Methionine metabolism in Arabidopsis seeds is strongly associated with stress-related metabolites and transcripts, suggesting that its content leads to desiccation stress during seed development.
Abstract
With an aim to elucidate novel metabolic and transcriptional interactions associated with methionine (Met) metabolism in seeds, we have produced transgenic Arabidopsis (Arabidopsis thaliana) seeds expressing a feedback-insensitive form of CYSTATHIONINE-γ-SYNTHASE, a key enzyme of Met synthesis. Metabolic profiling of these seeds revealed that, in addition to higher levels of Met, the levels of many other amino acids were elevated. The most pronounced changes were the higher levels of stress-related amino acids (isoleucine, leucine, valine, and proline), sugars, intermediates of the tricarboxylic acid cycle, and polyamines and lower levels of polyols, cysteine, and glutathione. These changes reflect stress responses and an altered mitochondrial energy metabolism. The transgenic seeds also had higher contents of total proteins and starch but lower water contents. In accordance with the metabolic profiles, microarray analysis identified a strong induction of genes involved in defense mechanisms against osmotic and drought conditions, including those mediated by the signaling cascades of ethylene and abscisic acid. These changes imply that stronger desiccation processes occur during seed development. The expression levels of transcripts controlling the levels of Met, sugars, and tricarboxylic acid cycle metabolites were also significantly elevated. Germination assays showed that the transgenic seeds had higher germination rates under salt and osmotic stresses and in the presence of ethylene substrate and abscisic acid. However, under oxidative conditions, the transgenic seeds displayed much lower germination rates. Altogether, the data provide new insights on the factors regulating Met metabolism in Arabidopsis seeds and on the mechanisms by which elevated Met levels affect seed composition and behavior.
Developing seeds can serve as an excellent system for studying metabolic regulation, including both metabolic and transcriptional parameters, since during their developmental stages seeds induce a massive synthesis of reserve compounds, including storage proteins, starch, and oil (Angelovici et al., 2010; Fait et al., 2011). Moreover, during their development and maturation, the seed’s metabolism is associated with temporally distinct metabolic switches that alter their metabolic and transcriptomic profiles (Fait et al., 2006; Angelovici et al., 2009). Seeds can also be used as an attractive target for manipulating the metabolism of amino acids, since they are important nutritional sources and their nutritional quality suffers from low levels of several essential amino acids (Angelovici et al., 2010; Gu et al., 2010).
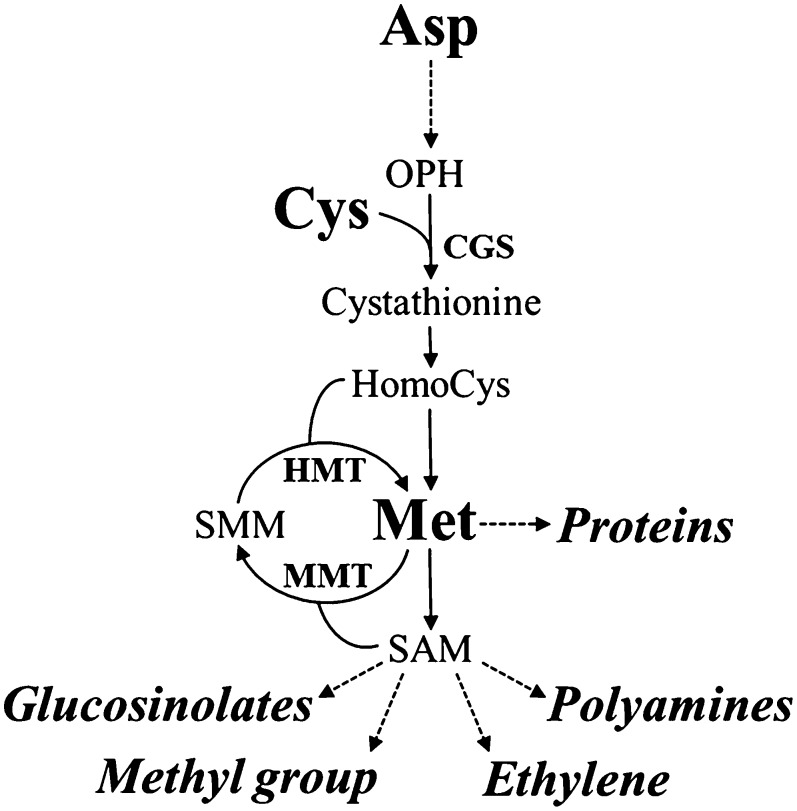
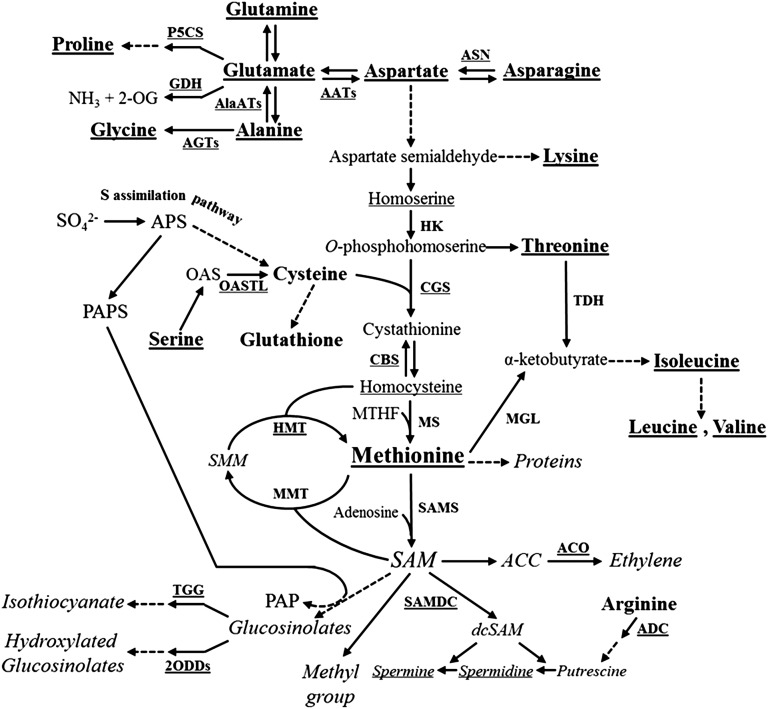
In these contexts, the essential sulfur-containing amino acid Met is an interesting candidate for such manipulations, since its levels are relatively low, limiting the nutritional values of seeds (Hesse et al., 2004; Amir et al., 2012; Galili and Amir, 2013). In addition, the complex Met metabolism presents an attractive system for studying the regulation of diverse biochemical pathways and Met interactions with other metabolites. Met is a fundamental metabolite in plant cells, since, in addition to its role as a protein constituent and its central role in the initiation of mRNA translation, it controls the levels of several essential metabolites through its important derivative, S-adenosyl-methionine (SAM). These include ethylene (ET), polyamines, and glucosinolates (Fig. 1). SAM, as a primary methyl group donor, also regulates important processes such as chlorophyll and cell wall synthesis (Roje, 2006). For these reasons, many studies were performed to assess the factors that regulate Met metabolism and its accumulation (Hesse et al., 2004; Amir et al., 2012; Galili and Amir, 2013). In vegetative tissues, Met is regulated mainly by the expression level of its first committed enzyme, CGS, which combines the carbon/amino skeleton derived from Asp with the sulfur moiety derived from Cys (Kim et al., 2002; Fig. 1). Recent studies performed in legumes and tobacco (Nicotiana tabacum) seeds show that seed-specific expression of feedback-insensitive mutated forms of Arabidopsis (Arabidopsis thaliana) CGS (AtCGS) leads to significantly higher levels of soluble Met (Hanafy et al., 2013; Matityahu et al., 2013; Song et al., 2013), indicating that Met can be synthesized de novo in these seeds via the Asp family pathway through CGS activity in a similar manner to that in vegetative tissues. However, studies performed on Arabidopsis and wheat (Triticum aestivum) seeds suggest the existence of an alternative pathway of Met synthesis in seeds, in which Met synthesized in vegetative tissues is converted to SMM, which is then transported through the phloem into the developing seeds and reconverted back to soluble Met by the activity of HMT (Bourgis et al., 1999; Lee et al., 2008; Fig. 1). Thus, the relative contribution of AtCGS to Met synthesis in Arabidopsis seeds has yet to be established.
Figure 1.
Met synthesis in higher plants. Key enzymes involved in Met synthesis through the Asp family and SMM pathways are presented. Main catabolic products of Met are presented in italics. Full arrows represent one metabolic step while dashed arrows represent several metabolic steps. OPH, O-Phosphohomoserine; SMM, S-methylmethionine; CGS, cystathionine-γ-synthase; HMT, homocysteine S-methyltransferase; MMT, SAM:Met S-methyltransferase.
The aim of this research was to study the regulatory roles of AtCGS in Arabidopsis seeds and to elucidate the effects of seed-specific expression of the feedback-insensitive mutated form of Arabidopsis CGS (AtD-CGS) on the genome-wide gene expression programs and primary metabolism. Since Met plays important metabolic roles in seeds, this manipulation also could shed light on how seeds tolerate the metabolic perturbations that occur in their central core pathways.
RESULTS
Creating Transgenic Arabidopsis Seeds Expressing AtD-CGS
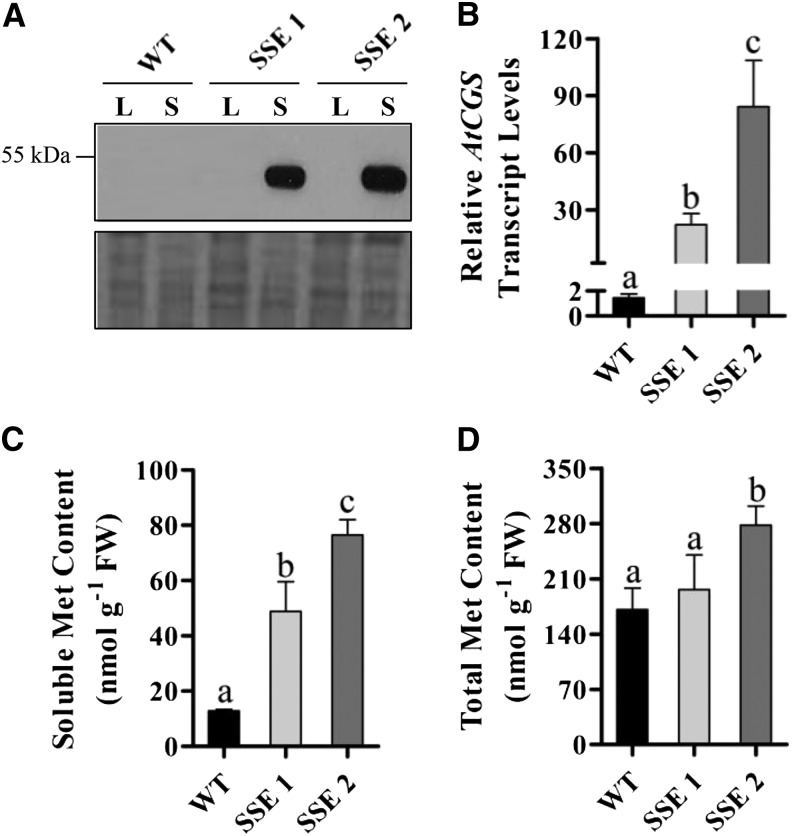
In order to assess the role of AtCGS in Met synthesis in Arabidopsis seeds and to elucidate novel interactions of Met metabolism with primary metabolites and gene expression programs in seeds, we generated transgenic Arabidopsis seeds expressing AtD-CGS (Hacham et al., 2006) under the control of the phaseolin promoter (Supplemental Fig. S1). This promoter is the most abundant seed storage protein in the common bean (Phaseolus vulgaris) and is stringently turned off during all vegetative stages of plant development (Sundaram et al., 2013). It was also demonstrated that this promoter is induced constitutively during the maturation and desiccation stages of seed development (Fait et al., 2011). Seeds from 20 kanamycin-resistant plants were screened by immunoblot, and the two transgenic lines, SSE 1 and SSE 2, exhibiting the highest expression levels of the AtD-CGS transgene (Fig. 2A) were chosen. Quantitative real-time (qRT)-PCR analysis showed that the expression levels of AtCGS in SSE 1 and SSE 2 genotypes increased 15- and 58-fold, respectively, compared with wild-type seeds (Fig. 2B). Similar morphology and growth rates were observed during the life cycle of the wild-type and SSE genotypes (Supplemental Fig. S2). Seeds from T3 homozygous plants were used for further analysis.
Figure 2.
