Recently discovered Arabidopsis peroxisomal mutants identify proteins involved in β-oxidation.
Abstract
Peroxisomes are highly dynamic and multifunctional organelles essential to development. Plant peroxisomes accommodate a multitude of metabolic reactions, many of which are related to the β-oxidation of fatty acids or fatty acid-related metabolites. Recently, several dozens of novel peroxisomal proteins have been identified from Arabidopsis (Arabidopsis thaliana) through in silico and experimental proteomic analyses followed by in vivo protein targeting validations. To determine the functions of these proteins, we interrogated their transfer DNA insertion mutants with a series of physiological, cytological, and biochemical assays to reveal peroxisomal deficiencies. Sugar dependence and 2,4-dichlorophenoxybutyric acid and 12-oxo-phytodienoic acid response assays uncovered statistically significant phenotypes in β-oxidation-related processes in mutants for 20 of 27 genes tested. Additional investigations uncovered a subset of these mutants with abnormal seed germination, accumulation of oil bodies, and delayed degradation of long-chain fatty acids during early seedling development. Mutants for seven genes exhibited deficiencies in multiple assays, strongly suggesting the involvement of their gene products in peroxisomal β-oxidation and initial seedling growth. Proteins identified included isoforms of enzymes related to β-oxidation, such as acyl-CoA thioesterase2, acyl-activating enzyme isoform1, and acyl-activating enzyme isoform5, and proteins with functions previously unknown to be associated with β-oxidation, such as Indigoidine synthase A, Senescence-associated protein/B12D-related protein1, Betaine aldehyde dehydrogenase, and Unknown protein5. This multipronged phenotypic screen allowed us to reveal β-oxidation proteins that have not been discovered by single assay-based mutant screens and enabled the functional dissection of different isoforms of multigene families involved in β-oxidation.
Peroxisomes are small (approximately 0.1–1.0 µm) single-membrane eukaryotic organelles that are essential for the development of animals and plants by mediating a multitude of conserved and lineage-specific metabolic functions (Beevers, 1979; van den Bosch et al., 1992; Kaur et al., 2009; Hu et al., 2012; Schrader et al., 2012). Plant peroxisomes house metabolic processes, including β-oxidation of fatty acids; hormone biosynthesis; photorespiration; the glyoxylate cycle; detoxification of reactive oxygen, nitrogen, and sulfur species; and metabolism of branched amino acids, urate, and polyamines (PAs; Beevers, 1979; Kaur et al., 2009; Hu et al., 2012). Peroxisomes also generate signaling molecules with regulatory roles in plant development (Weber, 2002; Corpas et al., 2013; Sandalio et al., 2013).
β-oxidation of fatty acids and related metabolites is a major function of peroxisomes throughout the lifecycle of a plant from seed germination to senescence. Mobilization of seed oil reserves during seed germination and postgerminative growth requires peroxisomal β-oxidation and the glyoxylate cycle. In this process, fatty acids are transported into the peroxisome, where they are activated into fatty acyl-CoAs and later, shortened by two carbons in each cycle of β-oxidation. The product, acetyl-CoA, is converted to four-carbon molecules by the glyoxylate cycle, and its products further undergo gluconeogenesis to provide energy for postgerminative development (Theodoulou and Eastmond, 2012). Using core β-oxidation enzymes as well as pathway-specific enzymes, 12-oxo-phytodienoic acid (OPDA), the jasmonic acid (JA) precursor that enters the peroxisome after being synthesized in the chloroplast, is converted to JA (Acosta and Farmer, 2010), and indole 3-butyric acid (IBA) is converted to the principal form of auxin, indole 3-acetic acid (IAA; Strader and Bartel, 2011). Other than the core β-oxidation pathway, which metabolizes straight-chain saturated fatty acids, auxiliary β-oxidation pathways also occur in the peroxisome to metabolize unsaturated fatty acids, in which case accessory enzymes are required (Goepfert and Poirier, 2007; Graham, 2008).
To assess the full composition of this versatile organelle in plants, both in silico analysis and experimental proteomics have been used to identify peroxisomal proteins. Bioinformatic analysis of the Arabidopsis (Arabidopsis thaliana) genome using Peroxisomal Target Signal type1 (PTS1) and PTS2 sequences predicted a total of over 400 proteins to be potentially peroxisomal (Lingner et al., 2011). Experimental proteomics of Arabidopsis, spinach (Spinacia oleracea), and soybean (Glycine max) peroxisomes using different tissue/cell types and development stages together identified several dozens of peroxisomal proteins that had not been found previously to associate with peroxisomes after in vivo targeting verification (Fukao et al., 2002, 2003; Arai et al., 2008a, 2008b; Eubel et al., 2008; Reumann et al., 2009; Babujee et al., 2010; Quan et al., 2013). After the identification of peroxisomal proteins from etiolated Arabidopsis seedlings through proteomics and in vivo protein-targeting analysis, we used reverse genetics to analyze the mutants of five newly identified proteins and revealed the role of a Cys protease, RESPONSE TO DROUGHT21A-LIKE1, in seed germination, β-oxidation, and stress response (Quan et al., 2013; Cassin-Ross and Hu, 2014). However, many other recently identified peroxisomal proteins have not been characterized with respect to their functions in peroxisomal physiology and plant development.
In this study, we interrogated the mutants of 27 recently identified peroxisomal genes with systematic phenotypic assays to analyze the function of the proteins in peroxisomal metabolism. Mutants for 20 of the tested genes showed statistically significant phenotypes in at least one of the assays. Additional analysis revealed that mutants for 7 of 20 genes displayed deficiencies in multiple assays, suggesting strongly that these seven proteins are involved in β-oxidation-related processes. This multifaceted screen enabled us to identify β-oxidation proteins that may not have been discovered otherwise by genetic screens based on single assays.
RESULTS
Identifying Mutants of Recently Discovered Arabidopsis Peroxisomal Genes
To investigate the role of recently identified and uncharacterized peroxisomal proteins in peroxisomal physiology, we took a reverse genetic approach by searching The Arabidopsis Information Resource database (http://www.Arabidopsis.org) for transfer DNA (T-DNA) insertion lines. Most of these proteins were identified from our own proteomic analyses of Arabidopsis peroxisomes from green leaves and etiolated seedlings (Reumann et al., 2009; Quan et al., 2013), and some were identified by other research groups. A few of the genes had been previously investigated in processes not directly related to β-oxidation, and a few others had been studied biochemically without mutant characterization. These included Calcium-dependent protein kinase1 (CPK1) and Hydroxy-acid oxidase isoform2 (HAOX2), which function in innate immunity (Coca and San Segundo, 2010; Rojas et al., 2012), 1,4-dihydroxy-2-naphthoyl (DHNA)-CoA thioesterase1 (DHNAT1), which is involved in DHNA-CoA hydrolysis in phylloquinone biosynthesis (Widhalm et al., 2012), Acetoacetyl CoA thiolase1 (AACT1), which is responsible for the condensation of two acetyl-CoA molecules to form acetoacetyl CoA (Jin et al., 2012), and Copper amine oxidase3 (CuAO3) in PA catabolism (Planas-Portell et al., 2013).
We obtained seed stocks of 53 putative mutant lines for 30 peroxisomal genes and genotyped these lines by PCR analysis of genomic DNA followed by sequencing of the PCR products. Despite numerous efforts, we were unable to confirm T-DNA insertion in six lines for three genes (Supplemental Table S1). In addition, homozygous plants could not be identified from seven lines (all in Columbia-0 [Col-0] background) for five peroxisomal proteins (i.e. NADP-dependent isocitrate dehydrogenase [ICDH], Uricase, Phosphogluconate dehydrogenase, Senescence-associated protein [B12D1], and Ala-glyoxylate aminotransferase2; Supplemental Table S1). We predicted that homozygous mutants for these five genes may be embryonic or seedling lethal in Col-0 background. Consistent with the notion that the null mutant of ICDH, an enzyme that regenerates NADPH, may be lethal, homozygous null mutants for two genes in the plastid-peroxisome dual-localized oxidative pentose-P pathway, which also generates NADPH, were found to be nonviable as well (Xiong et al., 2009; Bussell et al., 2013). These data support the idea that NADPH regeneration is essential for plant survival. Finally, the lethal phenotype seems to be ecotype specific for B12D1, because its null mutant was viable in Wassilewskija-4 (Ws-4) background.
