A key regulator of the actin cytoskeleton is shown to be associated with endosomal membranes.
Abstract
The actin cytoskeleton is a major regulator of cell morphogenesis and responses to biotic and abiotic stimuli. The organization and activities of the cytoskeleton are choreographed by hundreds of accessory proteins. Many actin-binding proteins are thought to be stimulus-response regulators that bind to signaling phospholipids and change their activity upon lipid binding. Whether these proteins associate with and/or are regulated by signaling lipids in plant cells remains poorly understood. Heterodimeric capping protein (CP) is a conserved and ubiquitous regulator of actin dynamics. It binds to the barbed end of filaments with high affinity and modulates filament assembly and disassembly reactions in vitro. Direct interaction of CP with phospholipids, including phosphatidic acid, results in uncapping of filament ends in vitro. Live-cell imaging and reverse-genetic analyses of cp mutants in Arabidopsis (Arabidopsis thaliana) recently provided compelling support for a model in which CP activity is negatively regulated by phosphatidic acid in vivo. Here, we used complementary biochemical, subcellular fractionation, and immunofluorescence microscopy approaches to elucidate CP-membrane association. We found that CP is moderately abundant in Arabidopsis tissues and present in a microsomal membrane fraction. Sucrose density gradient separation and immunoblotting with known compartment markers were used to demonstrate that CP is enriched on membrane-bound organelles such as the endoplasmic reticulum and Golgi. This association could facilitate cross talk between the actin cytoskeleton and a wide spectrum of essential cellular functions such as organelle motility and signal transduction.
The cellular levels of membrane-associated lipids undergo dynamic changes in response to developmental and environmental stimuli. Different species of phospholipids target specific proteins and this often affects the activity and/or subcellular localization of these lipid-binding proteins. One such membrane lipid, phosphatidic acid (PA), serves as a second messenger and regulates multiple developmental processes in plants, including seedling development, root hair growth and pattern formation, pollen tube growth, leaf senescence, and fruit ripening. PA levels also change during various stress responses, including high salinity and dehydration, pathogen attack, and cold tolerance (Testerink and Munnik, 2005, 2011; Wang, 2005; Li et al., 2009). In mammalian cells, PA is critical for vesicle trafficking events, such as vesicle budding from the Golgi apparatus, vesicle transport, exocytosis, endocytosis, and vesicle fusion (Liscovitch et al., 2000; Freyberg et al., 2003; Jenkins and Frohman, 2005).
The actin cytoskeleton and a plethora of actin-binding proteins (ABPs) are well-known targets and transducers of lipid signaling (Drøbak et al., 2004; Saarikangas et al., 2010; Pleskot et al., 2013). For example, several ABPs have the ability to bind phosphoinositide lipids, such as phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2]. The severing or actin filament depolymerizing proteins such as villin, cofilin, and profilin are inhibited when bound to PtdIns(4,5)P2. One ABP appears to be strongly regulated by another phospholipid; human gelsolin binds to lysophosphatidic acid and its filament severing and barbed-end capping activities are inhibited by this biologically active lipid (Meerschaert et al., 1998). Gelsolin is not, however, regulated by PA (Meerschaert et al., 1998), nor are profilin (Lassing and Lindberg, 1985), α-actinin (Fraley et al., 2003), or chicken CapZ (Schafer et al., 1996).
The heterodimeric capping protein (CP) from Arabidopsis (Arabidopsis thaliana) also binds to and its activity is inhibited by phospholipids, including both PtdIns(4,5)P2 and PA (Huang et al., 2003, 2006). PA and phospholipase D activity have been implicated in the actin-dependent tip growth of root hairs and pollen tubes (Ohashi et al., 2003; Potocký et al., 2003; Samaj et al., 2004; Monteiro et al., 2005a; Pleskot et al., 2010). Exogenous application of PA causes an elevation of actin filament levels in suspension cells, pollen, and Arabidopsis epidermal cells (Lee et al., 2003; Potocký et al., 2003; Huang et al., 2006; Li et al., 2012; Pleskot et al., 2013). Capping protein (CP) binds to the barbed end of actin filaments with high (nanomolar) affinity, dissociates quite slowly, and prevents the addition of actin subunits at this end (Huang et al., 2003, 2006; Kim et al., 2007). In the presence of phospholipids, AtCP is not able to bind to the barbed end of actin filaments (Huang et al., 2003, 2006). Furthermore, capped filament ends are uncapped by the addition of PA, allowing actin assembly from a pool of profilin-actin (Huang et al., 2006). Collectively, these data lead to a simple model whereby CP, working in concert with profilin-actin, serves to maintain tight regulation of actin assembly at filament barbed ends (Huang et al., 2006; Blanchoin et al., 2010; Henty-Ridilla et al., 2013; Pleskot et al., 2013). Furthermore, the availability of CP for filament ends can be modulated by fluxes in signaling lipids. Genetic evidence for this model was recently obtained by analyzing the dynamic behavior of actin filament ends in living Arabidopsis epidermal cells after treatment with exogenous PA (Li et al., 2012). Specifically, changes in the architecture of cortical actin arrays and dynamics of individual actin filaments that are induced by PA treatment were found to be attenuated in cp mutant cells (Li et al., 2012; Pleskot et al., 2013).
Structural characterization of chicken CapZ demonstrates that the α- and β-subunits of the heterodimer form a compact structure resembling a mushroom with pseudo-two-fold rotational symmetry (Yamashita et al., 2003). Actin- and phospholipid-binding sites are conserved on the C-terminal regions, sometimes referred to as tentacles, which comprise amphipathic α-helices (Cooper and Sept, 2008; Pleskot et al., 2012). Coarse-grained molecular dynamics (CG-MD) simulations recently revealed the mechanism of chicken and AtCP association with membranes (Pleskot et al., 2012). AtCP interacts specifically with lipid bilayers through interactions between PA and the amphipathic helix of the α-subunit tentacle. Extensive polar contacts between lipid headgroups and basic residues on CP (including K278, which is unique to plant CP), as well as partial embedding of nonpolar groups into the lipid bilayer, are observed (Pleskot et al., 2012). Moreover, a glutathione S-transferase fusion protein containing the C-terminal 38 amino acids from capping protein α subunit (CPA) is sufficient to bind PA-containing liposomes in vitro (Pleskot et al., 2012). Collectively, these findings lead us to predict that AtCP will behave like a membrane-associated protein in plant cells.
Additional evidence from animal and microbial cells supports the association of CP with biological membranes. In Acanthamoeba castellanii, CP is localized primarily to the hyaline ectoplasm in a region of the cytoplasm just under the plasma membrane that contains a high concentration of actin filaments (Cooper et al., 1984). Localization of CP with regions rich in actin filaments and with membranes was supported by subcellular fractionation experiments, in which CP was associated with a crude membrane fraction that included plasma membrane (Cooper et al., 1984). Further evidence demonstrates that CP localizes to cortical actin patches at sites of new cell wall growth in budding yeast (Saccharomyces cerevisiae), including the site of bud emergence. By contrast, CP did not colocalize with actin cables in S. cerevisiae (Amatruda and Cooper, 1992). CP may localize to these sites by direct interactions with membrane lipids, through binding the ends of actin filaments, or by association with another protein different from actin. In support of this hypothesis, GFP-CP fusion proteins demonstrate that sites of actin assembling in living cells contain both CP and the actin-related protein2/3 (Arp2/3) complex, and CP is located in two types of structures: (1) motile regions of the cell periphery, which reflect movement of the edge of the lamella during extension and ruffling; and (2) dynamic spots within the lamella (Schafer et al., 1998). CP has been colocalized to the F-actin patches in fission yeast (Schizosaccharomyces pombe; Kovar et al., 2005), which promotes Arp2/3-dependent nucleation and branching and limits the extent of filament elongation (Akin and Mullins, 2008). These findings lend additional support for a model whereby CP cooperates with the Arp2/3 complex to regulate actin dynamics (Nakano and Mabuchi, 2006). Activities and localization of other plant ABPs are linked to membranes. Membrane association has been linked to the assembly status of the ARP2/3 complex, an actin filament nucleator, in Arabidopsis (Kotchoni et al., 2009). SPIKE1 (SPK1), a Rho of plants (Rop)-guanine nucleotide exchange factor (GEF) and peripheral membrane protein, maintains the homeostasis of the early secretory pathway and signal integration during morphogenesis through specialized domains in the endoplasmic reticulum (ER; Zhang et al., 2010). Furthermore, Nck-associated protein1 (NAP1), a component of the suppressor of cAMP receptor/WASP-family verprolin homology protein (SCAR/WAVE) complex, strongly associates with membranes and is particularly enriched in ER membranes (Zhang et al., 2013a). Finally, a superfamily of plant ABPs, called NETWORKED proteins, was recently discovered; these link the actin cytoskeleton to various cellular membranes (Deeks et al., 2012; Hawkins et al., 2014; Wang et al., 2014).
In this work, we demonstrate that CP is a membrane-associated protein in Arabidopsis. To our knowledge, this is the first direct evidence for CP-membrane association in plants. This interaction likely targets CP to cellular compartments such as the ER and Golgi. This unique location may allow CP to remodel the actin cytoskeleton in the vicinity of endomembrane compartments and/or to respond rapidly to fluxes in signaling lipids.
RESULTS
Heterodimeric CP Is a Moderately Abundant Cellular Protein in Arabidopsis
CP is an α/β heterodimer encoded by two single genes in Arabidopsis (Huang et al., 2003). The α-subunit gene, CPA (NM_111425 and At3g05520), encodes a polypeptide that is 308 amino acids long and 35,038 D. The β-subunit gene, CPB (NM_105837 and At1g71790), encodes a polypeptide of 256 amino acids and 28,876 D. CP is an obligate heterodimer; for example, genetic ablation of either subunit in budding yeast (S. cerevisiae) leads to loss of the other subunit (Amatruda et al., 1992; Sizonenko et al., 1996; Kim et al., 2004). Similarly, knockdown mutants for either CP subunit in Arabidopsis result in a reduction in transcript levels for the other subunit (Li et al., 2012). We tested whether this was also the case for CP protein levels in Arabidopsis and sought to determine the abundance of CP in wild-type cells.
