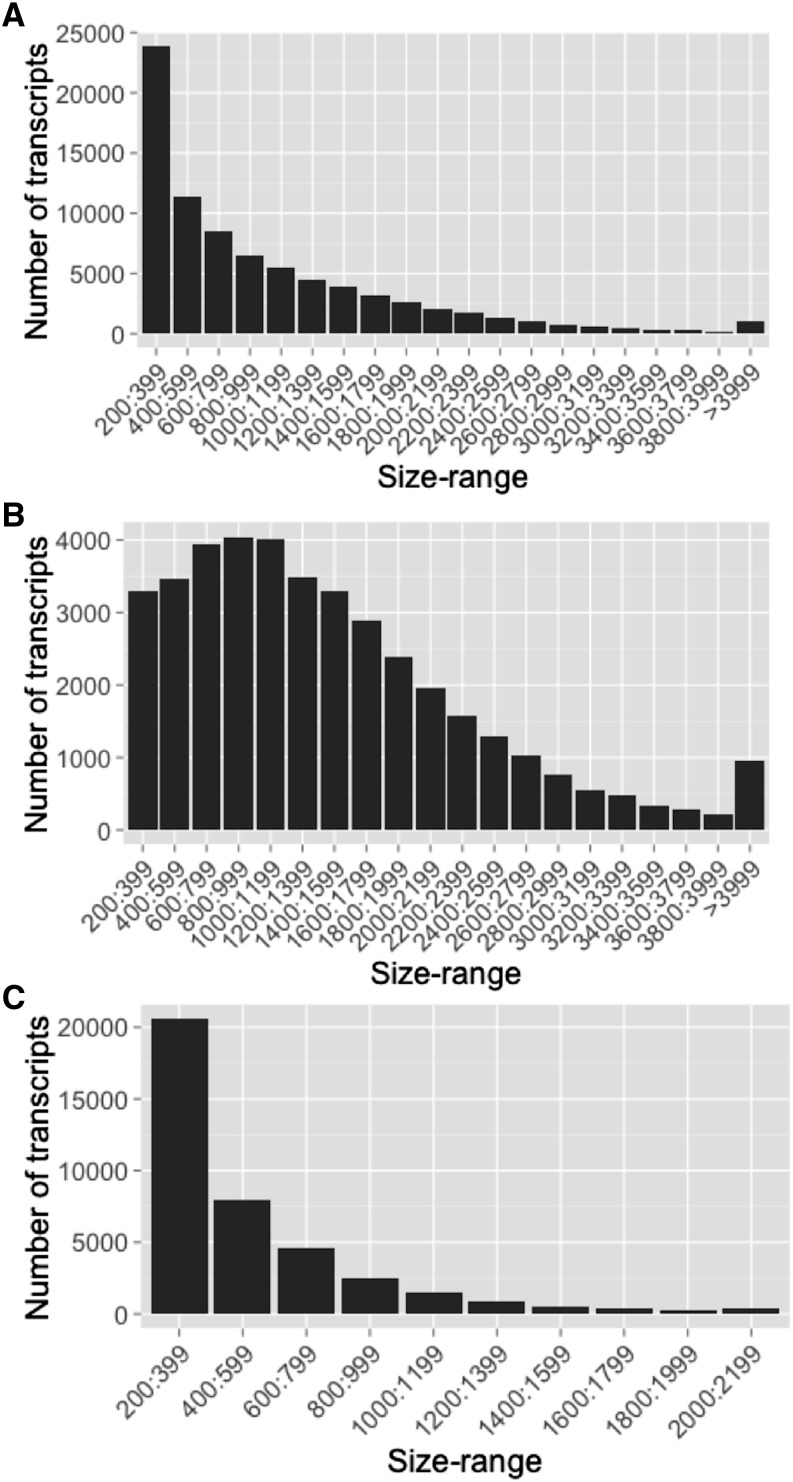
In organisms without a reference genome, massive Illumina short-read sequencing, in conjunction with efficient de novo transcriptome assembly, has become a feasible method for generating a reference transcriptome with sufficient depth coverage to carry out differential gene expression analysis (Wang et al., 2009; Surget-Groba and Montoya-Burgos, 2010; Schliesky et al., 2012). RNA-sequencing (RNA-seq) libraries were prepared from the seed, seedling, coiling stem strand, prehaustorial, haustorial, and flowering stages of dodder growing on both tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum) as hosts (Fig. 1, A and B). A total of 198,992,437 paired-end reads (100 bp) were obtained after sequencing libraries on the Illumina HiSeq2000 platform. Initial preprocessing of reads involved the removal of low-quality sequences, duplicated reads, and reads containing adapter/primer sequences. In order to minimize host tissue contamination, care was taken to remove any host tissue attached to dodder during tissue collection. Furthermore, preprocessed reads aligning to either host transcriptome, tomato or tobacco, were discarded prior to the assembly, as mRNA can translocate from host to dodder (Roney et al., 2007; David-Schwartz et al., 2008). A total of 127 million paired-end reads, obtained after processing and filtering, were used for transcriptome assembly utilizing the Trinity software package (Grabherr et al., 2011). The resulting Dodder_all_transcriptome, with 275,483 contigs (longer than 200 bp), N50 of 1.8 kb (N50 is defined as the largest contig length such that using equal or longer contigs produces half the bases of the transcriptome), and average length of 1.1 kb, was subsequently used for refinement and downstream differential expression analysis (Table I; Supplemental Data Set S1). The assembly generated a high number of transcripts particularly enriched for smaller sized contigs (approximately 28% of contigs are in the size range 200–400 bp; Supplemental Fig. S2).
Table I. Assembly statistics for dodder transcriptomes.
| Parameter | Dodder_all | Dodder_filtered | Dodder_final |
|---|---|---|---|
| No. of transcripts | 275,483 | 91,250 | 79,867 |
| No. of Trinity_components | 102,906 | 43,492 | 43,473 |
| Transcriptom_size (Mb) | 312 | 98 | 80 |
| N50 (bp) | 1,806 | 1,620 | 1,550 |
| Average_length (bp) | 1,132 | 1,069 | 1,005 |
| Median_length (bp) | 778 | 772 | 704 |
| Minimum_length (bp) | 201 | 201 | 201 |
| Maximum_length (bp) | 29,317 | 29,317 | 29,317 |
Likely misassembled transcripts and/or contigs not well supported by the reads used to assemble the transcriptome, based on mapping reads back to the contigs, were filtered out based on abundance estimation, and 91,250 transcripts were retained for further processing (Table I). In order to remove redundant and/or highly similar contigs, the filtered transcripts were then clustered at a sequence similarity threshold value of 95%. The resulting Dodder_final_transcriptome has 79,867 transcripts, with N50 of 1,550 bp and average contig size of 1,005 bp (Fig. 2A; Table I; Supplemental Data Set S2).
Figure 2.
Transcript size distribution showing the high proportion of small-sized transcripts in Dodder_final_transcriptome (A), the normal size distribution for BLASTX-annotated final transcripts (B), and the extremely high enrichment of small-sized transcripts for the unannotated final transcripts (C).
To investigate the efficacy of transcript refinement, the reads used to assemble the transcriptome were mapped to the raw transcriptome, filtered transcriptome, and final transcriptome as well as filtered out transcripts. Although the number of transcripts decreased significantly during the process of refinement of the assembled transcriptome, almost the same percentage of reads (approximately 80%) mapped to the retained transcripts at each step of refinement, indicating retention of most of the real transcripts throughout the process and, thus, good quality of the filtered transcriptome (Table II). Only 20% of reads mapped to discarded transcripts, of which only 7% were uniquely mapped, suggesting that filtered out transcripts are likely assembly artifacts and/or transcripts not well supported by the used Illumina reads.