Generation of transgenic Arabidopsis seeds with altered Met levels. A, Western-blot analysis of protein extracts from leaves (L) and mature dry seeds (S) of wild-type (WT) and SSE genotypes using antibodies against the hemagglutinin epitope tag. B, qRT-PCR analysis of AtCGS transcript levels in mature dry seeds of wild-type and SSE genotypes. Data shown are means ± se of three replicates. C, Soluble Met contents in mature dry seeds (fresh weight [FW]) of wild-type and SSE genotypes following GC-MS analysis. Data shown are means ± se of five replicates. D, Total Met contents in mature dry seeds (fresh weight) of wild-type and SSE genotypes following protein hydrolysis and GC-MS analysis. Data shown are means ± se of eight replicates. Significance was calculated using the two-way ANOVA test of P < 0.05 and identified by different letters.
Transgenic Seeds Expressing AtD-CGS Accumulate Higher Levels of Met and Other Amino Acids
Soluble amino acid analysis of wild-type and transgenic mature dry seeds indicated that Met levels were significantly increased 4- and 6-fold in SSE 1 and SSE 2, respectively (Fig. 2C). In accordance with the elevation in soluble Met contents, total Met levels measured after protein hydrolysis were elevated slightly in SSE 1 and significantly in SSE 2, up to 1.6-fold (Fig. 2D).
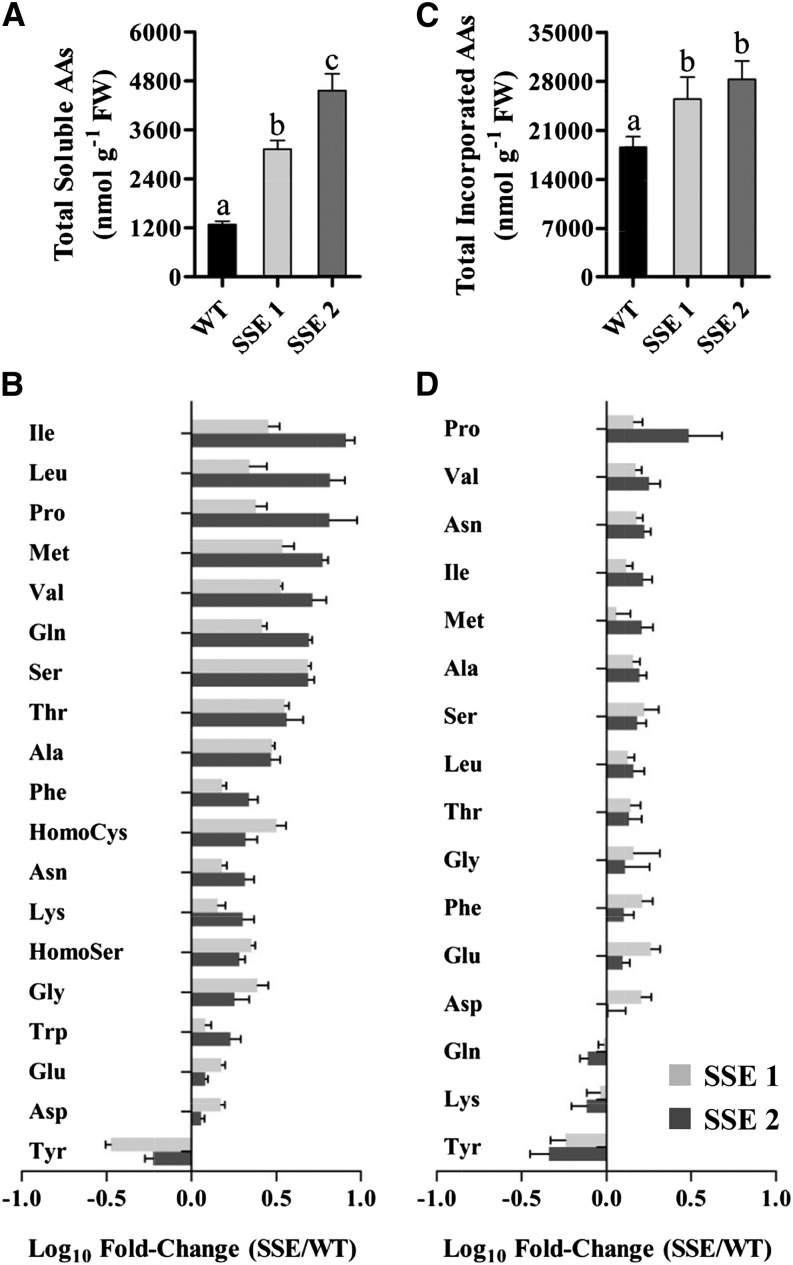
The elevations of soluble Met content in both transgenic lines were associated with significant increases in total soluble amino acid contents, up to 2.4- and 3.6-fold, respectively (Fig. 3A; Supplemental Table S1). Notably, the highest increases were detected in the levels of the stress-associated branched-chain amino acids (BCAAs) Ile, Leu, Val (up to 8.2-, 6.6-, and 5.2-fold, respectively), and Pro (up to 6.5-fold; Fig. 3B), whose levels were also found to be correlated with that of Met (Supplemental Fig. S3). These four amino acids are known to be involved in plant defense mechanisms against abiotic stresses and to play active roles in drought, salt, and osmotic stresses (Joshi et al., 2010). BCAAs are related to Met metabolism, since they can be produced from Met via METHIONINE-γ-LYASE (MGL), a catabolic enzyme of Met (Joshi et al., 2010). However, qRT-PCR analysis revealed that AtMGL expression levels were not altered significantly in the transgenic seeds (Supplemental Fig. S4). In addition, the expression level of Thr dehydratase, the main enzyme of BCAA synthesis (Joshi et al., 2010), did not exhibit any altered expression. Tyr is the only amino acid that exhibited reduced contents in its soluble and protein-bound forms, up to 3- and 2-fold reductions, respectively (Fig. 3, B and D). Phe, which exhibited significant elevations in both SSE lines, shares its biosynthesis pathway with Tyr (Maeda and Dudareva, 2012); thus, elevations in Phe might explain the reductions in Tyr contents. However, further studies are required to reveal the metabolic link between the aromatic amino acids and Met.
Figure 3.
GC-MS-based analysis of soluble and protein-incorporated amino acids. A, Total soluble amino acid contents in mature dry seeds (fresh weight [FW]) of wild-type (WT) and SSE genotypes. B, Changes in the contents of individual soluble amino acids in mature dry seeds of both SSE genotypes. Data shown in A and B are means ± se of five replicates. C, Total protein-incorporated amino acid contents in mature dry seeds (fresh weight) of wild-type and SSE genotypes following protein hydrolysis. D, Changes in the contents of individual protein-incorporated amino acids in mature dry seeds of both SSE genotypes. Data shown in C and D are means ± se of eight replicates. Significance was calculated using the two-way ANOVA test of P < 0.05 and identified by different letters. The bars in B and D represent the log10-fold change in amino acid content in seeds from both SSE genotypes compared to wild-type seeds. The bars at right (positive) indicate increased content, and the bars on the left (negative) indicate decreased content.
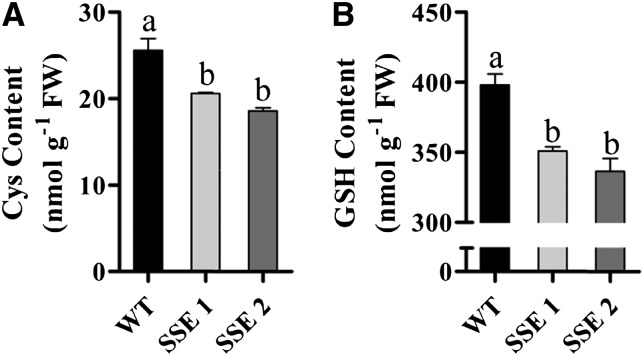
The level of Cys, the precursor for Met synthesis, decreased significantly in SSE 1 and SSE 2 by 19% and 27%, respectively (Fig. 4A). Since Cys is also used as the precursor for the synthesis of glutathione (GSH) and the level of GSH is limited by Cys content (Kopriva, 2006), the level of GSH was also determined. Similarly to Cys, the levels of GSH were reduced significantly by 12% in SSE 1 and by 16% in SSE 2 (Fig. 4B). Lower GSH contents may lead to oxidative stress. To examine this possibility further, we measured the level of malondialdehyde (MDA), which results from lipid peroxidation of polyunsaturated fatty acids and is used as a metabolic marker for oxidative stress (Bai et al., 2012). The results showed a significant increase in MDA contents in both SSE seeds (Supplemental Fig. S5), indicating that these seeds are exposed to greater oxidative conditions.
Figure 4.
HPLC analysis of Cys and GSH contents in mature dry seeds. A, Total Cys contents (fresh weight [FW]) of wild-type (WT) and SSE genotypes. B, Total GSH contents (fresh weight) of wild-type and SSE genotypes. Data shown are means ± se of four replicates. Significance was calculated using the two-way ANOVA test of P < 0.05 and identified by different letters.
Next, we measured the levels of protein-bound amino acids after protein hydrolysis. Figure 3C and Supplemental Table S1 show that the levels of protein-bound amino acids in seeds from SSE 1 and SSE 2 genotypes increased significantly up to 25% and 40%, respectively. In general, amino acids that exhibited high soluble contents also exhibited high total levels (Fig. 3D).
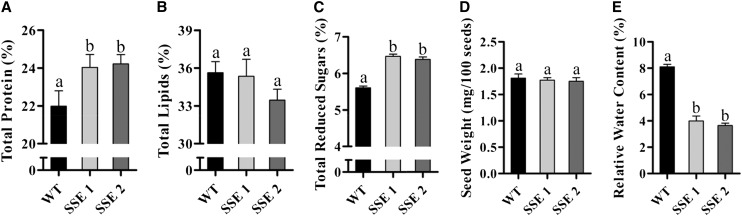
Transgenic Seeds Accumulate More Protein and Starch While Their Water Contents Are Reduced
The higher levels of amino acids in both SSE genotypes (Fig. 3) indicate that higher protein contents exist in these seeds. Indeed, Kjeldahl analysis revealed that seeds from both SSE genotypes exhibited an increase of 10% in their total protein contents compared with wild-type seeds (Fig. 5A). Higher protein levels may affect the levels of lipids and starch, as reported previously (Hernández-Sebastià et al., 2005; Matityahu et al., 2013). Measurement of the total lipids in SSE seeds revealed a slight decrease only in the SSE 2 line, although not statistically significant (Fig. 5B). However, the levels of total reducing sugars following carbohydrate hydrolysis that results mostly from starch hydrolysis were found to increase by 15% in both SSE genotypes compared with wild-type seeds (Fig. 5C).
Figure 5.
Chemical and physical characteristics of seeds from wild-type (WT) and SSE genotypes. A, Total protein contents according to the Kjeldahl method. B, Total lipid contents according to the Soxhlet method. C, Total reduced sugar contents after carbohydrate proteolysis according to the method of Sumner (1924). D, Weight of the seeds. E, Relative water contents of seeds. Data shown are means ± se of four replicates. Significance was calculated using the two-way ANOVA test of P < 0.05 and identified by different letters.
The higher levels of protein and starch that were observed in SSE seeds, in addition to the observation that these seeds exhibit similar morphology and dimensions to the wild-type seeds (Supplemental Fig. S2), suggest that the total seed weight or relative water content were altered. No significant differences in the weight of the transgenic seeds were detected (Fig. 5D); however, their relative water contents were decreased significantly by more than 2-fold compared with wild-type seeds (Fig. 5E). These results imply that stronger desiccation may occur during the late stages of seed maturation when the phaseolin promoter expresses the AtD-CGS.
