Abstract
Transcription activator-like effector nucleases successfully generate a heritable tomato mutant.
We report the successful use of transcription activator-like effector nucleases (TALENs) under the control of an estrogen-inducible promoter for targeted mutagenesis in tomato (Solanum lycopersicum) of the negative regulator of GA signaling PROCERA (PRO). TALEN expression was induced and plants were regenerated from cotyledons of seedlings derived from stable transgenic lines. Six of 40 regenerated plants carried pro alleles, and the mutations in the two lines examined were heritable. Homozygous pro segregants exhibited phenotypes consistent with increased GA response.
Tomato is an important agricultural crop in which significant resources are invested for breeding traits such as disease resistance and fruit shape and color (Foolad and Panthee, 2012). Mutations affecting these traits can be generated using random mutagens such as ethyl methanesulfonate and transfer DNA integration (Mathews et al., 2003; Menda et al., 2004); however, screening for the desired mutation is laborious and time consuming. A potentially more efficient method of gene disruption is targeted mutagenesis using sequence-specific nucleases, which create a double strand break in the target sequence. These breaks are then repaired either by the homologous recombination or nonhomologous end-joining pathway (Jasin and Rothstein, 2013). Nonhomologous end-joining is sometimes imprecise, resulting in deletions or insertions at the double strand break site.
TALENs are composed of a sequence-specific DNA-binding domain fused to the FokI nuclease domain (Christian et al., 2010; Zhang et al., 2010). Binding of TALEN monomers to the targeted gene allows the FokI nuclease domain to dimerize and cleave the DNA (Chen and Gao, 2013; Voytas, 2013). TALENs have been successfully used to make sequence-specific mutations in a variety of plant species, including Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), rice (Oryza sativa), Brachypodium spp., and barley (Hordeum vulgare; Cermak et al., 2011; Li et al., 2012; Christian et al., 2013; Shan et al., 2013a; Wendt et al., 2013; Zhang et al., 2013). More recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein9 (Cas9) system has also been demonstrated to be a promising genome-editing tool (Jiang et al., 2013; Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013b; Upadhyay et al., 2013). The CRISPR/Cas9 system has been shown to be an effective tool for targeted mutagenesis in transgenic tomato hairy roots (Ron et al., 2014). In an accompanying paper, CRISPR/Cas9 is shown to be useful for generating heritable mutations (Brooks et al., 2014).
DELLA proteins are negative regulators of GA signaling (Hauvermale et al., 2012). The loss of DELLA function results in increased GA response, which causes the mutants to be tall and slender with light green vegetation. Tomato has one DELLA gene called PROCERA (PRO). A pro mutant caused by a missense mutation has been characterized (Jones, 1987; Bassel et al., 2008; Jasinski et al., 2008; Carrera et al., 2012). Although the mutant has phenotypes corresponding to increased GA response, it is partially responsive to GA, suggesting that the mutant protein retains partial activity (Van Tuinen et al., 1999). We used TALENs to create new pro alleles that can be used to determine the role of PRO in GA signaling.
RESULTS AND DISCUSSION
TALEN pairs pTAL423/4 and pTAL425/6 were designed to target the PRO gene using TALE-NT (Supplemental Fig. S1; Supplemental Table S1; Doyle et al., 2012). Due to concerns about potential TALEN cytotoxicity (Christian et al., 2013), we generated transgenic tomato plants that express the TALEN pairs under the control of the estrogen-inducible XVE promoter (Zuo et al., 2000). One primary transgenic plant with TALEN pair pTAL423/4 (pTAL423/4 T0) and four with TALEN pair pTAL425/6 (pTAL425/6 T0) were generated.
Leaves of T0 plants were assayed for evidence of TALEN activity 7 d after spraying plants with β-estradiol. For this assay, PRO genomic DNA was PCR amplified and tested to determine if the Sm1I restriction site located at the TALEN cleavage site was mutated (Zhang et al., 2010). However, no Sm1I-resistant PRO amplicons were detected, suggesting that the TALENs were not active or that the mutation frequency was too low for detection without enrichment. Therefore, Sm1I-digested PRO amplicons were used as the template for a second round of PCR amplification, and one T0 plant (designated as pTAL425/6_1 T0) had Sm1I-resistant PRO amplicons. Mutations ranging in size from 2 to 168 bp were identified when these amplicons were sequenced (Supplemental Fig. S2).