Table II. Percentage of total and uniquely mapped reads to transcriptomes at different stages of refinement and to the filtered transcripts.
| Transcriptome | Total_mapped_reads | Uniquely_mapped_reads |
|---|---|---|
| % | ||
| Dodder_all | 80.36 | 39.25 |
| Dodder_filtered | 79.1 | 42.11 |
| Dodder_final | 78.57 | 53.40 |
| Filtered_out_transcripts | 19.75 | 6.78 |
To evaluate the quality of the Dodder_final_transcriptome, a full-length transcript analysis was performed by aligning transcripts to the UniProt database. Fifty-seven percent of unique hits in the protein database were represented by nearly full-length transcripts, having more than 70% alignment coverage, and 83% of proteins showed more than 50% alignment coverage (Supplemental Table S1). On the other hand, 73% of assembled transcripts with at least one BLAST hit showed more than 50% sequence identity to a matching database sequence, indicating the contiguity and quality of the assembled transcripts. In addition, the prediction of likely coding sequences from 79,867 filtered transcripts resulted in 49,463 putative open reading frames (ORFs)/coding sequences, of which more than half (25,345) were complete, further highlighting the quality of the filtered transcriptome. Those predicted ORFs were further clustered at the peptide level with a similarity threshold value of 95%, resulting in 44,758 ORFs/coding sequences (Supplemental Data Set S3).
Dodder Transcriptome Annotation
BLAST searches against the National Center for Biotechnology Information (NCBI) nonredundant database and the Arabidopsis (Arabidopsis thaliana) protein database (The Arabidopsis Information Resource 10 [TAIR10]) resulted in the annotation of 40,261 transcripts (50.4% of transcripts), of which 29,144 transcripts were assigned GO terms (Supplemental Data Set S4). BLASTX-annotated transcripts showed normal distribution for transcript size, whereas nonannotated transcripts were extremely enriched for small-sized transcripts, with more than 70% of unannotated transcripts being smaller than 600 bp (Fig. 2, B and C; Supplemental Table S2). Moreover, mapping reads back to annotated and nonannotated transcripts demonstrated that nonannotated transcripts were poorly supported by reads or represent lowly expressed sequences (Supplemental Table S2). Interestingly, no ORFs were predicted for 80% of nonannotated transcripts (31,359 of 39,606 nonannotated transcripts). In order to test if these nonannotated transcripts are real or assembly artifacts, we selected nine such nonannotated transcripts, five with a predicted ORF and four without an ORF, belonging to different size ranges, that appeared among top transcripts in differential expression analysis (described below) and detected the presence of all the selected transcripts by reverse transcription (RT)-PCR (Supplemental Fig. S3). Sequencing of the RT-PCR products confirmed that these sequences represent real dodder transcripts and are not assembly artifacts.
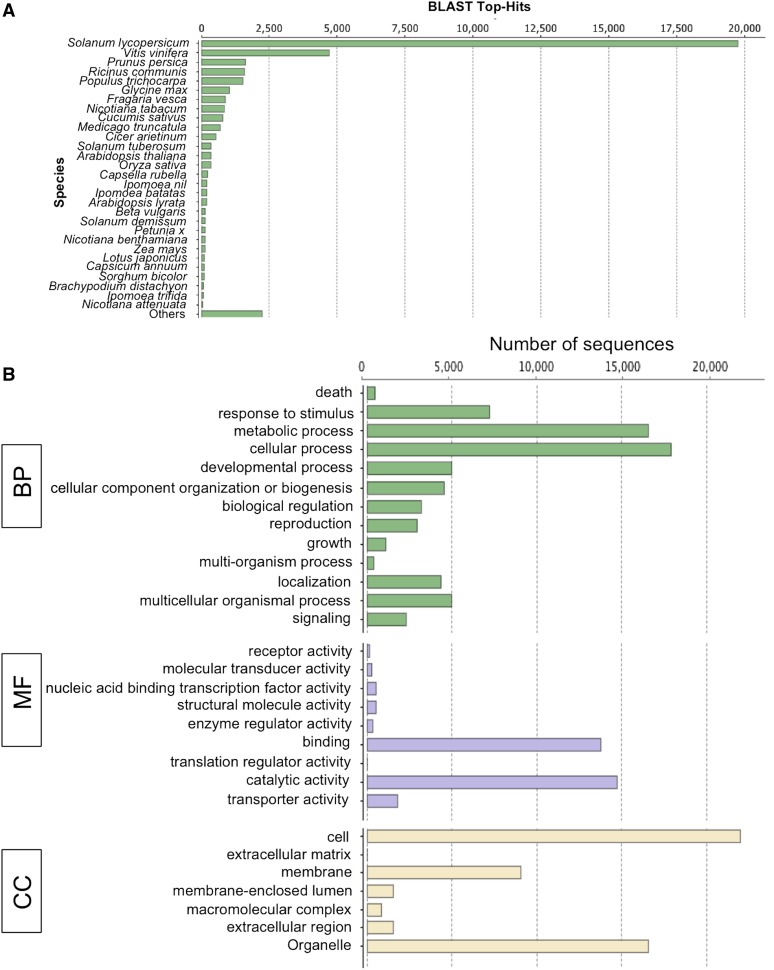
As part of Blast2GO, BLASTX against the nonredundant database provided insight into the taxonomic distribution of the transcripts (Fig. 3A). More than 55% of transcripts had top hits to sequences from members of the family Solanaceae (tomato, potato [Solanum tuberosum], Nicotiana spp., Petunia spp., and Capsicum spp.). This is consistent with the fact that the family Convolvulaceae is monophyletic and sister to Solanaceae (Stefanovic et al., 2002). Only a few top hits were found to be sequences from different species of Ipomoea, which also belongs to Convolvulaceae. This likely reflects a lack of Ipomoea spp. transcript sequences in the databases.
Figure 3.
Blast2GO annotation of Dodder_final_transcriptome. A, Top-hit species distribution of the Dodder_final_transcriptome showing the abundance of top hits to the sequences from members of the family Solanaceae. B, GO category distribution of dodder transcripts among level 1 GO categories: biological process (BP), molecular function (MF), and cellular component (CC). [See online article for color version of this figure.]
Among the 29,144 transcripts with at least one GO term assigned, 21,851 transcripts (27.3% of final transcripts), 21,961 transcripts (27.5% of final transcripts), and 23,287 transcripts (29.2% of final transcripts) were annotated in the biological process (GO:0008150), molecular function (GO:0003674), and cellular component (GO:0005575) categories, respectively. Overall and multilevel GO distribution within these broad GO categories is shown in Figure 3B and Supplemental Figure S4 (Supplemental Data Set S5). We have also classified transcripts among plant GOslims, a simplified version of the full GO ontologies (Supplemental Fig. S5). The overall and slim GO annotations for dodder transcripts are available as Supplemental Data Set S6. In addition, 12,675 transcripts were annotated as enzymes and were confirmed by the Enzyme Code number provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/), with transferases being the most abundant class of enzymes followed by hydrolases and oxidoreductases (Supplemental Fig. S6; Supplemental Data Set S7).