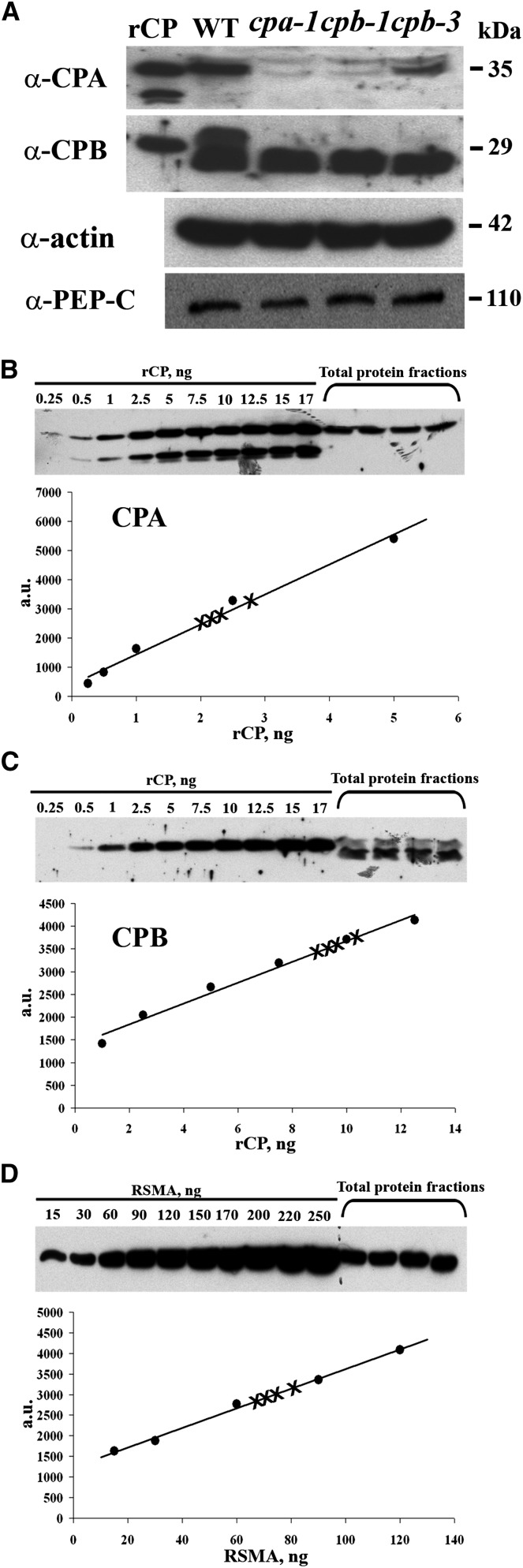
To assess the abundance of endogenous CP in Arabidopsis cellular extracts, we performed quantitative immunoblotting as previously established for actin, adenylate cyclase-associated protein1 (CAP1), profilin, and actin depolymerizing factor (ADF; Chaudhry et al., 2007). Here, recombinant AtCP was purified to generate standard curves for loading and detection limit determination, and we established the specificity of two affinity-purified antisera raised against CPA and CPB (Huang et al., 2003). As shown in Figure 1A, purified recombinant CPA and CPB subunits, as well as native polypeptides from cellular extracts with similar Mrs, were recognized by the respective affinity-purified polyclonal antibodies. Additional evidence for antibody specificity was obtained by probing cellular extracts from cpa and cpb homozygous knockdown plants (Li et al., 2012). Three independent transfer DNA (T-DNA) insertion lines were found to have markedly reduced CPA and CPB polypeptide levels (Fig. 1A). A second, lower Mr polypeptide is present and equally abundant in extracts of the wild type and all three cp mutants probed with anti-CPB; this likely represents a nonspecific cross reaction with another Arabidopsis protein. Interestingly, the insertion in CPA (cpa-1) led to reductions in both proteins of the heterodimer, and the cpb-1 and cpb-3 knockdown mutants had reduced levels of CPA and CPB (Fig. 1A). This is similar to the behavior of CPA and CPB transcripts in the respective mutant lines reported previously (Li et al., 2012). Thus, these two affinity-purified antibodies were appropriate for quantitative immunoblotting and subcellular localization studies.
Figure 1.
CP is a moderately abundant protein in total cellular extracts. A, On protein immunoblots, CPA and CPB antisera recognized polypeptides from purified rCP (10-ng load), as well as polypeptides of appropriate size from total cellular extracts of wild-type Arabidopsis Col-0 seedlings (50-µg load). Total cellular extracts (50 µg each) prepared from three homozygous T-DNA insertion mutant lines (cpa-1, cpb-1, and cpb-3; Li et al., 2012) had reduced levels of CPA and CPB polypeptides. Probing of identical membranes with anti-actin antibodies revealed that total actin levels were not reduced in the cp homozygous mutant lines compared with the wild type. The same blot was reprobed with anti-phosphoenolpyruvate carboxylase antibody, which recognized a band of 110 kD, and verifies the equal loading of samples. B and C, CPA and CPB protein levels were estimated by semiquantitative immunoblotting using total protein extracts from wild-type Arabidopsis seedlings and a standard curve comprising varying amounts of rCP. The Coomassie-stained gel images at the top show results from the blotting of a standard curve and four biological replicates of seedling extracts (75 µg each) with anti-CPA (B) and anti-CPB (C). The intensity of each band from the standard curve as a function of protein amount is plotted. The data were fit with a linear function and the correlation coefficients for these representative examples were 0.99 and 0.98 for CPA and CPB, respectively. In this example, CPA represents 0.0015% ± 0.0001% of total cellular protein, whereas CPB represents 0.0013% ± 0.0002%. D, Total actin levels were estimated by immunoblotting of seedling extracts prepared from the wild type and a standard curve comprising different amounts of purified rabbit skeletal muscle actin. The gel image shows the result from blotting of a standard curve and four biological replicates of seedling extracts (25 µg each). The intensity of each band from the standard curve as a function of protein amount is plotted. The data were fit with a linear function and the correlation coefficient for this representative example was 0.99. In this experiment, actin represents 0.37% ± 0.02% of total cellular protein. a.u., Arbitrary units; RSMA, rabbit skeletal muscle actin; WT, wild type.
The relative abundance of CP, with respect to actin and two other ABPs, in total cellular extracts from Arabidopsis seedlings was estimated by quantitative immunoblotting. At least four biological replicates of cell extracts were loaded on the same gel as a standard curve comprising known amounts of the recombinant protein. After transfer to nitrocellulose, probing with specific antisera, and detection with enhanced-chemiluminescence reagents, the intensity of the reactive bands was determined by densitometry and plotted as a function of protein amount. Representative examples for CPA and CPB, shown in Figure 1, B and C, respectively, demonstrate that the standard curves were linear over at least an order of magnitude in protein concentration and that each serum can detect nanogram quantities of recombinant capping protein (rCP). As a benchmark for the method, and to establish the relationship with CP, total cellular actin levels were also quantified (Fig. 1D). The CP determinations were repeated twice and the mean values (± sd) from eight biological replicates are reported in Table I. Actin was the most abundant protein of those examined, comprising 0.37% of total cellular protein from seedling extracts. This corresponds well with the concentration in rosette leaves (0.36%) determined previously (Chaudhry et al., 2007). The monomer-binding proteins, CAP1 and ADF, were also quite abundant with levels of approximately 0.05% of total cellular protein. Both subunits of CP were markedly less abundant than actin or the monomer-binding proteins, with estimated cellular levels of 0.0015% and 0.0013% of total protein for CPA and CPB, respectively.
Table I. CP is a moderately abundant cellular protein.
Values represent the mean percentage (± sd) of a particular ABP with respect to total protein. Number of samples is given in parentheses. Molar ratios of each ABP to total actin were determined by multiplying the percentage of protein by the ratio of molecular weights and normalizing to actin concentration.
| Protein | Total Protein | ABP:ActinMolar Ratio |
|---|---|---|
| Actin | 0.37 ± 0.02 (4) | — |
| CPA | 0.0015 ± 0.0001 (8) | 1:207 |
| CPB | 0.0013 ± 0.0002 (8) | 1:196 |
| CAP1 | 0.0522 ± 0.0002 (3) | 1:9 |
| ADF | 0.056 ± 0.002 (4) | 1:3 |
Additional information can be derived by transforming these data into a molar ratio of ABP abundance with respect to actin levels, as previously reported (Chaudhry et al., 2007). For the monomer-binding proteins CAP1 and ADF, this corresponds to a 1:9 and 1:3 relationship, respectively, between ABP and total cellular actin (Table I). This is in agreement with previous data from rosette leaves, in which CAP1 is present at 1:7 and ADF is present at 1:3 ABP:actin (Chaudhry et al., 2007). By contrast, the CPA subunit was present at 1:207 stoichiometry with total actin, and CPB was present at 1:196 (Table I). Analysis of CPA and CPB protein levels in cp knockdown mutants (Table II) showed a decreased stoichiometry of the CPA subunit, with total actin of 1:1922, 1:1889, and 1:2187 for cpa-1, cpb-1, and cpb-3, respectively. For the CPB subunit, stoichiometries with actin of 1:1029, 1:764, and 1:996 were determined for the cp mutant lines.
Table II. Quantitative protein analysis of AtCP in cp knockdown lines.
Values represent mean percentage (± sd) of a particular ABP with respect to total protein. Number of samples is given in parentheses. Molar ratios of each ABP to total actin were determined by multiplying the percentage of protein by the ratio of molecular weights and normalizing to actin concentration.
| Protein | Wild Type |
cpa-1 |
cpb-1 |
cpb-3 |
||||
|---|---|---|---|---|---|---|---|---|
| Total Protein | ABP:Actin Molar Ratio | Total Protein | ABP:Actin Molar Ratio | Total Protein | ABP:Actin Molar Ratio | Total Protein | ABP:Actin Molar Ratio | |
| Actin | 0.388 ± 0.011 (3) | — | 0.58 ± 0.02 (3) | — | 0.57 ± 0.02 (3) | — | 0.66 ± 0.03 (3) | — |
| CPA | 0.0016 ± 0.0002 (3) | 1:201 | 0.00025 ± 0.00002 (6)a | 1:1,922 | 0.00025 ± 0.00002 (6)a | 1:1,889 | 0.00025 ± 0.00002 (6)a | 1:2,187 |
| CPB | 0.0018 ± 0.0004 (3) | 1:260 | 0.00068 ± 0.00015 (3) | 1:1,029 | 0.0009 ± 0.0002 (3) | 1:764 | 0.0008 ± 0.0003 (3) | 1:996 |
This value represents the lower limit for detection of CPA protein on immunoblots.
We conclude that CP is a moderately abundant ABP in cellular extracts from Arabidopsis seedlings. However, this analysis does not tell us anything about CP concentration in different tissues or cell types or about its subcellular distribution.