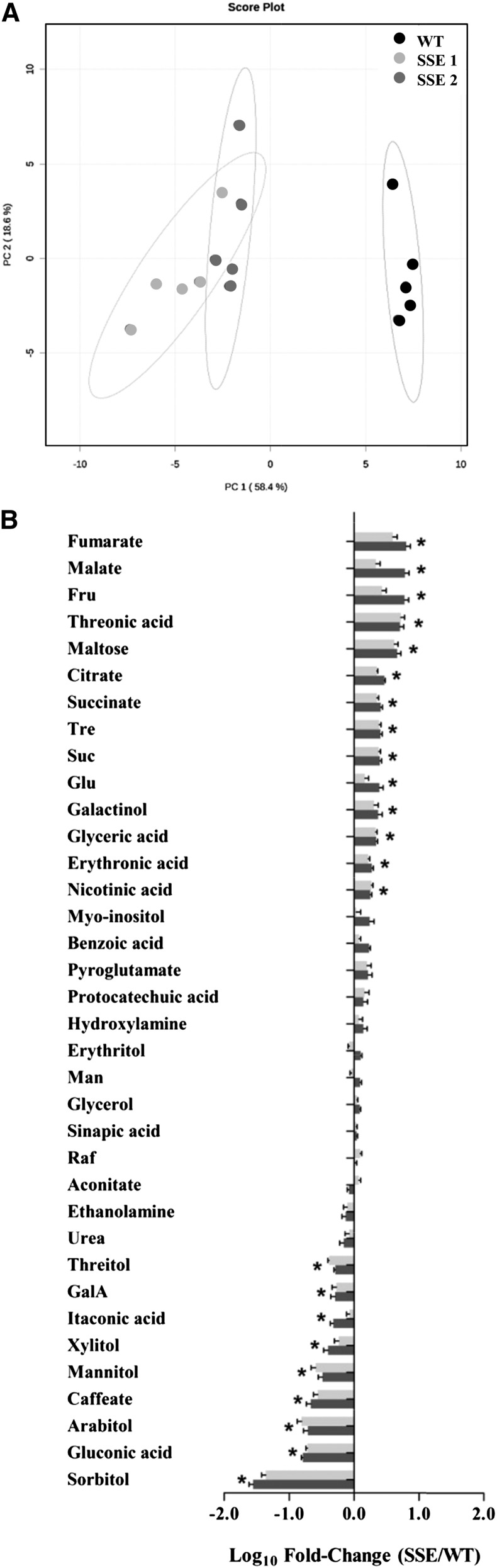
Global Analysis of Metabolic Profiles Using Gas Chromatography-Mass Spectrometry Reveals Distinct Stress-Associated Metabolic Responses in SSE Genotypes
The mode of interaction between metabolites can provide important insights into the modular behavior of biochemical processes and their regulation. To reveal these interactions, global metabolic profile analysis of maturing seeds of wild-type and SSE genotypes was performed using an established gas chromatography-mass spectrometry (GC-MS) protocol (Fait et al., 2006; Matityahu et al., 2013).
After analyzing the global trends of changes of metabolites in the seeds by principal component analysis, the wild-type and SSE genotypes exhibited clear distinguishable differences, implying relatively strong effects of the genetic manipulation on the primary metabolome of SSE seeds (Fig. 6A). The effect of the manipulation on the levels of individual metabolites of the central metabolism was investigated to show changes that are associated mostly with stress metabolic responses. These include (1) major increases in the abundance of intermediates of the tricarboxylic acid cycle, such as fumarate, malate, succinate, and citrate (6.4-, 6.1-, 4-, and 3-fold, respectively; Fig. 6B); (2) significant alterations in the metabolism of carbohydrates, including the accumulation of major sugars such as Fru, maltose, trehalose (Tre), Suc, Glu, and galactinol (6-, 4.6-, 2.6-, 2.5-, 2.5-, and 2.3-fold, respectively) and sugar acids such as threonic, glyceric, and erythronic acids (5.3-, 2.2-, and 1.9-fold, respectively; Fig. 6B); (3) dramatic reductions in the levels of sugar alcohols such as sorbitol, arabitol, mannitol, xylitol, and threitol (35.5-, 6.4-, 3.9-, 2.5-, and 2.4-fold, respectively) and in gluconic acid and GalA (6.2- and 2-fold, respectively; Fig. 6B); and (4) relatively minor alterations in the levels of organic acids, where only caffeate and itaconic acid exhibited significant decreases (4.6- and 2-fold, respectively), and in nicotinic acid levels, which were elevated up to 1.9-fold (Fig. 6B).
Figure 6.
Primary metabolome changes in transgenic seeds. A, Principal component analysis of GC-MS metabolite profiling data of wild-type (WT) and SSE genotypes. Variance explained by each component is indicated in brackets; principal component analysis was based in 56 annotated metabolites. B, Changes in the contents of individual metabolites in mature dry seeds of both SSE genotypes. Data shown in A and B are means ± se of five replicates. The bars in B represent the log10-fold change in metabolites contents in seeds from both SSE genotypes compared to wild-type seeds. The bars facing right (positive) indicate increased content, and the bars facing left (negative) indicate decreased content. Significance was calculated using the two-way ANOVA test of P < 0.05 and identified by asterisks.
Functional Classification of Altered Transcripts
The results show that the levels of stress-related amino acids, sugars, and metabolites of the tricarboxylic acid cycle were increased in SSE genotypes. This, in addition to the low water contents found in these seeds, suggests that SSE seeds undergo a situation that resembles osmotic or desiccation stresses. To assess this assumption further, a microarray analysis using Affymetrix ATH1 GeneChips was used (see “Materials and Methods”). ANOVA of the microarray results indicated that the expression of approximately 2,000 genes (1,103 up-regulated and 877 down-regulated) was changed significantly by at least 1.5-fold between the wild-type and SSE genotypes, with a false discovery rate of 0.05 (Supplemental Tables S2 and S3).
The genes were classified according to their physiological functions using the enrichment and functional annotation clustering tools embedded in PageMan and DAVID bioinformatic softwares (Usadel et al., 2006; Xia et al., 2009). (Full Arabidopsis Genome Initiative set lists and calculated ratios are presented in Supplemental Tables S2 and S3.) Enrichment analysis of the down-regulated transcripts did not show clear patterns except for a cluster of transcripts participating in ribosomal protein synthesis (Supplemental Table S3). Moreover, the fold changes observed in this group showed much lower values than those observed in the up-regulated group (Supplemental Tables S2 and S3). For these two reasons, we focused mainly on the list of up-regulated transcripts that shows three clusters of genes, which exhibited high fold changes in SSE genotypes compared with the wild type, as described below.
Microarray Analysis Reveals the Induction of Stress-Associated Transcriptional Phenotypes in SSE Seeds
The first cluster consists of highly induced transcripts corresponding to genes involved in stress-induced responses and is divided into three subclusters (Table I). The first subcluster (1.1 in Table I) is composed of stress-induced transcripts whose function in the reduction and/or repair of drought, osmotic, and salt stresses induced damage. This subcluster contains GLYCINE-RICH PROTEIN3 (At2g05520) displaying the highest induction (Table I), which was found previously to be up-regulated in response to drought, ET, and abscisic acid (ABA). The list also includes three genes responsive to dehydration, RD20 (At2g33380), RD21 (At1g47128), and RD19 (At4g39090); two genes having an early response to dehydration, ERD14 (At1g76180) and ERD10 (At1g20450); five senescence-associated genes, SAG21 (At4g02380), SAG14 (At5g20230), SAG2 (At5g60360), SAG29 (At5g13170), and SAG24 (At1g66580); three cold-regulated proteins, COR47 (At1g20440), COR15A (At2g42540), and COR413-PLASMA MEMBRANE1 (At2g15970); and the cold- and ABA-regulated gene of KINASE1 (At5g15960; Table I). All the above transcripts are well-characterized genes previously reported to participate in cellular responses to water deprivation and salinity in Arabidopsis (Kreps et al., 2002; Seki et al., 2002). Additionally, four lipid transfer proteins, LTP4 (At5g59310), LTP3 (At5g59320), LTP6 (At3g08770), and LTP1 (At2g38540), that showed increased expression, are known as cell wall-localized small proteins that participate in responses to water deprivation and salt stress (Arondel et al., 2000; Table I). The expression levels of FATTY ACID DESATURASE2 (At3g12120), whose function is essential for the proper maintenance of low cytosolic Na+ concentrations for salt tolerance during seed germination (Zhang et al., 2012), also increased, as well as the vegetative storage protein VSP2 (At5g24770), whose degradation serves as nitrogen and carbon sources that are available immediately for seed development and whose transcript is induced by osmotic and nutritional stresses (Liu et al., 2005; Table I). Finally, two HOMEOBOX proteins, HB12 (At3g61890) and HB7 (At2g46680), were up-regulated significantly (Table I). These two genes were reported previously to encode potential regulators of growth in response to water deficit (Olsson et al., 2004).
Table I. Gene cluster 1: highly expressed transcripts in SSE genotypes associated with stress, redox and detoxification, and cell wall modifications.
| Gene Symbola | Gene Namea | Gene Descriptiona | Pb | Fold Changec |
|---|---|---|---|---|
| Cluster 1.1: stress-induced transcriptsd | ||||
| At2g05520 | GRP3 | GLY-RICH PROTEIN3 | 2.89E-02 | 35.51 |
| At2g33380 | RD20 | RESPONSIVE TO DEHYDRATION20 | 2.02E-02 | 28.50 |
| At5g59310 | LTP4 | LIPID TRANSFER PROTEIN4 | 4.36E-02 | 28.28 |
| At5g24770 | VSP2 | VEGETATIVE STORAGE PROTEIN2 | 3.76E-02 | 23.31 |
| At1g47128 | RD21 | RESPONSIVE TO DEHYDRATION21 | 1.26E-02 | 19.80 |
| At5g59320 | LTP3 | LIPID TRANSFER PROTEIN3 | 4.41E-02 | 18.57 |
| At3g08770 | LTP6 | LIPID TRANSFER PROTEIN6 | 2.14E-04 | 17.69 |
| At4g02380 | SAG21 | SENESCENCE-ASSOCIATED GENE21 | 1.60E-02 | 14.39 |
| At5g15960 | KIN1 | KINASE1 | 4.71E-02 | 14.12 |
| At1g76180 | ERD14 | EARLY RESPONSE TO DEHYDRATION14 | 1.64E-02 | 11.15 |
| At1g20440 | COR47 | COLD REGULATED47 | 4.81E-02 | 9.88 |
| At2g38540 | LTP1 | LIPID TRANSFER PROTEIN1 | 4.56E-02 | 9.55 |
| At5g20230 | SAG14 | SENESCENCE-ASSOCIATED GENE14 | 4.45E-03 | 8.42 |
| At4g39090 | RD19 | RESPONSIVE TO DEHYDRATION19 | 2.19E-02 | 8.24 |
| At1g20450 | ERD10 | EARLY RESPONSE TO DEHYDRATION10 | 3.27E-02 | 5.90 |
| At3g12120 | FAD2 | FATTY ACID DEHYDROGENASE2 | 4.54E-05 | 5.65 |
| At5g60360 | SAG2 | SENESCENCE-ASSOCIATED GENE2 | 3.76E-02 | 5.48 |
| At3g61890 | HB12 | HOMEOBOX12 | 9.38E-03 | 5.26 |
| At2g46680 | HB7 | HOMEOBOX7 | 8.30E-03 | 4.82 |
| At2g42540 | COR15A | COLD REGULATED15A | 1.97E-02 | 3.81 |
| At2g15970 | COR413-PM1 | COLD REGULATED413 PLASMA MEMBRANE1 | 4.32E-02 | 3.21 |
| At5g13170 | SAG29 | SENESCENCE-ASSOCIATED GENE29 | 4.69E-02 | 2.02 |
| At1g66580 | SAG24 | SENESCENCE-ASSOCIATED GENE24 | 2.90E-02 | 1.68 |
| Cluster 1.2: redox- and detoxification-related transcriptsd | ||||
| At3g49110 | PRX33 | PEROXIDASE33 | 3.10E-02 | 33.97 |
| At3g15353 | MT3 | METALLOTHIONEIN3 | 3.56E-02 | 6.65 |
| At2g31570 | GPX2 | GLUTATHIONE PEROXIDASE2 | 3.25E-02 | 6.15 |
| At1g07890 | APX1 | ASCORBATE PEROXIDASE1 | 3.20E-02 | 5.73 |
| At1g53580 | GLXII-3 | GLYOXALASE II3 | 4.70E-02 | 5.61 |
| At1g07600 | MT1A | METALLOTHIONEIN1A | 1.40E-02 | 5.59 |
| At2g29940 | PDR3 | PLEOTROPIC DRUG RESISTANCE3 | 4.43E-03 | 5.35 |
| At2g47730 | GSTF8 | GLUTATHIONE S-TRANSFERASE8 | 5.76E-03 | 4.48 |
| At2g23150 | NRAMP3 | NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN3 | 2.83E-02 | 3.03 |
| At5g61640 | PMSR1 | PEPTIDE-MET SULFOXIDE REDUCTASE1 | 2.35E-04 | 1.92 |
| At5g07470 | PMSR3 | PEPTIDE-MET SULFOXIDE REDUCTASE3 | 3.73E-06 | 1.82 |
| Cluster 1.3: Cell wall-related transcriptsd | ||||
| At5g57560 | TCH4 | TOUCH4 | 4.88E-02 | 11.52 |
| At4g37990 | CAD8 | CINNAMYL-ALCOHOL DEHYDROGENASE8 | 1.62E-02 | 11.37 |
| At1g02790 | PGA4 | EXOPOLYGALACTURONASE4 | 2.44E-02 | 7.41 |
| At4g35010 | BGAL11 | β-GALACTOSIDASE11 | 2.45E-02 | 5.87 |
| At1g52400 | BGLU18 | β-GALACTOSIDASE18 | 2.14E-02 | 5.61 |
| At2g39700 | EXPA4 | EXPANSIN4 | 1.82E-02 | 4.86 |
| At3g60140 | BGLU30 | β-GALACTOSIDASE30 | 4.77E-02 | 4.41 |
| At2g22470 | AGP2 | ARABINOGALACTAN PROTEIN2 | 4.93E-02 | 4.36 |
| At3g20865 | AGP40 | ARABINOGALACTAN PROTEIN40 | 3.18E-02 | 4.22 |
| At5g40730 | AGP24 | ARABINOGALACTAN PROTEIN24 | 1.70E-02 | 4.09 |
| At1g14420 | AT59 | PECTATE LYASE-LIKE4 | 2.31E-02 | 4.08 |
| At4g34230 | CAD5 | CINNAMYL-ALCOHOL DEHYDROGENASE5 | 4.87E-02 | 4.01 |
| At3g01700 | AGP11 | ARABINOGALACTAN PROTEIN11 | 2.85E-02 | 3.55 |
| At5g11740 | AGP15 | ARABINOGALACTAN PROTEIN15 | 4.58E-02 | 3.43 |
| At3g13520 | AGP12 | ARABINOGALACTAN PROTEIN12 | 2.79E-02 | 3.14 |
Annotation for all the corresponding transcripts is based on The Arabidopsis Information Resource (TAIR) databases. bP values were calculated according to microarray correction of false discovery rate < 0.05. cFold change values represent the normalized signal intensity of SSE genotype samples over the wild-type control samples and correspond to an average value from two biological replicates from each genotype. Full probe lists are given in Supplemental Tables S2 and S3. dTranscripts were clustered according to enrichment analysis performed with the tools embedded in DAVID and PageMan softwares.