We attempted to produce heritable TALEN-induced mutations by spraying the shoot apex of pTAL425/6_1 T0 plants with β-estradiol daily for 1 week, but no TALEN-induced mutations were detected in the pTAL425/6_1 T1 seedlings. We then sprayed unopened and opened flowers with β-estradiol daily and injected immature and mature fruits with β-estradiol, but mutations were not detected in seedlings from the treated plants. In addition, when pTAL425/6_1 T1 seeds were germinated in the presence of β-estradiol, no mutations were detected in the T2 seedlings.
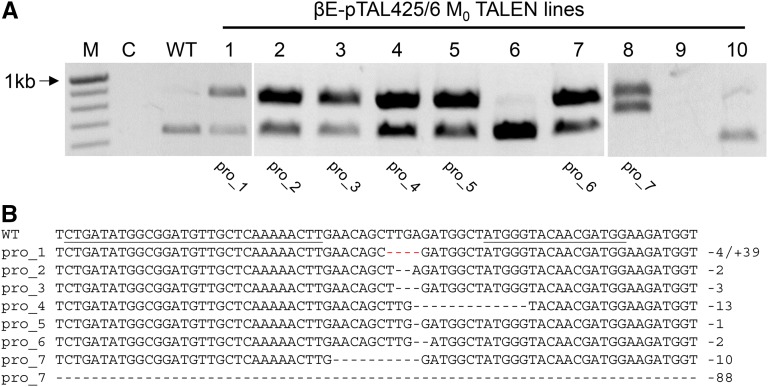
Since the TALENs were active in somatic tissue, we attempted to regenerate plants bearing mutations from cotyledons (Supplemental Fig. S3). pTAL425/6_1 T1 cotyledons were immersed in β-estradiol solution weekly, and plants (βE-pTAL425/6 M0) were regenerated. Six and three plants were regenerated from tissue that received either one or two β-estradiol treatments, respectively, but none carried TALEN-induced mutations. Forty βE-pTAL425/6 M0 plants were regenerated from tissue that had received three treatments, and seven of these plants (designated as pro_1–pro_7 M0) carried mutations (Fig. 1A). Since restriction enzyme assays do not detect all TALEN-induced mutations (Suzuki et al., 2014; Veres et al., 2014), additional plants may have carried mutations. While pro_1 to pro_6 each carried one mutant allele, pro_7 carried two different mutant alleles and exhibited characteristic pro mutant phenotypes, including long internode length and smooth leaf margins. Sequencing indicated that each allele was unique (Fig. 1B). The mutations in pro_2 to pro_7 were deletions ranging in size from 1 to 88 bp; pro_1 has a 39-bp insertion and a 4-bp deletion (Supplemental Fig. S4). Seven of the deletions cause frame shifts that would result in the production of a truncated PRO protein.
Figure 1.
Characterization of TALEN-induced mutations. A, PCR screen for TALEN-induced mutations in βE-pTAL425/6 M0 plants. Wild-type (WT) PRO amplicons were cleaved by Sm1I, producing 647- and 225-bp products, but the mutant amplicons in lines 1 to 5, 7, and 8 are not cleaved. Note that line 9 failed to amplify. The mutant line designations below the image correspond to the DNA sequences in B. M indicates the marker lane, and C indicates the no-template control. B, DNA sequence alignment of TALEN-induced mutations. Left and right TALEN-binding sequences are underlined in the wild type. The sizes of deletions (−) and insertions (+) are indicated to the right of the sequences. The red dashes indicate the location of a 39-bp insertion in pro_1.
