Abstract
Despite our increasing knowledge of the molecular events that induce the glycolysis pathway in effector T cells, very little is known about the transcriptional mechanisms that dampen the glycolysis program in quiescent cell populations such as memory T cells. Here, we show that the transcription factor Bcl-6 directly repressed genes involved in the glycolysis pathway, including Slc2a1, Slc2a3, Pkm2 and Hk2, in TH1 cells exposed to low amounts of interleukin 2 (IL-2). Thus, Bcl-6 plays an opposing role to the IL-2-sensitive glycolytic transcriptional program that c-Myc and HIF-1α promote in effector T cells. Additionally, the Th1-lineage-specifying factor T-bet functionally antagonized the Bcl-6-dependent repression of genes in the glycolysis pathway, implicating the molecular balance between these two factors in metabolic gene program regulation.
Introduction
Bcl-6 is necessary for the development of a number of immune cell types including germinal center (GC) B cells and CD4+ T follicular helper (TFH) cells1–3. Bcl-6 has also been implicated in promoting memory cell formation in both CD4+ and CD8+ T cells4–8. In both TFH and memory cell development, an important role for Bcl-6 is to inhibit the expression of Blimp-1, a transcriptional regulatory protein required for the terminal differentiation of effector cell populations2,6,9,10. Although this is one critical activity for Bcl-6 in several immune cell populations, to date, it has been unclear what additional gene pathways are regulated by Bcl-6 to promote the functional characteristics of different T cell populations11.
Recent studies have uncovered that an important functional difference between effector and memory cell populations is their metabolic states12–14. Elegant studies in CD8+ T cells have shown that genes in the glycolysis pathway are induced in effector cells, resulting in a switch to aerobic glycolysis for energy production15–17. This is thought to be essential for the rapid proliferative burst of activated T cells and for promoting aspects of effector cell functions15,18. In contrast, the glycolysis pathway is downregulated in CD8+ memory T cells19, which causes the cells to utilize mitochondrial fatty acid oxidation as a predominant form of cellular metabolism resulting in a shift to a catabolic state16,20. Notably, experimentally inhibiting glycolysis promotes CD8+ T cell memory formation, while artificially activating the glycolysis pathway causes the cells to preferentially adopt an effector state14,21. These experiments suggest that the utilization of different metabolic pathways in T cells actively contributes to differentiation outcomes.
The transcriptional regulatory events that induce glycolytic genes to promote effector cell differentiation have been elucidated13,19. T cell receptor (TCR) signaling and CD28 co-stimulation activate c-Myc, which is required for the initial upregulation of glucose transporters and rate-limiting glycolysis enzymes15. Additionally, interleukin-2 (IL-2)-signaling promotes the sustained expression of glycolysis genes in effector CD8+ T cells, with the hypoxia factor HIF-1α required for this activity17. In contrast to our expanding knowledge of the events that induce the glycolysis pathway in effector T cells, little information is known about the transcriptional regulatory events that downregulate, or alternatively prevent, the expression of these genes to enhance memory cell formation.
Recent research has indicated that the TFH and memory T cell gene expression programs have several common features, suggesting a close relationship between these two populations22. One similarity between TFH and memory T cells is the role for the transcriptional repressor Bcl-6 in determining aspects of each gene program4,23. It appears that the abundance of Bcl-6 is partially responsible for defining the unique characteristics for the two gene programs, with the highest amounts of Bcl-6 promoting TFH differentiation whereas moderate Bcl-6 expression is needed for memory cell development5,7,24. One environmental signal that regulates Bcl-6 expression in T cells is IL-2. Strong IL-2-signaling inhibits Bcl-6 expression whereas low environmental IL-2 conditions promote the expression of Bcl-67,9,25–27. Since IL-2-signaling also regulates the metabolic state of CD8+ T cells17, there is an inverse correlation between the capacity of IL-2-signaling to functionally regulate Bcl-6 expression and the expression of glycolysis genes.