The second subcluster (1.2 in Table I) includes genes that are involved in redox- and detoxification-related processes. The greatest increase in induction (34-fold) was detected for a transcript encoding the peroxidase PRX33 (At3g49110; Table I). A recent study shows that this peroxidase belongs to class III cell wall peroxidases, which play key roles in the generation of a hydrogen peroxide (H2O2) oxidative burst required for triggering biotic and abiotic stress responses in Arabidopsis (O’Brien et al., 2012). In addition, the levels of GLUTATHIONE PEROXIDASE2 (At2g31570) and ASCORBATE PEROXIDASE1 (At1g07890), two cytosolic H2O2-scavenging enzymes involved in the protection against oxidative damage generated by reactive oxygen species (Davletova et al., 2005), were up-regulated (Table I). Significant elevation was also observed in the expression level of GLUTATHIONE S-TRANSFERASE8 (At2g47730), a member of a group of enzymes playing crucial roles in oxidative stress tolerance and in plant responses against abiotic stresses (Wagner et al., 2002). The levels of GLYOXALASE II (At1g53580), which takes part in the GSH-dependent glyoxalase detoxification system (Maiti et al., 1997), also increased significantly (Table I). In addition, PEPTIDE-METHIONINE SULFOXIDE REDUCTASE1 (PMSR1; At5g61640) and PMSR3 (At5g07470), two transcripts participating in the active repair of oxidatively damaged Met residues in protein backbones (Bechtold et al., 2004), showed an approximately 2-fold increased expression (Table I). We also identified four transcripts involved in metal handling and detoxification processes, METALLOTHIONEIN3 (At3g15353), METALLOTHIONEIN1A (At1g07600), PLEOTROPIC DRUG RESISTANCE3 (At2g29940), and NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN3 (At2g23150; Table I), which were reported previously to play active roles in redox homeostasis (Benatti et al., 2014). We assume that the induction of these genes is most likely mediated by the low GSH contents found in the transgenic seeds (Fig. 4), since GSH plays a significant role in redox homeostasis and protection from oxidative stress (Noctor et al., 2012).
The third subcluster (1.3 in Table I) contains cell wall-related transcripts. Modifications in cell wall components occur in response to many stresses (Cosgrove, 2000). In SSE seeds, the expression levels of cell wall-related transcripts increased significantly, including several arabinogalactan proteins, β-galactosidases, and enzymes participating in the metabolism of cell wall components, such as TOUCH4 (At5g57560), CINNAMYL-ALCOHOL DEHYDROGENASE8 (At4g37990), EXOPOLYGALACTURONASE4 (At1g02790), EXPANSIN4 (At2g39700), PECTATE LYASE-LIKE4 (At1g14420), and CINNAMYL-ALCOHOL DEHYDROGENASE5 (At4g34230; Table I). These transcripts were reported previously to play key roles in the modification and loosening of cell wall components during environmental stresses and desiccation (for review, see Seifert and Roberts, 2007).
Stress-Associated Transcriptional Phenotypes Are Correlated with ET and ABA Metabolism and with Their Signal Transduction
Plants utilize ET and ABA as hormones to regulate multiple developmental processes and coordinate responses to abiotic stresses. Our analysis reveals that the expression of the two genes encoding the last enzyme of ET synthesis, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) OXIDASE, ACO4 (At1g05010) and ACO2 (At1g62380), increased up to 6.4- and 2.2-fold, respectively, suggesting the enhancement of ET synthesis in SSE seeds (Table II). Three ET response factors (ERFs), RELATED TO AP2-12 (RAP2.12; At1g53910), RAP2.2 (At3g14230), and RAP2.4 (At1g78080), exhibited approximately 2-fold increases (Table II). These ERFs are often involved in plant stress responses, regulating downstream ET-responsive genes (Cheng et al., 2012). Increased expression was also observed for two ET transcription factors, ETHYLENE INSENSITIVE3 (At3g20770) and ENHANCED ETHYLENE RESPONSE5 (At2g19560; Table II). The first is a well-characterized transcription factor that initiates downstream transcriptional cascades for ET responses, and the second is involved in ET signaling and response (Christians et al., 2009). Interestingly, another two transcription factors that function as active repressors of ET biosynthesis, ETHYLENE OVERPRODUCER-LIKE1 (At4g02680) and ERF7 (At3g20310; Christians et al., 2009), were down-regulated by 1.61-fold (Table II). All of the above strongly suggest the enhancement of ET responses in SSE genotypes.
Table II. Gene cluster 2: transcripts associated with ET and ABA metabolism and signaling.
| Gene Symbola | Gene Namea | Gene Descriptiona | Pb | Fold Changec |
|---|---|---|---|---|
| Ethylene metabolism and signalingd | ||||
| At1g05010 | ACO4 | ACC OXIDASE4 | 3.49E-02 | 6.44 |
| At1g62380 | ACO2 | ACC OXIDASE2 | 2.29E-02 | 2.24 |
| At1g53910 | RAP2.12 | RELATED TO AP2-12 | 6.41E-05 | 2.07 |
| At2g19560 | EER5 | ENHANCED ETHYLENE RESPONSE5 | 4.72E-04 | 2.05 |
| At3g14230 | RAP2.2 | RELATED TO AP2-2 | 4.61E-03 | 1.87 |
| At1g78080 | RAP2.4 | RELATED TO AP2-4 | 1.23E-02 | 1.65 |
| At3g20770 | EIN3 | ETHYLENE INSENSITIVE3 | 8.65E-06 | 1.56 |
| At4g02680 | EOL1 | ETHYLENE OVERPRODUCER-LIKE1 | 3.51E-02 | −1.53 |
| At3g20310 | ERF7 | ETHYLENE-RESPONSIVE BINDING FACTOR3 | 2.08E-04 | −1.61 |
| ABA metabolism and signalingd | ||||
| At5g59220 | HAI1 | HIGHLY ABA-INDUCED PP2C GENE1 | 1.80E-02 | 4.54 |
| At2g26980 | SnRK3.17 | SNF1-RELATED PROTEIN KINASE3.17 | 1.47E-06 | 3.81 |
| At1g16540 | ABA3 | ABA DEFICIENT3 | 1.03E-05 | 3.10 |
| At4g26080 | ABI1 | ABA INSENSITIVE1 | 0.25E-03 | 2.05 |
| At2g36270 | ABI5 | ABA INSENSITIVE5 | 9.52E-05 | 1.96 |
| At4g33950 | SnRK2.6 | SNF1-RELATED PROTEIN KINASE2.6 | 1.06E-02 | 1.94 |
| At4g24400 | SnRK3.13 | SNF1-RELATED PROTEIN KINASE3.13 | 6.25E-04 | 1.85 |
| At1g10940 | SnRK2.4 | SNF1-RELATED PROTEIN KINASE2.4 | 8.91E-04 | 1.69 |
| At1g01360 | RCAR1 | REGULATORY COMPONENT OF ABA RECEPTOR1 | 3.03E-03 | 1.55 |
| At1g07430 | HAI2 | HIGHLY ABA-INDUCED PP2C GENE2 | 5.45E-03 | −1.53 |
| At2g13540 | ABH1 | ABA HYPERSENSITIVE1 | 4.75E-04 | −1.57 |
| At1g78390 | NCED9 | 9-CIS-EPOXYCAROTENOID DIOXYGENASE9 | 2.36E-02 | −1.83 |
| At2g29090 | CYP707A2 | (+)-ABA 8’-HYDROXYLASE/OXYGEN BINDING | 4.42E-02 | −2.36 |
Annotation for all the corresponding transcripts is based on the TAIR databases. bP values were calculated according to microarray correction of false discovery rate < 0.05. cFold change values represent the normalized signal intensity of SSE genotype samples over the wild-type control samples and correspond to an average value from two biological replicates from each genotype. Full probe lists are given in Supplemental Tables S2 and S3. dTranscripts were clustered according to enrichment analysis performed with the tools embedded in DAVID and PageMan softwares.
Many of the up-regulated stress-associated genes are also associated with ABA. The levels of ABA were most probably altered in SSE seeds, since ABA3 (At1g16540), which is the last enzyme of ABA biosynthesis (Nonogaki et al., 2014), increased up to 3.1-fold (Table II), while CYP707A2 (At2g29090), which encodes 8′-hydrolase, a key enzyme involved in ABA catabolism (Nonogaki et al., 2014), exhibited a 2.3-fold decrease (Table II). Additionally, the expression level of NCED9 (At1g78390), a 9-cis-epoxycarotenoid dioxygenase, which is a rate-limiting enzyme in the biosynthesis of ABA in seeds, showed a 1.83-fold decrease. Although these results are somewhat conflicting, the data point to a higher ABA signaling cascade in SSE genotypes. Three negative PROTEIN PHOSPHATASES TYPE 2C (PP2C)-ABA regulators, HIGHLY ABSCISIC ACID-INDUCED PP2C GENE1 (HAI1; At5g59220), ABSCISIC ACID INSENSITIVE1 (ABI1; At4g26080), and HAI2 (At1g07430), were identified. The first two increased up to 4.54- and 2.05-fold, while the third exhibited a 1.53-fold decreased expression (Table II). HAI1 in particular was suggested to attenuate ABA signaling controlling Pro and osmoregulatory solute accumulation (Bhaskara et al., 2012). The expression level of the ABA receptor REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR1 (RCAR1; At1g01360) also increased up to 1.55-fold, although it was previously demonstrated that HAI1 is only slightly regulated by this receptor (Bhaskara et al., 2012). Generally, the interaction between HAI-PP2Cs and the RCAR receptors blocks their negative regulation, thus allowing the activation of ABA downstream targets. Among these targets are the SUCROSE NONFERMENTING-RELATED KINASES2 (SnRK2s), which are activated by ABA or osmotic stress and positively regulate the ABA response (Umezawa et al., 2009). In our study, four SnRKs (At2g26980, At4g33950, At4g24400, and At1g10940) were up-regulated significantly (Table II). In addition, the expression level of ABI5 (At2g36270), which was identified as a transcriptional regulator mediating ABA responses in seed germination and development (Finkelstein, 2013), was also elevated (Table II), while ABSCISIC ACID HYPERSENSITIVE1 (At2g13540), an mRNA cap-binding protein that negatively modulates early ABA signal transduction, showed decreased expression (Table II). Taken together, we assume that the signaling cascades of ABA increased in SSE seeds apparently due to the lower water contents and enhanced desiccation found in these seeds (Fig. 5).
Up-Regulation of Genes Involved in Met Metabolism and the Synthesis of Other Amino Acids
Microarray analysis revealed major increases in the expression of genes related to the metabolism of amino acids that were grouped as the third cluster (Table III). The expression levels of several genes related to Met and Cys metabolism were up-regulated (Table III). These include CBSX3 (At5g10860), which encodes a CYSTATHIONINE-β-SYNTHASE that catalyzes the first reversed step of the transsulfuration pathway from homo-Cys back to cystathionine (Fig. 7), the last enzyme in Cys biosynthesis, O-ACETYL-SERINE(THIOL)LYASE (OASTL; At4g14880; Fig. 7), and a Cys-lyase, CORONATINE INDUCED1 (At4g23600), which was reported previously to be involved in a range of processes, including the generation of precursors for the synthesis of ET and polyamines and which exhibits similar activity to CYSTATHIONINE-β-SYNTHASE (Jones et al., 2003). The results also show that the expression levels of HMT1 (At3g25900) and HMT3 (At2g27740), which catalyze the conversion of SMM to Met through the SMM cycle (Fig. 7), increased 3.2- and 1.5-fold, respectively (Table III).