In total, 40 homozygous T-DNA insertion mutants for 27 genes were obtained; at least two mutant alleles were available for 12 genes (Table I; Supplemental Fig. S1). We performed reverse transcription (RT) -PCR analysis to detect the level of full-length transcripts of the target genes and discovered that 30 mutants for 23 peroxisomal genes were possible null alleles (Supplemental Fig. S2). In addition, the expression levels of acyl-CoA thioesterase2 (ACH2), 2,4-dienoyl-CoA reductase (DECR), Epoxide hydrolase isoform3 (EH3), and two genes encoding unknown proteins (unknown protein5 [UP5] and UP7) were decreased in ach2-3, decr-3, eh3-1, up5-1, and up7-2, respectively, suggesting that these mutants were knockdown alleles. However, the expression levels of ICDH, calcium binding EF-hand (CEF), acyl transferase2 (ATF2), DHNAT1, and small thioesterase5 (sT5) were increased in icdh1, cef1, atf2-1, dhnat1-2, and st5-2, respectively, indicating that the target genes were overexpressed in these lines (Supplemental Fig. S2). All of these viable homozygous lines, including both loss-of-function mutants and overexpression lines, were used in subsequent analyses.
Table I. Confirmed homozygous peroxisomal mutants analyzed in this study.
Based on the level of the full-length target transcripts, mutants were considered to be knockout (KO), knockdown (KD), or overexpressor (OE). β-oxidation assays were Suc dependence (SC), 2,4-DB (DB), OPDA, detection of oil bodies using Nile Red (NR), or fatty acids quantification (FA). ABA, Seed germination in the presence of ABA; D/L, requires light for complete germination; N/A, not applicable; X, mutant exhibits a phenotype; —, mutant has no phenotype.
| Gene Locus | Acronym | Annotation | PTS | Allele | Line | Ecotype | Level of Target Transcripts | β-Oxidation Processes | D/L | ABA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KO | KD | OE | SC | DB | OPDA | NR | FA | |||||||||
| At1g01710 | ACH2 | Acyl-CoA thioesterase2 | SKL | ach2-1 | SALK_134567 | Col-0 | X | — | X | X | — | X | — | X | ||
| ach2-2 | GK_705E01 | Col-0 | X | — | X | X | — | X | — | X | ||||||
| ach2-3 | SALK_126817C | Col-0 | X | — | — | X | X | X | X | X | ||||||
| At1g16730 | UP6 | Unknown protein6 | CRL | up6-1 | SALK_122395 | Col-0 | X | — | X | X | — | N/A | X | X | ||
| At1g19570 | DHAR1 | Dehydroascorbate reductase1 | dhar1-1 | SALK_005382 | Col-0 | X | — | X | — | — | N/A | X | X | |||
| dhar1-2 | SALK_029966C | Col-0 | X | — | X | — | — | N/A | X | X | ||||||
| At1g20560 | AAE1 | Acyl-activating enzyme isoform1 | SKL | aae1-1a | SALK_041152 | Col-0 | X | — | — | X | X | X | X | X | ||
| At1g48320 | DHNAT1/sT1 | DHNA-CoA thioesterase1/small thioesterase1 | AKL | dhnat1-1b | CS846230 | Col-0 | X | — | X | X | — | N/A | X | X | ||
| dhnat1-2 | SALK_066477C | Col-0 | X | — | — | — | — | N/A | X | X | ||||||
| At1g50510 | INDA | Indigoidine synthase A | RIx5HL | inda1 | GK_681B01 | Col-0 | X | X | — | — | X | X | X | X | ||
| At1g54340 | ICDH/IDHP1 | NADP-dependent isocitrate deshydrogenase | SRL | icdh1 | SALK_034151 | Col-0 | X | X | X | — | — | N/A | X | X | ||
| At1g64850 | CEF | Calcium binding EF-hand | SRL | cef-1 | CS806024 | Col-3 | X | — | — | — | N/A | N/A | N/A | N/A | ||
| At1g77540 | ATF2 | Acetyltransferase2 | SSI | atf2-1 | FLAG_206E09 | Ws-4 | X | — | — | — | N/A | N/A | N/A | N/A | ||
| At2g29590 | sT5 | Small thioesterase5 | SKL | st5-1 | SALK_089257 | Col-0 | X | — | — | X | — | N/A | X | X | ||
| st5-2 | SALK_092512 | Col-0 | X | — | — | X | — | N/A | X | X | ||||||
| At2g42490 | CuAO3 | Copper amine oxidase3 | SKL | cuao3-1 | SALK_095214 | Col-0 | X | — | — | X | — | N/A | X | — | ||
| cuao3-2 | SALK_096065C | Col-0 | X | — | — | — | — | N/A | X | — | ||||||
| At3g12800 | SDRb/DECR | Short-chain dehydrogenase/reductase isoform b/2,4-dienoyl-CoA reductase | SKL | decr-2 | CS877641 | Col-0 | X | — | X | — | — | N/A | X | X | ||
| decr-3 | CS871159 | Col-3 | X | — | X | — | — | N/A | X | X | ||||||
| At3g14150 | HAOX2 | Hydroxy-acid oxidase isoform2 | SML | haox2-1b | SALK_102409C | Col-0 | X | — | — | — | N/A | N/A | N/A | N/A | ||
| At3g48140 | B12D1 | Senescence-associated protein/B12D-related protein | b12d1-1 | FLAG_548C12 | Ws-4 | X | — | X | — | X | X | X | — | |||
| At3g48170 | BADH | Betaine Aldehyde deshydrogenase | SKL | badh1 | CS822971 | Col-0 | X | — | X | — | X | X | X | X | ||
| At3g55640 | CADC | Ca2+-dependent carrier | cadc1 | CS832927 | Col-0 | X | — | X | — | — | N/A | X | X | |||
| cadc2 | GK_237H07 | Col-0 | X | — | X | — | — | N/A | X | X | ||||||
| At3g56460 | ZnDH | Zinc-binding dehydrogenase | SKL | zndh1 | SALK_056640 | Col-0 | X | — | — | — | N/A | N/A | N/A | N/A | ||
| zndh2 | SALK_082243 | Col-0 | X | — | X | — | N/A | N/A | N/A | N/A | ||||||
| At4g02340 | EH3 | Epoxide hydrolase isoform3 | ASL | eh3-1 | SALK_106529C | Col-0 | X | — | — | X | — | N/A | X | X | ||
| eh3-2 | SALK_149885C | Col-0 | X | — | — | — | — | N/A | X | X | ||||||
| At4g04320 | MCD | Malonyl-CoA decarboxylase | SRL | mcd1 | SALK_069574C | Col-0 | X | — | — | — | N/A | N/A | N/A | N/A | ||
| mcd2 | GK_859E12 | Col-0 | X | — | — | — | N/A | N/A | N/A | N/A | ||||||
| At4g09320 | NDPK1 | Nucleoside diphosphate kinase type1 | ndpk1-1 | SALK_089749 | Col-0 | X | — | — | — | N/A | N/A | N/A | N/A | |||
| At5g04870 | CPK1 | Calcium-dependent protein kinase1 | cpk1-1b | SALK_007698 | Col-0 | X | — | X | X | — | N/A | — | — | |||
| cpk1-2b | SALK_080155 | Col-0 | X | — | X | — | — | N/A | — | — | ||||||
| At5g11910 | ELT1 | Esterase/lipase/thioesterase family isoform1 | SRI | elt1-1 | FLAG_632F03 | Ws-4 | X | — | X | — | — | N/A | X | X | ||
| At5g16370 | AAE5 | Acyl-activating enzyme5 | SRM | aae5-1a | SALK_009731 | Col-0 | X | — | X | — | X | X | X | |||
| At5g44250 | UP5 | Unknown protein5 | SRL | up5-1 | SALK_107281 | Col-0 | X | X | X | — | X | X | X | X | ||
| At5g47720 | ACAT1.3/AACT1 | Acetoacetyl-CoA thiolase1.3 | SAL | aact1-2b | SALK_008505 | Col-0 | X | — | X | — | — | N/A | X | X | ||
| At5g58220 | TTL1 | Transthyretin-like protein1 | RLx5HL | ttl3 | SALK_137289 | Col-0 | X | — | — | — | N/A | N/A | N/A | N/A | ||
| At5g65400 | UP7 | Ser Hydrolase1 | SLM | up7-1 | SALK_062218C | Col-0 | X | X | — | — | — | N/A | X | X | ||
| up7-2 | SALK_067081 | Col-0 | X | — | — | — | — | N/A | X | — | ||||||
Except for CPK1 and AACT1, 25 peroxisomal proteins analyzed in this study were initially identified from the leaf peroxisome proteome (Supplemental Table S2), indicating that they may be involved in photorespiration or general plant growth. Photorespiration is a major peroxisomal function in green tissue, where peroxisomes, together with chloroplasts and mitochondria, convert phosphoglycolate produced by the oxygenase activity of Rubisco into glycerate, a molecule that is reused in the chloroplastic Calvin-Benson cycle (Foyer et al., 2009; Peterhansel et al., 2010). Apart from the control mutant peroxin14 (pex14), which is defective in peroxisomal protein import, none of the mutants showed obvious difference in appearance from the wild type while growing under ambient air (CO2 = 400 µL L−1) or decreased levels of CO2 (80 µL L−1), the latter of which was expected to enhance the growth defect of photorespiratory mutants because of the higher demand for photorespiration under low CO2 (Supplemental Fig. S3). We concluded that these proteins are not essential for plant growth or photorespiration or that they play redundant roles with other proteins in these processes under our laboratory conditions. Because the majority of the peroxisomal proteome is dedicated to β-oxidation-related processes (Hu et al., 2012), we next used parallel physiological assays to assess the efficiency of peroxisomal β-oxidation in these mutants.