Here, we discovered that Bcl-6 repressed the IL-2-sensitive expression of genes encoding glucose transporters and rate-limiting enzymes involved in glycolysis. In a comparison of previously published microarray datasets, we observed an overlap in the identity of genes that were reciprocally regulated by HIF-1α versus Bcl-6, including numerous genes in the glycolysis pathway. The expression of the genes in this overlapping subset was also sensitive to IL-2-signaling. This led us to hypothesize that Bcl-6 might serve as a key repressor for the genes in the glycolytic pathway that are sensitive to IL-2-signaling and are differentially expressed between effector and memory T cell states. We found that Bcl-6 directly repressed numerous genes in the glycolysis pathway, effectively functioning in direct opposition to the gene programs activated by HIF-1α and c-Myc. Additionally, the TH1-lineage-specifying transcription factor T-bet functionally inhibited the ability of Bcl-6 to repress genes involved in glycolysis. This suggests that the molecular balance between T-bet and Bcl-6 influences the expression of the metabolic gene program.
Results
Overlap between genes regulated by Bcl-6 and HIF-1α
To start to address the question of which regulatory pathways Bcl-6 represses to promote the normal differentiation and activity of unique immune cells, we compared gene expression patterns between wild-type and Bcl-6-deficient bone marrow derived myeloid cells from a published microarray study using GEO2R28. This analysis revealed that numerous genes involved in glycolysis, including rate-limiting enzymes and glucose transporters, were upregulated in Bcl-6-deficient cells (Supplementary Table 1). These findings suggested that Bcl-6 plays a role in functionally repressing genes encoding components of the glycolysis pathway, at least in some circumstances.
There is a reciprocal expression pattern between Bcl-6 and HIF-1α in T cells responding to IL-2. Bcl-6 expression was inhibited in the presence of high concentrations of IL-2 (Figs. 1 and 2)9,25,26, whereas HIF-1α is enhanced by IL-2-signaling15,17. Therefore, we hypothesized that Bcl-6 might play an opposing role to HIF-1α in the IL-2-sensitive regulation of glycolytic target genes. To begin to examine this possibility, we compared the genes from the Bcl-6 GEO2R analysis with the genes previously identified to be IL-2-sensitive and HIF-1α-dependent in a microarray study analyzing wild-type versus HIF-1α-deficient effector CD8+ T cells17. There was a substantial overlap in the identity of the genes functionally activated by HIF-1α in CD8+ T cells and the genes that are functionally repressed by Bcl-6 in myeloid cells (Supplementary Table 1). The overlapping HIF-1α and Bcl-6 subset included genes from the glycolytic pathway, such as Slc2a3 and Slc2a1 (the genes that encode the glucose transporters Glut3 and Glut1), Hk2, and Aldoc. It also included gene products that encode important hydrolases that modify proteins, such as Plod2, as well as P4ha and Egln family members. This overlap suggested that Bcl-6 might functionally oppose the IL-2-sensitive, HIF-1α-dependent gene program.
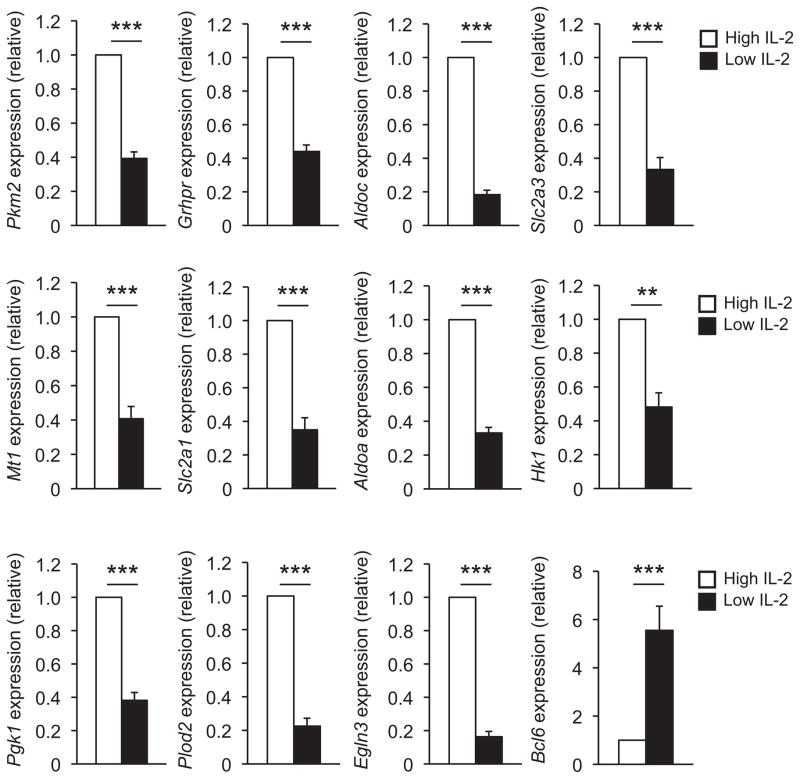
Figure 1. IL-2 signaling regulates the expression of glycolysis pathway genes in CD8+ TC1 cells.