CP Localizes to Punctate Structures That Overlap Partially with the Actin Cytoskeleton in Cells
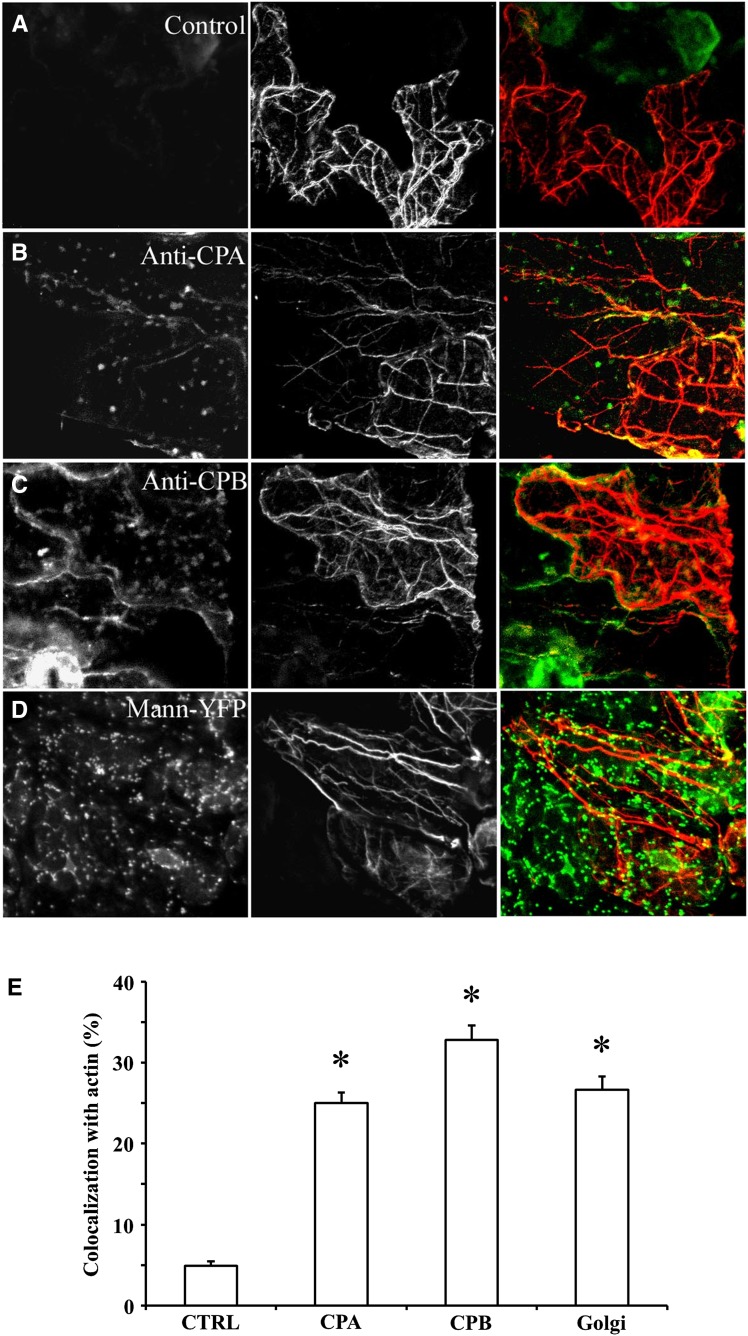
To further understand the relationship between CP and the actin cytoskeleton, we determined its subcellular distribution by immunolocalization. Our expectation was that CP would at least partially colocalize with actin filaments or bundles. The two affinity-purified antibodies, raised against recombinant CPA and CPB, respectively, were used in combination with a mouse monoclonal anti-actin IgM on fixed and freeze-fractured rosette leaves of Arabidopsis. In epidermal pavement cells, actin filaments were arrayed into a randomly oriented set of individual filaments, mostly located in the cortical cytoplasm (Fig. 2, middle image). A second population of actin filaments comprised large bundles that were present in the cortical cytoplasm, but also ramified through the central vacuole. Both CPA (Fig. 2B) and CPB (Fig. 2C) antisera recognized numerous puncta of heterogenous sizes that were distributed randomly throughout the cytoplasm. In epidermal pavement cells, the largest CPA- and CPB-labeled particles had a mean diameter (± sd) of 1.01 ± 0.13 and 0.98 ± 0.12 µm (n = hundreds of puncta from more than 30 cells). Some of these puncta appeared to colocalize with or align along actin filament cables on color overlays of the two fluorescence channels (Fig. 2, B and C, right image). To assess the extent of overlap, we quantified colocalization of signals. Individual regions of interest (ROIs) were selected from maximum intensity z-series projection images of cells that were double labeled with anti-CPA or anti-CPB and anti-actin. After thresholding to remove background, the percentage of pixels positive for both CPA or CPB and actin was measured. Figure 2E shows the results from this quantitative colocalization analysis. CPA and CPB puncta had 25.0% ± 1.3% (mean ± sem; n = 64 ROIs from 16 cells) and 32.8% ± 1.8% (n = 63 ROIs from 15 cells) overlap with actin filaments in epidermal pavement cells. These values seem somewhat low; nevertheless, they were significantly different from controls in which CPA or CPB primary antibody was excluded (4.9% ± 0.5% colocalization, n = 33 optical sections from 10 cells; P < 0.0001 with a paired Student’s t test). We also performed a cross correlation analysis on the colocalization data according to the methods of Costes et al. (2004). The Pearson correlation coefficient (PCC) value was determined for numerous ROIs on individual optical sections and mean values for all pairwise combinations (± sem) were calculated. The PCC for CPA and CPB localization with actin was 0.51 ± 0.12 and 0.55 ± 0.16, respectively, whereas the control without primary CP antibody had a PCC value of 0.25 ± 0.13. The PCC values for CP colocalization with actin were significantly different from the controls (Student’s t test, P < 0.0001). These data indicate moderate colocalization between the two signals (Costes et al., 2004) and are similar to values obtained for the ARP2/3 complex associated with actin filaments (PCC = 0.61; Zhang et al., 2013b). Thus, a modest amount of CP is present in large particles that associate with actin filaments or cables in epidermal pavement cells.
Figure 2.
CP is present on cytoplasmic puncta that display only modest colocalization with actin filaments or cables in epidermal pavement cells. Seedlings of wild-type Arabidopsis plants (20 DAG) were fixed and prepared by the freeze-shattering method prior to incubation with affinity-purified CPA or CPB polyclonal antisera, as well as with a mouse monoclonal IgM against actin. Epidermal pavement cells were examined by confocal laser scanning microscopy and images shown are z-series projections. A, The left image shows a control with secondary antibody only (i.e. no CP primary antibody). The middle image shows actin labeling and the right image is a color overlay of the control (green) and actin (red) images. B, A representative epidermal pavement cell that is double labeled for CPA (left) and actin (middle). The right image is a color overlay of CPA (green) and actin (red). CPA is present on cytoplasmic puncta or foci of varying size and intensity. A small subset of these colocalize (right, yellow) with actin filaments or cables. C, A representative epidermal cell that is double labeled for CPB (left) and actin (middle). The right image is a color overlay of CPB (green) and actin (red). Similar to CPA, CPB is present on puncta that sometimes colocalize (yellow) with actin cables. D, Colocalization of Golgi and actin filaments. Arabidopsis seedlings expressing the Golgi marker mannosidase-YFP were prepared and immunolabeled as above with the actin monoclonal antibody. The left image shows mannosidase-YFP fluorescence and the middle image is actin. The right image is a color overlay of mannosidase-YFP (green) and actin (red), showing a substantial overlap (yellow) of Golgi on the actin cables (yellow). E, Quantitative analysis of CPA, CPB, and mannosidase-YFP association with actin filaments and cables. See “Materials and Methods” for details. The mean values (± sem) from analysis of more than 25 ROIs per treatment are plotted. Compared with controls, in which the CP primary antibody was excluded, the extent of colocalization between CPA, CPB, or mannosidase-YFP with actin was significant (*P < 0.01). CTRL, Control; Mann, mannosidase.
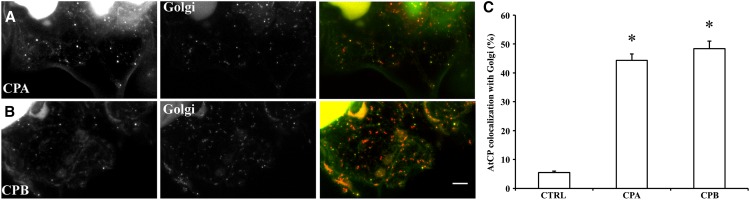
To better understand whether this colocalization analysis could reveal the association of a membrane-bound compartment with the actin cytoskeleton, we performed immunolocalization of the filament network on an Arabidopsis line expressing a Golgi marker, the transmembrane domain from soybean (Glycine max) α-1,2-mannosidase fused to yellow fluorescent protein (YFP; Nelson et al., 2007). The plant cell Golgi apparatus has long been recognized to associate with and locomote along actin filament cables (Satiat-Jeunemaitre et al., 1996; Boevink et al., 1998; Nebenführ et al., 1999) and depends upon Myosin XI motors for its movement (Avisar et al., 2008; Peremyslov et al., 2008; Prokhnevsky et al., 2008). Mannosidase-YFP decorated numerous, large puncta that were present throughout the cytoplasm of epidermal pavement cells (Fig. 2D, left image). The average size of these compartments was 1.83 ± 0.09 µm (n = hundreds of Golgi from seven cells). Many of these compartments were arrayed along actin cables in two-color overlays (Fig. 2D, right image). Quantitative assessment of colocalization revealed that 26.6% ± 1.7% of the Golgi signal overlapped with actin filaments or cables and this was significantly different from controls (P < 0.0001; Fig. 2E). Similarly, the PCC value for mannosidase-actin colocalization was 0.45 ± 0.09 (n = 52 ROIs); this was significantly different (P < 0.0001) from the value of 0.26 ± 0.15 (n = 25 ROIs) for controls without actin primary antibody. These results indicate that it is possible to use quantitative colocalization to describe the association of a membrane-bound organelle with the actin cytoskeleton. We hypothesize that the majority of CP is present on a cytoplasmic compartment or organelle, a fraction of which associates with actin filaments.
Subcellular Fractionation Reveals That CP Associates with Membrane Fractions
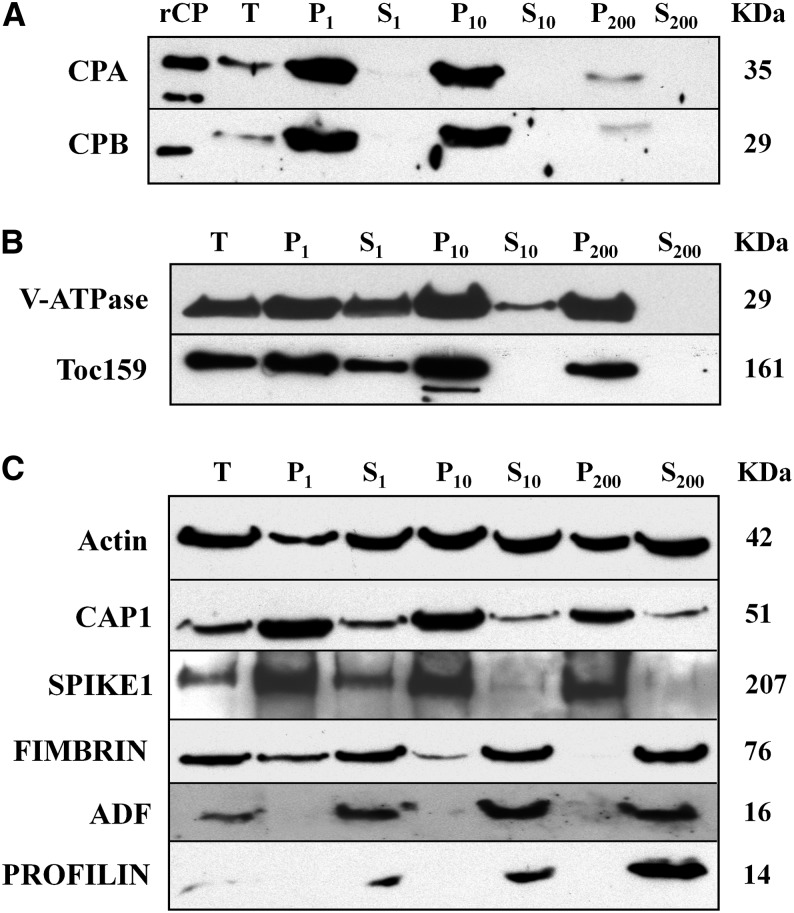
Given the heterogenous size, random distribution, and density of the CP-labeled puncta, we speculated that a substantial amount of CP is present on a membrane-bound compartment. To assess the membrane association of CP and to identify which compartment(s) might contain this protein, we separated cellular organelles from Arabidopsis seedlings by differential centrifugation and Suc density gradient sedimentation. In differential centrifugation experiments, filtered homogenates of 20 d after germination (DAG) seedlings were subjected to consecutive sedimentation at 1,000g, 10,000g, and 200,000g. The resulting pellet (P) and supernatant (S) fractions were analyzed by protein gel immunoblotting with CPA and CPB antibodies (Fig. 3A). As controls, we probed blots with antibodies against the vacuolar proton pump ATPase (V-ATPase), and the chloroplast outer envelope translocon component translocase of chloroplast (Toc159) (Fig. 3B). We also analyzed the distribution of actin and several cytoskeletal proteins, including CAP1, SPK1, fimbrin, ADF, and profilin (Fig. 3C).
Figure 3.
CP is present in membrane fractions after differential centrifugation of cellular extracts. Analysis of CP and several other ABPs during differential centrifugation of extracts prepared from 20 DAG Arabidopsis Col-0 seedlings. The individual lanes represent the pellet (P) and supernatant (S) fractions obtained after total cellular extracts (T) were subjected to differential centrifugation at 1,000g, 10,000g, and 200,000g, respectively. Lanes were loaded with equal amounts of protein (75 µg), separated by SDS-PAGE, and immunoblotted with antibodies against CP, V-ATPase, AtToc159, and various ABPs. The molecular weight in kilodaltons for each polypeptide is given at right. A, CPA and CPB were most abundant in the pellet fractions and were virtually undetectable in the soluble fractions. rCP loaded in the first lane verifies the size of the native protein in extracts. B, Antibodies against the tonoplast marker V-ATPase and the chloroplast outer envelope protein Toc159, were used as positive controls for differential centrifugation of membrane-associated proteins. C, Actin and several cytoskeletal-associated proteins also partitioned with membranes or organellar fractions. Antibodies were used to detect the following: actin; CAP1; the ROP-GEF, SPK1; an actin filament cross linking protein, FIMBRIN; and, two actin monomer-binding proteins, ADF and PROFILIN. Actin partitioned almost equally between soluble and pellet fractions, whereas CAP1 and SPK1 were mainly in pellet fractions. By contrast, FIMBRIN, ADF, and PROFILIN were predominantly soluble proteins.