Transcript Expression Patterns across Different Stages of Dodder
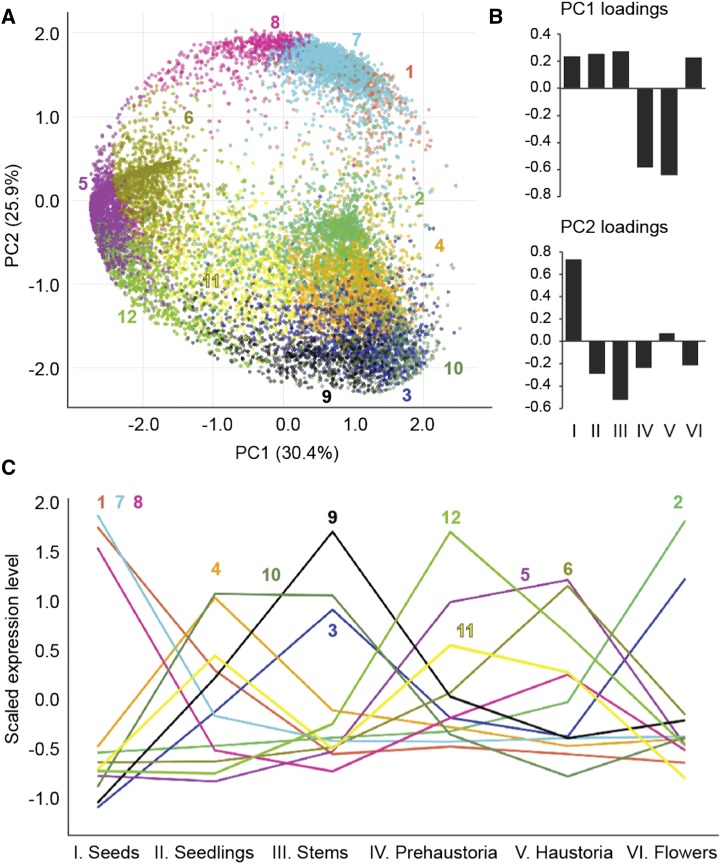
Illumina reads (combined from both tomato and tobacco plants as well as from either host plant separately) from six different developmental stages, seeds, seedlings, stems/strands, prehaustoria, haustoria, and flowers, were aligned to the assembled transcripts. A multidimensional scaling plot showed clustering together of stage-specific biological replicates, despite being from different host plants, suggesting that gene expression at specific stages of dodder development was not greatly influenced by the host (Supplemental Fig. S7). Therefore, in order to investigate gene expression dynamics across different stages of dodder development, a principal component analysis (PCA) with self-organizing maps (SOM) was performed using normalized read counts (Supplemental Data Set S8) obtained from mapping reads combined from both host plants (Wehrens and Buydens, 2007; Chitwood et al., 2013). Principal component 1, representing haustorial- and prehaustorial-stage specific gene expression patterns, explained 30% of variation in our data set (Fig. 4, A and B). This suggests that haustoria formation and subsequent infection of host plants involve a major transcriptional transition in dodder. In addition, principal component 2, representing seed-specific gene expression patterns, explained 26% of variation in our data set, highlighting the uniqueness of this life cycle stage.
Figure 4.
PCA with SOM clustering of gene expression across tissues. A, The expression profile of each transcript is represented in PCA space with SOM node memberships indicated by different colors and numbers. A total of 12 clusters, showing expression patterns specific to one or more stages of the dodder life cycle, were defined. B, Loadings of principal components (PC)1 and PC2 with variance explained. Roman numerals indicate different sample tissues, as defined in C. PC1 represented the haustoria- and prehaustoria-specific gene expression pattern, whereas PC2 represented the seed-specific gene expression pattern. C, Scaled expression patterns of each SOM cluster across tissues. Colors and numbers are as depicted in A.
The SOM clustering analysis dissected multiple different clusters of transcripts showing expression patterns specific to one or more developmental stages (Fig. 4). Clusters 12 and 6 showed expression patterns specific to the prehaustorial and haustorial stages, respectively, whereas cluster 5 showed a pattern common to prehaustorial and haustorial stages. GO enrichment analysis showed that the prehaustoria-specific cluster 12 was enriched for the GO terms Cell Wall and Hydrolase activity, whereas haustoria-specific cluster 6 was enriched for the GO terms Transporter Activity, Secondary Metabolic Process, Response to External Stimuli, Secondary Shoot Formation, and Polar Auxin Transport. Enriched GO terms for haustorial and prehaustorial stage-specific cluster 5 were Response to Biotic Stimulus, Response to Stress, Cell Death, Regulation of Plant-Type Hypersensitive Response, Transport, Receptor Activity, and Kinase Activity (Supplemental Data Set S9).
In addition, clusters specific to seed, seedling, and flower stages were also detected; notable are seedling-specific cluster 4, stem- and flower-specific cluster 3, and flower-specific cluster 2. Seedling-specific cluster 4 showed enrichment of GO terms relating to Photosynthesis, Postembryonic Development, Chlorophyll Biosynthetic Process, Carotenoid Biosynthetic Process, and Leaf Morphogenesis, among others. Consistent with the developmental stages, enriched GO categories for flower-specific clusters 2 and 3 included Flower Development, Specification of Floral Organ Identity, Petal Development, Ovule Development, and Pollination. This gene expression pattern for dodder seedlings and flowers suggests that PCA- and SOM-dissected transcript clustering is a valid method for capturing developmental transitions in gene expression at different stages of parasite development.
Differential Transcript Expression and GO Enrichment Analysis
Three independent differential expression analyses were performed using reads originating from dodder grown on tomato, reads originating from dodder grown on tobacco, and all the reads combined together. The number of differentially expressed transcripts was comparable for reads originating from tomato and tobacco host plants but was substantially higher for the combined data set, due to an increased number of replicates from the two hosts for all tissues except for seeds and seedlings (Table III). Hence, we primarily focused on investigating differentially expressed transcripts from the combined data set (Supplemental Data Set S10).
Table III. Number of differentially expressed transcripts for each tissue pair comparison for three independent analyses.
| Tissue Comparison | Tomato_host | Tobacco_host | Combined |
|---|---|---|---|
| Flowers_vs_haustoria | 10,578 | 10,115 | 17,177 |
| Flowers_vs_prehaustoria | 8,164 | 6,732 | 13,242 |
| Flowers_vs_seedlings | 12,212 | 11,214 | 13,359 |
| Flowers_vs_seeds | 24,598 | 25,688 | 26,978 |
| Flowers_vs_stems | 5,951 | 4,335 | 7,524 |
| Haustoria_vs_prehaustoria | 474 | 3,743 | 4,375 |
| Haustoria_vs_seedlings | 11,027 | 11,779 | 13,604 |
| Haustoria_vs_seeds | 23,300 | 22,779 | 25,456 |
| Haustoria_vs_stems | 9,930 | 9,479 | 16,066 |
| Prehaustoria_vs_seedlings | 7,138 | 7,887 | 9,109 |
| Prehaustoria_vs_seeds | 24,466 | 26,026 | 27,634 |
| Prehaustoria_vs_stems | 5,736 | 4,756 | 10,005 |
| Seedlings_vs_seeds | 24,659 | 24,659 | 21,499 |
| Seedlings_vs_stems | 7,192 | 6,309 | 7,505 |
| Seeds_vs_stems | 25,264 | 23,765 | 24,996 |
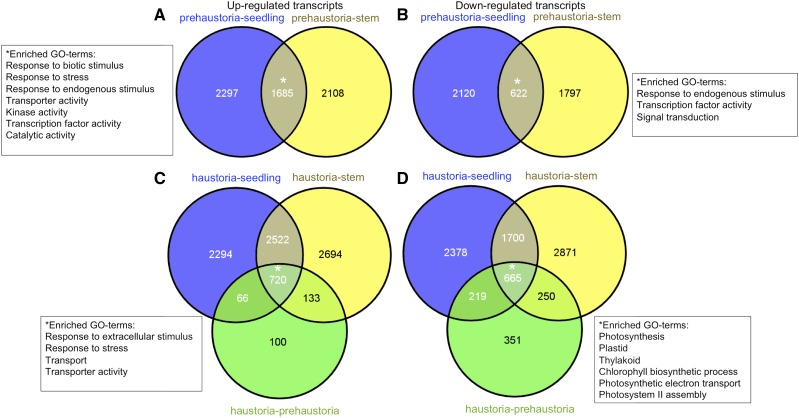
To investigate gene expression changes associated with the process of dodder parasitism, first we compared differentially expressed transcripts in prehaustoria with reference tissues, stems and seedlings. By using fold change ≥ 2 and false discovery rate (FDR) < 0.05 as a cutoff to identify differentially regulated transcripts, 1,685 and 622 transcripts were shared among up-regulated and down-regulated transcripts, respectively, in prehaustoria as compared with stems and seedlings (Fig. 5, A and B; Supplemental Data Set S11). GOslim categories enriched in up-regulated transcripts included Response to Biotic Stimulus, Response to Stress, Response to Endogenous Stimulus, Transporter Activity, Kinase Activity, Transcription Factor Activity, Catalytic Activity, and Cell Wall. Transcripts under those enriched GO terms included genes encoding disease, defense, and drug response signals; cell wall-loosening enzymes and modulators such as pectin lyase, pectin methylesterase, polygalacturonase, cellulase, and members of both the expansin A and B families; kinases, such as Leu-rich repeat kinases, receptor-like kinases, and mitogen-activated protein kinases; the strigolactone biosynthetic MORE AXILLARY GROWTH (MAX1), MAX3, and MAX4 enzymes; multiple transporters such as sugar transporter1, amino acid transporter, ATP-binding cassette-type transporter, ammonium transporter, phosphate transporter, nitrate transporter, and potassium transporter; and transcription factors like class II homeobox transcription factors KNOTTED-like in Arabidopsis (KNAT3) and KNAT4, AGAMOUS-LIKE, BEL1-like homeodomain1 (BLH1), WRKY, and YABBY. Additional overall GO terms for up-regulated genes include Secondary Shoot Formation, with underlying MAX genes; Auxin Polar Transport, with underlying gene AUXIN RESISTANT1 (AUX1) and a gene encoding auxin efflux carrier family protein; Mucilage Extrusion and Mucilage Metabolic Process, with underlying genes encoding subtilase family protein; and Abscisic Acid Transport. Among the enriched GO terms for down-regulated transcripts were Response to Endogenous Stimulus, Auxin Mediated Signaling Pathway, Cell Wall, Transcription Factor Activity, Tropism, and Anatomical Structural Morphogenesis, with underlying transcripts involved in auxin response, such as multiple INDOLE-3-ACETIC ACID INDUCIBLEs (IAAs) and AUXIN RESPONSE FACTORs (ARFs), and SMALL AUXIN UPREGULATED (SAUR)-like genes. A few members of the expansin A family, different from those up-regulated in prehaustoria, were also found in the list of down-regulated genes.