Mutants for Four Genes Were Sugar Dependent in Postgerminative Growth
Before the establishment of photosynthesis, peroxisomal fatty acid β-oxidation is required to fuel early seedling growth. As a result, many β-oxidation mutants arrest or develop slowly after germination, a phenotype that can be rescued by adding a carbon source, such as Suc (Hu et al., 2012), to the growth medium. To check for Suc dependence, we grew mutants for 7 d on one-half-strength Linsmaier and Skoog (LS) medium in dark or light conditions with or without Suc and quantified hypocotyl or root lengths. Only differences between mutants and wild-type plants with a P value < 0.01 were considered statistically significant after an unpaired Student’s t test.
As expected, pex14 displayed sugar-dependent growth in both dark and light conditions (Fig. 1). Although not showing sugar dependence, dehydroascorbate reductase1 (dhar1), icdh, atf2, b12d1, and cpk1 had statistically shorter or longer hypocotyls and/or roots than wild-type plants, regardless of the presence of Suc in dark or light conditions (Fig. 1). Because the size of these mutants was indistinguishable from that of the wild type as adults (Supplemental Fig. S3), we reasoned that the developmental differences observed at the seedling stage were possibly overcome later.
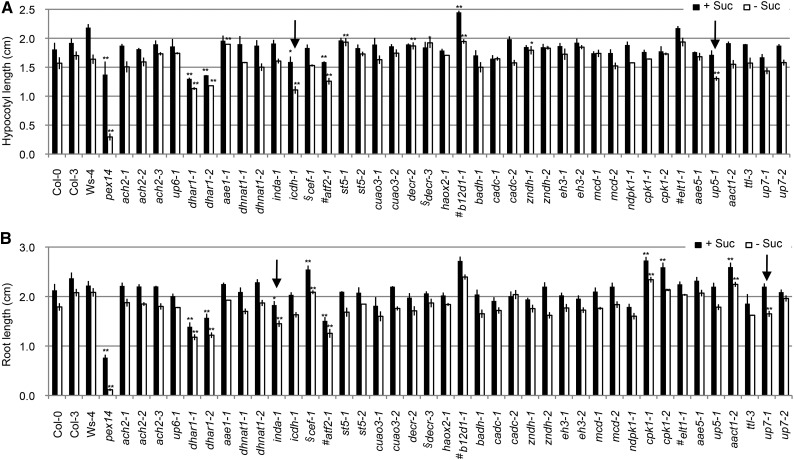
Figure 1.
Suc dependence assay. Shown are hypocotyl lengths of 7-d etiolated seedlings (A) and root lengths of 7-d light-grown seedlings (B), both of which were grown on one-half-strength LS medium with or without 1% Suc. Data represent means ± se of three independent experiments. For each experiment, n ≥ 30. Student’s t test. *, Statistically significant difference of P < 0.01 from the wild type (Col-0, Col-3, or Ws-4); **, statistically significant difference of P < 0.001 from the wild type (Col-0, Col-3, or Ws-4); #, mutants in Ws-4 background; §, mutants in Col-3 background; arrows, mutants that show sugar dependence.
In the dark, the absence of Suc resulted in an approximately 15% decrease in the hypocotyl length of wild-type Col-0 and most mutant seedlings, whereas the decrease was approximately 26% for icdh1 and 21% for up5-1 (Fig. 1A; Supplemental Table S3). In light-grown seedlings, the absence of Suc caused an approximately 16% reduction of the primary root length in Col-0 and most mutants, whereas this reduction was approximately 24% for both indigoidine synthase A (inda1) and up7-1 (Fig. 1B; Supplemental Table S3). These results indicated possible roles of ICDH and UP5 as well as INDA and UP7 in lipid mobilization during postgerminative growth in dark-grown and light-grown conditions, respectively.
Mutants for 14 Genes Showed Abnormal 2,4-Dichlorophenoxybutyric Acid Response
IBA is converted to IAA through peroxisomal β-oxidation (Strader and Bartel, 2011). To check whether any mutants showed reduced response to the inhibitory effect of IBA on primary root elongation, root lengths of 7-d seedlings grown on one-half-strength LS medium supplemented with IBA or IAA were measured. For the wild types and mutants, control treatment with 100 nm IAA resulted in similar degrees of decreases in the primary root length coupled with excessive root hair growth (Supplemental Fig. S4). The pex14 mutant seedlings exhibited strong IBA resistance, but all of the other mutants showed IBA response similar to that of the wild type (Supplemental Figs. S4 and S5), with no statistically significant differences observed at tested IBA concentrations (Student’s t test, P < 0.01).
Given that 2,4-dichlorophenoxybutyric acid (2,4-DB) is converted to the synthetic auxin analog 2,4-dichlorophenoxy acetic acid (2,4-D) by β-oxidation (Hayashi et al., 2000), we studied the response of the mutants to 2,4-DB and 2,4-D. Wild-type plants and the tested mutants showed similar levels of response to 100 nm 2,4-D (i.e. shortened primary root length and excessive root hairs; Supplemental Fig. S4). Wild-type seedlings showed a heterogeneous response when growing on 2,4-DB (Supplemental Fig. S6). Therefore, we instead scored the percentage of 2,4-DB-resistant seedlings, which were defined as having root length statistically the same as that of untreated seedlings. Treatment with 0.4, 0.8, and 1 µm 2,4-DB resulted in a decrease of the primary root length in approximately 30%, 65%, and 67%, respectively, of wild-type Col-0 seedlings but only approximately 8%, 24%, and 26%, respectively, of wild-type Ws-4 seedlings (Fig. 2). Because wild-type Ws-4 and Col-0 plants showed similar sensitivity to 2,4-D (Supplemental Fig. S4), we speculated that Ws-4 plants were less sensitive to 2,4-DB and therefore, assessed the response of mutants in Ws-4 background with higher concentrations of 2,4-DB; 66% and 95% of wild-type Ws-4 seedlings were sensitive to 1.5 and 2 µm 2,4-DB, respectively (Supplemental Fig. S7).
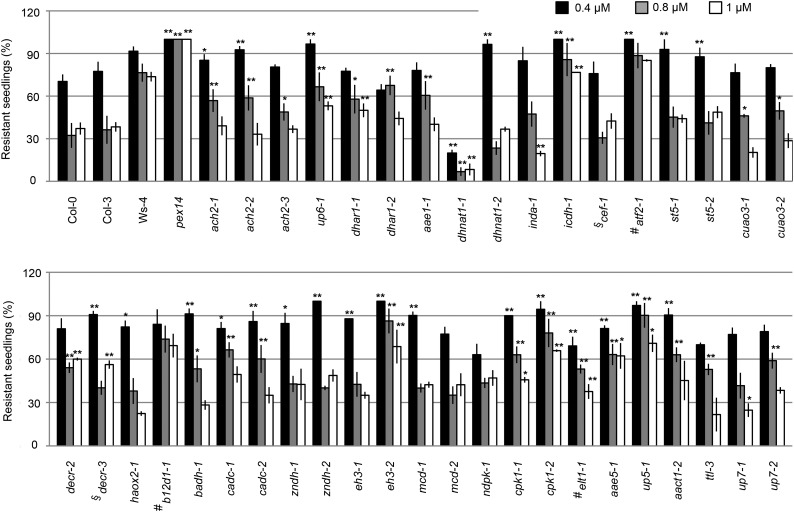
Figure 2.
2,4-DB response assay. Percentage of 7-d light-grown seedlings displaying resistance to 2,4-DB after growing on one-half-strength LS medium supplemented with 0.5% Suc and 0.4, 0.8, or 1 µm 2,4-DB is presented. Resistance was defined as having a root length with no statistically significant difference from that on medium without 2,4-DB. Data represent means ± se of three independent experiments. For each experiment, n ≥ 30. Student’s t test. *, Statistically significant difference of P < 0.01 from the wild type (Col-0, Col-3, or Ws-4); **, statistically significant difference of P < 0.001 from the wild type (Col-0, Col-3, or Ws-4); #, mutants in Ws-4 background; §, mutants in Col-3 background.