With the exception of the low-speed pellet (P1), which contains mainly cellular debris and cell wall material, CPA and CPB were found primarily in the insoluble, membrane-containing fractions (P10 and P200; Fig. 3A). A substantial amount of CP was detected in the P10 fraction, which is enriched for organelles such as chloroplasts, mitochondria, and nuclei. By comparison, only small amounts of CPs were present in the microsomal fraction (P200), which contains vesicles and membranes of the endomembrane system. Notably, little or no CP was detected in the S200 cytosolic fraction. A similar distribution was observed for the chloroplast envelope protein, Toc159, which was most abundant in all pellet fractions and preferentially in P10 (Fig. 3B). The V-ATPase antibody also detected a polypeptide that was abundant in pellet fractions, but nearly equally abundant in P10 and P200. Of the cytoskeletal proteins, both CAP1 and SPK1 showed a similar distribution to CP; however, each of these was more prevalent in P200 and had some cytosolic signal (S200; Fig. 3C). By contrast, considerably more fimbrin antigen, an F-actin bundling protein, was detected in the soluble (S10 and S200) fractions and the monomer-binding proteins ADF and profilin were almost completely soluble (Fig. 3C).
Because individual actin filaments and higher order structures like bundles or cables can also sediment under these conditions, it was important to assess the distribution of actin during differential centrifugation. Actin appeared to be equally abundant in all soluble and pellet fractions (Fig. 3C), in contrast with the membrane markers (V-ATPase and Toc159) and CP. These results suggest that CP may associate with a membrane-bound compartment, independent of its binding to actin filaments. Similar results were reported for the plant Arp2/3 complex, which is a peripheral membrane protein present in microsomal fractions (Dyachok et al., 2008; Kotchoni et al., 2009). Furthermore, SPK1 is a peripheral membrane-associated protein that accumulates at the ER (Zhang et al., 2010). Little colocalization of NAP1, a component of the SCAR/WAVE complex, was found with actin, whereas a big pool of NAP1 was associated with the surface of ER (Zhang et al., 2013a).
To get a better sense about the association of CP and actin with the microsomal (P200) fraction, we extended our quantitative immunoblotting analyses to these samples and determined the relative abundance of each protein (Table III). As observed for total cellular extracts, actin is relatively abundant in the P200 fraction, representing 0.25% of total microsomal protein. The monomer-binding protein CAP1 was less abundant at 0.01% of total protein. In addition, CP subunits were present at 0.0007% and 0.0008% of total protein for CPA and CPB, respectively. Expressed as molar ratios with total actin, CAP1 was present at 1:28, whereas CPA and CPB were 1:290 and 1:201, respectively. These amounts are slightly less than those found in total cell extracts but still quite prevalent. The presence of both a monomer-binding protein (CAP1) and a filament end-binding protein (CP) in the microsomal fraction could indicate the presence of both G- and F-actin on these membranes or contamination of this fraction with cytoskeletal elements. Alternatively, CP and CAP1 could associate directly with membranes or membrane proteins independent of their association with actin.
Table III. CP is present in the microsomal membrane fraction.
Values represent mean percentage (±sd) of a particular ABP with respect to total protein. Number of samples is given in parentheses. Molar ratios of each ABP to total actin were determined by multiplying the percentage of protein by the ratio of molecular weights and normalizing to actin concentration.
| Protein | Total Protein | ABP:ActinMolar Ratio |
|---|---|---|
| Actin | 0.245 ± 0.014 (3) | — |
| CPA | 0.00071 ± 0.00006 (3) | 1:291 |
| CPB | 0.00084 ± 0.00006 (3) | 1:201 |
| CAP1 | 0.0105 ± 0.0003 (3) | 1:28 |
CP Behaves Like an Integral Membrane-Associated Protein
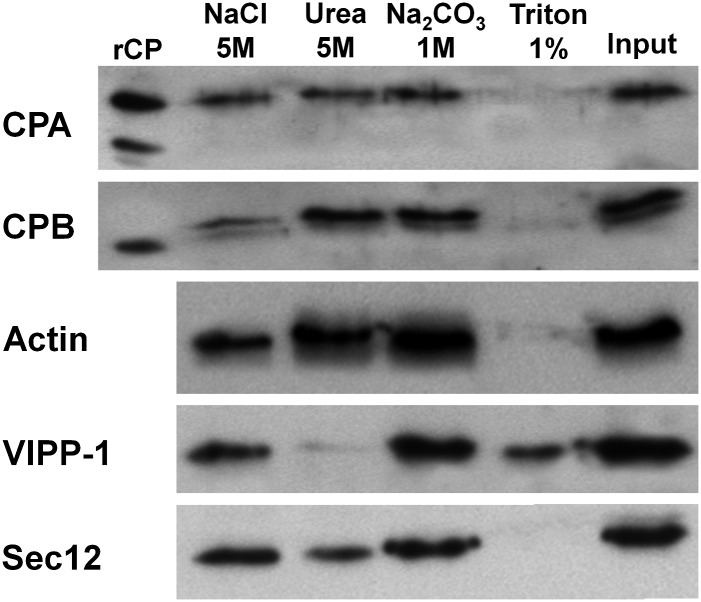
To determine the nature of CP association with the microsomal fraction, we analyzed the P200 fraction from Arabidopsis seedlings by extraction with high salt, chaotrope, alkaline pH, and nonionic detergent. The P200 fraction was divided into equal amounts and resuspended in buffer containing the different agents to discriminate between peripheral and integral membrane proteins. If CP is a peripheral membrane protein that associates with other membrane proteins or phospholipid head groups, it should be eluted partially or fully by treatments with increased ionic strength (5 m NaCl), by mild chaotropic salt conditions (5 m urea) or with alkaline conditions (1 m Na2CO3, pH 10.9). If CP behaves like an integral membrane protein, which is embedded in the phospholipid bilayer, it should be removed from the microsomal pellet by treatment with a nonionic detergent (1% (v/v) Triton X-100). All reactions were incubated for 30 min at 4°C and centrifuged at 200,000g to give supernatant (S2200) and pellet (P2200) fractions. The resulting pellets (and supernatants; data not shown) were blotted for the presence of CP and actin (Fig. 4). Antibodies against well-characterized proteins guanine-nucleotide exchange factor (Sec12; Bar-Peled and Raikhel, 1997) and vesicle-inducing protein in plastids-1 (VIPP-1) were used as controls for integral and peripheral membrane proteins, respectively.
Figure 4.
CP behaves like an integral membrane-associated protein. The supernatant S1 fraction was centrifuged at 200,000g to give a P200 microsomal membrane fraction, which was resuspended and divided into five equal fractions in buffer containing either 5 m NaCl, 5 m urea, 1 m Na2CO3, pH 10.9, or 1% (v/v) Triton X-100 and incubated on a shaker for 30 min at 4°C. The resulting suspension was recentrifuged for 60 min at 200,000g, providing pellet and soluble fractions. Shown here are the pellet fractions that were blotted and probed with CPA and CPB antibodies, as well as with actin, VIPP-1, and Sec12 antibodies as controls for peripheral and integral membrane-associated proteins, respectively. Similar experiments were performed four independent times.
A major proportion of CP antigen dissociated from the membranes and very little was present in the P200 fraction after treatment with 1% Triton X-100 (Fig. 4). No significant amount of CP was released from the membrane fraction after treatments with the chaotrope (5 m urea), whereas a small proportion of CPB was released in the presence of 5 m NaCl (Fig. 4). Alkaline conditions transform the structure of membrane compartments, turning closed compartments into sheets (Zheng et al., 2003). This has the effect of releasing soluble proteins that are trapped inside membranous vesicles. Only a minor amount of CP was released from membranes in the presence of 1 m Na2CO3, pH 10.9 (Fig. 4). These data indicate that CP behaves somewhat like an integral membrane protein.
For controls (Fig. 4), we observed that the integral protein Sec12 was also solubilized from the membrane with Triton X-100 (Bar-Peled and Raikhel, 1997). By contrast, the peripheral membrane protein VIPP-1 was not released from membranes with salt treatment (5 m NaCl), or with alkaline conditions. However, urea and detergent did elute VIPP-1 from the membrane, showing the peripheral but tight association with microsomal membranes. Actin was released from the membrane mainly with Triton X-100, although a small proportion was released from the membrane under high-salt treatment.
Collectively, these findings provide evidence that CP behaves like a protein integrated in the phospholipid bilayer, rather than an extrinsic protein associated peripherally with membranes. Because of the partial release with high salt treatment, we cannot completely rule out that CP behaves like a peripheral protein that is tightly associated with membranes. This is consistent with CG-MD simulations showing that the C terminus of the α-subunit of AtCP associates with PA-containing membranes via extensive polar and nonpolar contacts, and that part of this amphipathic helix partially inserts into the lipid bilayer (Pleskot et al., 2012).
CP Is Located on the Cytoplasmic Side of Microsomes
To confirm that CP is not simply trapped inside of membranes from the microsomal fraction and to further reveal its association with membranes, we treated microsomes with proteinase K (PK). Results from this experimental approach indicated that CP was present on the outside of the microsomes, because no CPA or CPB was detected when P200 microsomes were treated with PK (Fig. 5). Experimental controls showed that samples not digested with PK, but treated equivalently in all other respects, suffered little appreciable proteolysis. Controls for other peripheral proteins, actin and VIPP-1, showed the same behavior as CP (Fig. 5). These data support the conclusion that CP associates with the cytoplasmic face of microsomal membranes.
Figure 5.
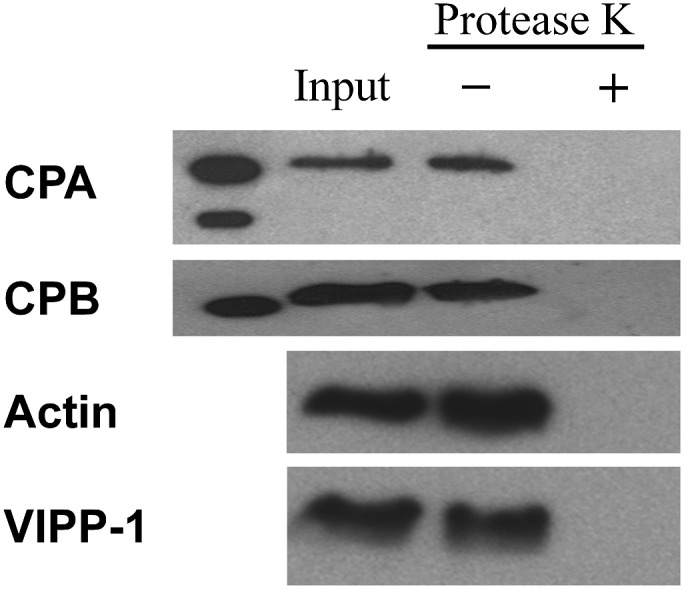
CP localizes on the cytoplasmic side of the membrane. The P200 fraction containing CP was incubated with and without PK. Immunoblots of the resulting samples were performed with antibodies against CPA and CPB, anti-actin, and anti-VIPP1, The P200 fraction prior to addition of protease was used as a loading control. rCP was loaded in the first lane as a molecular weight marker for CP.
CP Cofractionates with ER and Golgi
The above analyses establish CP as a membrane-associated protein. To further investigate which cellular membranes or organelle/compartments contain CP, we employed two distinct approaches: Suc density gradient fractionation of the microsomal fraction and confocal microscopy of epidermal cells with organelle markers.