Figure 5.
Differentially expressed transcripts and enriched GO terms in prehaustoria and haustoria. A and B, Number of genes up-regulated (A) and down-regulated (B) at prehaustorial stage with associated enriched GO terms relative to stems and seedlings. C and D, Number of genes up-regulated (C) and down-regulated (D) at haustorial stage with associated enriched GO terms relative to prehaustoria, stems, and seedlings. [See online article for color version of this figure.]
To examine gene expression changes associated with the progression of parasitism from prehaustorial to haustorial stage, we compared expressed transcripts in haustoria with those in prehaustoria, stems, and seedlings (Supplemental Data Set S12). In the differentially expressed genes, the enrichment of GOslim terms Photosynthesis, Plastid, and Thylakoid and multiple Metabolic Process and overall GO terms Chlorophyll Biosynthetic Process, Photosynthetic Electron Transport, and Photosystem II Assembly was noted for 665 common down-regulated transcripts in haustoria, with underlying genes related to photosystem subunit and reaction center proteins (Fig. 5D). A total of 720 common transcripts up-regulated in haustoria as compared with prehaustoria, stems, and seedlings showed enrichment of the GOslim terms Transport and Transporter Activity, Response to Extracellular Stimulus, Response to Stress, and Catalytic Activity and overall GO terms Secondary Shoot Formation, Polar Auxin Transport, Gibberellin Biosynthetic and Metabolic Processes, and Mucilage Metabolic Process and Extrusion. These categories included underlying transcripts encoding multiple transporters and stress- and stimuli-responsive signals, such as pleiotropic drug resistance protein, basic pathogenesis-related proteins, and heat shock proteins, as noted for the prehaustorial stage. In addition, strigolactone biosynthetic enzymes, the GA biosynthetic enzymes GIBBERELLIN-OXIDASE8(GA2OX8), GA20OX1, GA20OX2 and GA20OX5, and transcription factors such as WRKY, YABBY, and GRAS families were also among the up-regulated genes in the haustorial stage (Fig. 5C). Thus, except for the down-regulation of photosynthetic genes, differentially expressed transcripts in haustoria compared with prehaustoria, stems and seedlings shared a significant proportion of enriched GO terms with those differentially expressed in prehaustoria compared with stems and seedlings. This suggests that trends in the expression of genes regulating early parasitism-related processes that begin in the prehaustorial stages are exaggerated as parasitism proceeds to the haustorial stage.
GO categories for common transcripts up-regulated or down-regulated at each developmental stage compared with all other stages were consistent with the developmental stage, suggesting the robustness of our differential gene expression analysis (Supplemental Data Set S13). Notable are the enrichment of GO terms Flower Development, Stamen, Petal, Embryo Sac Development, and Pollination for common up-regulated transcripts in flowers as compared with all other tissues; enrichment of Photosynthesis, Transport, Transporter, and Structural Morphogenesis GO terms for transcripts down-regulated in seeds; and enrichment of GO terms related to Photosynthesis, Photosystem Assembly, Chlorophyll Biosynthetic Process, Gibberellin Biosynthetic Process, Auxin-Mediated Signaling Pathway, and Meristem Growth for common transcripts up-regulated in seedlings as compared with other tissues.
DISCUSSION
This study provides, to our knowledge, the first comprehensive view of plant parasitism at the transcriptome level and identifies genes and gene categories underlying interactions between the obligate shoot parasite dodder and its host. We use high replication across several developmental stages from dodder grown on two hosts to generate a comprehensive transcriptome and two different methods of analysis to identify key genes involved in parasite development. Responses to stimuli and transporter gene categories were found among those highly up-regulated in the infective stages containing prehaustoria and haustoria. Photosynthetic genes were among the down-regulated transcripts at the haustorial stage. Additionally, other major categories associated with the infection process were transcripts coding for cell wall-modifying enzymes and genes involved in phytohormone (such as auxin, GA, and strigolactone) biosynthesis, transport, and responses.
Transcriptome Assembly and Annotation
In order to build a comprehensive transcriptome, we used RNA-seq reads not only from the stages important for investigating dodder parasitism (stem, haustorial, and prehaustorial stages) but also from other significant stages of dodder development (seed, seedling, and flower). Thus, the transcriptome allows a comprehensive look at fundamental principles of dodder development and metabolism in addition to the process of parasitism.
Initially, the assembled transcriptome was highly enriched for small-sized transcripts, similar to other de novo-assembled plant transcriptomes, which could in part be due to the transcript assembly algorithm, where the reads are decomposed into k-mers that may report hypothetical and misassembled transcripts (Zhao et al., 2011; Gruenheit et al., 2012). Filtering out transcripts based on abundance estimation and clustering by sequence identity would have likely removed these assembly artifacts, including transcript variants that were poorly supported by the reads. Nonetheless, only half of the filtered and clustered transcripts were functionally annotated, which is similar to many other de novo-assembled plant transcriptomes, such as tobacco, pepper (Capsicum frutescens), and sweet potato (Ipomoea batatas; Tao et al., 2012; Liu et al., 2013; Nakasugi et al., 2013). RT-PCR followed by sequencing confirmed the existence of the nonannotated transcripts, 80% of which had no predicted protein-coding regions, suggesting that most of these nonannotated transcripts may constitute poorly expressed assembled intergenic noncoding RNAs besides potential dodder-specific transcripts and 3′ or 5′ untranslated regions. In plants, the functions of intergenic noncoding RNAs are poorly characterized, except for the involvement of such RNAs in biotic and abiotic stress responses in Arabidopsis (Liu et al., 2012). A number of those potential noncoding RNAs were represented among transcripts showing differential expression in haustorial and prehaustorial stages compared with seedlings and stems, suggesting their possible involvement in the process of parasitism (Supplemental Data Set S14).
Genes and Gene Categories Associated with Dodder Infection and Development
Based upon PCA and SOM clustering in combination with differential expression analysis, our study identifies transcripts specific to individual stages of dodder development and highlights ones associated with key sequential events in the process of Cuscuta spp. parasitism, including increased responses to biotic and external stimuli, cell wall modifications, increased transporter activities, and reduced photosynthesis.