As shown in Figure 2 and Supplemental Figure S7, dhnat1-1 and esterase/lipase/thioesterase family isoform1 (elt1-1) had statistically lower percentages of resistant seedlings compared with wild-type plants and were, therefore, considered hypersensitive to 2,4-DB. In contrast, ach2, up6-1, dhar1, icdh1, b12d1, betaine aldehyde dehydrogenase1 (badh1), Ca2+-dependent carrier (cadc), eh3-2, cpk1, acyl-activating enzyme5 (aae5-1), up5-1, and aact1-2 showed statistically higher percentages of resistant seedlings than the wild type under at least two concentrations of 2,4-DB (Student’s t test, P < 0.01) and were considered as having reduced 2,4-DB response. Taken together, these results suggested that DHNAT1, ELT1, ACH2, UP6, DHAR1, ICDH, B12D1, BADH, CADC, EH3, CPK1, AAE5, UP5, and AACT1 may be involved in 2,4-DB metabolism. The fact that ICDH was overexpressed in icdh1 implied that ICDH may play a negative role in 2,4-DB catabolism.
Mutants of Nine Genes Showed Deficiency in OPDA Metabolism
Methyl-jasmonic acid (MeJA) inhibits primary root elongation (Staswick et al., 1992). We reasoned that the JA precursor OPDA, which is converted to JA through peroxisomal β-oxidation, would similarly inhibit primary root elongation. Based on this assumption, we previously developed a simple assay to identify peroxisomal proteins involved in OPDA metabolism (Cassin-Ross and Hu, 2014). The same assay was applied in this study to assess the function of the tested proteins in OPDA metabolism.
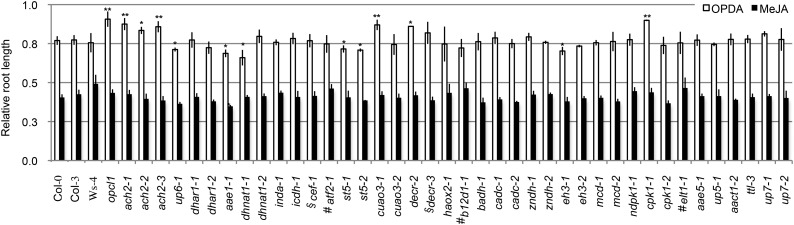
Compared with the wild type, the control mutant OPC-8:0 CoA ligase1 (opcl1), which is defective in 3-oxo-2-(2′-[Z]-pentenyl)cyclopentane-1-octanoic acid (OPC)8:0 CoA Ligase1 in JA biosynthesis (Koo et al., 2006), showed resistance to the inhibition of OPDA on primary root elongation, whereas MeJA exerted a similar effect on opcl1 and wild-type plants, suggesting that opcl1 was specifically resistant to OPDA (Fig. 3). Except for aae1-1, all other mutants responded to MeJA similarly to the wild type (i.e. approximately 60% inhibition of root elongation; Fig. 3). The primary root of ach2, copper amine oxidase3 (cuao3-1), decr-2, and cpk1-1 showed reduced OPDA response similar to opcl1, whereas up6-1, aae1-1, dhnat1-1, st5, and eh3-1 seemed to show enhanced response to OPDA (Student’s t test, P < 0.01; Fig. 3). These results suggested that ACH2, CUAO3, DECR, UP6, AAE1, DHNAT1, ST5, EH3, and CPK1 may affect the catabolism of OPDA.
Figure 3.
Root response to OPDA. Relative root lengths (treated versus untreated) of 7-d seedlings grown on one-half-strength LS medium supplemented with 250 nm OPDA or 10 µm MeJA are shown. Data represent means ± se of three independent experiments. For each experiment, n = 50. Student’s t test. *, Statistically significant difference of P < 0.01 from the wild type (Col-0, Col-3, or Ws-4); **, statistically significant difference of P < 0.001 from the wild type (Col-0, Col-3, or Ws-4); #, mutants in Ws-4 background; §, mutants in Col-3 background.
Most Proteins with β-Oxidation-Related Phenotypes Are Involved in Seed Germination
Our Suc dependence and 2,4-DB/OPDA response assays together identified 31 mutants for 20 peroxisomal genes that showed a phenotype in at least one of the assays; 10 of the genes had at least two alleles with similar phenotypes. These data suggested the potential roles of AACT1, AAE1, AAE5, ACH2, B12D1, BADH, CADC, CPK1, CUAO3, DECR, DHAR1, DHNAT1, EH3, ELT1, INDA, ICDH, ST5, UP5, UP6, and UP7 in β-oxidation. Because fatty acid β-oxidation plays a key role in seed germination and early postgerminative growth (Theodoulou and Eastmond, 2012), we determined whether these 20 proteins are involved in seed germination. Positive controls used in this experiment and subsequent assays were previously characterized null mutants acyl-CoA oxidase4-1 (acx4-1) and 3-ketoacyl CoA thiolase2-3 (kat2-3), which are impaired in genes encoding the core β-oxidation enzymes ACX4 and KAT2, respectively (Germain et al., 2001; Rylott et al., 2003).
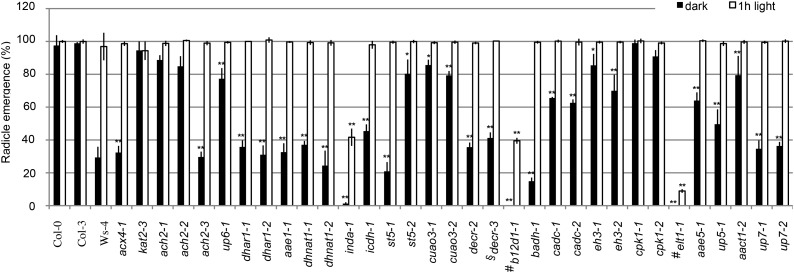
Fresh seeds from all 30 mutants germinated normally as quantified by radicle emergence from seeds grown on plain agar (0.8% [w/v] agar) or one-half-strength LS medium (Supplemental Fig. S8). We then tested seed germination in response to factors known to influence this process. Light is a positive regulator of seed germination (Lau and Deng, 2010), and therefore, we compared the germination rate of mutant seeds that were placed in total darkness with that of mutant seeds that were subjected to 1 h of light treatment before germination. Quantification of radicle emergence from 5-d seedlings revealed that, whereas wild-type Col-0, Columbia-3 (Col-3), and the positive control kat2-3 showed no light dependence, wild-type Ws-4 seemed to depend more on light treatment for germination (Fig. 4). Together with acx4-1, mutants for 19 of 20 genes tested showed statistically lower germination rates compared with their respective wild-type controls when germinated in total darkness (Student’s t test, P < 0.01), and the 1 h of light pretreatment could rescue the germination potential for all of these mutants, except for inda-1, b12d1-1, and elt1-1 (Fig. 4).
Figure 4.
Seed germination in the mutants. Percentage of radicle emergence from seeds on plain agar (0.8% [w/v] agar) in the dark without or with 1 h of light pretreatment is presented. Data represent means ± se of three independent experiments. For each experiment, n = 50. Student’s t test. *, Statistically significant difference of P < 0.01 from the wild type (Col-0, Col-3, or Ws-4); **, statistically significant difference of P < 0.001 from the wild type (Col-0, Col-3, or Ws-4); #, mutants in Ws-4 background; §, mutants in Col-3 background.
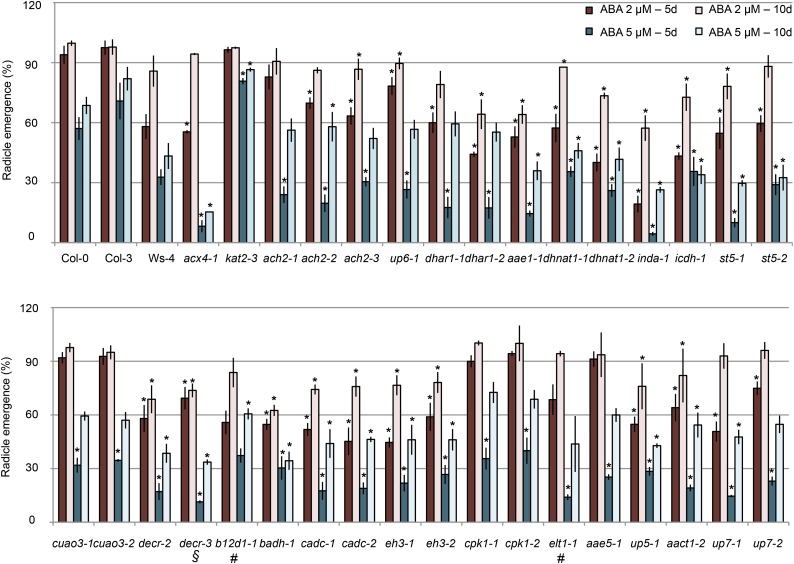
We also quantified seed germination rate on medium supplemented with the phytohormone abscisic acid (ABA), which acts synergistically with OPDA to inhibit germination (Vanstraelen and Benková, 2012). Radicle emergence was quantified on seeds sown on plain agar supplemented with 2 or 5 µm ABA (Fig. 5). As previously reported, kat2-3 exhibited insensitivity to ABA, regardless of the concentration (Jiang et al., 2011). However, germination rate of acx4-1 was statistically lower than that in the wild type, and similarly, most of the 5- or 10-d mutant seedlings showed statistically significant hypersensitivity to 2 µm ABA compared with the wild type (Student’s t test, P < 0.001). At 5 µm ABA, all of the 5-d seeds except b12d1-1 showed hypersensitive response, whereas at 10 d, the germination rates for ach2, up6, dhar1, cuao3, cpk1, and aae5 were comparable with the germination rate of the wild type (Fig. 5).