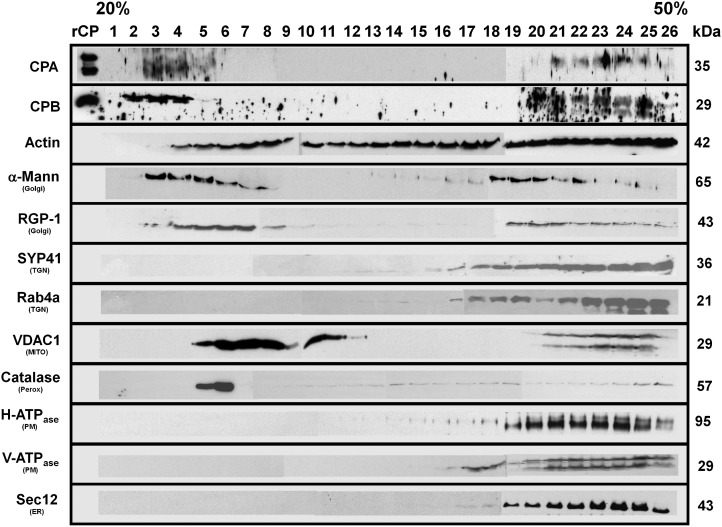
To further separate endomembranes and organelles from the microsomal pellet, the P200 fraction was subjected to isopycnic ultracentrifugation on 20% to 50% (w/v) linear Suc gradients and the results analyzed by immunoblotting (Fig. 6). A selection of previously characterized organelle/compartment markers was used as controls (full details and sources of antibodies are provided in Supplemental Table S1). This included antibodies against the following: CPA and CPB; the mitochondrial voltage-dependent anion channel, VDAC1; the peroxisomal marker, catalase; the ER marker, Sec12; the Golgi enzymes, α-1,2-mannosidase and reversibly-glycosylated protein1 (RGP1); a SNARE protein associated with the trans-Golgi network, Syntaxin of Plants41 (SYP41); the secretory vesicle-associated GTPase, Ras-related GTP-binding protein A4b (RabA4b); the plasma membrane proton-translocating adenosine triphosphate synthase (H+-ATPase); and the vacuolar H+-ATPase, V-ATPase. A representative experiment is shown in Figure 6 and this assay was repeated three times on independent Suc density gradients with similar results. The behavior of compartment markers is consistent with the results of Oliviusson et al. (2006), whose methods were used herein for Suc gradient separations.
Figure 6.
CP is coenriched with several membrane-bound compartments in the microsomal fraction. Microsomal (P200) membrane fractions were separated on an isopycnic 20% to 50% (w/v) linear Suc gradient. Equal volumes of protein fractions collected from the gradient were separated on SDS-PAGE gels, blotted, and probed with antibodies against the following: CPA and CPB; actin; cis-Golgi, α-1,2-mannosidase; trans-Golgi, RGP1; plasma membrane, H+-ATPase; ER, Sec12; tonoplast, V-ATPase; mitochondrial outer membrane porin 1, VDAC1; trans-Golgi network, AtSYP41 and RabA4; and peroxisome, catalase. Protein names and sizes are indicated on the left and right, respectively. The entire gradient, fractions 1 to 26, required several gels and membranes for probing with each antibody. Separation between the individual blots or membranes comprising the full gradient is not shown on the figure, for clarity of presentation. Mann, Mannosidase; MITO, mitochondria; Perox, peroxisome; PM, plasma membrane; TGN, trans-Golgi network.
CP was present in two discrete regions of the Suc density gradient: a major peak at low density, around fractions 2 to 5; and a somewhat less abundant peak at high density, between fractions 20 and 25 (Fig. 6). By contrast, CP was not detected in the middle of the gradient (fractions 6–18). The low-density fraction of CP overlapped best with the Golgi compartment as revealed by the α-1,2-mannosidase and RGP1 protein in fractions 3 to 7 and 17 to 24. The high-density CP fraction corresponded with the migration of several endomembrane markers, including the ER, plasma membrane, and tonoplast (Fig. 6), making it difficult to rule out these compartments. On the other hand, the CP peaks were clearly separated from those of VDAC1 and catalase, showing that CP-enriched fractions did not cosediment with the mitochondria- or peroxisome-enriched fractions.
We also tested the behavior of actin in the Suc density gradient fractions (Fig. 6). Actin was ubiquitous throughout virtually the entire gradient, from fractions 4 to 26, indicating that it is present on numerous membrane compartments. As with the microsomal fractionation described above, this analysis does not reveal whether the actin is present as monomers or filaments. An alternative interpretation of these results is that individual and/or bundles of actin filaments, with varying sizes, migrate at different densities throughout the gradient. Collectively, our subcellular fractionation results demonstrate that CP in plant cells is present on several subcellular compartments, probably the Golgi and the ER.
To further evaluate the CP-Golgi association, we analyzed an Arabidopsis line expressing the mannosidase-YFP marker by immunolocalization (Fig. 7) and Suc density gradient separations (Supplemental Fig. S1). The quantitative imaging experiments showed 44.3% ± 2.2% and 48.4% ± 2.6% colocalization with cis-Golgi for CPA and CPB, respectively, whereas the control without primary CP antibody had 5.4% ± 0.5% colocalization (Fig. 7C). The mean PCC values (± sem) calculated from the same ROI on individual images were 0.78 ± 0.13 (n = 59), 0.80 ± 0.10 (n = 40), and 0.26 ± 0.15 (n = 25). The PCC values for CP-mannosidase colocalization were significantly different from controls (Student’s t test, P < 0.0001), indicating a very strong association of CP with the cis-Golgi marker (Costes et al., 2004). The fractionation experiments demonstrated comigration in the low-density fractions of CP and α-mannosidase when proteins were detected with anti-CPB and anti-GFP, respectively (Supplemental Fig. S1A). Use of a trans-Golgi marker (Dhugga et al., 1997) also revealed partial overlap between fractions containing CP and RGP1 protein (Supplemental Fig. S1A). Specificity of the anti-GFP antibody was demonstrated by probing membrane fractions from ecotype Columbia-0 of Arabidopsis (Col-0) not expressing a fluorescent fusion protein (Supplemental Fig. S1B). Collectively, these results confirm that some proportion of cellular CP associates with Golgi.
Figure 7.
CP colocalizes with a cis-Golgi marker. A and B, Colocalization of CP with Golgi. Arabidopsis seedlings expressing the Golgi marker, mannosidase-YFP, were prepared and immunolabeled with CP polyclonal antibodies. The left image shows a representative image from an epidermal pavement cell labeled with CPA (A) and CPB (B), respectively. Middle images correspond to mannosidase-YFP fluorescence from the same cells. The right images show merged images depicting colocalization. C, Quantitative analysis of colocalization between CPA and CPB with mannosidase-YFP. See “Materials and Methods” for details. The mean values (± sem) from analysis of >41 ROIs within at least seven epidermal pavement cells per treatment are plotted. As a control, the primary anti-CPB antibody was left out and samples were processed in identical fashion. The extent of colocalization between both CP subunits and mannosidase-YFP was significantly different from the negative control (*P < 0.01). CTRL, Control. Bar = 10 μm.
DISCUSSION
Phospholipids are important regulatory molecules in eukaryotic cells and have diverse roles in various cellular events, including intracellular signaling responses, membrane trafficking, and modulating cytoskeletal organization (Saarikangas et al., 2010). Although numerous ABPs are regulated by phospholipids in vitro (Saarikangas et al., 2010), evidence for the existence and mechanism of regulation in vivo is limited. CP is one such ABP that, along with phospholipase D, may serve as a hub for positive feedback between lipid signaling events and cortical cytoskeletal organization (Pleskot et al., 2013).
Our quantification of the total amount of CP showed that more than enough CP was present to bind all available actin filament barbed ends in the cell. The observed stoichiometry with total actin is in the same range as reported for CP in mammalian neutrophils and platelets, as well as in Acanthamoeba or Dictyostelium spp. cells, which have CP concentrations of 1 to 5 µm and stoichiometries with total actin of 1:90 to 1:400 (Cooper et al., 1984; DiNubile et al., 1995; Barkalow et al., 1996; Eddy et al., 1997; Pollard et al., 2000). In S. cerevisiae, CP is also present at micromolar levels but total actin is much less abundant, making the ratio of CP to total actin of approximately 1:4 (Kim et al., 2004). Both F-actin levels and the number of filament barbed ends are estimated to be rather low in plant cells (reviewed by Staiger and Blanchoin, 2006).
To examine the role of CP in vivo, we localized this protein in cells and examined its subcellular fractionation properties. CP was localized primarily in the cytoplasm as numerous puncta that were distributed randomly. Immunolocalization demonstrated that about 30% of AtCP colocalizes with actin bundles. Why there is more CP available to bind with the cytoskeleton than barbed ends is not clear, but some of the CP molecules in cells not bound to actin filaments may associate with other cellular fractions, including membrane-bound compartments. Our immunolocalization results clearly show that CP colocalized with the Golgi apparatus in Arabidopsis epidermal pavement cells. These results indicate that fluorescent spots are sites colocalized with actin filaments and with membrane subcellular components. Furthermore, colocalization experiments using Arabidopsis plants expressing mannosidase-YFP revealed 50% colocalization with Golgi by antibody staining with CP sera, a result that was supported by AtCP comigration with a cis-Golgi marker and partial comigration with a trans-Golgi marker on Suc density gradients.
Cytoplasmic CP puncta have been observed but not well characterized in S. cerevisiae (Amatruda and Cooper, 1992), cultured myocytes and fibroblasts (Schafer et al., 1994), cardiac muscle (Hart and Cooper, 1999), and Drosophila spp. bristles (Frank et al., 2006). In stably transformed Potorous tridactylus K1 cell line fibroblasts, GFP-CPβ2 marks large, motile puncta in the peripheral cytoplasm that depend on actin for movement (Schafer et al., 1998). Similarly, enhanced GFP-CPβ1 is present on cytoplasmic punctate structures in lamellipodia in Xenopus laevis cell line XTC fibroblasts after 2 h of transient expression (Miyoshi et al., 2006). In addition, previous research has shown that CP localizes in the hyaline ectoplasm, a region of the cytoplasm just under the plasma membrane that contains a high concentration of actin filaments. These experiments show that CP is associated with a region of cells rich in actin filaments and with a membrane fraction that itself contains actin filaments (Cooper et al., 1984).
In addition to immunolocalization in cells, we provide further evidence that plant CP is associated with cellular endomembranes. Specifically, differential centrifugation of cellular fractions showed that AtCP was present in the microsomal membrane fraction. Further fractionation and immunoblotting of microsomes separated on Suc density gradients show that CP may be associated with Golgi and/or ER. To our knowledge, we provide the first direct experimental evidence that confirms AtCP binds directly to cellular organelles in plants. Thus, AtCP may assume a role in sensing and transducing membrane signaling lipids into changes in actin cytoskeleton dynamics.
Additional support for the CP-membrane localization was provided by the investigations of Pleskot et al. (2012), using molecular docking and CG-MD simulations. They uncovered a specific mode of high-affinity interaction between membranes containing PA/phosphatidylcholine and plant CP. In this mechanism, the C-terminal amphipathic helix of plant CPα-subunit partially intercalates into the lipid bilayer through specific polar and nonpolar interactions. The mechanism and specific residues on the CPα C terminus were found to be unique to the plant kingdom.
CP from Arabidopsis is regulated by interaction with both PA and PtdIns(4,5)P2 (Huang et al., 2003, 2006; Li et al., 2012). PA is markedly more abundant in plant membranes than is PtdIns(4,5)P2 Thus, PA binding may be physiologically relevant for CP activity. Moreover, PA levels change rapidly in response to water deficit, wounding, and microbial attack (Li et al., 2009; Testerink and Munnik, 2011). Interestingly, PA also mediates functions such as recruitment of effector (peripheral) proteins to membranes mediated by a growing number of modular membrane-targeting domains that specifically recognize their cognate lipid ligands, to form protein-protein and lipid-protein interactions during cell signaling and membrane trafficking (Cho and Stahelin, 2005).