Recruitment of Genes Relating to Responses to Stimuli and Cell Wall-Modifying Enzymes for Induction of Plant Parasitism
Our study uses the transcriptome to identify the involvement of genes related to responses to stimuli and plant defense in the process of parasitism. Many such genes, such as basic pathogenesis-related proteins, heat shock proteins, and drug resistance proteins among others, showed increased expression in prehaustorial and haustorial stages compared with stems and seedlings. Genes associated with stress responses were also found to be up-regulated in roots of a root parasite after contact with a host plant (Torres et al., 2005; Honaas et al., 2013). Thus, it can be hypothesized that parasitic plants recruit defense-related genes for host recognition. Considering that a host may demonstrate active defense responses to dodder attack (Runyon et al., 2010), it also appears that the parasite is under biotic stress and, hence, may recruit its own defense mechanism to overcome the counterattack.
Our study also showed the increased expression of many genes encoding cell wall-modifying enzymes, such as pectin lyase, pectin methylesterase, and cellulase, besides expansins in dodder infective stages. This, in combination with classical histological and immunocytochemical studies indicating the prominence of cell wall-loosening complexes at the dodder-host plant interface, suggests the importance of cell wall modifications in the induction and penetrance of host tissue by haustoria as well as the rapid expansion of haustorial tissues (Nagar et al., 1984; Vaughn, 2002, 2003). Expansin cell wall modulators were identified as core regulators of parasitism in the root parasites Striga spp. and T. versicolor (O’Malley and Lynn, 2000; Vaughn, 2002; Honaas et al., 2013). However, expansin regulation of plant parasitism might be a more complex phenomenon in the stem parasite Cuscuta spp., as different genes encoding expansins were represented among both the up-regulated and down-regulated genes.
Increased Transporter Activity and Reduced Photosynthesis with Progression of Plant Parasitism
After the establishment of vascular connections with the host plant, dodder becomes a very strong sink, acquiring nutrients and solutes from the host plant and significantly reducing the biomass of the host plant (Shen et al., 2005). Our investigation showed increased expression of genes associated with the transport of not only sugars but also of other nutrients, such as amino acids, phosphate, nitrate, and ammonia, in the haustorial stage of dodder (Supplemental Data Set S15), indicating that dodder acquires many nutrients in addition to water and assimilated carbon from its host. The symplastic versus apoplastic mode of nutrient acquisition from the host plant by dodder is debated in the literature (Haupt et al., 2001; Birschwilks et al., 2006; Shen et al., 2006). Our gene expression analysis highlights the fact that the transport mechanism includes apoplastic and transmembrane transport processes in addition to symplastic transport.
The process of dodder becoming a strong sink after infecting host plants is associated with reduced chlorophyll biosynthesis and photosynthesis, suggesting that after successful parasitism, dodder acquires its nutrients from the host plant mostly through haustorial transport, reducing photosynthesis to a minimal level, if any. Lack of true leaves in all Cuscuta spp. further supports this observation (Fig. 1C). However, some studies showed that stems of different Cuscuta spp., including dodder, photosynthesize to varying degrees, consistent with other lineages of parasitic plants (Choudhury and Sahu, 1999; Revill et al., 2005; Wickett et al., 2011). Our differential gene expression analysis also showed high expression of photosynthetic genes in dodder seedlings. Despite the retention of a functional photosynthetic apparatus in Cuscuta reflexa, 99% of carbon used by Cuscuta spp. comes from the host plant, suggesting little or no photosynthesis in dodder stems after host plant infestation (Jeschke et al., 1994; Hibberd et al., 1998).
Phytohormone Activity in Dodder Development and Parasitism
This study also demonstrates a significant involvement of the phytohormones auxin, GA, and strigolactone in dodder parasitism. Polar auxin accumulation has been postulated as a universal feature of early plant organogenesis, and localized auxin accumulation has been shown to be required for early haustorium development in T. versicolor (Benková et al., 2003; Tomilov et al., 2005). An increased activity of genes underlying polar auxin transport in dodder infective stages likely establishes auxin maxima that induce haustoria formation. Besides auxin, genes encoding GA biosynthetic and metabolic enzymes and the strigolactone biosynthetic enzymes MAX1, MAX3, and MAX4 were among the up-regulated genes in dodder prehaustoria and haustoria. However, the functional significance of GA and strigolactone for haustoria induction and parasitism needs to be investigated. GA, in combination with auxin, has been shown to regulate the growth of excised Cuscuta spp. shoot tips in vitro (Maheshwari et al., 1980). In addition, interaction between strigolactone and auxin is also known to regulate bud outgrowth and shoot branching in Arabidopsis (Brewer et al., 2009; Hayward et al., 2009). The expression of genes relating to secondary shoot formation in the development of prehaustorium further supports the involvement of auxin and strigolactone in dodder parasitism. It also provides additional support for the recruitment of shoot developmental processes in the evolution of dodder haustoria, as opposed to these structures simply being modified adventitious roots, as suggested previously (Kuijt and Toth, 1976; Lee, 2007). Therefore, the significance of auxin and its interaction with GA and/or strigolactone to regulate haustoria induction and, thus, dodder parasitism will be an interesting question to investigate in the future.
CONCLUSION
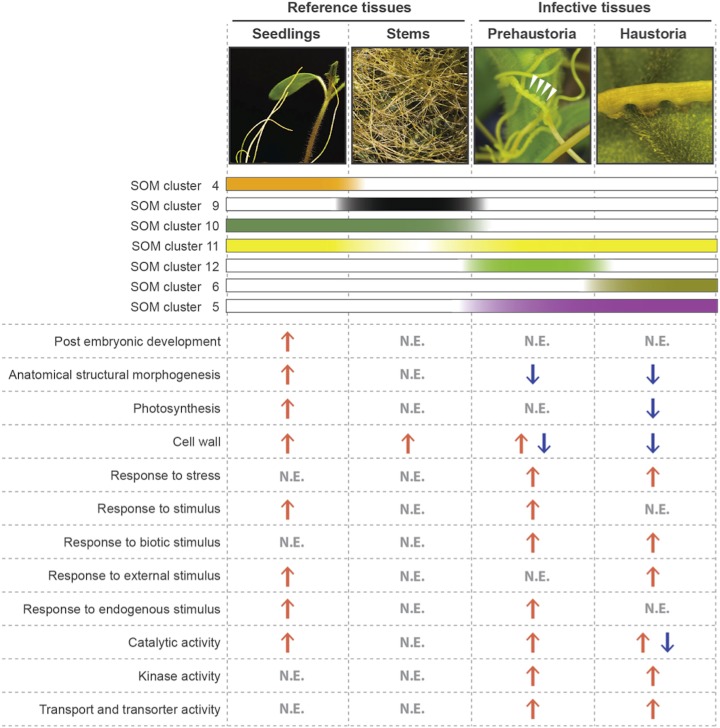
This study generates a robust and well-annotated transcriptome for the stem parasitic weed dodder and then outlines the sequential molecular events underlying dodder infestation of host plants (Fig. 6). Utilizing both SOM transcript clusters and differential gene expression analysis, the unique prehaustorial and haustorial stages of dodder showed statistically robust enrichment of a few key gene categories likely to be involved in the process of parasitism. Based on the significant changes in gene expression at the various stages of parasite development, the proposed progression of events leading to parasite establishment involve a mechanical stimulus generated from initial contact with the host plant that induces haustoria formation and penetration into the host stem. This is facilitated by the recruitment of defense- and stress-responsive genes for host recognition and the action of cell wall-modifying enzymes. Once vascular connections between dodder and the host plant are established, the host-parasite relationship relies on the transfer of nutrients and solutes from host to parasite, employing multiple transporters and reducing photosynthesis. This comprehensive transcriptomic study not only provides molecular insight into dodder development and parasitism but also an opportunity to identify potential genes that could be utilized for controlling destructive plant parasitic weeds. In the future, a gene coexpression network analysis might help us identify core regulators of the highlighted molecular processes involved in dodder parasitism, such as transcription factors regulating stress responses, cell wall-modifying enzymes, and transporters. These can then be exploited for targeted knockdown in dodder using a host-mediated RNA interference strategy to attain more complete control of parasitic weeds and to develop parasitic weed-resistant crop varieties.
Figure 6.