Primary CD8+ T cells were cultured in TC1 polarizing conditions (IL-12 and anti-IL-4) and exposed to either high or low environmental IL-2 conditions (250 U/ml or 10 U/ml, respectively). RNA was isolated from the low (black bar) and high (white bar) IL-2 treated cells and transcript abundance for the indicated genes were determined by quantitative RT-PCR. Relative transcript expression was first normalized to the Rps18 (ribosomal protein S18) control and then the sample values were compared relative to the high IL-2 concentration (set to 1) for each independent experiment. At least three or four independent experiments were performed for all genes analyzed. All error bars represent standard error of the mean (SEM) *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
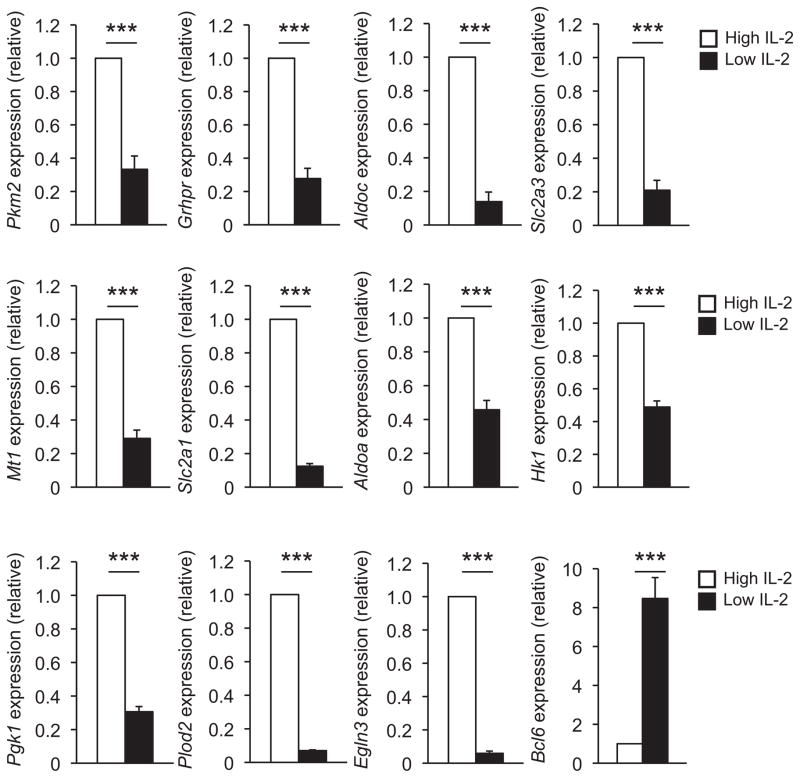
Figure 2. Glycolysis pathway genes are also regulated by environmental IL-2 conditions in CD4+ TH1 cells.