The subcellular distribution of PA pools in plants is poorly characterized, although a new fluorescent reporter that shows great promise for future work indicates abundant PA at the plasma membrane of pollen tubes (Potocký et al., 2014). Furthermore, native PA binding proteins accumulate at the plasma membrane (Zhang et al., 2004), whereas in mammalian cells, PA is abundant in not only the plasma membrane, but also the ER, Golgi, and endosomal membranes (Rizzo et al., 2000; Baillie et al., 2002; Loewen et al., 2004). Because the filament end-capping activity of AtCP is negatively regulated by membrane phospholipids in vivo (Li et al., 2012), we speculate that the membrane-bound CP may represent an inactive pool of ABP. Alternatively, membrane-bound CP may position this key regulator of actin dynamics near sites of filament assembly and turnover.
There are two possibilities for how ABPs may be regulated by phospholipids: (1) direct interaction with phosphoinositides regulates the activity and/or subcellular localization of ABPs; or (2) phosphoinositides control the localization of scaffolding proteins that relay between the actin cytoskeleton and plasma membrane or intracellular membrane organelles. In this regard, various ABPs have been demonstrated to be membrane-associated proteins (Saarikangas et al., 2010). In this study, in addition to CP-membrane association, we show that CAP1 from Arabidopsis (Chaudhry et al., 2007) is highly enriched on microsomal membranes.
Several other plant ABPs or regulators of actin organization are found on membranes or cellular organelles. The xyloglucan galactosyltransferase KATAMARI1 (KAM1/MUR3) is a membrane-associated ABP that mediates actin organization and function in proper endomembrane organization as well as cell elongation; KAM1/MUR3 is located specifically on Golgi membranes, where it associates with membranes as an integral membrane protein (Tamura et al., 2005). Another membrane-associated ABP family is the formins, which nucleate unbranched filaments and processively elongate filaments at the barbed end. The majority of plant group I formins contain a transmembrane domain at the N terminus in their primary amino acid sequence (Cvrcková, 2000; Deeks et al., 2002), which indicates that these formins are likely to be membrane-bound proteins in Arabidopsis. This prediction was verified for Arabidopsis Formin1 (AtFH1), AtFH6, and AtFH5 (Banno and Chua, 2000; Cheung and Wu, 2004; Favery et al., 2004; Ingouff et al., 2005). The subcellular localization of AtFH1 was shown by fluorescent protein fusion to be targeted to the plasma membrane and colocalized with a known plasma membrane protein, aquaporin, plasma membrane intrinsic protein PIP2;1 (Martinière et al., 2011). AtFH6 has been shown to be uniformly distributed throughout the plasma membrane and to generate actin cables that serve as tracks for vesicle trafficking for extensive plasma membrane and cell wall biogenesis (Favery et al., 2004). Arabidopsis group Ie formins AtFH4 and AtFH8 not only associate with the cell membrane, but also accumulate to specific subcellular domains along the cell perimeter (i.e. AtFH4 localized to cell-to-cell contact points in mesophyll cells of leaf and cotyledon; Deeks et al., 2005).
Because of their often large size, plant cells require a transport network that orchestrates the movement of endomembranes and other macromolecular complexes through the cytoplasm and delivers them to their subcellular destinations. Plant Myosin XI is a molecular motor that is implicated in organelle transport along the actin cytoskeleton. Myosin XI-K has a critical role in this process through its association with endomembrane vesicles (Peremyslov et al., 2012). Furthermore, subcellular localization and fractionation experiments showed that the nature of myosin-associated vesicles is organ specific and cell type specific: (1) in leaves, a large proportion of these vesicles aligned and cofractionated with a motile ER subdomain; and (2) in roots, non-ER vesicles were the dominant myosin cargo. Furthermore, Myosin XI-K had a polar localization at the tips of growing, but not mature, root hairs, suggesting that myosins contribute to vesicle transport during tip growth (Peremyslov et al., 2012). The physical association of Myosin XI-K with endomembranes was explored by fractionation experiments using leaf extracts from Arabidopsis plants and an XI-K-specific antibody (Peremyslov et al., 2012). On isopycnic Suc gradients, Myosin XI-K migrated with peaks of the ER marker and Golgi markers Sec21 and NAG. The distribution of the trans-Golgi/secretory vesicle marker RabA4b also corresponded broadly to that of the myosin. These results suggest that most of the Myosin XI-K in leaf cells is associated with the ER-, organelle-, and secretory vesicle-derived membranes. A distinct plant-specific transport vesicle compartment in Arabidopsis was recently identified and is associated with Myosin XI and a novel cargo adaptor MyoB1 (Peremyslov et al., 2013).
In many eukaryotic cells, actin polymerization is involved in generating forces for organelle movement and remodels or transports membranes during trafficking events (i.e. endocytosis, vesicle formation where actin polymerization might assist invagination formation, pinching off vesicles, and/or driving vesicles away from membrane; Kaksonen et al., 2005). Most of these examples require the ARP2/3 complex, which nucleates new actin filaments and generates branched actin networks. This complex is also membrane associated in nonplant systems (Beltzner and Pollard, 2008) as well as in plants, because a large fraction of the ARP2/3 pool was found to be strongly associated with cell membranes in Arabidopsis (Zhang et al., 2013b). ARP2/3-membrane association correlates with the assembly status and subunit composition of the complex (Kotchoni et al., 2009), and may be regulated by its lipid-binding specificity (Fiserová et al., 2006; Maisch et al., 2009). Association of ARP2/3 complex with membranes is expected because ARP2/3 has a wide variety of organelle-based functions in eukaryotic cells as an actomyosin-based transporter of ARP2/3-containing organelles (Fehrenbacher et al., 2005; Kaksonen et al., 2005), and because of observations of punctate ARP2/3 localization in mammalian cells linked to endomembrane dynamics (Welch et al., 1997; Strasser et al., 2004; Shao et al., 2006). However, demonstrating similar functions for plant ARP2/3 complex requires further experimentation.
The ARP2/3 complex interacts with nucleation promoting factor proteins, such as WAVE/SCAR, in order to be activated and converted into an efficient actin filament nucleator (for review, see Higgs and Pollard, 2001; Welch and Mullins, 2002). Moreover, WAVE/SCAR and ARP2/3 complexes are part of a conserved Rho-of-Plants (ROP) small GTPase signal transduction cascade that integrates actin and microtubule organization with trafficking through the secretory pathway (Bloch et al., 2005; Fu et al., 2005; Lavy et al., 2007; Yalovsky et al., 2008; Szymanski, 2009), and controls actin-dependent morphogenesis in many tissues and developmental contexts (Smith and Oppenheimer, 2005; Szymanski, 2005; Yalovsky et al., 2008).
Several core subunits of the WAVE/SCAR regulatory complex (W/SRC), NAP1 and SCAR2, were found to be peripheral membrane-associated proteins on the ER (Zhang et al., 2010, 2013a). The association of NAP1 with membranes was relatively strong, because no NAP1 solubilization was observed after treatment with high concentrations of salt or the nonionic detergent Triton X100. Furthermore, NAP1 cofractionates with ER membranes (Zhang et al., 2013a). Based on live-cell imaging with fluorescent fusion proteins, the W/SRC subunits SCAR1 and BRICK1 have been reported to localize at the plasma membrane (Dyachok et al., 2008, 2011). SCAR2, like the abundant NAP1, overlapped with an ER marker (Sec12) in Suc gradients, and SEC12, SCAR2, and NAP1 were shifted to less dense Suc fractions when ER-associated ribosomes were destabilized by chelating free Mg2+ (Zhang et al., 2013a). Furthermore, a positive regulator of W/SRC, the DOCK family guanine nucleotide-exchange factor SPK1, is an Arabidopsis protein that strongly associates with cell membranes. SPK1 localizes to the surface of the ER, as suggested by localization and cell fractionation data, and most prominent at ER exit site subdomains (Zhang et al., 2010).
Knowledge from this study demonstrating CP-membrane association in plants, along with an ever-expanding list of membrane-cytoskeletal linkages supported by plant ABPs (Deeks et al., 2012; Wang et al., 2014), suggest that F-actin polymerization driving endomembrane compartment movement as well as vesicle formation and trafficking events between the ER and the Golgi apparatus in plants might be orchestrated and tightly regulated by a cytoskeletal protein network.
MATERIALS AND METHODS
Plant Growth Conditions
The T-DNA insertion lines for AtCPA (cpa-1; SALK_080009) and AtCPB (cpb-1; SALK_014783 and cpb-3; SALK_101017) were obtained from the Arabidopsis Biological Resources Center (Ohio State University), genotyped to identify homozygous mutant plants, and backcrossed to the wild type at least twice prior to use in experiments. Description of the plant growth and cytoskeletal phenotypes associated with these cp knockdown lines are described elsewhere (Li et al., 2012, 2014; Pleskot et al., 2013). For all experiments herein, Arabidopsis (Arabidopsis thaliana) Col-0 was used as wild-type plant material. Wild-type and cp homozygous mutant seedlings were grown aseptically on one-half-strength Murashige and Skoog medium (Sigma-Aldrich) containing 1% (w/v) agar and 1% (w/v) Suc. The growth condition was 16-h light at 100 μmol m−2 s−1 and 8-h dark at 25°C, and seedlings were harvested at 20 DAG for preparation of total cell extracts and subcellular fractionation experiments.
Recombinant Protein Purification
A construct for bacterial expression of CPA and CPB from the same plasmid and identical promoters was described previously (Huang et al., 2003). rCP was isolated from soluble bacterial (Escherichia coli BL21[DE3]) extracts and purified to >90% homogeneity by chromatography on DEAE-Sephacel, hydroxylapatite, and Q-Sepharose (Huang et al., 2003). CP concentration was determined with the Bradford assay (Bio-Rad) using bovine serum albumin as a standard. For loading controls and generation of standard curves in quantitative immunoblotting experiments, recombinant AtCAP1 (Chaudhry et al., 2007) and AtADF1 (Carlier et al., 1997) were purified according to published methods. Protein concentrations were determined by spectrometry at 280 nm with an extinction coefficient of 33,671 m−1 cm−1 for CAP1 (Chaudhry et al., 2007) and at 277 nm assuming an A277 of 0.89 for a 1-mg/mL solution of AtADF1 (Didry et al., 1998). Rabbit skeletal muscle actin was purified according to Spudich and Watt (1971) and was gel filtered on Sephacryl S-300 by the methods of Pollard (1984). Actin concentration was determined by spectrometry assuming an A290 of 0.63 for a 1-mg/mL solution.
Quantitative Immunoblotting
The cellular abundance of CPA, CPB, actin, CAP1, and ADF from wild-type Arabidopsis seedling extracts, as well as from cpa-1, cpb-1, and cpb-3 T-DNA insertion mutant lines (Li et al., 2012), was estimated by quantitative immunoblotting, roughly as described by Wu and Pollard (2005) and Chaudhry et al., (2007). A linear standard curve was generated by loading various amounts of each recombinant purified protein on the same gel as the seedling samples. Total protein extracts from 20 DAG seedlings were prepared by grinding the plant material with liquid nitrogen in a mortar and pestle, obtaining a thin powder, which was loaded into homogenization buffer containing 20 mm HEPES/KOH, pH 7.2, 50 mm KOAc, 2 mm Mg(OAc)2, 250 mm sorbitol, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 1% (v/v) protease inhibitor cocktail (2 mm O-phenanthroline, 0.5 mg/mL leupeptin, 2 mg/mL aprotinin, and 1 mg/mL pepstatin). The extracts were clarified by centrifugation at 15,000g for 2 min, and total protein concentration was determined by the Bradford assay. To estimate the amount of CP in microsomal membrane fractions, we obtained the P200 fraction by differential centrifugation, as described in the section below. For determination of actin, CAP, and ADF concentrations, 25 μg of total protein was loaded, whereas 75 μg of total protein was loaded for CP determinations on the same SDS-PAGE as the standard curve samples. Proteins separated by SDS-PAGE were transferred to nitrocellulose membranes and probed with appropriate antibodies. The primary polyclonal antibodies used were anti-AtCPA and anti-AtCPB (Huang et al., 2003), anti-AtCAP1 (Chaudhry et al., 2007), anti-maize (Zea mays) pollen actin (Gibbon et al., 1999), and anti-AtADF2 (Chaudhry et al., 2007) at dilutions given in Supplemental Table S1. For loading control, we used anti-phosphoenolpyruvate carboxylase (Rockland Immunochemicals). Horseradish peroxidase-coupled secondary antibody (Sigma-Aldrich) was diluted 1:50,000 and detection was with SuperSignal West Pico Chemoluminescent substrate (Thermo Scientific). Images of developed blots were captured on autoradiographic film and scanned, prior to analysis of band intensity with ImageJ. At least three biological replicates of total cellular extract were prepared and tested with each antisera and recombinant protein. With these conditions, the linear range for detection was as follows: 0.25 to 5 ng for CPA, 0.5 to 12.5 ng for CPB, 2 to 20 ng for CAP1, 5 to 25 ng for ADF, and 15 to 120 ng for actin (Fig. 1). Actin and ABP cellular abundance were expressed as a percentage of total cellular protein, and the ratio of actin to ABP was estimated using these percentages after normalizing for Mr of each protein (Tables I–III).