Figure 5.
Seed germination in response to ABA. Percentage of radicle emergence from seeds grown on plain agar supplemented with 0, 2, or 5 µm ABA for 5 and 10 d is presented. Data represent relative means (treated versus untreated) ± se of three independent experiments. For each experiment, n = 100. Student’s t test. *, Statistically significant difference of P < 0.001 from the wild type (Col-0, Col-3, or Ws-4); #, mutants in Ws-4 background; §, mutants in Col-3 background.
Based on results from these two germination assays, we concluded that most of these 20 proteins have potential positive roles in initial seed germination and that the role for INDA and B12D1 seemed more prominent given the stronger phenotypes of their mutants. To determine whether the seed germination phenotypes correlated with gene expression pattern, we analyzed the expressions of these 20 genes during seed maturation and germination. Using data from the publicly available Botany Array Resource database (Toufighi et al., 2005), we generated a heat map using log2-normalized data of the relative expression for 18 of these genes (data for INDA and UP7 were unavailable). During some stages of seed maturation and germination, UP6, DECR, EH3, AACT1, and AAE5 were up-regulated, whereas DHAR1, ACH2, sT5, DHNAT1, AAE1, CuAO3, ICDH, BADH, ELT1, and B12D1 were repressed to various levels. Consistent with the relatively strong germination phenotype of the b12d1 mutant, the expression of B12D1 is markedly increased during the seed germination process (Supplemental Fig. S9; Supplemental Table S4).
Mutants for Seven Peroxisomal Proteins Retain Oil Bodies and Accumulate Fatty Acids in Early Seedling Development
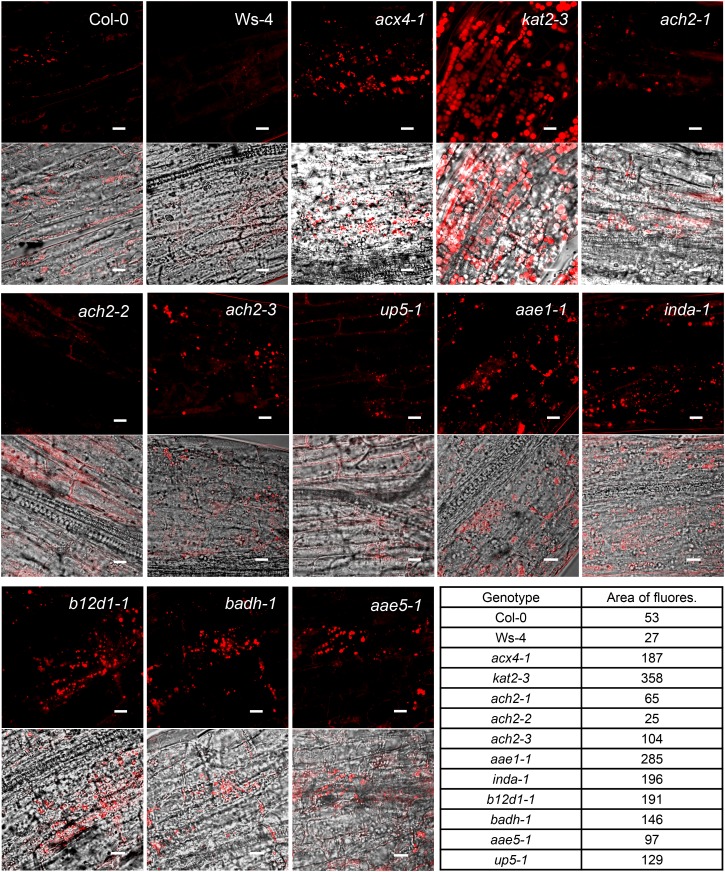
Previous studies have shown that the inability to break down triacylglycerol (TAG) leads to the prolonged presence of oil bodies in β-oxidation mutants (Graham, 2008). To assess lipid mobilization defects in the peroxisomal mutants, we used the lipophilic stain Nile Red (Greenspan et al., 1985) to detect the presence of lipid bodies in hypocotyls of 5-d and 7-d etiolated seedlings. Fluorescence microscopy revealed that, 5 d after germination, both wild-type and mutant hypocotyls contained abundant oil bodies (Supplemental Fig. S10), which were markedly diminished at 7 d in the wild type and most mutants (Supplemental Fig. S11). In contrast, the two positive controls (acx4-1 and kat2-3) and seven mutants (ach2-3, aae1-1, inda-1, b12d1-1, badh-1, aae5-1, and up5-1) still retained a significant number of oil bodies 7 d after germination (Fig. 6).
Figure 6.
Confocal microscopic visualization of oil bodies in the hypocotyl of 7-d etiolated seedlings. Seedlings were grown on 0.8% (w/v) agar plate. Nile Red fluorescence images (top) and merged images (bottom) of bright-field and Nile Red fluorescence are shown. Area of fluorescence is the arbitrary value (assigned by Image J) within the 75-µm2 area of the Nile Red fluorescent images shown. Bars = 10 µm.
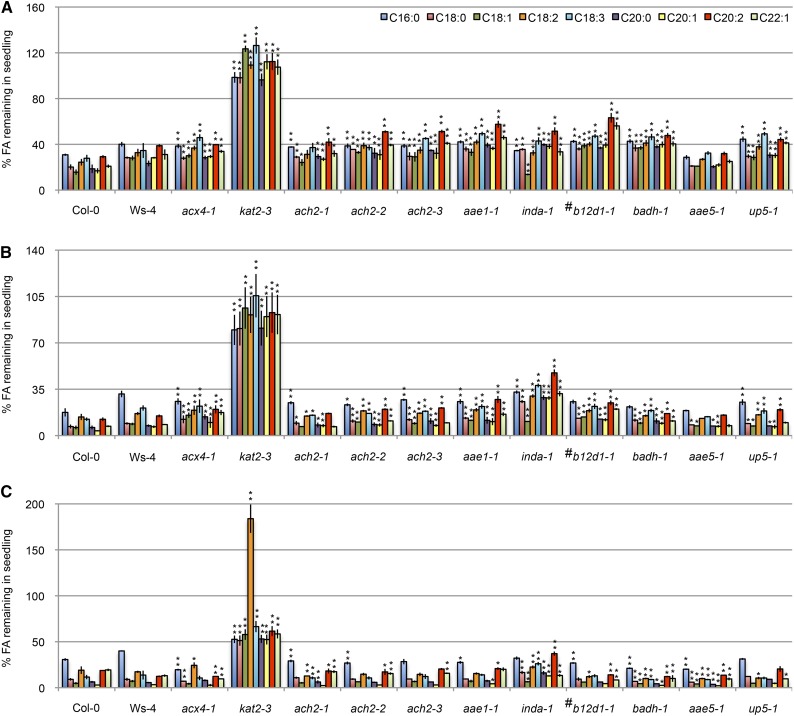
To confirm the oil body accumulation phenotype in these seven mutants, we quantified long-chain fatty acid species and compared their levels in 3-d, 5-d, and 7-d etiolated seedlings with seeds. At 3 and 5 d after germination, the amount of C20:1, which is a marker for TAG (Lemieux et al., 1990), remained at a statistically higher level in virtually all of the mutants than in wild-type seedlings (Fig. 7, A and B). Similarly, higher accumulation of other long-chain fatty acids (i.e. C16:0, C18:0, C18:1, C18:2, C18:3, C20:2, and C20:3) was observed in nearly all of the mutants at 3 d and all of the mutants at 5 d (Student’s t test, P < 0.01; Fig. 7, A and B). Consistent with the strongest retention of oil bodies found in this study (Fig. 6), kat2-3 showed the highest accumulation of all long-chain fatty acids analyzed (Fig. 7). At 7 d after germination, the level of long-chain fatty acids was still higher in kat2-3 and inda1 but comparable with or even lower in other mutants compared with the wild type (Fig. 7C). These data support the notion that ach2-3, aae1-1, inda-1, b12d1-1, badh-1, aae5-1, and up5-1 had a reduced rate of β-oxidation, which caused delayed degradation of the fatty acid substrates.
Figure 7.
Fatty acid (FA) quantification in seedlings. Percentage of FAs remaining is shown for 3-d (A), 5-d (B), and 7-d (C) etiolated seedlings grown on 0.8% (w/v) agar plates compared with the level in seeds. The amount of FA in dry seeds was set as 100%. Data represent means ± se of three independent experiments. For each experiment, n ≥ 50. *, Statistically significant difference of P < 0.01 from the wild type (Col-0, Col-3, or Ws-4); **, statistically significant difference of P < 0.001 from the wild type (Col-0, Col-3, or Ws-4); #, mutants in Ws-4 background.