Primary CD4+ T cells were cultured in TH1 polarizing conditions (IL-12 and anti-IL-4) with either low (black bars) or high (white bars) IL-2 concentrations as in Fig. 1. Transcript abundance for the indicated genes were determined by quantitative RT-PCR and represented as in Fig. 1. At least three or four independent experiments were performed for all genes analyzed. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
IL-2-dependent regulation of glycolytic genes in T cells
We next hypothesized that environmental IL-2 conditions may serve as a conserved stimulus that functionally regulates the expression of the overlapping subset of HIF-1α and Bcl-6 genes in TH1 cells and CD8+ TC1 cells. Consistent with previous results in CD8+ T cells, numerous genes in the glycolysis pathway were preferentially expressed in high versus low environmental IL-2 conditions in CD8+ TC1 cells (Fig. 1 and Supplementary Figs. 1 and 2a). This included Slc2a3 and Slc2a1, as well as enzymes important in the glycolytic pathway including Aldoa, Aldoc, Pkm2, Hk2, and Grhpr. Indeed, there was a global induction of the key components that regulate the glycolysis and associated pathways in high environmental IL-2 conditions, whereas their expression was severely diminished in low IL-2 conditions (Fig. 1 and Supplementary Figs. 1 and 2a). Several other genes in the overlapping HIF-1α and Bcl-6 subset followed the same IL-2-sensitive gene expression pattern as those in the glycolytic pathway in CD8+ TC1 cells (Fig. 1 and Supplementary Fig. 2a). Similar to the results in CD8+ TC1 cells, genes involved in the glycolytic and associated pathways were preferentially expressed in CD4+ TH1 cells exposed to high environmental IL-2 conditions in comparison to the low IL-2 (Fig. 2 and Supplementary Figs. 2b and 3). Notably, Bcl-6 expression inversely correlated with the expression of the glycolytic pathway genes in both TH1 and TC1 cells, with Bcl-6 expression robustly induced in low environmental IL-2 conditions (Figs. 1and 2)9,27.
Bcl-6 directly regulates genes important in glycolysis
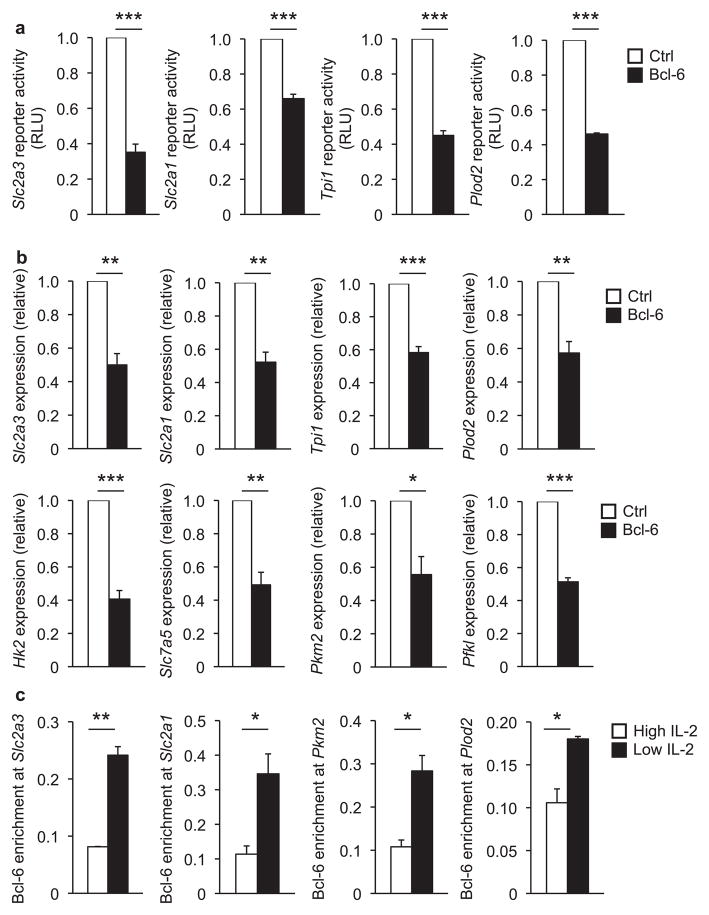
The inverse correlation between the expression of Bcl-6 and the glycolysis pathway genes in CD4+ TH1 and CD8+ TC1 cells led us to investigate whether Bcl-6 may be involved in the direct repression of this gene program. To start to test this hypothesis, we cloned promoter-reporter constructs for glycolysis pathway genes and additional genes from the overlapping HIF-1α and Bcl-6 subset. Notably, there was a reduction in the promoter activities of Slc2a3, Slc2a1, and Tpi1 as well as Plod2 and P4ha2 in response to Bcl-6 expression (Fig. 3a and Supplementary Fig. 4a). As a control, Bcl-6 expression alone did not repress the activity of the pGL3-promoter vector or several other promoter-reporter constructs (Supplementary Fig. 4b)29. These data suggest that Bcl-6 is capable of repressing the promoter activities of a subset of genes involved in glycolysis and the IL-2-sensitive regulatory pathways that are controlled by HIF-1α.