Subcellular Fractionation
Two grams (fresh weight) of wild-type Arabidopsis seedlings were homogenized for 5 min with a hand-held mixer (Polytron; Brinkmann Instruments) on ice in 10 mL of precooled homogenization buffer. The homogenate was filtered through two layers of Miracloth and subjected to differential centrifugation. The first spin, performed for 25 min at 1,000g, removed cell debris and cell walls and resulted in pellet (P1) and supernatant (S1) fractions. The supernatant S1 was removed to a fresh tube and centrifuged for 25 min at 10,000g, yielding the P10 and S10 fractions. The pellet (P10) was retained and S10 was transferred to a fresh tube, centrifuged for 25 min at 200,000g to yield P200, which is a membrane-enriched microsomal fraction, and S200, the soluble cytosolic protein fraction (Kotchoni et al., 2009).
The microsomal fraction was further separated on isopycnic Suc density gradients. For most experiments, however, the 10,000g step of differential centrifugation was eliminated. Organelles and membrane-bound compartments in the P200 were resuspended in suspension buffer containing the following: 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, 1 mm PMSF, and 1% protease inhibitor cocktail. The resuspended microsomal fraction was subjected to centrifugation for 16 h at 200,000g on a linear 20% to 50% (w/w) Suc density gradient prepared in centrifugation buffer (10 mm Tris-HCl, pH 7.6, 2 mm EDTA, 1 mm dithiothreitol, 1 mm PMSF, and 1% protease inhibitor cocktail). The resulting Suc gradient was divided into fractions of 0.2 mL, Laemmli sample buffer (Laemmli, 1970) was added, and samples were boiled for 5 min. Equal volumes of each fraction were separated by SDS-PAGE, transferred to nitrocellulose, and probed with CP, actin, ABP, and various organelle marker antibodies (Supplemental Table S1).
Assays for Integral or Peripheral Membrane Proteins
To determine how CP is associated with the membrane fraction, experiments were performed to evaluate whether CP behaves like an integral or peripheral membrane protein. The P200 fraction was resuspended in buffer containing 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, 1 mm PMSF, and 1% (v/v) protease inhibitor cocktail. This sample was treated separately with either 5 m NaCl, 1 m Na2CO3, 5 m urea, or 1% (v/v) Triton X-100, respectively, and incubated on a rotator for 60 min at 4°C. The resulting suspension was centrifuged for 60 min at 200,000g, providing pellet and solubilized fractions. Proteins contained in each fraction were precipitated overnight by the addition of TCA to a final concentration of 20% (v/v). Each protein sample was centrifuged at 15,000g, washed twice with cold acetone, and resuspended in an equal volume of buffer. Solubilized and pellet-associated proteins were separated on 12.5% (w/v) SDS-PAGE gels, transferred to nitrocellulose, and probed with anti-CP, anti-actin, anti-Sec12, and anti-VIPP-1 antibodies (Supplemental Table S1).
Protease Digestion Assay
To determine whether CP is present on the external or internal face of membrane fractions, protease K treatments were performed. The P200 fraction was dissolved in homogenization buffer without EGTA, PMSF, or protease inhibitor cocktail. This fraction was incubated with PK (Promega), at a final concentration of 70 μg/mL for 1.5 h at 4°C, under rotation. Protease digestion was terminated by adding PMSF to a final concentration of 5 mm and further incubation for 10 min at room temperature. Membranes were collected by centrifugation after protease treatment and resuspended in sample buffer. Control samples for protease digestion (without addition of PK) were treated in the same way as samples containing PK. Samples were separated on 12.5% (w/v) SDS-PAGE gels, transferred to nitrocellulose, and probed with anti-CP, anti-actin, and anti-VIPP1 (Supplemental Table S1).
Immunofluorescence and Confocal Microscopy
The subcellular localization of CP was analyzed by immunofluorescence microscopy with the freeze-fracture technique (Wasteneys et al., 1997; Szymanski et al., 1999; Qiu et al., 2002) using rosette leaves from Arabidopsis. Quantitative colocalization of CP and actin, or CP and specific compartment markers, was performed on material that was double-labeled with two antisera or by CP immunolabeling of Arabidopsis lines expressing a fluorescent fusion protein for cis-Golgi (Nelson et al., 2007). Three-week-old seedlings were fixed in 2% (v/v) formaldehyde and 0.5% (v/v) glutaraldehyde in PEM buffer (100 mm PIPES, 10 mm EGTA, and 4 mm MgCl2) for 1 h. Samples were washed with PEMT (100 mm PIPES, 10 mm EGTA, 5 mm MgCl2, and 0.1% (v/v) Triton X-100) three times for 10 min each. The excess buffer was absorbed from samples with filter paper placed on glass microscope slides, and covered with a second slide. The sandwich of two slides and sample was submerged into liquid nitrogen, allowed to freeze, and placed between two aluminum blocks previously cooled to −80°C. Gentle pressure was applied over the sample with the aluminum blocks. After separating the two glass slides, the freeze-fractured samples were incubated in permeabilization buffer (phosphate-buffered saline and 1% Triton X-100) for 2 h and then washed three times with PBST-G buffer (phosphate-buffered saline, 50 mm Gly, and 0.1% [v/v] Triton X-100). The samples were incubated overnight at 4°C with affinity-purified anti-AtCP (1:5 dilution) and anti-actin monoclonal antibody (JLA-20; 1:400 dilution). After washing, samples were incubated for 3 h at 37°C in fluorescein isothiocyanate-conjugated anti-rabbit sera (1:400; Sigma-Aldrich) and rhodamine-conjugated anti-mouse serum (1:400; Sigma-Aldrich) in PBST. Controls included the elimination of one primary antisera, or use of CPA or CPB preimmune serum from the same animals used to generate the affinity-purified antibody (Huang et al., 2003). Samples were mounted and imaged with a laser scanning confocal microscope (Bio-Rad 2100), using the excitation light from an argon ion (488 nm) and an He-Ne (543 nm) laser. Images of the cortical cytoplasm from the outer periclinal face of epidermal pavement cells were obtained by collecting 17 to 25 optical sections at 0.3-µm steps and generating a maximum intensity projection of the z-series stack.
Immunofluorescence Image Analysis
MetaMorph software (version 6) was used for image analysis and to quantify the extent of AtCP and actin colocalization. ROIs were selected from each image in order to maximize the area of cells analyzed. Individual images were thresholded, based on the antibody control images, and typical threshold values were 30 to 80. The extent of colocalization was determined between the two channels as previously described (Zhang et al., 2010).
The Pearson correlation coefficient (PCC) is a quantitative measurement that estimates the degree of overlap between fluorescence signals obtained in two channels (Barlow et al., 2010; Zinchuk et al., 2013). PCC values range from 1.0 (which indicates complete colocalization of two structures) to 0 (which indicates no significant correlation) to −1 (which indicates complete separation of two signals). Comparison with an experimental control condition provides a baseline for the experiment. Here, a PCC value was calculated for each antibody pairing from individual ROIs on z-series maximum intensity projections using ImageJ software (version 1.47) for the analysis (French et al., 2008). Background correction values were identical for all images (Costes et al., 2004). The PCCs were averaged, and the sem was calculated. Statistical analyses were performed using SPSS software (version 14.0; SPSS). A paired Student’s t test was used to compare mean PCC scores (McDonald and Dunn 2013).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT3G05520 (AAF64531) and AT1G71790 (EFH63646).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CP comigrates with a cis-Golgi fraction on sucrose density gradients.
Supplemental Table S1. Cytoskeletal and compartment markers antibodies used in differential centrifugation, Suc gradients, and immunofluorescence experiments.
Supplementary Material
Acknowledgments
We thank Sebastian Bednarek (University of Wisconsin, Madison), Félix Kessler (University of Neuchatel), Norbert Rolland (Commissariat à l’Energie Atomique), Natasha Raikhel (University of California, Riverside), Erik Nielsen (University of Michigan), Laurent Blanchoin (Commissariat à l’Energie Atomique), and Liwen Jiang (Chinese University of Hong Kong) for providing antisera, as well as Andreas Nebenführ (University of Tennessee, Knoxville) for the Arabidopsis line expressing mannosidase-YFP used in this study. The JLA-20 monoclonal anti-actin was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and maintained by the University of Iowa.