Table III. Gene cluster 3: transcripts associated with amino acid metabolism.
| Gene Symbola | Gene Namea | Gene Descriptiona | Pb | Fold Changec |
|---|---|---|---|---|
| At3g01120 | CGS | CYSTATHIONINE γ-SYNTHASE | 2.24E-04 | 21.72 |
| At2g25450 | 2ODD | 2-OXOACID-DEPENDENT DIOXYGENASE | 2.67E-02 | 19.70 |
| At2g38400 | AGT3 | ALA-GLYOXYLATE TRANSAMINASE3 | 1.02E-02 | 6.74 |
| At4g23600 | CORI3 | CORONATINE INDUCED1 | 3.05E-02 | 4.96 |
| At2g39800 | P5CS1 | Δ-1-PYRROLINE-5-CARBOXYLATE SYNTHASE | 2.40E-06 | 3.89 |
| At5g25980 | TGG2 | THIOGLUCOSIDE GLUCOHYDROLASE2 | 1.21E-05 | 3.78 |
| At3g25900 | HMT1 | HOMOCYSTEINE S-METHYLTRANSFERASE1 | 1.30E-02 | 3.25 |
| At5g10860 | CBSX3 | CYSTATHIONINE-β-SYNTHASE3 | 2.72E-07 | 3.14 |
| At4g34710 | ADC2 | ARG DECARBOXYLASE2 | 4.08E-02 | 2.69 |
| At5g07440 | GDH2 | GLU DEHYDROGENASE2 | 1.82E-02 | 2.58 |
| At5g19550 | AAT2 | ASP AMINOTRANSFERASE2 | 3.01E-03 | 2.36 |
| At1g17290 | AlaAT1 | L-ALA:2-OXOGLUTARATE AMINOTRANSFERASE1 | 8.15E-03 | 2.10 |
| At4g14880 | OASTL | O-ACETYL-SERINE(THIOL)LYASE | 4.64E-04 | 1.97 |
| At3g22200 | GABA-T | γ-AMINOBUTYRATE TRANSAMINASE | 4.17E-02 | 1.92 |
| At4g31990 | AAT5 | ASP AMINOTRANSFERASE5 | 9.95E-04 | 1.75 |
| At3g47340 | ASN1 | ASN SYNTHETASE1 | 1.65E-03 | 1.73 |
| At3g02470 | SAMDC | S-ADENOSYL-MET DECARBOXYLASE | 3.10E-02 | 1.59 |
| At3g22740 | HMT3 | HOMOCYSTEINE S-METHYLTRANSFERASE3 | 1.13E-03 | 1.53 |
Annotation for all the corresponding transcripts is based on the TAIR databases. bP values were calculated according to microarray correction of false discovery rate < 0.05. cFold change values represent the normalized signal intensity of SSE genotype samples over the wild-type control samples and correspond to an average value from two biological replicates from each genotype. Full probe lists are given in Supplemental Tables S2 and S3.
Figure 7.
Schematic representation of Met-associated altered transcripts and metabolites in SSE seeds. Key enzymes are presented in small caps, boldface; amino acids are presented in all initial capital letter and lower case, boldface; and Met catabolic products are presented in italics. Underlined enzymes and metabolites represent up-regulated genes and metabolites, respectively. Full arrows represent one metabolic step while dashed arrows represent several metabolic steps. AATs, Aspartate aminotransferases; ADC, arginine decarboxylase; AGTs, alanine-glyoxylate transaminases; AlaATs, alanine aminotransferases; APS, adenosine 5′-phosphosulfate; ASN, asparagine synthetase; CBS, cystathionine-β-synthase; dcSAM, decarboxylated S-adenosylmethionine; GDH, glutamate dehydrogenase; HK, homoserine kinase; MS, methionine synthase; MTHF, methyltetrahydrofolate; OAS, O-acetylserine; PAP, adenosine 3′,5′-bisphosphate; PAPS, 3′-phosphoadenosine-5′-phosphosulfate; SAMS, SAM synthase; TDH, threonine dehydratase; TGG, thioglucoside glucohydrolase; 2ODDs, 2-oxoacid dependent dioxygenases; 2-OG, 2-oxoglutarate.
In addition, the analysis revealed altered expression levels of several key genes that participate in secondary metabolism derived from Met. Two transcripts (At2g25450 and At5g25980) involved in the metabolism of glucosinolates were highly expressed, exhibiting 19.7- and 3.8-fold increases, respectively (Table III). The first is a 2-oxoacid-dependent dioxygenase involved in the hydroxylation of glucosinolates as part of their secondary modifications, and the second is a β-THIOGLUCOSIDE GLUCOHYDROLASE2 (TGG; At5g25980), which degrades glucosinolates to produce toxins, such as isothiocyanate, that deter herbivores (Fig. 7; Hansen et al., 2008). The expression levels of SAMDC (At3g02470) and ARG DECARBOXYLASE2 (At4g34710), two rate-limiting enzymes that catalyze the first steps of polyamine biosynthesis, were also increased (Table III; Fig. 7). To reveal whether the levels of polyamines were indeed altered in the SSE seeds, we separated polyamines extracted from the transgenic and wild-type seeds by thin-layer chromatography (TLC). The results show that the levels of putrescine decreased significantly in SSE 2, while the levels of spermidine and spermine increased in both SSE genotypes (Supplemental Fig. S6). Since decarboxylated SAM provides the propylamino group for the synthesis of spermidine and spermine derived from putrescine (Fig. 7), we assume that the elevations in SAMDC expression resulted in higher levels of decarboxylated SAM, which led to the observed increases in spermidine and spermine contents simultaneously with decreases in putrescine.
Notably, although the levels of Met, Cys, and GSH were altered, as well as the levels of enzymes involved in Met catabolism, the expression levels of genes involved in the sulfur assimilation pathway (apart from OASTL) were not changed significantly. This can be explained by the observation that this pathway is regulated partly by the activity of enzymes and not by the expression levels of its genes (Takahashi et al., 2011) or that changes in Met metabolism do not affect the expression levels of genes involved in the sulfur assimilation pathway.
Several transcripts associated with Glu metabolism, which is related to the Asp family, were also altered. The expression levels of two aspartate aminotransferases, AAT2 (At5g19550) and AAT5 (At4g31990), and ASPARAGINE SYNTHETASE1 (ASN1; At3g47340), which convert Asp to Glu and Asn, respectively (Fig. 7), were up-regulated (Table III). The levels of three catabolic enzymes of Glu were also elevated. The first is GLU DEHYDROGENASE2 (At5g07440), which converts Glu to 2-oxoglutarate and ammonia and was characterized as a stress-responsive enzyme induced by exogenous ammonium, senescence, reactive oxygen species, and salt (Skopelitis et al., 2006). The second is γ-AMINOBUTYRATE TRANSAMINASE (GABA-T; At3g22200), which converts γ-aminobutyrate that is produced from Glu to succinic semialdehyde, restoring the tricarboxylic acid cycle succinate pool. It was demonstrated recently that this gene participates in salt stress tolerance in Arabidopsis (Renault et al., 2010). The third is Δ-1-PYRROLINE-5-CARBOXYLATE SYNTHETASE (P5CS; At2g39800), the main regulatory enzyme of the Pro synthesis pathway.
In addition to Met and Asp metabolism, the expression of genes involved in other amino acid metabolisms was also elevated. Two transcripts related to Ala catabolism were up-regulated, ALANINE-GLYOXYLATE TRANSAMINASE3 (AGT3; At2g38400) and l- ALANINE:2-OXOGLUTARATE AMINOTRANSFERASE1 (AlaAT1; At1g17290; Table III; Fig. 7). AGT3 converts Ala to glyoxylate, thus producing pyruvate and Gly, while AlaAT1 converts Ala to α-ketoglutarate, thus producing pyruvate and Glu (Liepman and Olsen, 2003). Interestingly, the products of these transamination reactions provide substrates for the tricarboxylic acid cycle. Finally, in accordance with the accumulations of Pro in the SSE genotypes (Fig. 3), its main regulatory synthetic enzyme, P5CS, was up-regulated up to 3.9-fold (Table III).
In addition to changes in the transcript levels of genes associated with amino acid metabolism, three transcripts encoding seed storage proteins (At1g03890, At4g27140, and At4g27150) were also up-regulated significantly (Supplemental Table S2). This implies that larger amounts of these proteins are being synthesized in the SSE seeds. These results correlate with the higher total protein contents measured in these seeds and with previous studies indicating that Met may affect proteome composition (Holowach et al., 1984).
Up-Regulation of Key Synthetic Enzymes Leads to an Elevation in the Contents of Tricarboxylic Acid Cycle Intermediates and Sugars
The microarray analysis also revealed changes in transcripts involved in core metabolism pathways. Increased expression of several transcripts coding for key enzymes in the tricarboxylic acid cycle was observed. These include ACONITASE (At2g43090), SUCCINATE DEHYDROGENASES (At3g27380 and At5g66760), MALATE DEHYDROGENASE1 (At2g22780), and CITRATE SYNTHASE2 (At3g58750; Supplemental Table S4). Furthermore, a higher expression level was also found in genes participating in the pyruvate dehydrogenase complex, which links the glycolysis to the tricarboxylic acid cycle (Supplemental Table S4).
In addition, several transcripts involved in sugar metabolism were also up-regulated. These include two SUC SYNTHASES, SUS1 (At5g20830) and SUS2 (At5g49190), key enzymes participating in Suc synthesis that exhibited 3- and 7-fold increases, respectively (Supplemental Table S5). Five transcripts involved in Tre metabolism (At5g65140, At4g24040, At1g23870, At1g60140, and At1g68020), as well as RAF SYNTHASE6 (At5g20250), were all up-regulated in SSE seeds, supporting the observed changes in carbohydrate metabolism (Fig. 6; Supplemental Table S5). Interestingly, even higher up-regulations were detected in several key carbohydrate transporters, such as ALKALINE/NEUTRAL INVERTASE C (At3g06500), SUGAR TRANSPORT PROTEIN13 (At5g26340), GLUCOSE-6-PHOSPHATE TRANSMEMBRANE TRANSPORTER2 (At1g61800), SUCROSE-PROTON SYMPORTER2 (At1g22710), CARBOHYDRATE TRANSMEMBRANE TRANSPORTER1 (At5g27350), and SUGAR TRANSPORT PROTEIN11 (At5g23270; Supplemental Table S5), indicating the allocation of sugars in SSE seeds.
Germination Rates of Transgenic Seeds under Stress and Hormonal Treatments
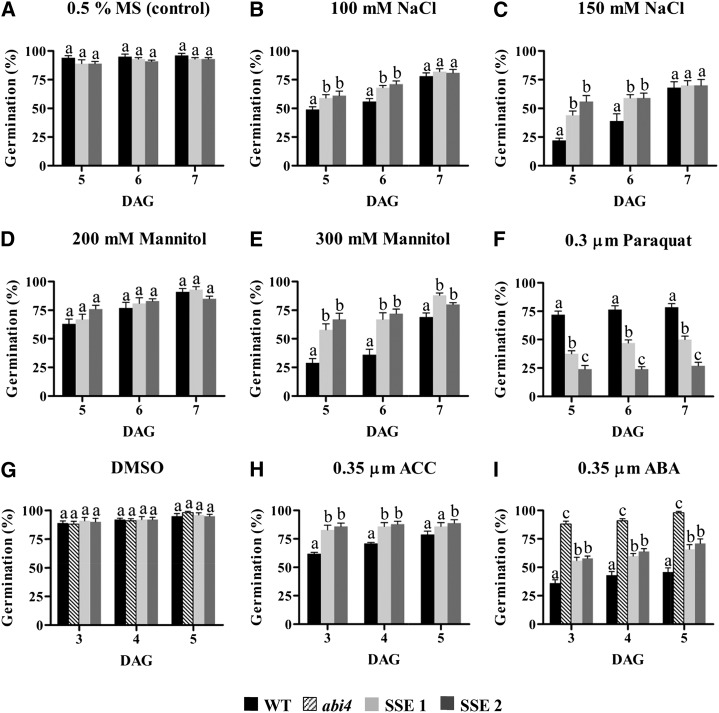
The changes in cellular constituents, which include higher contents of stress-related metabolites and transcripts, total proteins, and starch and lower contents of water and GSH, may affect the seed germination rate. Therefore, a series of germination assays was conducted to evaluate SSE seed germination rate.