Figure 3. Bcl-6 directly represses genes in the glycolytic pathway.
(a) EL4 T cells were transfected with the indicated promoter-reporter constructs in combination with either a Bcl-6 expression vector (black bars) or an empty vector control (white bars). Luciferase promoter-reporter values were normalized to a renilla control and expressed relative to the control sample (set to 1) for each experiment. (b) Primary CD4+ T cells cultured in TH1 conditions with high IL-2 concentrations were transfected with either a control (white bars) or Bcl-6 (black bars) expression vector. Relative transcript abundance was determined by quantitative RT-PCR analysis using the PrimePCR system customized with primers specific to the indicated genes. Samples were first normalized to the expression of Rps18 and then compared to the control sample (set to 1) in each independent experiment. (c) ChIP experiments were performed with TH1 polarized cells maintained in either low (black bars) or high (white bars) IL-2 conditions. Chromatin samples were immunoprecipitated with either an antibody specific to Bcl-6 or a nonspecific IgG antibody control. The indicated promoter regions were monitored by qPCR with promoter-specific primers. The Bcl-6-precipitated samples were first normalized to a standardized aliquot of the input chromatin for each condition, followed by subtraction of the IgG antibody control as the nonspecific background of the experiment to determine the Bcl-6 enrichment relative to the percent input. At least two (c) or three (a, b) independent experiments were performed. (a–c) Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
We next transfected either a control or Bcl-6 expression vector into primary TH1 cells that were differentiated in high environmental IL-2 conditions and analyzed the endogenous expression of glycolysis pathway genes. This experimental system tests whether increasing Bcl-6 expression alone is sufficient to repress the glycolysis pathway genes in conditions where HIF-1α and c-Myc would otherwise strongly promote their expression. Numerous genes in the glycolysis pathway, including the rate-limiting enzymes Hk2 and Pkm2, were repressed by the expression of Bcl-6 in primary TH1 cells maintained in high environmental IL-2 conditions (Fig. 3b and Supplementary Fig. 5). These data suggest that Bcl-6 expression dominantly represses genes involved in the glycolysis pathway even when the cellular conditions are favorable for their expression.
If Bcl-6 participates in the direct repression of glycolytic target genes in the context of T cells, then there would be an inverse correlation between the expression of these genes and Bcl-6 association with these loci. To examine this prediction, we performed chromatin immunoprecipitation (ChIP) analyses assessing Bcl-6 association with the promoter regions for several glycolysis pathway genes in primary TH1 cells exposed to either high or low environmental IL-2 conditions. In ChIP experiments, Bcl-6 associated with the Slc2a3, Slc2a1, Aldoc and Pkm2 promoters in low IL-2 conditions, coinciding with the repression of these genes (Fig. 3c and Supplementary Fig. 4c). In contrast, when TH1 cells were exposed to high environmental IL-2 conditions, Bcl-6 association with these promoters was diminished, correlating with the upregulation of gene expression. A similar inverse correlation of Bcl-6 binding with gene expression was observed for Plod2 and P4ha2 (Fig. 3c and Supplementary Fig. 4c). Collectively, the data indicate that Bcl-6 associates with a subset of genes important in the glycolysis pathway in TH1 cells and is functionally important for repressing their expression.