DISCUSSION
Multipronged Phenotypic Screen Allows the Identification of Additional Peroxisomal Proteins Potentially Involved in β-Oxidation
Forward genetic screens for mutants with sugar dependence or resistance to 2,4-DB/IBA identified key enzymes in β-oxidation and proteins involved in peroxisome biogenesis (Hu et al., 2012). However, these screens rely on single assays and tend to isolate mutants with strong visual phenotypes. In this study, we used multiple assays to simultaneously screen mutants of 27 uncharacterized peroxisomal genes in a quantitative manner. Mutants of 20 genes showed statistically significant phenotypes in β-oxidation-related processes, and additional investigations discovered a subset of them with abnormal seed germination, accumulation of oil bodies, and delayed degradation of long-chain fatty acids during early seedling development. Mutants for seven genes exhibited deficiencies in multiple β-oxidation-based assays, strongly suggesting the involvement of these proteins in peroxisomal β-oxidation. ACH2, AAE1, and AAE5 are isoforms of enzymes known to be related to β-oxidation, whereas INDA, B12D1, BADH, and UP5 belong to functional categories previously unknown to be associated with β-oxidation; their roles in this pathway could be direct or indirect. This screen provided a complementary approach to previous genetic screens in the identification of β-oxidation proteins that may not have been uncovered by single assay-based screens. In addition, although β-oxidation occurs throughout the lifecycle of a plant, many mutants in this pathway do not show obvious phenotypes at mature stages (Hu et al., 2012). Therefore, using assays aimed at dissecting β-oxidation in initial plant development, this study was able to unmask phenotypes for several mutants and will help us understand the β-oxidation network in more depth.
ACH2, an ACH Involved in the Hydrolysis of Long-Chain Fatty Acyl-CoAs
Acyl-CoA thioesterase enzymes (ACOTs) hydrolyze fatty acyl-CoAs, yielding free fatty acids and CoA. The complex role of these enzymes in lipid metabolism has been previously documented, particularly in animals (Hunt et al., 2012). Mammalian ACOT8, the closest homolog to the two Arabidopsis peroxisomal ACOTs (ACH1 and ACH2), showed high activities toward a broad range of acyl-CoA substrates and was strongly inhibited by CoA, suggesting that ACOT8 is involved in the regulation of the intracellular levels of acyl-CoAs, free fatty acids, and CoA (Hunt et al., 2002; Ofman et al., 2002).
ACH2 was the first ACH to be cloned from plants, and its recombinant protein showed high levels of ACH activity against both medium-chain and long-chain fatty acyl-CoAs, with the highest activity toward long-chain unsaturated fatty acyl-CoAs (Tilton et al., 2000, 2004). We have shown in this study that loss-of-function ach2 mutants have higher accumulation of long-chain fatty acids in germinated seedlings and partial resistance to 2,4-DB and OPDA, supporting the positive role for ACH2 in β-oxidation. Interestingly, the ACH2 knockdown allele ach2-3 displayed a stronger seed germination phenotype than the null alleles, possibly because of functional compensation among multigene family members in the null alleles. Our study has expanded the list of potential substrates for ACH2, suggesting that it may be involved in the regulation of the intracellular level of acyl-CoAs, acid-CoAs (OPDA and 2,4-DB), free fatty acids, acids, and CoA. However, the high expression of ACH2 in mature tissues, such as young leaves and flowers, the low expression of ACH2 in germinating seedlings (Tilton et al., 2004), the relatively weak phenotypes observed in ach2 mutants in seed germination, and the fact that ach2 mutant was never isolated from previous forward genetic screens together suggest that ACH2 may not play a major role during early seedling development.
Two Additional Isoforms of Acyl-Activating Enzymes Involved in Early Seedling Development
Arabidopsis AAE1 and AAE5 belong to a 14-member plant-specific clade in the acyl-activating enzymes superfamily (Shockey et al., 2003). Some proteins from this clade have been reported to contain acid-CoA ligase activities. Benzoyloxyglucosinolate1 possesses benzoyl-CoA ligase activity (Kliebenstein et al., 2007), and AAE7 and AAE11 have acyl-CoA synthetase activity toward short and medium chains, respectively (Shockey et al., 2003). However, the function and/or substrate specificity of AAE1 and AAE5 and those of many other AAEs are still unknown.
It was reported that AAE1 is not involved in the activation of short-chain to medium-chain acid substrates ranging in length from C2 (acetate) to C14 (myristate; Shockey et al., 2003). We have shown in this study the accumulation of oil bodies and delayed degradation of long-chain fatty acids in aae1-1 etiolated seedlings. In addition, aae1-1 is hypersensitive to ABA and OPDA, two hormones required for dormancy. These results suggest that, during seed germination and early seedling development, AAE1 is involved in the peroxisomal activation of long-chain fatty acids before these substrates enter the β-oxidation cycle.
In our study, the loss-of-function mutant of AAE5 showed a weaker phenotype than that of AAE1, where aae5-1 exhibited partial resistance to 2,4-DB, weak accumulation of oil bodies, and delayed degradation of long-chain fatty acids in etiolated seedlings. These results suggest that AAE5 may be involved in the activation of long-chain fatty acids in Arabidopsis, but its role is not as prominent as AAE1 in germination and early seedling development. AAE1 is ubiquitously expressed throughout the plant, whereas the expression of AAE5 is limited to developing seeds and roots (Shockey et al., 2003). In light of its high expression in developing seeds, a tissue in which the flow of fatty acids can be adjusted through β-oxidation (Poirier et al., 1999), we speculate that AAE5 may play a stronger role in activating long-chain fatty acids in developing seeds.
Taken together, our assays enabled us to distinguish the functions of AAE1 and AAE5, two proteins that encode enzymes with the same biochemical activity and had been considered functionally redundant.
A BADH Involved in β-Oxidation
BADH belongs to subfamily 10 of the aldehyde dehydrogenase family, which is composed of amino-aldehyde dehydrogenases (EC 1.2.1.19), 4-aminobutyraldehyde dehydrogenases, 4-guanidinobutyr-aldehyde dehydrogenases (EC 1.2.1.54), and BADHs (EC 1.2.1.8; Brocker et al., 2013). Based on findings that BADH is induced by ABA, dehydration, salt, chilling, and oxidative stresses and that BADH oxidizes betaine aldehyde, 4-aminobutyraldehyde, and 3-aminopropionaldehyde to their corresponding carboxylic acids, it has been suggested that BADH may serve as a detoxification enzyme essential for the control of the level of aminoaldehydes that are produced in response to abiotic stresses (Missihoun et al., 2011). A recent study suggested that BADH, in conjunction with CuAO2 and CuAO3, functions in PA homeostasis (Planas-Portell et al., 2013). Similar to BADH, the expression of CuAO3 is also increased in response to ABA, MeJA, salicylic acid, and flagellin, suggesting that PA catabolism may be involved in plant stress response (Planas-Portell et al., 2013). In addition, PAs are essential for a wide range of physiological processes, including seed germination (Pieruzzi et al., 2011).
We have shown that plants lacking BADH are hypersensitive to the inhibitory effect of ABA on germination, accumulate higher amounts of long-chain fatty acids and oil bodies days after germination, and exhibit 2,4-DB resistance. These data point to a positive role for BADH in β-oxidation and seed germination. The role of BADH in β-oxidation remains to be determined at the biochemical level.
An Indigoidine Synthase in Seed Germination
INDA is involved in the biosynthesis of indigoidine, a blue pigment first described in the bacterium Erwinia chrysanthemi, which is implicated in protecting bacteria against oxidative stresses generated by reactive oxygen species produced during plant defense (Reverchon et al., 2002). Other than being present in most bacteria, INDA is found in the yeast Candida elegans and Arabidopsis (Finn et al., 2014). Based on the low expression level of this gene in Arabidopsis (Supplemental Fig. S2), we predict that INDA may play a regulatory rather than a housekeeping role and that its expression may be under tight temporal/spatial control. Among all of the mutants tested in this study, plants lacking INDA exhibited the strongest phenotype across most assays. Although the biochemical pathway in which this protein is involved remains speculative, INDA seems to have a profound effect on β-oxidation.
B12D1 in β-Oxidation
The expression of the barley (Hordeum vulgare) B12D gene (HvB12Dg1) is high in the aleurone layer and the embryo of developing seeds, diminishes toward seed maturity, and reappears in germinating seeds (Aalen et al., 1994). Consistently, the expression of HvB12Dg1 was shown to be regulated by two key hormones that control seed germination: up-regulation by gibberellic acid and down-regulation by ABA (Steinum and Berner, 1998). Similarly, Arabidopsis B12D1 is preferentially expressed during seed maturation and germination, causing poor seed germination in its absence. In cereal seeds, proteins localized in aleurone and embryo are involved in the synthesis and accumulation of lipid bodies, desiccation tolerance, and dormancy (Aalen et al., 1994), but the precise function of HvB12D1 is still unclear. We have shown that plants lacking B12D1 are resistant to 2,4-DB and accumulate oil bodies and long-chain fatty acids in germinated seedlings, suggesting the involvement of B12D1 in β-oxidation-related processes during germination. The biochemical function of B12D1 remains to be determined.