Under normal culture conditions, no differences in germination rates between wild-type and SSE genotypes were detected, both reaching close to 100% germination on day 5 (Fig. 8A). However, under mild (100 mm NaCl) and severe (150 mm NaCl) salt stresses, both SSE genotypes exhibited higher germination rates than wild-type seeds (Fig. 8, B and C). Notably, 7 d after germination under acute salt stress, the wild-type seeds closed the gap and exhibited the same germination rates as both SSE genotypes, reaching slightly lower than 80% germination (Fig. 8, B and C). Differences between germination rates of wild-type and SSE seeds were also observed on mannitol, which mimics osmotic stress. Under mild osmotic conditions (200 mm mannitol), no changes were observed (Fig. 8D); however, under higher concentrations (300 mm mannitol), the SSE seeds germinated better than the wild-type seeds, reaching approximately 85% germination (Fig. 8E).
Figure 8.
Germination assays under normal and stress conditions. Germination rates of wild-type (WT) and transgenic seeds in medium containing 0.5% MS medium (control; A), 100 mm NaCl (B), 150 mm NaCl (C), 200 mm mannitol (D), 300 mm mannitol (E), 0.3 μm paraquat (F), DMSO (G), 0.35 μm ACC (H), and 0.35 μm ABA (I) are shown. DAG, Days after germination. The experiment was repeated twice, and the same patterns were observed. Data shown are means ± se of six replicates, each composed of 20 seeds from each genotype. Significance was calculated using the two-way ANOVA test of P < 0.05 and identified by different letters.
Since the levels of GSH were reduced in SSE seeds, the germination rates under oxidative stress conditions (0.3 μm paraquat) were also examined. Analysis of the germination rate after 7 d showed that SSE 1 and SSE 2 reached 50% and 23% germination, respectively, while wild-type seeds reached 79% (Fig. 8F), implying that higher levels of AtD-CGS affect the ability of the transgenic seeds to cope with oxidative damages.
Microarray analysis revealed the up-regulation of genes associated with the metabolism and response of ET and ABA. Since ET is produced from Met (Fig. 7), we measured its levels in the immature green seeds in separated siliques at two different developmental stages, maturation and desiccation; however, its levels were not altered significantly between the transgenic seeds and the wild type. Most probably, seeds do not emit significant levels of ET; thus, it is difficult to measure such changes. Therefore, to assess whether the levels of ET were altered in SSE seeds compared with the wild type, the seeds were germinated in the presence of 0.35 μm ACC, which is the substrate for ACO, the last enzyme of the ET pathway (Fig. 7). The results show that in the presence of ACC, both SSE genotypes germinated faster than wild-type seeds and seemed to cope better with elevated contents of ACC (Fig. 8H).
We also determined the ability of these seeds to germinate in the presence of 0.35 μm ABA. The ABA-insensitive mutant abi4 was used as a control (Finkelstein, 2013). In the presence of ABA, the abi4 mutant germinated much faster than the wild-type and the transgenic seeds. Both SSE seeds exhibited higher germination rates than wild-type seeds (Fig. 8I).
DISCUSSION
Effects of Seed-Specific Expression of AtD-CGS on Met Metabolism
The results of this study show that higher expression levels of AtD-CGS elevate the levels of soluble and total Met, indicating that Arabidopsis seeds, like legume and tobacco seeds (Hanafy et al., 2013; Matityahu et al., 2013; Song et al., 2013), are able to synthesize Met de novo through the Asp family pathway. However, further investigation is required to reveal the relative contribution of the two pathways of Met synthesis (Asp and SMM) in Arabidopsis seeds, since significant increases in the expression of HMT1 and HMT3 in the SMM cycle were detected. This suggests possible cross talk between the SMM cycle and the Asp family pathway.
Our results indicate that the transgenic seeds try to minimize the effects caused by higher expression levels of AtD-CGS and/or higher Met content by reducing the flux toward Met and elevating its catabolism. Several observations support these assumptions. (1) The higher levels of intermediates in the Asp family and Met pathways, homo-Ser and homo-Cys, indicating the existence of bottlenecks within the pathway that may reduce flux toward Met synthesis. Notably, the levels of these two intermediates were also increased in transgenic tobacco seeds expressing AtD-CGS (Matityahu et al., 2013). (2) The higher expression levels of CYSTATHIONINE-β-SYNTHASE and Cys-lyase that catalyze the reverse step of the transsulfuration pathway from homo-Cys back to cystathionine (Fig. 7). (3) The up-regulation of genes that convert Asp to Asn and Glu, and two Glu catabolic enzymes, thus reducing the availability of the carbon/amino skeleton substrate of Met synthesis. Moreover, up-regulation of P5CS, the first enzyme in the Pro synthesis pathway that uses Glu as a substrate, might pull the flux from Asp toward Glu and Pro. (4) The higher expression levels of genes in the glucosinolate, polyamine, and ET biosynthesis pathways suggest that Met is catabolized to these metabolites. In addition, the higher levels of BCAAs that also can be produced from Met suggest that Met catabolism contributes to their accumulation. Since the expression levels of MGL and Thr dehydratase, the main enzymes that produce the BCAAs (Joshi et al., 2010), were not altered significantly in SSE seeds, the accumulation of BCAAs most probably occurred via the enhanced activities of enzymes in their biosynthetic pathways or from higher flux toward their biosynthesis pathways.
Taken together, the results indicate that, in addition to enhanced catabolism of Met, the pathways of Asp and Met synthesis are attenuated in order to minimize the effect of the uncontrolled CGS activity. These processes were activated most probably by the transgenic seeds due to the large impact of that manipulation on the seed metabolic and transcriptomic profiles, which also led to a strong desiccation stress phenotype. Although the elevated expression levels of genes involved in Asp and Glu metabolism imply that a flux toward Asp was reduced and the catabolism of Glu was enhanced, the levels of these two amino acids were increased. This is apparently connected to the total elevation of amino acids found in SSE seeds; the reasons underlying these elevations are not clear yet. However, whereas the expression of the key Pro synthesis enzyme was enhanced, the expression levels of other key synthetic enzymes were not up-regulated and the expression levels of their catabolic enzymes were not reduced. Taking into account that the level of proteins increased in SSE seeds, the results imply that the elevation of amino acid levels is not due to protein degradation, as was suggested previously (Araújo et al., 2011). The higher levels of amino acids might result from enhanced transport from leaves, but further studies are required to assess this hypothesis. The higher levels of soluble amino acids triggered their incorporation into seed storage proteins, leading to the elevation of total protein contents. Similar results were also found in two lines of transgenic soybean (Glycine max) and tobacco seeds expressing the AtD-CGS (Matityahu et al., 2013; Song et al., 2013). However, such elevations were not observed in transgenic seeds with elevated levels of other amino acids. For example, transgenic soybean seeds with higher levels of total Trp and Arabidopsis seeds having higher levels of Lys did not show increases in total amino acid levels (Zhu and Galili, 2003; Kita et al., 2010). The higher levels of soluble amino acids and total proteins are most probably related more to higher Met content than the higher expression level of AtCGS, since immature soybean seeds grown in vitro and supplemented with exogenous Met showed increased total protein contents (Holowach et al., 1984).
The increase in total protein contents in SSE seeds raises the question of the nature of these proteins. The finding that transcripts participating in ribosomal protein synthesis were reduced significantly suggests that the protein synthesis rate decreased, although three transcripts involved in the synthesis of seed storage proteins were up-regulated. Accordingly, analysis of the seed proteomic profiles may shed light on these conflicting findings, since it is known that Met can influence the seed protein profile differentially, as demonstrated in soybean seeds (Holowach et al., 1984).
High Expression Levels of AtD-CGS Induce Typical Stress-Associated Metabolic Responses
The most pronounced changes detected in the metabolic and transcriptomic profiles of SSE seeds reveal significant elevations in the levels of stress-associated metabolites and transcripts. Among the metabolites that were elevated significantly are amino acids (BCAAs and Pro), tricarboxylic acid cycle intermediates (citrate, fumarate, malate, and succinate), the major plant sugars (Fru, maltose, Tre, Suc, Glu, and galactinol), and MDA, which is a common metabolic marker for oxidative stress.
The increased expression of the key gene for Pro synthesis, together with the accumulation of Pro, was shown previously to be strongly associated with plant responses to abiotic stress and environmental cues (for review, see Verbruggen and Hermans, 2008). In addition to higher Pro in SSE seeds, the level of soluble Pro was also increased in AtD-CGS-expressing tobacco seeds, where it was the only amino acid whose soluble level increased (Matityahu et al., 2013). This suggests a strong relationship between Met metabolism and Pro in seeds. One possible explanation for this connection might be that Pro is able to balance cell redox status (Verbruggen and Hermans, 2008); therefore, it is accumulated to cope with the altered redox state that might occur in SSE seeds due to the lower GSH contents. Another option is that Pro is accumulated as part of the metabolic stress phenotype in SSE seeds (Alla et al., 2012), and it is not directly related to Met metabolism. Additional investigations are required to clarify the putative relations between these two amino acids.
The mechanisms underlying the association between higher expression levels of AtD-CGS and the induction of the tricarboxylic acid cycle metabolism have yet to be elucidated, but they might be related to the higher expression levels of genes observed in this pathway. In addition, the list of up-regulated genes also includes the pyruvate dehydrogenase complex, which links the glycolysis to the tricarboxylic acid cycle by converting pyruvate into acetyl-CoA, thereby contributing to the higher levels of tricarboxylic acid metabolites. Higher levels of these metabolites can also be derived from the BCAAs, whose catabolic products feed directly into the tricarboxylic acid cycle (Araújo et al., 2011), from higher expression levels of GABA-T that degrades γ-aminobutyrate, which is then converted to succinate in the tricarboxylic acid cycle, and/or from the catabolism of Ala, whose two transcripts of its catabolism were up-regulated, leading to the production of pyruvate and α-ketoglutarate. These findings suggest that the tricarboxylic acid cycle is fed by several metabolites. However, we cannot exclude the possibility that the elevations in these tricarboxylic acid cycle metabolites result from perturbations or regulatory points operating inside the cycle. Altogether, these observations indicate that the energy status of SSE seeds was altered, similar to the alterations occurring during stress (Angelovici et al., 2009).
Another mechanism used by plants in their response to drought stress is the accumulation of sugars. It has been demonstrated that certain sugars (such as Suc, Tre, and Raf) play a central role in the protection from drought stress in a wide range of organisms by helping to maintain the structure of the cytoplasm and to stabilize certain proteins when the amount of water is reduced (for review, see Ingram and Bartels, 1996). Galactinol, which increased up to 2.3-fold and is used for Raf synthesis, also plays an important role in abiotic stress tolerance (Elsayed et al., 2014). The higher levels of sugars in SSE seeds are apparently not the result of starch degradation, since the levels of starch increased in these seeds. Moreover, the up-regulation of key sugar synthesis enzymes also suggests that the higher levels of sugars originated from the enhancement of their biosynthesis pathways.
Unlike sugars, the levels of polyols were reduced dramatically in SSE seeds. Polyols are synthesized in leaves and then translocated via the phloem to the sink tissues, where they can accumulate and function as osmoprotectants, especially during osmotic stress. This is due to their chemical nature, which allows them not to interfere with cell metabolism and to enter into the tricarboxylic acid cycle as a carbon and energy source (Williamson et al., 2002). It was demonstrated that, in seeds, polyols accumulate at the desiccation stage (Fait et al., 2006; Angelovici et al., 2010). Thus, one option to explain the reduction of polyols in SSE seeds is that the desiccation process in these seeds was altered, resulting in their lower relative contents. Another explanation could be that their catabolic rate increased, leading to higher levels of tricarboxylic acid metabolites.
Altogether, the results point at major changes in endogenous metabolic processes that occur in SSE seeds during the transition from late reserve accumulation to desiccation when the phaseolin promoter is active (Fait et al., 2011). This assumption is based on previous results showing that amino acids and sugars accumulate specifically during seed desiccation in Arabidopsis wild-type seeds (Fait et al., 2006; Angelovici et al., 2010). Our findings also indicate that, unlike Lys metabolism, which had only minor effects on the metabolome and lower contents of intermediates of the tricarboxylic acid cycle (Angelovici et al., 2009), Met metabolism has a relatively large impact on primary metabolites, particularly those involved in osmotic and desiccation stresses.
Lower GSH Contents in SSE Genotypes Lead to Susceptibility to Oxidative Stress
The levels of Cys and its product, GSH, were reduced significantly in both SSE genotypes; however, these reductions were more pronounced in seeds from the SSE 2 line than those observed in the SSE 1 line, suggesting that higher expression of AtD-CGS increases the flux of Cys toward Met synthesis at the expense of GSH. Similar results were also observed in tobacco seeds expressing AtD-CGS (Matityahu et al., 2013). GSH is a key player in cellular redox homeostasis and signaling under optimal and stress conditions, particularly oxidative stress, which accompanies many abiotic stresses (Noctor et al., 2012). Therefore, we assume that the reduction of GSH may lead to oxidative stress. Indeed, a marker for oxidative stress (MDA) increased significantly in SSE seeds. The low level of GSH may also trigger the expression of genes encoding several H2O2-scavenging enzymes, such as peroxidases and metal-detoxifying enzymes. These enzymes are up-regulated under direct and indirect oxidative stresses and participate in plant responses against oxidative conditions (Caverzan et al., 2012). An inverse relation between GSH and the expression levels of peroxidase was reported previously, since lower levels of GSH increased the expression levels of these genes (Caverzan et al., 2012), while higher levels of GSH caused a reduction in the expression levels of 10 peroxidases (Hacham et al., 2014). Although the reduction in GSH contents triggered the expression of these genes, the germination rates of the SSE seeds were lower under oxidative conditions mimicked by the presence of paraquat, indicating that higher oxidative stress occurs in SSE seeds.