Bcl-6 interacts with glycolysis genes in many cell types
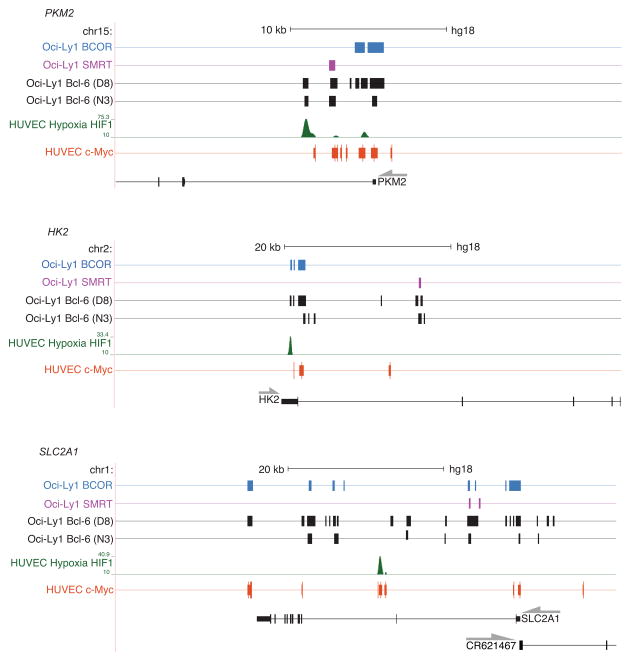
ChIP-seq studies have been performed to examine the genomic localization of Bcl-6 in B cells and Th9 cells to define the mechanisms that Bcl-6 utilizes to repress target gene expression30–33. These comprehensive datasets provide extensive information about the genomic localization of Bcl-6 and its co-repressor complexes in different cellular settings. We next compared our ChIP-PCR results with the previously published Bcl-6 ChIP-seq datasets from other lymphocyte subsets30–33. We visualized the data from the published ChIP-seq studies using the UCSC Genome Browser and focused on the Bcl-6 peaks found in proximity to the glycolysis pathway genes (Fig. 4 and Supplementary Fig. 6). Notably, Bcl-6 peaks were identified within the regulatory regions for Slc2a3, Slc2a1 and Pkm2 in B cells (Fig. 4 and Supplementary Fig. 6). Additionally, Slc2a1 and Pkm2 were identified within the list of genes that contain IL-2-sensitive, overlapping Bcl-6 and STAT transcription factor ChIP-seq peaks in TH9 cells33. Together, these data suggest that Bcl-6 associates with the loci for genes involved in the glycolysis pathway in both T and B cells in several different settings.
Figure 4. Genomic distribution of Bcl-6, HIF-1α, and c-Myc surrounding the loci for glycolysis pathway genes.
Shown are images derived from the UCSC genome browser displaying ChIP-seq tracks for BCOR30 (blue), SMRT30 (purple), two independent Bcl-6 antibodies30 (black), c-Myc ChIP-seq peaks from ENCODE52 (orange), and HIF-1α in hypoxic conditions35 (green). The boxes represent the BED file location of the significant ChIP-seq peaks for each experiment. Genes are displayed below the browser image, with the grey arrow indicating the direction of transcription. The cell types and antibodies for each ChIP-seq experiment are indicated to the left of the tracks. See Methods section for the GSE accession numbers for the individual ChIP-seq datasets30,31,35.
Given the large number of genes that are functionally repressed by Bcl-6 overexpression in primary TH1 cells, we next assessed how wide-spread the association of Bcl-6 was with the loci for the genes that were functionally repressed in the Bcl-6 overexpression experiments. The ChIP-seq datasets from B cells30–32 revealed Bcl-6 peaks at most of the genes that were repressed by Bcl-6 expression in the primary TH1 cell experiments including Hk2, Tpi1, Aldoa, Pfkl, Pfkm, Pck2, and Grhpr (Fig. 4 and Supplementary Fig. 6). Many of the Bcl-6 peaks also contained overlapping BCOR peaks, and less often SMRT peaks, suggesting that Bcl-6 may at least in part be preferentially utilizing a BTB-domain-mediated BCOR repression mechanism to inhibit their expression30. Collectively, these data suggest that Bcl-6 likely plays a direct role in the repression of an extensive network of the glycolytic gene program.