CONCLUSION
Our systematic and quantitative analysis of mutants of recently discovered Arabidopsis peroxisomal genes identified additional peroxisomal proteins with varying degrees of contribution to β-oxidation. This study has taken a step forward toward completely dissecting the plant β-oxidation network and it will provide a framework for future investigations to integrate genetics and physiology with biochemical and metabolic assays to identify substrates for all enzymes in β-oxidation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotypes Col-0, Col-3, and Ws-4 as well as pex14 (SALK_007441), opcl1 (SALK_140659), acx4-1 (SALK_000879), and kat2-3 (SALK_024922) mutants were used as controls. Seed stocks were obtained from the Arabidopsis Biological Resource Center, the Nottingham Arabidopsis Stock Centre, and the Institut National de la Recherche Agronomique-Versailles Genomic Resource Center (Table I; Supplemental Table S1). Surface-sterilized seeds were plated on one-half-strength LS medium (Caisson Laboratories, Inc.) solidified with 0.8% (w/v) agar and supplemented with 1% (w/v) Suc. Plants were grown at 22°C under continuous illumination at 75 µmol photons m−2 s−1. Homozygous lines were identified by PCR analysis of genomic DNA from 7-d seedlings. The location of the T-DNA insertion was confirmed by sequencing of the PCR products. Primers used are listed in Supplemental Table S5.
RNA Analysis
For RT-PCR analysis of the mutants, total RNA was isolated from 7-d seedlings grown on medium with 1% (w/v) Suc as described previously (Mallory et al., 2001). After being treated with DNaseI (QIAGEN), 1 µg of each RNA sample was reverse transcribed using the SuperScript III Reverse Transcriptase Kit (Invitrogen). The expression level of the target gene was analyzed by PCR using 50 ng of complementary DNA for 25 (for UBIQUITIN10) or 35 (for target gene) cycles. The gene-specific primers used are summarized in Supplemental Table S5.
Physiological Assays
For germination assays, seeds harvested from plants that had been grown simultaneously were sown on 0.8% (w/v) agar plates (i.e. plain agar plate) or one-half-strength LS medium solidified with 0.8% (w/v) agar. The plates were kept at 4°C for 4 d before being transferred to a growth chamber with continuous low light intensity (plates covered with mesh) or darkness. An additional set of plates was exposed to light for 1 h before being placed in the dark in the growth chamber. After 5 d, radicle emergence was scored. For seed germination in response to ABA, seeds were sown on plain agar plates supplemented with ABA (Sigma-Aldrich). After 4 d of stratification at 4°C, plates were transferred to the growth chamber and grown in the light for 5 or 10 d before radicle emergence was scored. All of the data are representative of at least three independent experiments. For each experiment, n = 50.
For Suc dependence analysis, seeds were placed on plates supplemented with or without 1% (w/v) Suc and stratified at 4°C for 2 d. After 7 d in the growth chamber with continuous low-intensity light or darkness, the plates were scanned using an EPSON scanner (Epson Perfection 4870 PHOTO). Hypocotyl lengths of dark-grown seedlings and roots of light-grown seedlings were measured using ImageJ (imagej.nih.gov/ij/). To study IAA, IBA, 2,4-D, 2,4-DB, and OPDA responses, seeds were sown on plates containing 0.5% (w/v) Suc and various concentrations of IAA (BioWORLD), IBA (Sigma-Aldrich), 2,4-D (Sigma-Aldrich), 2,4-DB (Sigma-Aldrich), OPDA (Cayman Chemical), or MeJA (Sigma-Aldrich) as described in the figures. The plates were then stored at 4°C for 2 d and placed in the growth chamber with continuous low-intensity light. Root length was quantified at 7 d. All of the data are representative of at least three independent experiments. For each experiment, n = 50.
To detect photorespiratory phenotypes, 2-week seedlings were transferred from plates to soil and placed in a growth cabinet with a controlled environment at a light intensity of 115 µmol photons m−2 s−1, 20°C, 16-h-light/8-h-dark photoperiod, and CO2 concentration of 80 (low CO2 concentration) or 400 µL L−1 (ambient air). After 2 weeks, plants were photographed with a COOLPIX 8800 VR camera (Nikon).
Visualization of Oil Bodies
Etiolated seedlings grown on 0.8% (w/v) agar for 5 or 7 d were stained for 5 min with 1 µg mL−1 of aqueous solution of Nile Red (Molecular Probes) as previously described (Greenspan et al., 1985; Linka et al., 2008). Images were recorded using an Olympus FluoView 1000 Spectral-Based Laser Scanning Confocal Microscope System (excitation wavelength = 450–500 nm; emission wavelength > 528 nm) with the 60× objective. All of the data are representative of at least three independent experiments. For each experiment, n = 12.
Fatty Acid Analysis
Amounts of fatty acids in seeds and seedlings were analyzed as fatty acid methyl esters (FAMEs) according to a published protocol (Li et al., 2006). Briefly, 50 seedlings or 100 seeds were placed in a tube with a screw cap. Added to the tube were 2 mL of 5% (v/v) H2SO4 in methanol, 300 µL of Toluene, and 10 to 60 µg of Tri15:0TAG (amount used depends on the tissue) as the internal standard. The tubes were capped and vortexed to ensure submergence of the sample; later, they were heated for 90 min at 85 °C. The resulting FAMEs were extracted using hexane and 0.9% NaCl, dried down under nitrogen gas, and resuspended in the appropriate volume of hexane. FAMEs were analyzed by gas chromatography with a flame ionization detector. Samples were separated on a DB-23 capillary column (30 m × 0.25 mm i.d., 0.25-µm film thickness; J&W Scientific), and helium was used as carrier gas at a constant flow of 1.5 mL min−1. For gas chromatography, oven temperature was maintained at 140°C for 3 min followed by a 5°C per minute increase until the oven reached 230°C and a final 3 min at 230°C. Injector and detector were maintained at 250°C throughout the analysis.
Statistical Analysis
To reveal statistical differences between the mutants and the wild type, all data generated from the assays were subjected to an unpaired Student’s t test. We considered differences between mutants and wild-type plants with P < 0.01 as significant.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematics of peroxisomal genes analyzed in this study.
Supplemental Figure S2. RT-PCR analysis of 7-d mutant seedlings.
Supplemental Figure S3. Images of 4-week-old plants grown in ambient air or under low CO2.
Supplemental Figure S4. IAA and 2,4-D responses.
Supplemental Figure S5. IBA response of peroxisomal mutants.
Supplemental Figure S6. Responsiveness of Col-0 and pex14 to 2,4-DB.
Supplemental Figure S7. Responsiveness of mutants in Ws-4 background to 2,4-DB.
Supplemental Figure S8. Seed germination potential of peroxisomal mutants.
Supplemental Figure S9. Heat map showing relative expression levels of candidate genes in seed maturation and germination.
Supplemental Figure S10. Visualization of hypocotyl oil bodies in 5-d etiolated seedlings.
Supplemental Figure S11. Visualization of hypocotyl oil bodies in 7-d etiolated seedlings.
Supplemental Table S1. Lines without T-DNA insertion and lines for which homozygotes could not be identified.
Supplemental Table S2. Identification of peroxisomal proteins analyzed in this study from previous proteomic studies.
Supplemental Table S3. Measurements for the Suc dependence assays.
Supplemental Table S4. Log2 expression values downloaded from the Bio-Analytic Resource expression browser for heat map generation.
Supplemental Table S5. Primers used for genotyping and RT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Navneet Kaur for helpful discussions throughout this project and critical reading of the article; Dr. Henrik Tjellström for help with fatty acid measurements; Drs. Dirk Colbry and Ian Dworkin for expert assistance in generating heat maps; Dr. Melinda Frame for helping with confocal microscopy; Drs. Gregg Howe and Gilles Basset for providing opcl1 and dhnat1-1 seeds; and the Arabidopsis Biological Resource Center, the Nottingham Arabidopsis Stock Centre, and the Institut National de la Recherche Agronomique-Versailles Genomic Resource Center for providing most of the mutant seeds.