Glossary
- CP
capping protein
- PA
phosphatidic acid
- ABP
actin-binding protein
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- CG-MD
coarse-grained molecular dynamics
- ER
endoplasmic reticulum
- T-DNA
transfer DNA
- rCP
recombinant capping protein
- ROI
region of interest
- PCC
Pearson correlation coefficient
- YFP
yellow fluorescent protein
- DAG
d after germination
- PK
proteinase K
- H+-ATPase
proton-translocating adenosine triphosphate synthase
- Col-0
ecotype Columbia-0 of Arabidopsis
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This work was supported by the Physical Biosciences Program of the U.S. Department of Energy, Office of Basic Energy Sciences (contract no. DE–FG02–09ER15526 to C.J.S.). Work in the laboratory of D.B.S. was sponsored by the U.S. National Science Foundation (grant nos. MCB–0640872 and MCB–1121893).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Akin O, Mullins RD. (2008) Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Cooper JA. (1992) Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol 117: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Gattermeir DJ, Karpova TS, Cooper JA. (1992) Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J Cell Biol 119: 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. (2008) Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Huston E, Scotland G, Hodgkin M, Gall I, Peden AH, MacKenzie C, Houslay ES, Currie R, Pettitt TR, et al. (2002) TAPAS-1, a novel microdomain within the unique N-terminal region of the PDE4A1 cAMP-specific phosphodiesterase that allows rapid, Ca2+-triggered membrane association with selectivity for interaction with phosphatidic acid. J Biol Chem 277: 28298–28309 [DOI] [PubMed] [Google Scholar]
- Banno H, Chua NH. (2000) Characterization of the Arabidopsis formin-like protein AFH1 and its interacting protein. Plant Cell Physiol 41: 617–626 [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. (1997) Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol 114: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkalow K, Witke W, Kwiatkowski DJ, Hartwig JH. (1996) Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J Cell Biol 134: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AL, Macleod A, Noppen S, Sanderson J, Guérin CJ. (2010) Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of Pearson’s correlation coefficient. Microsc Microanal 16: 710–724 [DOI] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. (2008) Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem 283: 7135–7144 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Henty JL, Khurana P, Staiger CJ. (2010) Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr Opin Plant Biol 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, Sadot E, Yalovsky S. (2005) Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol Biol Cell 16: 1913–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. (1998) Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua N-H, Pantaloni D. (1997) Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol 136: 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F, Guérin C, von Witsch M, Blanchoin L, Staiger CJ. (2007) Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol Biol Cell 18: 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. (2004) Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. (2005) Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct 34: 119–151 [DOI] [PubMed] [Google Scholar]
- Cooper JA, Blum JD, Pollard TD. (1984) Acanthamoeba castellanii capping protein: properties, mechanism of action, immunologic cross-reactivity, and localization. J Cell Biol 99: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Sept D. (2008) New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol 267: 183–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. (2004) Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86: 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrcková F. (2000) Are plant formins integral membrane proteins? Genome Biol 1: RESEARCH001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Calcutt JR, Ingle EKS, Hawkins TJ, Chapman S, Richardson AC, Mentlak DA, Dixon MR, Cartwright F, Smertenko AP, et al. (2012) A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr Biol 22: 1595–1600 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Cvrcková F, Machesky LM, Mikitová V, Ketelaar T, Zársky V, Davies B, Hussey PJ. (2005) Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol 168: 529–540 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ, Davies B. (2002) Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci 7: 492–498 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM. (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA 94: 7679–7684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didry D, Carlier MF, Pantaloni D. (1998) Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J Biol Chem 273: 25602–25611 [DOI] [PubMed] [Google Scholar]
- DiNubile MJ, Cassimeris L, Joyce M, Zigmond SH. (1995) Actin filament barbed-end capping activity in neutrophil lysates: the role of capping protein-beta 2. Mol Biol Cell 6: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak BK, Franklin-Tong VE, Staiger CJ. (2004) Tansley review: the role of the actin cytoskeleton in plant cell signaling. New Phytol 163: 13–30 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Shao MR, Vaughn K, Bowling A, Facette M, Djakovic S, Clark L, Smith L. (2008) Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol Plant 1: 990–1006 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao F, He J, Huq E, Blancaflor EB. (2011) SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23: 3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy RJ, Han J, Condeelis JS. (1997) Capping protein terminates but does not initiate chemoattractant-induced actin assembly in Dictyostelium. J Cell Biol 139: 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Chelysheva LA, Lebris M, Jammes F, Marmagne A, De Almeida-Engler J, Lecomte P, Vaury C, Arkowitz RA, Abad P. (2004) Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher KL, Boldogh IR, Pon LA. (2005) A role for Jsn1p in recruiting the Arp2/3 complex to mitochondria in budding yeast. Mol Biol Cell 16: 5094–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserová J, Schwarzerová K, Petrásek J, Opatrný Z. (2006) ARP2 and ARP3 are localized to sites of actin filament nucleation in tobacco BY-2 cells. Protoplasma 227: 119–128 [DOI] [PubMed] [Google Scholar]
- Fraley TS, Tran TC, Corgan AM, Nash CA, Hao J, Critchley DR, Greenwood JA. (2003) Phosphoinositide binding inhibits α-actinin bundling activity. J Biol Chem 278: 24039–24045 [DOI] [PubMed] [Google Scholar]
- Frank CA, Kennedy MJ, Goold CP, Marek KW, Davis GW. (2006) Mechanisms underlying the rapid induction and sustained expression of synaptic homeostasis. Neuron 52: 663–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Freyberg Z, Siddhanta A, Shields D. (2003) “Slip, sliding away”: phospholipase D and the Golgi apparatus. Trends Cell Biol 13: 540–546 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Kovar DR, Staiger CJ. (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MC, Cooper JA. (1999) Vertebrate isoforms of actin capping protein β have distinct functions In vivo. J Cell Biol 147: 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins TJ, Deeks MJ, Wang P, Hussey PJ. (2014) The evolution of the actin binding NET superfamily. Front Plant Sci 5: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Li J, Blanchoin L, Staiger CJ. (2013) Actin dynamics in the cortical array of plant cells. Curr Opin Plant Biol 16: 678–687 [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. (2001) Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70: 649–676 [DOI] [PubMed] [Google Scholar]
- Huang S, Blanchoin L, Kovar DR, Staiger CJ. (2003) Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J Biol Chem 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S, Gao L, Blanchoin L, Staiger CJ. (2006) Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol Biol Cell 17: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Fitz Gerald JN, Guérin C, Robert H, Sørensen MB, Van Damme D, Geelen D, Blanchoin L, Berger F. (2005) Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat Cell Biol 7: 374–380 [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. (2005) Phospholipase D: a lipid centric review. Cell Mol Life Sci 62: 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. (2005) A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123: 305–320 [DOI] [PubMed] [Google Scholar]
- Kim K, McCully ME, Bhattacharya N, Butler B, Sept D, Cooper JA. (2007) Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: implications for how capping protein binds the actin filament. J Biol Chem 282: 5871–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Yamashita A, Wear MA, Maéda Y, Cooper JA. (2004) Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J Cell Biol 164: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni SO, Zakharova T, Mallery EL, Le J, El-Assal Sel-D, Szymanski DB. (2009) The association of the Arabidopsis actin-related protein2/3 complex with cell membranes is linked to its assembly status but not its activation. Plant Physiol 151: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. (2005) Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell 16: 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. (1985) Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature 314: 472–474 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S. (2007) A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol 17: 947–952 [DOI] [PubMed] [Google Scholar]
- Lee S, Park J, Lee Y. (2003) Phosphatidic acid induces actin polymerization by activating protein kinases in soybean cells. Mol Cells 15: 313–319 [PubMed] [Google Scholar]
- Li J, Henty-Ridilla JL, Huang S, Wang X, Blanchoin L, Staiger CJ. (2012) Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 24: 3742–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Staiger BH, Henty-Ridilla JL, Abu-Abied M, Sadot E, Blanchoin L, Staiger CJ. (2014) The availability of filament ends modulates actin stochastic dynamics in live plant cells. Mol Biol Cell 25: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hong Y, Wang X. (2009) Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 1791: 927–935 [DOI] [PubMed] [Google Scholar]
- Liscovitch M, Czarny M, Fiucci G, Tang X. (2000) Phospholipase D: molecular and cell biology of a novel gene family. Biochem J 345: 401–415 [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Maisch J, Fiserová J, Fischer L, Nick P. (2009) Tobacco Arp3 is localized to actin-nucleating sites in vivo. J Exp Bot 60: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Gayral P, Hawes C, Runions J. (2011) Building bridges: formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J 66: 354–365 [DOI] [PubMed] [Google Scholar]
- McDonald JH, Dunn KW. (2013) Statistical tests for measures of colocalization in biological microscopy. J Microsc 252: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerschaert K, De Corte V, De Ville Y, Vandekerckhove J, Gettemans J. (1998) Gelsolin and functionally similar actin-binding proteins are regulated by lysophosphatidic acid. EMBO J 17: 5923–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. (2006) Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol 175: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro D, Liu Q, Lisboa S, Scherer GEF, Quader H, Malhó R. (2005a) Phosphoinositides and phosphatidic acid regulate pollen tube growth and reorientation through modulation of [Ca2+]c and membrane secretion. J Exp Bot 56: 1665–1674 [DOI] [PubMed] [Google Scholar]
- Nakano K, Mabuchi I. (2006) Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol Biol Cell 17: 1933–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA. (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol 121: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T. (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Oliviusson P, Heinzerling O, Hillmer S, Hinz G, Tse YC, Jiang L, Robinson DG. (2006) Plant retromer, localized to the prevacuolar compartment and microvesicles in Arabidopsis, may interact with vacuolar sorting receptors. Plant Cell 18: 1239–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Klocko AL, Fowler JE, Dolja VV. (2012) Arabidopsis Myosin XI-K localizes to the motile endomembrane vesicles associated with F-actin. Front Plant Sci 3: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Morgun EA, Kurth EG, Makarova KS, Koonin EV, Dolja VV. (2013) Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25: 3022–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol 146: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskot R, Li J, Zárský V, Potocký M, Staiger CJ. (2013) Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci 18: 496–504 [DOI] [PubMed] [Google Scholar]
- Pleskot R, Pejchar P, Žárský V, Staiger CJ, Potocký M. (2012) Structural insights into the inhibition of actin-capping protein by interactions with phosphatidic acid and phosphatidylinositol (4,5)-bisphosphate. PLoS Comput Biol 8: e1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskot R, Potocký M, Pejchar P, Linek J, Bezvoda R, Martinec J, Valentová O, Novotná Z, Zárský V. (2010) Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J 62: 494–507 [DOI] [PubMed] [Google Scholar]
- Pollard TD. (1984) Polymerization of ADP-actin. J Cell Biol 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29: 545–576 [DOI] [PubMed] [Google Scholar]
- Potocký M, Eliás M, Profotová B, Novotná Z, Valentová O, Zárský V. (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130 [DOI] [PubMed] [Google Scholar]
- Potocký M, Pleskot R, Pejchar P, Vitale N, Kost B, Zárský V. (2014) Live-cell imaging of phosphatidic acid dynamics in pollen tubes visualized by Spo20p-derived biosensor. New Phytol 203: 483–494 [DOI] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV. (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105: 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB. (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Watkins SC, Romero G. (2000) The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem 275: 23911–23918 [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P. (2010) Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev 90: 259–289 [DOI] [PubMed] [Google Scholar]
- Samaj J, Baluska F, Menzel D. (2004) New signalling molecules regulating root hair tip growth. Trends Plant Sci 9: 217–220 [DOI] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre B, Steele C, Hawes C. (1996) Golgi membrane dynamics are cytoskeleton dependent: a study on Golgi stack movement induced by brefeldin A. Protoplasma 191: 21–33 [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. (1996) Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol 135: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Korshunova YO, Schroer TA, Cooper JA. (1994) Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. J Cell Biol 127: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Welch MD, Machesky LM, Bridgman PC, Meyer SM, Cooper JA. (1998) Visualization and molecular analysis of actin assembly in living cells. J Cell Biol 143: 1919–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D, Forge A, Munro PMG, Bailly M. (2006) Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil Cytoskeleton 63: 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizonenko GI, Karpova TS, Gattermeir DJ, Cooper JA. (1996) Mutational analysis of capping protein function in Saccharomyces cerevisiae. Mol Biol Cell 7: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem 246: 4866–4871 [PubMed] [Google Scholar]
- Staiger CJ, Blanchoin L. (2006) Actin dynamics: old friends with new stories. Curr Opin Plant Biol 9: 554–562 [DOI] [PubMed] [Google Scholar]
- Strasser GA, Rahim NA, VanderWaal KE, Gertler FB, Lanier LM. (2004) Arp2/3 is a negative regulator of growth cone translocation. Neuron 43: 81–94 [DOI] [PubMed] [Google Scholar]
- Szymanski DB. (2005) Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr Opin Plant Biol 8: 103–112 [DOI] [PubMed] [Google Scholar]
- Szymanski DB. (2009) Plant cells taking shape: new insights into cytoplasmic control. Curr Opin Plant Biol 12: 735–744 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM. (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. (2005) KATAMARI1/MURUS3 is a novel golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C, Munnik T. (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10: 368–375 [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T. (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot 62: 2349–2361 [DOI] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C, Cummins I, Deeks MJ, Sparkes I, Hawes C, Hussey PJ. (2014) The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr Biol 24: 1397–1405 [DOI] [PubMed] [Google Scholar]
- Wang X. (2005) Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol 139: 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO, Willingale-Theune J, Menzel D. (1997) Freeze shattering: a simple and effective method for permeabilizing higher plant cell walls. J Microsc 188: 51–61 [DOI] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. (1997) The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. (2002) Cellular control of actin nucleation. Annu Rev Cell Dev Biol 18: 247–288 [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. (2005) Counting cytokinesis proteins globally and locally in fission yeast. Science 310: 310–314 [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Maéda K, Maéda Y. (2003) Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J 22: 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kotchoni SO, Samuels AL, Szymanski DB. (2010) SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr Biol 20: 2144–2149 [DOI] [PubMed] [Google Scholar]
- Zhang C, Mallery E, Reagan S, Boyko VP, Kotchoni SO, Szymanski DB. (2013a) The endoplasmic reticulum is a reservoir for WAVE/SCAR regulatory complex signaling in the Arabidopsis leaf. Plant Physiol 162: 689–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Mallery EL, Szymanski DB. (2013b) ARP2/3 localization in Arabidopsis leaf pavement cells: a diversity of intracellular pools and cytoskeletal interactions. Front Plant Sci 4: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X. (2004) Phospholipase D α 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J. (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V, Wu Y, Grossenbacher-Zinchuk O. (2013) Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci Rep 3: 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.