Seed-Specific Expression of AtD-CGS Stimulates Transcriptomic Defense Responses
Our transcriptomic analysis revealed that the induced alteration of Met metabolism in the SSE genotypes had significant effects on gene expression programs. The most profound effect was the up-regulation of genes principally controlling drought, salt, and oxidative stress defense mechanisms associated with abiotic responses. These include genes involved in stress-induced responses and redox, detoxification, and cell wall-related transcripts. In addition, genes associated with ET and ABA metabolism and their signaling were also up-regulated. The majority of these genes are well-known transcripts induced under various environmental cues and responsive to ET and ABA (Kreps et al., 2002; Seki et al., 2002). Indeed, many of these transcripts were reported previously to contain either GCC/DRE/CRT boxes, to which ERFs bind specifically under abiotic stresses, or ABA-responsive elements (CACGTG[T/C/G]; Kaplan et al., 2006; Cheng et al., 2012). This implies that most of the transcriptomic stress responses observed in the SSE genotypes are regulated primarily through the ET and/or ABA signaling cascades.
The stress-related transcriptomic and metabolic behavior of SSE seeds urged us to examine their germination performance under nonstress conditions and following exposure to abiotic stresses. The germination rates of the transgenic seeds under nonstress conditions were not altered significantly compared with those of the wild type. This result is unlike those reported for tobacco seeds expressing AtD-CGS, whose germination rates were significantly lower than those of the wild type (Matityahu et al., 2013). Assessment of the germination rates of SSE seeds under osmotic and salt stresses showed that, under these stresses, SSE seeds germinate better than wild-type seeds. In addition, these seeds coped better with high doses of ET and ABA. The results are unexpected due to the inhibitory role of ABA in seed germination. One possible explanation can be that the SSE seeds may have higher levels of ET, which was found to play a crucial role in overcoming seed dormancy and can alleviate germination inhibition induced by salinity in many seeds (Lin et al., 2013). Moreover, a negative interaction between ET and ABA, in which ET can overcome the inhibitory action of ABA in seeds, was reported (Linkies et al., 2009). Another possible explanation for the ability of these seeds to germinate better than wild-type seeds under stress is the higher stress-related metabolites such as sugars, BCAAs, polyamines, and different transcripts found in these seeds. One group of up-regulated genes that might be involved is the SnRKs, which were shown to protect seeds during the desiccation stage (Bhaskara et al., 2012).
Taken together based on the results described above, we suggest three options, or their combination, to explain the link between high expression levels of AtD-CGS and the stress phenotypes that were observed in SSE seeds. (1) The lower water contents found in SSE seeds. The elevations of total proteins and total carbohydrates (mainly starch) were similar to the reduction in water content (calculated by the percentage of the seed’s fresh weight). This suggests that the elevations of storage compounds are at the expense of water content. We hypothesize that the reduction in water contents led to stronger desiccation during seed development, which in turn triggered changes in ABA and its downstream stress-related signaling cascades, leading to elevations of stress-related metabolites and transcripts. (2) ET metabolism. Although we were not able to detect ET levels in the developing seeds, we suspect that its levels increased, since we found that several key genes involved in its synthesis and downstream targets were up-regulated. We previously showed a strong correlation between higher Met content and higher ET levels in tomato (Solanum lycopersicum; Katz et al., 2006), suggesting that this also might occur in SSE seeds. The higher germination rates of SSE seeds in the presence of ACC, the substrate for ET synthesis, also supports this assumption. (3) The reduction in GSH contents. Low GSH contents might induce oxidative stress in SSE seeds. Although the three options described above are strongly reflected through our observations, we cannot exclude additional factors that contributed or triggered the stress phenotypes observed in SSE seeds.
CONCLUSION
By using metabolic and transcriptomic profiles, this study exposed novel metabolic and transcriptional interactions associated with Met metabolism in seeds. The results illustrate distinct stress behaviors of these interactions and suggest that the increased expression level of AtCGS in SSE genotypes during the last stages of seed development, when the phaseolin promoter is active, strongly stimulates phenotypes that resemble drought stress and enhances the desiccation stage. As a result, metabolic pathways and gene expression programs associated with these stresses were induced. This implies that Met metabolism has a strong influence on the seed’s metabolome during development. Apparently, due to this stress phenotype, the results imply that several regulatory mechanisms operate in order to reduce the flux toward Met synthesis and to promote Met catabolism. However, despite these metabolic and transcriptomic changes, the SSE seeds germinated as well as wild-type seeds under nonstress conditions, suggesting that seeds can tolerate perturbations even when their core metabolic pathways are strongly altered.
From a nutritional prospective, although Arabidopsis is not used as a crop plant, the results indicate a way to improve the nutritional quality of crop plants related to Arabidopsis, such as false flax (Camelina sativa). These improvements include significantly higher levels of Met and higher levels of total storage components, proteins and carbohydrates.
Despite the stress phenotype, the higher expression level of AtD-CGS brings several benefits to the transgenic seeds: the higher levels of Met and its associated metabolites (polyamines, glucosinolates, and ET) provide protection especially during stress, and the higher contents of proteins and carbohydrates, two important seed storage components, provide essential nutritional energy to the embryo during germination. From an evolutionary point of view, these benefits raise an interesting question regarding the reasons why AtCGS expression is subjected to such tight regulation (Amir et al., 2012; Galili and Amir, 2013) and why very low levels of Met are found in tissues of various plant species (Amir et al., 2012). One possible explanation is that, without such tight regulation, the seeds might experience difficulties in germinating under oxidative conditions that are likely to occur in SSE seeds, apparently due to lower GSH content.
Altogether, these data provide new insights into the factors participating in the regulation of Met and the mechanisms mediating the effects of elevated Met levels on seed composition and behavior.
MATERIALS AND METHODS
Generation of Transgenic Arabidopsis Seeds Expressing AtD-CGS in a Seed-Specific Manner
Arabidopsis (Arabidopsis thaliana) plants of ecotype Columbia-0 were used to generate lines expressing AtD-CGS in a seed-specific manner. The complementary DNA of AtD-CGS lacking the regulatory 30-amino acid domain leading to the accumulation of high Met levels was inserted previously into a pCE vector (Hacham et al., 2006). This fragment was cut from the pCE vector using SphI and introduced with that enzyme into the pCEPH vector, which contains the phaseolin promoter (Shaul and Galili, 1992). This promoter is the most abundant seed storage protein in the common bean (Phaseolus vulgaris) and is stringently turned off during all vegetative stages of plant development (Sundaram et al., 2013). It was demonstrated that this promoter is induced constitutively during the maturation and desiccation stages of seed development (Fait et al., 2011). The fragment containing the phaseolin promoter-AtD-CGS without its stop codon was then digested from the vector using SmaI and introduced with the same enzyme into the binary Ti plasmid pZP111 (9,735 bp) carrying three in-frame copies of hemagglutinin epitope tag and an octopine synthase terminator (Hacham et al., 2006; Supplemental Fig. S1). This plasmid was introduced into Agrobacterium tumefaciens EHA105 cells, and Arabidopsis transformation was performed by the floral dip method. Transformed seeds were selected on medium supplemented with kanamycin (50 mg mL−1; Duchefa) and carbenicillin (85 mg mL−1; Duchefa). Twenty independent transformation events were selected and planted in an enriched soil medium. Seeds were collected from the T1 generation and then analyzed for a 3:1 segregation based on the presence of the transgene and the observed resistant phenotype to kanamycin. Segregated T1 lines were then grown again, and the T2 generation was examined for nonsegregating lines to produce T2 and further T3 homozygous lines. The two T3 homozygous lines showing the highest levels of AtD-CGS protein accumulation by western-blot analysis were chosen for further analysis.
Plant Material and Seed Collection
In all experiments, wild-type and transgenic seeds were grown on petri dishes with 0.5% Murashige and Skoog (MS) medium (Duchefa), 0.5% Suc (J.D. Baker), and 7 mg mL−1 plant agar (Duchefa). The young seedlings were grown for 10 d and then planted on fertilized soil at 22°C ± 1°C under a 16/8-h light/dark cycle at 100 μmol m–2 s–1 with 50% to 70% relative humidity. Mature dry seed pools were collected from all genotypes at the end of the desiccation period, each collected from at least five different plants. Following collection, the seeds were allowed to dry fully for 3 d in a vacuum desiccator and stored under dry conditions at 4°C until further analyses.
Protein and mRNA Analyses
Wild-type and transgenic seeds (10 mg) expressing AtD-CGS from T3 homozygous lines were analyzed by western blot using antibodies against the epitope tag of hemagglutinin, as described previously (Matityahu et al., 2013). For the expression level analysis of AtCGS and AtMGL, total RNA was extracted from 50 mg of mature dry seeds from the wild-type and SSE genotypes. All RNA samples were extracted and analyzed as described previously (Matityahu et al., 2013). To normalize variance among samples, the PP2A-A3 transcript level was used as an endogenous control (Czechowski et al., 2005). The values presented are means of three biological replicates, each with three technical replicates. All primers used for qRT-PCR analyses are listed in Supplemental Table S6.
Microarray Analysis
Total RNA was extracted from mature dry seeds as described above, three biological replicates for the wild-type genotype and two biological replicates for each transgenic line, and then amplified using two-cycle Affymetrix labeling according to standard Affymetrix protocols. Hybridization, labeling, scanning, and data extraction were performed according to standard Affymetrix protocols. Transcriptome analysis was carried out using Partek Genome Suite software (http://www.partek.com). Preprocessing was carried out using the robust microarray averaging algorithm (Irizarry et al., 2003), and two-way ANOVA was performed. False discovery rate was applied to correct multiple comparisons. Differentially expressed genes were chosen according to a false discovery rate of 0.05 and a 1.5-fold change between SSE genotypes and the wild-type genotype (full probe set, Arabidopsis Genome Initiative list, and calculated ratios are presented in Supplemental Tables S2 and S3). Genes that responded significantly were clustered and classified according to their physiological functions using the enrichment and functional annotation clustering tools embedded in PageMan (http://mapman.mpimp-golm.mpg.de/pageman/; Usadel et al., 2005) and DAVID (http://david.abcc.ncifcrf.gov/; Huang et al., 2009).
Extraction, Derivatization, and Analysis of Seed Primary Metabolites Using GC-MS and HPLC Analyses
Mature dry seeds from wild-type and SSE genotypes were collected separately from six distinct biological pools, and 10 mg of mature dry seeds was treated and derivatized as described previously (Matityahu et al., 2013). The single-ion mass method was used for soluble and protein-incorporated amino acid determination with the RXI-5-Sil MS capillary column (RESTEK; 30 m, 0.25-mm i.d., and 0.25-μm thickness), while the total-ion-count method was used for metabolic profiling and separation using the VF-5ms capillary column (Agilent; 30 m + 10 m EZ-guard, 0.25-mm i.d., and 0.25-μm thicknesses). All analyses were carried out on a GC-MS system (Agilent 7890A) coupled with a mass selective detector (Agilent 5975c) and a Gerstel multipurpose sampler (MPS2; Matityahu et al., 2013).
For Cys and GSH determination, 100 mg of mature dry seeds was collected, ground using a mortar and pestle, and then extracted and analyzed by HPLC as described previously (Matityahu et al., 2013).
Morphological and Chemical Characterization of Seeds
Immature green seeds and mature dry seeds were observed with a Zeiss Axio Imager A1 light microscope and photographed with a Zeiss Axiocam MRc 5 camera using the AxioVision program, version 4.6.3.0 SP1. Using the measurement tools embedded in this software, 100 seeds from all genotypes were analyzed for their width and length. Seven-week-old plants and mature plants at the start of the desiccation processes were also observed for their morphology and photographed using a Nikon D3200 SLR camera.
Total seed weight was determined by analyzing the weight of exactly 100 seeds from each genotype. Relative water contents were calculated by the weight loss of 100 mg of mature dry seeds from all genotypes following oven incubation at 100°C for 24 h.
For total protein and lipid determinations, 100 mg of mature dry seeds was collected from four distinct biological pools and treated according to the Kjeldahl and Soxhlet methods, as described previously (Song et al., 2013). The same amounts were used to calorimetrically measure total reducing sugars after carbohydrate and starch hydrolysis using the method of Sumner (1924).