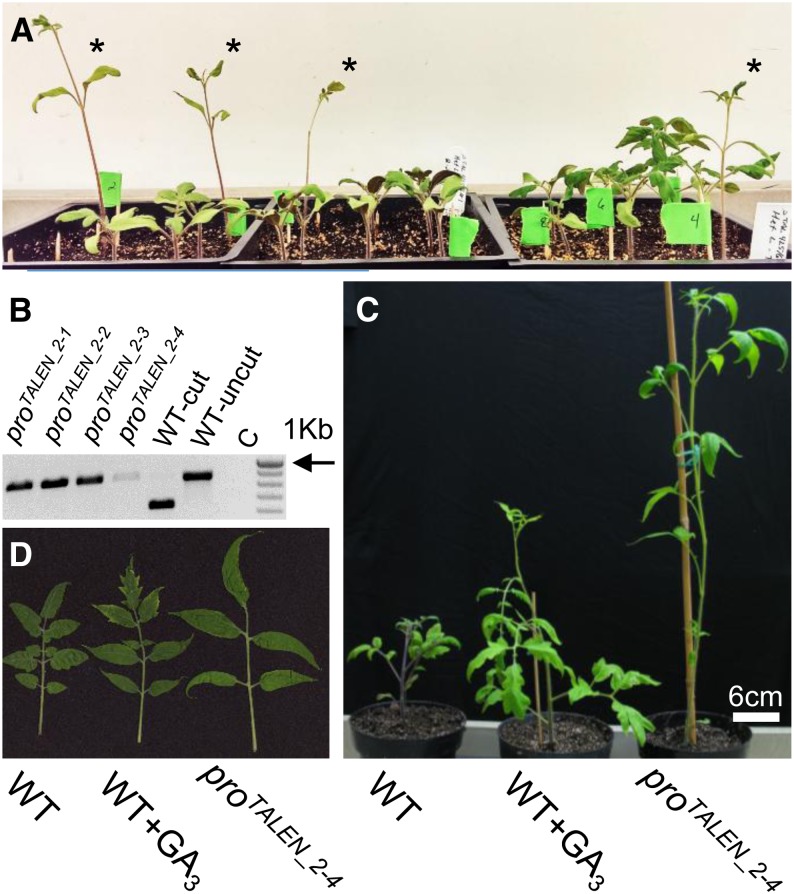
We then determined if the mutant alleles in the pro_2 and pro_1 lines (proTALEN_2 and proTALEN_1, respectively) are heritable. The previously described pro allele is recessive (Jones, 1987); thus, if proTALEN_2 and proTALEN_1 are heritable, we expected one-quarter of the seedlings derived from the heterozygous pro_2 and pro_1 plants to exhibit a mutant phenotype. Consistent with the mutations being heritable, four of 20 pro_2 seedlings (Fig. 2) and three of 20 pro_1 seedlings (Supplemental Fig. S5) were tall, and genotyping indicated that they were homozygous mutants. In further experiments, 55 of 293 pro_2 M1 seedlings were tall, and the resulting χ2 P value for a 3:1 segregation ratio was 0.014, which is consistent with Mendelian inheritance of the proTALEN_2 allele. In addition, we tested if the transgene encoding the TALEN nuclease segregated away from the TALEN-induced pro mutation and found that proTALEN_2-2 and proTALEN_2-4 M1 mutants did not have the transfer DNA insertion (data not shown).
Figure 2.
A, Three-week-old seedlings segregating for the proTALEN_2 allele. Homozygous proTALEN_2 mutants are indicated by the asterisks. B, Genotyping of the homozygous proTALEN_2 seedlings marked in A by PCR amplification and digestion with Sm1l. C indicates the no-template control. C, Comparison of 8-week-old wild-type (WT) and WT+GA3 plants with homozygous proTALEN_2 plants. D, Third youngest fully expanded leaves of the plants shown in C.