Glossary
- ABA
abscisic acid
- Col-0
Columbia-0
- Col-3
Columbia-3
- FAME
fatty acid methyl ester
- IAA
indole 3-acetic acid
- IBA
indole 3-butyric acid
- JA
jasmonic acid
- LS
Linsmaier and Skoog
- MeJA
methyl-jasmonic acid
- OPDA
12-oxo-phytodienoic acid
- PA
polyamine
- RT
reverse transcription
- TAG
triacylglycerol
- Ws-4
Wassilewskija-4
- 2,4-D
2,4-dichlorophenoxy acetic acid
- 2,4-DB
2,4-dichlorophenoxybutyric acid
- T-DNA:
transfer DNA
Footnotes
This work was supported by the National Science Foundation Arabidopsis 2010 Program (Molecular and Cellular Biology grant no. 0618335) and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant no. DE–FG02–91ER20021 to J.H.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aalen RB, Opsahl-Ferstad HG, Linnestad C, Olsen OA. (1994) Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. Plant J 5: 385–396 [DOI] [PubMed] [Google Scholar]
- Acosta IF, Farmer EE. (2010) Jasmonates. Arabidopsis Book 8: e0129, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y, Hayashi M, Nishimura M. (2008a) Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol 49: 526–539 [DOI] [PubMed] [Google Scholar]
- Arai Y, Hayashi M, Nishimura M. (2008b) Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid beta-oxidation in soybean and Arabidopsis. Plant Cell 20: 3227–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babujee L, Wurtz V, Ma C, Lueder F, Soni P, van Dorsselaer A, Reumann S. (2010) The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin K1) biosynthesis in plant peroxisomes. J Exp Bot 61: 1441–1453 [DOI] [PubMed] [Google Scholar]
- Beevers H. (1979) Microbodies in higher plants. Annu Rev Plant Physiol 30: 159–193 [Google Scholar]
- Brocker C, Vasiliou M, Carpenter S, Carpenter C, Zhang Y, Wang X, Kotchoni SO, Wood AJ, Kirch HH, Kopečný D, et al. (2013) Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237: 189–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell JD, Keech O, Fenske R, Smith SM. (2013) Requirement for the plastidial oxidative pentose phosphate pathway for nitrate assimilation in Arabidopsis. Plant J 75: 578–591 [DOI] [PubMed] [Google Scholar]
- Cassin-Ross G, Hu J. (2014) A simple assay to identify peroxisomal proteins involved in 12-oxo-phytodienoic acid metabolism. Plant Signal Behav 9: e29464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca M, San Segundo B. (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63: 526–540 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Palma JM, del Río LA. (2013) Peroxisomes as cell generators of reactive nitrogen species (RNS) signal molecules. Subcell Biochem 69: 283–298 [DOI] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH. (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Hara-Nishimura I, Nishimura M. (2003) Novel glyoxysomal protein kinase, GPK1, identified by proteomic analysis of glyoxysomes in etiolated cotyledons of Arabidopsis thaliana. Plant Cell Physiol 44: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M. (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696 [DOI] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM. (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28: 1–12 [DOI] [PubMed] [Google Scholar]
- Goepfert S, Poirier Y. (2007) Beta-oxidation in fatty acid degradation and beyond. Curr Opin Plant Biol 10: 245–251 [DOI] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M. (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt MC, Siponen MI, Alexson SE. (2012) The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochim Biophys Acta 1822: 1397–1410 [DOI] [PubMed] [Google Scholar]
- Hunt MC, Solaas K, Kase BF, Alexson SE. (2002) Characterization of an acyl-coA thioesterase that functions as a major regulator of peroxisomal lipid metabolism. J Biol Chem 277: 1128–1138 [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhang XF, Wang XF, Zhang DP. (2011) Arabidopsis 3-ketoacyl-CoA thiolase-2 (KAT2), an enzyme of fatty acid β-oxidation, is involved in ABA signal transduction. Plant Cell Physiol 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Jin H, Song Z, Nikolau BJ. (2012) Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. Plant J 70: 1015–1032 [DOI] [PubMed] [Google Scholar]
- Kaur N, Reumann S, Hu J. (2009) Peroxisome biogenesis and function. Arabidopsis Book 7: e0123, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, D’Auria JC, Behere AS, Kim JH, Gunderson KL, Breen JN, Lee G, Gershenzon J, Last RL, Jander G. (2007) Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana. Plant J 51: 1062–1076 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Chung HS, Kobayashi Y, Howe GA. (2006) Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lemieux B, Miquel M, Somerville C, Browse J. (1990) Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet 80: 234–240 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lingner T, Kataya AR, Antonicelli GE, Benichou A, Nilssen K, Chen XY, Siemsen T, Morgenstern B, Meinicke P, Reumann S. (2011) Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23: 1556–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N, Theodoulou FL, Haslam RP, Linka M, Napier JA, Neuhaus HE, Weber AP. (2008) Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20: 3241–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Ely L, Smith TH, Marathe R, Anandalakshmi R, Fagard M, Vaucheret H, Pruss G, Bowman L, Vance VB. (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missihoun TD, Schmitz J, Klug R, Kirch HH, Bartels D. (2011) Betaine aldehyde dehydrogenase genes from Arabidopsis with different sub-cellular localization affect stress responses. Planta 233: 369–382 [DOI] [PubMed] [Google Scholar]
- Ofman R, el Mrabet L, Dacremont G, Spijer D, Wanders RJ. (2002) Demonstration of dimethylnonanoyl-CoA thioesterase activity in rat liver peroxisomes followed by purification and molecular cloning of the thioesterase involved. Biochem Biophys Res Commun 290: 629–634 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, Kurkcuoglu S, Kreuzaler F. (2010) Photorespiration. Arabidopsis Book 8: e130, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieruzzi FP, Dias LL, Balbuena TS, Santa-Catarina C, dos Santos AL, Floh EI. (2011) Polyamines, IAA and ABA during germination in two recalcitrant seeds: Araucaria angustifolia (Gymnosperm) and Ocotea odorifera (Angiosperm). Ann Bot (Lond) 108: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Portell J, Gallart M, Tiburcio AF, Altabella T. (2013) Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Ventre G, Caldelari D. (1999) Increased flow of fatty acids toward beta-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121: 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Yang P, Cassin-Ross G, Kaur N, Switzenberg R, Aung K, Li J, Hu J. (2013) Proteome analysis of peroxisomes from etiolated Arabidopsis seedlings identifies a peroxisomal protease involved in β-oxidation and development. Plant Physiol 163: 1518–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, Linka N, Switzenberg R, Wilkerson CG, Weber AP, et al. (2009) In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S, Rouanet C, Expert D, Nasser W. (2002) Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J Bacteriol 184: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Wang K, Ryu CM, Kaundal A, Mysore KS. (2012) Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA. (2003) Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid beta-oxidation is essential for embryo development. J Biol Chem 278: 21370–21377 [DOI] [PubMed] [Google Scholar]
- Sandalio LM, Rodríguez-Serrano M, Romero-Puertas MC, del Río LA. (2013) Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules. Subcell Biochem 69: 231–255 [DOI] [PubMed] [Google Scholar]
- Schrader M, Bonekamp NA, Islinger M. (2012) Fission and proliferation of peroxisomes. Biochim Biophys Acta 1822: 1343–1357 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse J. (2003) Arabidopsis contains a large superfamily of acyl-activating enzymes: Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol 132: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinum TM, Berner HS, Rap S, Salehian Z, Aalen RB. (1998) Differential regulation of the barley (Hordeum vulgare) transcripts B12D and B22E in mature aleuron layers. Physiologia Plantarum 102: 337–345 [Google Scholar]
- Strader LC, Bartel B. (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou FL, Eastmond PJ. (2012) Seed storage oil catabolism: a story of give and take. Curr Opin Plant Biol 15: 322–328 [DOI] [PubMed] [Google Scholar]
- Tilton G, Shockey J, Browse J. (2000) Two families of acyl-CoA thioesterases in Arabidopsis. Biochem Soc Trans 28: 946–947 [PubMed] [Google Scholar]
- Tilton GB, Shockey JM, Browse J. (2004) Biochemical and molecular characterization of ACH2, an acyl-CoA thioesterase from Arabidopsis thaliana. J Biol Chem 279: 7487–7494 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RB, Wanders RJ, Tager JM. (1992) Biochemistry of peroxisomes. Annu Rev Biochem 61: 157–197 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E. (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28: 463–487 [DOI] [PubMed] [Google Scholar]
- Weber H. (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7: 217–224 [DOI] [PubMed] [Google Scholar]
- Widhalm JR, Ducluzeau AL, Buller NE, Elowsky CG, Olsen LJ, Basset GJC. (2012) Phylloquinone (vitamin K(1) ) biosynthesis in plants: two peroxisomal thioesterases of Lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-CoA. Plant J 71: 205–215 [DOI] [PubMed] [Google Scholar]
- Xiong Y, DeFraia C, Williams D, Zhang X, Mou Z. (2009) Characterization of Arabidopsis 6-phosphogluconolactonase T-DNA insertion mutants reveals an essential role for the oxidative section of the plastidic pentose phosphate pathway in plant growth and development. Plant Cell Physiol 50: 1277–1291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.