Germination Assays under Normal and Stress Conditions
For abiotic and hormone treatment germination assays, 20 seeds from each genotype were grown separately in petri dishes under normal or stress conditions in six biological replicates. Germination rates were calculated on days 5, 6, and 7 after germination, and the experiments were repeated twice to observe the same patterns. For the ABA treatment assay, we also included the ABA-insensitive abi4 mutant as a control.
Salt stress (100 and 150 mm NaCl), osmotic stress (200 and 300 mm mannitol), oxidative stress (0.3 μm paraquat), ET treatment (0.35 μm ACC), and ABA treatment (0.35 μm ABA) were applied to the medium, while the control for all experiments was 0.5% MS medium. Since the ABA was dissolved with dimethyl sulfoxide (DMSO), a similar final concentration of DMSO was added to the MS medium of the control (Fig. 7G). All the above materials were manufactured by Sigma-Aldrich.
Polyamine Extraction and Analysis Using TLC
Free polyamines were extracted from 100 mg of mature dry seeds of the wild-type and SSE genotypes according to Ye et al. (1997). TLC separation was performed on Kieselgel-60 plates (Merck), and photomicrographs were taken with transmitted UV light. Individual dansylated polyamine bands were identified by comparing the RF values of dansylated putrescine, spermidine, and spermine standards (Sigma-Aldrich). Band quantification and relative fluorescence intensity calculation were done using QuantityOne software (Bio-Rad).
Determination of Lipid Peroxidation by MDA Assay
The thiobarbituric acid test was used to determine MDA as an end product of lipid peroxidation. Mature dry seeds (100 mg) from the wild-type and SSE genotypes were treated, and MDA contents were determined as reported previously (Bai et al., 2012).
Statistical Analyses
All statistical analyses were performed and bar graphs were compiled using GraphPad Prism 5.01 scientific software (http://www.graphpad.com/). In all experiments throughout this study, significance was calculated using the two-way ANOVA test of P < 0.05 and examined between each SSE genotype and the wild type and between SSE 1 and SSE 2. Principal component analysis and correlation and pattern analyses of GC-MS data were done using MetaboAnalyst 2.0, a comprehensive tool suite for metabolomic data analysis (http://metaboanalyst.ca/; Xia et al., 2009, 2012), following data log10 transformation and pareto scaling (mean centered and divided by the square root of sd of each variable) manipulations.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The plant transformation binary vector construct for seed-specific expression of AtD-CGS.
Supplemental Figure S2. Morphological and phenotypic characteristics of wild-type and SSE seeds.
Supplemental Figure S3. Correlation and pattern analyses of GC-MS metabolic profiling data.
Supplemental Figure S4. Transcript analysis of MGL.
Supplemental Figure S5. Lipid peroxidation determination by MDA assay.
Supplemental Figure S6. Polyamine contents in wild-type and SSE seeds.
Supplemental Table S1. Soluble and protein-incorporated amino acid contents in mature dry seeds of wild-type and SSE genotypes.
Supplemental Table S2. Differentially up-regulated transcripts in SSE seeds compared with wild-type seeds.
Supplemental Table S3. Differentially down-regulated transcripts in SSE seeds compared with wild-type seeds.
Supplemental Table S4. Clustering of transcripts associated with tricarboxylic acid cycle metabolism.
Supplemental Table S5. Clustering of transcripts associated with carbohydrate metabolism.
Supplemental Table S6. List of primers used for qRT-PCR analyses.
Supplementary Material
Acknowledgments
We thank Dr. Yael Hacham for fruitful assistance in several analyses and Dr. Gidi Baum for critical reading of the article.
Glossary
- SAM
S-adenosyl-methionine
- ET
ethylene
- SMM
S-methyl-methionine
- qRT
quantitative real-time
- BCAA
branched-chain amino acid
- GSH
glutathione
- MDA
malondialdehyde
- GC-MS
gas chromatography-mass spectrometry
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- ACC
1-aminocyclopropane-1-carboxylic acid
- TLC
thin-layer chromatography
- MS
Murashige and Skoog
- DMSO
dimethyl sulfoxide
- TAIR
The Arabidopsis Information Resource
Footnotes
This work was supported by the Israel Science Foundation (grant no. 231–09).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alla MMN, Khedr AHA, Serag MM, Abu-Alnaga AZ, Nada RM. (2012) Regulation of metabolomics in Atriplex halimus growth under salt and drought stress. Plant Growth Regul 67: 281–304 [Google Scholar]
- Amir R, Han T, Ma F. (2012) Bioengineering approaches to improve the nutritional values of seeds by increasing their methionine content. Mol Breed 29: 915–924 [Google Scholar]
- Angelovici R, Fait A, Zhu X, Szymanski J, Feldmesser E, Fernie AR, Galili G. (2009) Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol 151: 2058–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A. (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Arondel V, XII, Vergnolle C, II, Cantrel C, Kader J. (2000) Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci 157: 1–12 [DOI] [PubMed] [Google Scholar]
- Bai B, Sikron N, Gendler T, Kazachkova Y, Barak S, Grafi G, Khozin-Goldberg I, Fait A. (2012) Ecotypic variability in the metabolic response of seeds to diurnal hydration-dehydration cycles and its relationship to seed vigor. Plant Cell Physiol 53: 38–52 [DOI] [PubMed] [Google Scholar]
- Bechtold U, Murphy DJ, Mullineaux PM. (2004) Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights. Plant Cell 16: 908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti MR, Yookongkaew N, Meetam M, Guo WJ, Punyasuk N, AbuQamar S, Goldsbrough P. (2014) Metallothionein deficiency impacts copper accumulation and redistribution in leaves and seeds of Arabidopsis. New Phytol 202: 940–951 [DOI] [PubMed] [Google Scholar]
- Bhaskara GB, Nguyen TT, Verslues PE. (2012) Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 160: 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL, et al. (1999) S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11: 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caverzan A, Passaia G, Rosa SB, Ribeiro CW, Lazzarotto F, Margis-Pinheiro M. (2012) Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet Mol Biol (Suppl 35: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Hsieh EJ, Chen JH, Chen HY, Lin TP. (2012) Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol 158: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD. (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38: 109–124 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed AI, Rafudeen MS, Golldack D. (2014) Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol 16: 1–8 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Nesi AN, Angelovici R, Lehmann M, Pham PA, Song L, Haslam RP, Napier JA, Galili G, Fernie AR. (2011) Targeted enhancement of glutamate-to-γ-aminobutyrate conversion in Arabidopsis seeds affects carbon-nitrogen balance and storage reserves in a development-dependent manner. Plant Physiol 157: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166, 10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G, Amir R. (2013) Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol J 11: 211–222 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Koussevitzky S, Kirma M, Amir R. (2014) Glutathione application affects the transcript profile of genes in Arabidopsis seedling. J Plant Physiol 171: 1444–1451 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Schuster G, Amir R. (2006) An in vivo internal deletion in the N-terminus region of Arabidopsis cystathionine gamma-synthase results in CGS expression that is insensitive to methionine. Plant J 45: 955–967 [DOI] [PubMed] [Google Scholar]
- Hanafy MS, Rahman SM, Nakamoto Y, Fujiwara T, Naito S, Wakasa K, Ishimoto M. (2013) Differential response of methionine metabolism in two grain legumes, soybean and azuki bean, expressing a mutated form of Arabidopsis cystathionine γ-synthase. J Plant Physiol 170: 338–345 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kerwin RE, Ober JA, Lambrix VM, Mitchell-Olds T, Gershenzon J, Halkier BA, Kliebenstein DJ. (2008) A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol 148: 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Sebastià C, Marsolais F, Saravitz C, Israel D, Dewey RE, Huber SC. (2005) Free amino acid profiles suggest a possible role for asparagine in the control of storage-product accumulation in developing seeds of low- and high-protein soybean lines. J Exp Bot 56: 1951–1963 [DOI] [PubMed] [Google Scholar]
- Hesse H, Kreft O, Maimann S, Zeh M, Hoefgen R. (2004) Current understanding of the regulation of methionine biosynthesis in plants. J Exp Bot 55: 1799–1808 [DOI] [PubMed] [Google Scholar]
- Holowach LP, Thompson JF, Madison JT. (1984) Storage protein composition of soybean cotyledons grown in vitro in media of various sulfate concentrations in the presence and absence of exogenous l-methionine. Plant Physiol 74: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jones PR, Manabe T, Awazuhara M, Saito K. (2003) A new member of plant CS-lyases: a cystine lyase from Arabidopsis thaliana. J Biol Chem 278: 10291–10296 [DOI] [PubMed] [Google Scholar]
- Joshi V, Joung JG, Fei Z, Jander G. (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39: 933–947 [DOI] [PubMed] [Google Scholar]
- Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H. (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz YS, Galili G, Amir R. (2006) Regulatory role of cystathionine-gamma-synthase and de novo synthesis of methionine in ethylene production during tomato fruit ripening. Plant Mol Biol 61: 255–268 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee M, Chalam R, Martin MN, Leustek T, Boerjan W. (2002) Constitutive overexpression of cystathionine gamma-synthase in Arabidopsis leads to accumulation of soluble methionine and S-methylmethionine. Plant Physiol 128: 95–107 [PMC free article] [PubMed] [Google Scholar]
- Kita Y, Nakamoto Y, Takahashi M, Kitamura K, Wakasa K, Ishimoto M. (2010) Manipulation of amino acid composition in soybean seeds by the combination of deregulated tryptophan biosynthesis and storage protein deficiency. Plant Cell Rep 29: 87–95 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot (Lond) 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Huang T, Toro-Ramos T, Fraga M, Last RL, Jander G. (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J 54: 310–320 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ. (2003) Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol 131: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yang L, Paul M, Zu Y, Tang Z. (2013) Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiol Biochem 73: 211–218 [DOI] [PubMed] [Google Scholar]
- Linkies A, Müller K, Morris K, Turecková V, Wenk M, Cadman CS, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, et al. (2009) Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ahn JE, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, Pittendrigh B, Murdock LL, Koiwa H, Zhu-Salzman K. (2005) Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol 139: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Maiti MK, Krishnasamy S, Owen HA, Makaroff CA. (1997) Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol Biol 35: 471–481 [DOI] [PubMed] [Google Scholar]
- Matityahu I, Godo I, Hacham Y, Amir R. (2013) Tobacco seeds expressing feedback-insensitive cystathionine gamma-synthase exhibit elevated content of methionine and altered primary metabolic profile. BMC Plant Biol 13: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Nonogaki M, Sall K, Nambara E, Nonogaki H. (2014) Amplification of ABA biosynthesis and signaling through a positive feedback mechanism in seeds. Plant J 78: 527–539 [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Daudi A, Butt VS, Bolwell GP. (2012) Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236: 765–779 [DOI] [PubMed] [Google Scholar]
- Olsson AS, Engström P, Söderman E. (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55: 663–677 [DOI] [PubMed] [Google Scholar]
- Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C. (2010) The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol 10: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S. (2006) S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67: 1686–1698 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Shaul O, Galili G. (1992) Threonine overproduction in transgenic tobacco plants expressing a mutant desensitized aspartate kinase of Escherichia coli. Plant Physiol 100: 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, Kouvarakis A, Papadakis AK, Stephanou EG, Roubelakis-Angelakis KA. (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18: 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Hou W, Godo I, Wu C, Yu Y, Matityahu I, Hacham Y, Sun S, Han T, Amir R. (2013) Soybean seeds expressing feedback-insensitive cystathionine γ-synthase exhibit a higher content of methionine. J Exp Bot 64: 1917–1926 [DOI] [PubMed] [Google Scholar]
- Sumner JB. (1924) The estimation of sugar in diabetic urine, using dinitrosalicylic acid. J Biol Chem 62: 287–290 [Google Scholar]
- Sundaram S, Kertbundit S, Shakirov EV, Iyer LM, Jurícek M, Hall TC. (2013) Gene networks and chromatin and transcriptional regulation of the phaseolin promoter in Arabidopsis. Plant Cell 25: 2601–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. (2008) Proline accumulation in plants: a review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F. (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Williamson DJ, Jennings BD, Guo WW, Pharr DM. (2002) Sugar alcohols, salt stress, and fungal resistance: polyols–multifunctional plant protection? J Am Soc Hortic Sci 127: 467–473 [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. (2012) MetaboAnalyst 2.0: a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40: W127–W133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37: W652–W660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Muller HH, Zhang J, Gressel J. (1997) Constitutively elevated levels of putrescine and putrescine-generating enzymes correlated with oxidant stress resistance in Conyza bonariensis and wheat. Plant Physiol 115: 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu H, Sun J, Li B, Zhu Q, Chen S, Zhang H. (2012) Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 7: e30355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Galili G. (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.