Progression of dodder parasitism from seedling to infective stages involves increased responses to stress and stimuli, increased transport and transporter activity, and decreased photosynthesis. The schematic diagrams show enriched GO terms for dodder parasitic, prehaustorial, and haustorial stages and reference tissues, seedlings and stems, based upon combining transcript clustering and differential gene expression analysis. Red upward arrows indicate increased expression of genes underlying the GO term, and blue downward arrows indicate decreased expression of genes underlying the GO term at the relevant dodder stage. N.E. stands for not enriched. Also shown are the relevant SOM clusters for each of these dodder stages. The colors of the SOM cluster bars indicate the stage specificity of the corresponding clusters.
MATERIALS AND METHODS
Dodder Germination, Host Plant Infection, and Tissue Collection
Dodder (Cuscuta pentagona) seeds were germinated after treating with concentrated sulfuric acid and bleach followed by cleaning several times with water (Alakonya et al., 2012). Treated seeds were germinated on moist filter paper in magenta boxes, and germinated seedlings were used to achieve synchronized infection of the host plant alfalfa (Medicago sativa). Later, strands/twines from infected alfalfa plants were coiled around tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum) host plants before being left to establish for 4 weeks (Alakonya et al., 2012). For scanning electron microscopy analysis, tissue of dodder infecting tomato stem and leaf was fixed as described (Bharathan et al., 2002), and electronic images were obtained with a Hitachi S-3500 N device (Hitachi Science Systems).
Treated seeds, 3-d-old seedlings, noninfective stems of dodder (grown more than 30 cm away from host plants), infective stems with prehaustoria, host-penetrated stems with haustoria, and flowers of dodder grown on host plants were used to make RNA-seq libraries. Prehaustorial stems were collected by unwinding the dodder stems from the host plants as soon as prehaustoria were visible and before host plant penetration. Haustorial dodder stems were collected after host stem penetration by making vertical slices of the host stem in order to capture most of the penetrated dodder tissues. While dissecting haustorial strands, visible traces of host tissues attached to haustoria were removed. Dodder flowers with traces of dodder stem tissues attached to them were also collected from infected host plants. Plants were grown in the greenhouse at 22°C with 70% relative humidity and a daylength of 16 h.
RNA-seq Library Preparation and Sequencing
RNA-seq libraries were prepared from collected tissues (i.e. seeds, seedlings, coiling stem strands, prehaustoria, haustoria, and flowers) using a custom high-throughput method for the Illumina RNA-seq library (Kumar et al., 2012). Libraries were prepared from four replicates of seeds and seedlings as well as from four replicates from both host plants for each stem, prehaustoria, haustoria, and flower. These RNA-seq libraries were sequenced at the University of California Berkeley Genomics Sequencing Laboratory on a single lane of the HiSEquation 2000 platform (Illumina), and reads were generated in 100-bp paired-end format.
Preprocessing of Illumina Reads
Preprocessing of reads involved Q20-quality trimming (removal of low-quality reads with average Phred quality score < 20 and trimming of low-quality bases from the 3′ ends of the reads) and removal of adapter/primer contamination and duplicated reads using custom Perl scripts. The preprocessed reads were sorted into individual samples based on barcodes using fastx_barcode_splitter, and barcodes were trimmed using fastx_trimmer from Fastx_toolkit.
In order to remove host tissue contamination, paired barcode-trimmed combined reads were mapped to the tomato (ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.3_release/ITAG2.3_cdna.fasta) and tobacco (http://www.plantgdb.org/download/Download/Sequence/ESTcontig/Nicotiana_tabacum/current_version/Nicotiana_tabacum.mRNA.PUT.fasta.bz2) transcriptomes separately using BWA (Burrows-Wheeler Aligner) software in paired-read mapping mode with parameters -k 1 -l 25 -n 0.04 -e 15 -i 10 (Li and Durbin, 2009). Reads that did not map to host transcriptomes were extracted using SAMtools and used for subsequent transcriptome assembly (Li et al., 2009).
De Novo Transcriptome Assembly
The Trinity software package (version r2013-02-25) was used for efficient and robust de novo assembly of a transcriptome from the RNA-seq data (Grabherr et al., 2011). Transcriptome assembly was performed at the Lonestar Linux Cluster at the Texas Advance Computing Center (University of Texas) using 24 large-memory computing nodes and 1 TB JELLYFISH memory. The command line used for assembly was Trinity.pl–seqType fq–JM 1000G–left reads-1.fq–right reads-2.fq–min_contig_length 200–CPU 24–bflyHeapSpaceMax 7G. Subsequently, assembly statistics and downstream analysis were performed in the iPlant atmosphere and Discovery computing atmosphere (Goff et al., 2011).
Refinement and Assessment of Assembly
In order to filter out transcriptional artifacts, misassembled transcripts, and poorly supported transcripts, original reads were mapped to assembled transcripts using Bowtie2 with parameters -a–rdg 6,5–rfg 6,5–score-min L,-0.6,-0.4, followed by SAMtools usage to generate a bam alignment file (Li et al., 2009; Langmead and Salzberg, 2012). Subsequently, express software was used to calculate abundance estimation for each transcript in terms of FPKM (fragments per kilobase per transcript per million mapped reads), and transcripts with one or more FPKM were retained for downstream analysis (Roberts and Pachter, 2013).
In order to handle the issue of highly similar/redundant contigs/ORFs, CD-HIT suite, a clustering program based on similarity threshold, was used (Huang et al., 2010). For clustering transcripts, CD-HIT-EST with a sequence similarity threshold of 95% and word size of eight was used, whereas to cluster ORFs, CD-HIT with a sequence similarity threshold of 95% and word size of five was used.
Reads were mapped to transcripts at each stage of transcriptome refinement as well as to annotated and unannotated transcripts using BWA using the same parameters as described above (Li and Durbin, 2009). A custom Perl script was used to extract uniquely mapped reads, followed by the use of SAMtools flagstat to determine the number of total mapped reads and uniquely mapped reads (Li et al., 2009).
For full-length transcript analysis, transcripts were compared against the UniProt database (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz) using parameters -evalue 1e-20 -num_threads 6 -max_target_seqs 1 -outfmt 6, and then Perl script from the Trinity suite was used to examine the percentage of the UniProt target being aligned to the best-matching assembled transcript (Altschul et al., 1997; Haas et al., 2013).
Functional Annotation of the Transcriptome
The contigs from the final transcriptome were compared with the NCBI nonredundant database using BLASTX with an e-value threshold of 1e-3 (Altschul et al., 1997). The BLASTX output, generated in xml format, was used for Blas2GO analysis to annotate the contigs with GO terms describing biological processes, molecular functions, and cellular components (Götz et al., 2008). The e-value filter for GO annotation was 1e-6. After the Blast2GO mapping process, proper GO terms and Enzyme Code numbers from the KEGG pathway were generated. ANNEX and GOslim, which are integrated in the Blast2GO software, were used to enrich the annotation. Sequence description was also generated from Blast2GO, with an arbitrary nomenclature based upon degrees of similarities identified in the nonredundant database according to e-value and identity to BLASTed genes.
Sequence comparison against TAIR10 was also performed using BLASTX at the e-value cutoff of 0.001.
Differential Expression Analysis
RNA-seq by expectation maximization (RSEM), which allows for an assessment of transcript abundances based on the mapping of RNA-seq reads to the assembled transcriptome, was used for transcript abundance estimation of the de novo-assembled transcripts (Li and Dewey, 2011). Reads from individual libraries of each tissue were mapped to final transcripts using default RSEM parameters using script run_RSEM_align_n_estimate.pl, followed by joining RSEM-estimated abundance values for each sample using merge_RSEM_frag_counts_single_table.pl. Finally, differential expression analysis was carried out using run_DE_analysis.pl, which involves the Bioconductor package EdgeR in the R statistical environment (Robinson and Oshlack, 2010; R Development Core Team, 2011). Removal of transcripts with very low estimated counts (40 for the combined data set and 24 for the individual host plant data set) followed by normalization of RSEM-estimated abundance value preceded pairwise comparison for each tissue pair using EdgeR. All these Perl scripts are bundled with the Trinity software suit (Haas et al., 2013).