Homozygous proTALEN_2 plants are phenocopies of wild-type plants sprayed with GA3 (Fig. 2, C and D). In contrast to control wild-type plants, wild-type tomato plants sprayed with 50 μm GA3 daily and homozygous proTALEN_2 plants were taller and had lighter green leaves with smoother margins. These phenotypes are similar to those of the previously characterized pro mutant (Jones, 1987; Bassel et al., 2008; Jasinski et al., 2008; Carrera et al., 2012). The new pro alleles described here, and others that can be created through targeted mutagenesis, should prove useful in dissecting the role of PRO in GA signaling.
This report and the accompanying report by Brooks et al. (2014) demonstrate that both TALENs and CRISPR/Cas9 are effective tools for creating heritable mutations in tomato. The greater ease of synthesizing CRISPR/Cas9 and its higher mutagenesis frequency relative to TALEN might make it the preferred technology. However, CRISPR/Cas9 has been reported to have a higher frequency of off-target cleavage than TALENs (Fu et al., 2013; Suzuki et al., 2014; Veres et al., 2014), but progress is being made in addressing this deficiency (Fu et al., 2014).
MATERIALS AND METHODS
Materials and methods are described in Supplemental Materials and Methods S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Locations of TALEN target sites.
Supplemental Figure S2. Sequences of TALEN-induced mutations generated in somatic cells.
Supplemental Figure S3. Strategy for regenerating pTAL425/6 TALEN-induced mutants.
Supplemental Figure S4. The coding region of PRO showing the sequences of proTALEN_1 and one of the pro_7 alleles.
Supplemental Figure S5. Six-week-old seedling segregating for the proTALEN_1 allele.
Supplemental Table S1. TALEN Repeat Variable Diresidues sequences and TALEN target sequences.
Supplemental Materials and Methods S1. Materials and methods used.
Supplementary Material
Glossary
- TALEN
transcription activator-like nuclease
- CRISPR
clustered regularly interspaced short palindromic repeats
Footnotes
This work was supported by the United States-Israel Binational Agricultural Research and Development Fund (grant no. IS–4429–11C to N.E.O. and D.W.) and the National Science Foundation (grant no. DBI 0923827 to D.F.V.).
The online version of this article contains Web-only data.
References
- Bassel GW, Mullen RT, Bewley JD. (2008) Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): effects on the seed and vegetative plant. J Exp Bot 59: 585–593 [DOI] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J. (2014) Efficient gene editing in tomato in the first generation using the clustered regularly intespaced short palindromic repeats/Cas9 system. Plant Physiol 166: 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Ruiz-Rivero O, Peres LEP, Atares A, Garcia-Martinez JL. (2012) Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol 160: 1581–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia N V, Bogdanove AJ, Voytas DF. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Gao C. (2013) TALENs: customizable molecular DNA scissors for genome engineering of plants. J Genet Genomics 40: 271–279 [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Qi Y, Zhang Y, Voytas DF. (2013) Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 (Bethesda) 3: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. (2012) TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40: W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foolad MR, Panthee DR. (2012) Marker-assisted selection in tomato breeding. CRC Crit Rev Plant Sci 31: 93–123 [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM. (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Tattersall A, Piazza P, Hay A, Martinez-Garcia JF, Schmitz G, Theres K, McCormick S, Tsiantis M. (2008) PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J 56: 603–612 [DOI] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MG. (1987) Gibberellins and the procera mutant of tomato. Planta 172: 280–284 [DOI] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al. (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, et al. (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, Zhang K, Liu J, Voytas DF, Zheng X, et al. (2013a) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant 6: 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, et al. (2013b) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31: 686–688 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, et al. (2014) Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 15: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay SK, Kumar J, Alok A, Tuli R. (2013) RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3: 2233–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tuinen A, Peters AHLJ, Kendrick RE, Zeevaart JAD, Koornneef M. (1999) Characterization of the procera mutant of tomato and the interaction of gibberellins with end-of-day far-red light treatments. Physiol Plant 106: 121–128 [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Talkowski ME, Musunuru K. (2014) Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF. (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64: 327–350 [DOI] [PubMed] [Google Scholar]
- Wendt T, Holm PB, Starker CG, Christian M, Voytas DF, Brinch-Pedersen H, Holme IB. (2013) TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol 83: 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF. (2013) Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161: 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.