PCA with SOM Clustering
Normalized RSEM-estimated counts (Supplemental Data Set S8) were used for a gene expression clustering method (Chitwood et al., 2013). After selecting genes in the upper 95% quartile of coefficient of variation for expression across samples, scaled expression values within tissues were used to cluster these genes for multilevel four-by-three hexagonal SOM (Wehrens and Buydens, 2007). One hundred training interactions were used during clustering, over which the α learning rate decreased from 0.008 to 0.007. The final assignment of genes to winning units forms the basis of the gene clusters. The outcome of SOM clustering was visualized in PCA space where principal component values were calculated based on gene expression across samples (R stats package, prcomp function).
GO Enrichment Analysis
GO enrichment analysis of the differentially expressed genes as well as genes detected in specific clusters generated by PCA and SOM was performed using the GOSeq Bioconductor package and GO terms and GOslim terms generated by Blast2GO (Young et al., 2010).
RT-PCR Analysis
RNA from dodder stems was extracted using the RNeasy Plant kit (Qiagen) according to the manufacturer’s protocol. Resulting RNA was treated with DNase I (Qiagen). Complementary DNA from total RNA was prepared using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s protocol. Two microliters of the complementary DNA was used for amplification by PCR using the primer pairs listed in Supplemental Table S3.
The quality filtered, barcode-sorted, and trimmed short read data set, which was used for transcriptome assembly, was deposited to the NCBI Short Read Archive under accession numbers SRR965929, SRR965963, SRR966236, SRR966405, SRR966412, SRR966513, SRR966542, SRR966549, SRR966619 to SRR966622, SRR967154, SRR967164, SRR967181 to SRR967190, SRR967275 to SRR967289, and SRR967291. The all assembled transcripts (Dodder_all_transcriptome) have been deposited at GenBank/EMBL/DNA Data Bank of Japan under accession number GAON00000000. The version described in this article is the first version, GAON01000000.
Supplemental Data
The following materials are available in the online version of this article and deposited in the DRYAD repository: http://datadryad.org/resource/doi:10.5061/dryad.7fr20.
Supplemental Figure S1. Flow chart showing steps in dodder transcriptome assembly and annotation as well as downstream transcript clustering and differential expression analysis.
Supplemental Figure S2. Transcript size distribution for Dodder_all_transcriptome.
Supplemental Figure S3. Expression of nonannotated transcripts as detected by RT-PCR in dodder stems.
Supplemental Figure S4. Pie charts for multilevel GO distribution of annotated transcripts in three categories: biological processes, cellular components, and molecular function.
Supplemental Figure S5. Histogram representation of GOslim classification in three categories: biological processes, molecular function, and cellular components.
Supplemental Figure S6. Distribution of transcripts annotated as enzymes among different enzyme classes.
Supplemental Figure S7. Multidimensional scaling plot of all replicates of each dodder tissue used for transcriptome assembly and, subsequently, transcript clustering and differential expression analysis.
Supplemental Table S1. Distribution of the percentage length coverage for the top-matching UniProt database entries.
Supplemental Table S2. Size statistics and percentage of read mapping to annotated and unannotated transcripts.
Supplemental Table S3. Primers used in RT-PCR analysis.
Supplemental Data Set S1. Sequences of all transcripts of Dodder_all_transcriptome can be downloaded as a FASTA file at http://de.iplantcollaborative.org/dl/e00e9ea8-6aad-439b-a1fa-49dd3939693d.
Supplemental Data Set S2. Sequences of all transcripts of Dodder_final_transcriptome, obtained after filtering and clustering of transcripts, can be downloaded as a FASTA file at http://de.iplantcollaborative.org/dl/a85e3682-4315-43e8-9f9d-597908616b4a.
Supplemental Data Set S3. Sequences of all predicted ORFs from Dodder_all_transcriptome can be downloaded as a FASTA file at http://de.iplantcollaborative.org/dl/f3eb4e4a-05b8-4db4-94f2-87fb77c1622a.
Supplemental Data Set S4. Combined annotation of dodder transcripts obtained from BLASTX against nonredundant and TAIR10 databases.
Supplemental Data Set S5. GO chart data for number of annotated transcripts in each GO category along with GO level, score, and parent GO terms for each category under biological process, molecular function, and cellular component.
Supplemental Data Set S6. GO identifiers for all annotated dodder transcripts.
Supplemental Data Set S7. Enzyme code distribution from KEGG for all annotated dodder transcripts.
Supplemental Data Set S8. Normalized RSEM-estimated counts for all replicates of each dodder tissue used for transcript clustering and differential expression analysis.
Supplemental Data Set S9. Enriched GO categories, both overall GO and GOslim, for all clusters generated from PCA with SOM.
Supplemental Data Set S10. Differentially expressed transcripts (log fold change ≥ 1, FDR < 0.05) for each pairwise comparison for all dodder tissues.
Supplemental Data Set S11. Shared up-regulated and down-regulated transcripts at prehaustorial stage compared with seedlings and stems and associated enriched GO categories.
Supplemental Data Set S12. Shared up-regulated and down-regulated transcripts at haustorial stage compared with prehaustoria, seedlings, and stems and associated enriched GO categories.
Supplemental Data Set S13. GO terms enriched for shared transcripts up-regulated or down-regulated for each dodder developmental stage compared with all other stages.
Supplemental Data Set S14. List of nonannotated transcripts showing differential expression (log fold change ≥ 1, FDR < 0.05) in prehaustorial and haustorial stages compared with seedlings and stems.
Supplemental Data Set S15. Transcripts underlying the GO terms transport (GO:0006810) and transporter (GO:0005215) represented among up-regulated genes in prehaustorial stage compared with seedlings and stems and in haustorial stage compared with prehaustoria, seedlings, and stems.
Supplementary Material
Acknowledgments
We thank Dr. Jessica Budke for helpful comments on the manuscript.
Glossary
- GO
Gene Ontology
- ORF
open reading frame
- TAIR10
The Arabidopsis Information Resource 10
- RT
reverse transcription
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PCA
principal component analysis
- SOM
self-organizing maps
- FDR
false discovery rate
- RSEM
RNA-seq by expectation maximization
Footnotes
This work was supported by the U.S. Department of Agriculture/National Institute of Food and Agriculture (grant no. 2013–02345 to N.R.S.), the National Science Foundation (grant no. IOS–0820854 to N.R.S.), the Japan Society for the Promotion of Science (to Y.I.), and the United States-Israel Binational Agricultural Research and Development Fund (postdoctoral fellowship no. FI–463–12 to M.F.).
Present address: Institute of Plant Sciences, Volcani Center, Bet Dagan, Israel.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alakonya A, Kumar R, Koenig D, Kimura S, Townsley B, Runo S, Garces HM, Kang J, Yanez A, David-Schwartz R, et al (2012) Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell 24: 3153–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly R, Cholakh H, Joel DM, Leibman D, Steinitz B, Zelcer A, Naglis A, Yarden O, Gal-On A. (2009) Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol J 7: 487–498 [DOI] [PubMed] [Google Scholar]
- Bandaranayake PC, Yoder JI. (2013) Trans-specific gene silencing of acetyl-CoA carboxylase in a root-parasitic plant. Mol Plant Microbe Interact 26: 575–584 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. (2002) Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Birschwilks M, Haupt S, Hofius D, Neumann S. (2006) Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J Exp Bot 57: 911–921 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Maloof JN, Sinha NR. (2013) Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant Physiol 162: 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury NK, Sahu D. (1999) Photosynthesis in Cuscuta reflexa: a total plant parasite. Photosynthetica 36: 1–9 [Google Scholar]
- Cook JC, Charudattan R, Zimmerman TW, Rosskopf EN, Stall WM, MacDonald GE. (2009) Effects of Alternaria destruens, glyphosate, and ammonium sulfate individually and integrated for control of dodder (Cuscuta pentagona). Weed Technol 23: 550–555 [Google Scholar]
- David-Schwartz R, Runo S, Townsley B, Machuka J, Sinha N. (2008) Long-distance transport of mRNA via parenchyma cells and phloem across the host-parasite junction in Cuscuta. New Phytol 179: 1133–1141 [DOI] [PubMed] [Google Scholar]
- Dawson JH, Musselman LJ, Wolswinkel P, Doerr I. (1994) Biology and control of Cuscuta. Rev Weed Sci 6: 265–317 [Google Scholar]
- Ejeta G. (2007) Breeding for Striga resistance in sorghum: exploitation of an intricate host-parasite biology. Crop Sci 47: S216–S227 [Google Scholar]
- Furuhashi T, Furuhashi K, Weckwerth W. (2011) The parasitic mechanism of the holostemparasitic plant Cuscuta. J Plant Interact 6: 207–219 [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, et al. (2011) The iPlant Collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheit N, Deusch O, Esser C, Becker M, Voelckel C, Lockhart P. (2012) Cutoffs and k-mers: implications from a transcriptome study in allopolyploid plants. BMC Genomics 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney AL, Slate J, Press MC, Scholes JD. (2006) A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol 169: 199–208 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Oparka KJ, Sauer N, Neumann S. (2001) Macromolecular trafficking between Nicotiana tabacum and the holoparasite Cuscuta reflexa. J Exp Bot 52: 173–177 [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Bungard RA, Press MC, Jeschke WD, Scholes JD, Quick WP. (1998) Localization of photosynthetic metabolism in the parasitic angiosperm Cuscuta reflexa. Planta 205: 506–513 [Google Scholar]
- Honaas LA, Wafula EK, Yang Z, Der JP, Wickett NJ, Altman NS, Taylor CG, Yoder JI, Timko MP, Westwood JH, et al (2013) Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol 13: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26: 680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke WD, Raeth N, Baeumel P, Czygan FC, Proksch P. (1994) Modelling of the flows and partitioning of carbon and nitrogen in the holoparasite Cuscuta reflexa Roxb. and its host Lupinus albus L. II. Flows between host and parasite within the parasitized host. J Exp Bot 45: 801–812 [Google Scholar]
- Kuijt J (1983) Tissue Compatibility and the Haustoria of Parasitic Angiosperms. Baylor University, Waco, TX [Google Scholar]
- Kuijt J, Toth R. (1976) Ultrastructure of angiosperm haustoria: a review. Ann Bot (Lond) 40: 1121–1130 [Google Scholar]
- Kumar R, Ichihashi Y, Kimura S, Chitwood DH, Headland LR, Peng J, Maloof JN, Sinha NR. (2012) A high-throughput method for Illumina RNA-Seq library preparation. Front Plant Sci 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB. (2007) Structure and development of the upper haustorium in the parasitic flowering plant Cuscuta japonica (Convolvulaceae). Am J Bot 94: 737–745 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li W, Wu Y, Chen C, Lei J. (2013) De novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PLoS ONE 8: e48156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari R, Shailini C, Veluthambi K, Mahadevan S. (1980) Interaction of gibberellic acid and indole-3-acetic acid in the growth of excised Cuscuta shoot tips in vitro. Plant Physiol 65: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik CP, Singh MB. (1979) Physiological and biochemical aspects of parasitism in Cuscuta: a review. Annu Rev Plant Sci 1: 67–112 [Google Scholar]
- Mishra J. (2009) Biology and management of Cuscuta species. Indian J Weed Sci 41: 1–11 [Google Scholar]
- Musselman LJ, Yoder JI, Westwood JH. (2001) Parasitic plants major problem to food crops. Science 293: 1434. [DOI] [PubMed] [Google Scholar]
- Nadler-Hassar T, Shaner DL, Nissen S, Westra P, Rubin B. (2009) Are herbicide-resistant crops the answer to controlling Cuscuta? Pest Manag Sci 65: 811–816 [DOI] [PubMed] [Google Scholar]
- Nagar R, Singh M, Sanwal GG. (1984) Cell wall degrading enzymes in Cuscuta reflexa and its hosts. J Exp Bot 35: 1104–1112 [Google Scholar]
- Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM. (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS ONE 8: e59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Lynn DG. (2000) Expansin message regulation in parasitic angiosperms: marking time in development. Plant Cell 12: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Revill MJ, Stanley S, Hibberd JM. (2005) Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta. J Exp Bot 56: 2477–2486 [DOI] [PubMed] [Google Scholar]
- Roberts A, Pachter L. (2013) Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10: 71–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roney JK, Khatibi PA, Westwood JH. (2007) Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol 143: 1037–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runo S, Alakonya A, Machuka J, Sinha N. (2011) RNA interference as a resistance mechanism against crop parasites in Africa: a ‘Trojan horse’ approach. Pest Manag Sci 67: 129–136 [DOI] [PubMed] [Google Scholar]
- Runyon JB, Mescher MC, Felton GW, De Moraes CM. (2010) Parasitism by Cuscuta pentagona sequentially induces JA and SA defence pathways in tomato. Plant Cell Environ 33: 290–303 [DOI] [PubMed] [Google Scholar]
- Sandler HA. (2010) Managing Cuscuta gronovii (swamp dodder) in cranberry requires an integrated approach. Sustainability 2: 660–683 [Google Scholar]
- Schliesky S, Gowik U, Weber AP, Bräutigam A. (2012) RNA-Seq assembly: are we there yet? Front Plant Sci 3: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Ye W, Hong L, Cao H, Wang Z. (2005) Influence of the obligate parasite Cuscuta campestris on growth and biomass allocation of its host Mikania micrantha. J Exp Bot 56: 1277–1284 [DOI] [PubMed] [Google Scholar]
- Shen H, Ye W, Hong L, Huang H, Wang Z, Deng X, Yang Q, Xu Z. (2006) Progress in parasitic plant biology: host selection and nutrient transfer. Plant Biol (Stuttg) 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Krueger L, Olmstead RG. (2002) Monophyly of the Convolvulaceae and circumscription of their major lineages based on DNA sequences of multiple chloroplast loci. Am J Bot 89: 1510–1522 [DOI] [PubMed] [Google Scholar]
- Surget-Groba Y, Montoya-Burgos JI. (2010) Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res 20: 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Gu YH, Wang HY, Zheng W, Li X, Zhao CW, Zhang YZ. (2012) Digital gene expression analysis based on integrated de novo transcriptome assembly of sweet potato [Ipomoea batatas (L.) Lam]. PLoS ONE 7: e36234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilov AA, Tomilova NB, Abdallah I, Yoder JI. (2005) Localized hormone fluxes and early haustorium development in the hemiparasitic plant Triphysaria versicolor. Plant Physiol 138: 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MJ, Tomilov AA, Tomilova N, Reagan RL, Yoder JI. (2005) Pscroph, a parasitic plant EST database enriched for parasite associated transcripts. BMC Plant Biol 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC. (2002) Attachment of the parasitic weed dodder to the host. Protoplasma 219: 227–237 [DOI] [PubMed] [Google Scholar]
- Vaughn KC. (2003) Dodder hyphae invade the host: a structural and immunocytochemical characterization. Protoplasma 220: 189–200 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Buydens LMC. (2007) Self- and super-organizing maps in R: the Kohonen package. J Stat Softw 21: 1–19 [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. (2010) The evolution of parasitism in plants. Trends Plant Sci 15: 227–235 [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Honaas LA, Wafula EK, Das M, Huang K, Wu B, Landherr L, Timko MP, Yoder J, Westwood JH, et al (2011) Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol 21: 2098–2104 [DOI] [PubMed] [Google Scholar]
- Yoder JI, Gunathilake P, Wu B, Tomilova N, Tomilov AA. (2009) Engineering host resistance against parasitic weeds with RNA interference. Pest Manag Sci 65: 460–466 [DOI] [PubMed] [Google Scholar]
- Yoder JI, Scholes JD. (2010) Host plant resistance to parasitic weeds: recent progress and bottlenecks. Curr Opin Plant Biol 13: 478–484 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K. (2012) Plants that attack plants: molecular elucidation of plant parasitism. Curr Opin Plant Biol 15: 708–713 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. (2010) Gene Ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. (2011) Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics (Suppl 14) 12: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.