HIF-1α and c-Myc bind to glycolytic genes in TH1 cells
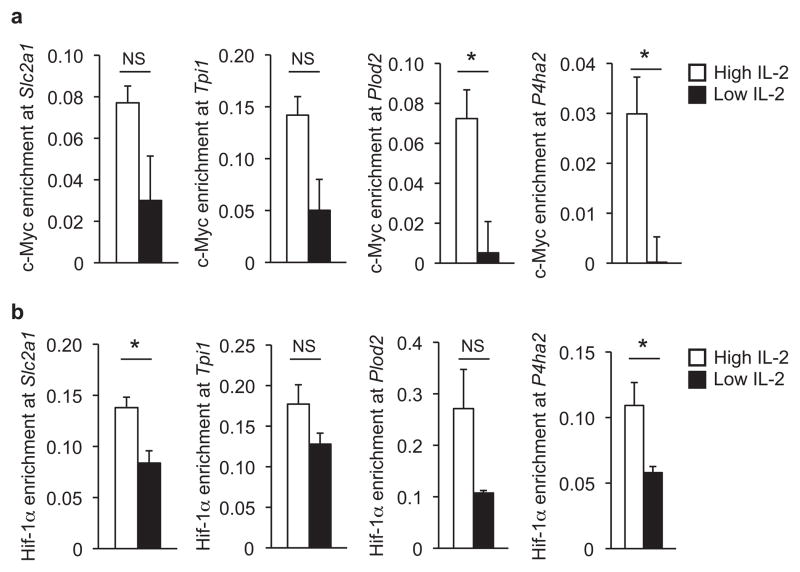
The inverse correlation between the IL-2-sensitive expression of HIF-1α and c-Myc versus Bcl-6, and the overlapping Bcl-6 and HIF-1α microarray subset, suggested that the Bcl-6 gene program might oppose the direct HIF-1α and c-Myc gene programs. To test this possibility, we first performed a ChIP analysis in primary TH1 cells exposed to either high or low environmental IL-2 conditions and examined HIF-1α and c-Myc association with a subset of the Bcl-6 repressed promoters. c-Myc and HIF-1α were enriched at the Slc2a1, Plod2 and P4ha2 promoters in TH1 cells maintained in high environmental IL-2 conditions, correlating with the expression of these genes (Fig. 5). In contrast, c-Myc and HIF-1α association with these promoters were reduced in low environmental IL-2 conditions. c-Myc was also enriched at the Slc2a3 and Tpi1 promoters in TH1 cells exposed to high environmental IL-2 conditions (Fig. 5 and Supplementary Fig. 4d).
Figure 5. The association of c-Myc and HIF-1α inversely correlates with Bcl-6 binding at the promoters for genes involved in glycolysis.
(a, b) ChIP experiments were performed with TH1 polarized cells maintained in either low (black bars) or high (white bars) IL-2 conditions. Chromatin samples were immunoprecipitated with either an antibody specific to (a) c-Myc, (b) HIF-1α or (a, b) a nonspecific IgG antibody control. The indicated promoter regions were monitored by qPCR. The c-Myc or HIF-1α-precipitated samples were normalized and graphically represented as described in Fig. 3c. (a, b) Three independent experiments were performed and error bars represent the SEM. The P value for (a) Tpi1 was 0.0577 and for (b) Plod2 was 0.0981, which were not quite statistically significant (NS). (a, b) NS, not significant; *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
We next extended the analysis of the glycolysis pathway gene loci by visualizing c-Myc ChIP-seq datasets from the ENCODE consortium34 and a published HIF-1α ChIP-seq study35. There was a substantial overlap in the c-Myc and Bcl-6 peaks at the loci for the glycolysis pathway genes, with many Bcl-6 peaks localized to the same promoter regions as the c-Myc peaks (Fig. 4 and Supplementary Fig. 6). Additionally, numerous genes that were identified in the overlapping HIF-1α and Bcl-6 subset contained hypoxia-inducible HIF-1α peaks. In some instances, the HIF-1α peaks appeared to overlap with the Bcl-6 peaks (Fig. 4 and Supplementary Fig. 6; e.g. see Pkm2, Aldoa, and Pfkfb3), whereas in others they were localized to different regions (Fig. 4 and Supplementary Fig. 6; e.g. see Slc2a1 and Slc7a5). There was also a subset of genes that only contained Bcl-6 peaks without any detectable HIF-1α peaks (Supplementary Fig. 6; e.g. see B4galt1 and Pfkm). Many of these genes were not identified in the initial overlapping HIF-1α and Bcl-6 subset, but were rather found to be repressed by Bcl-6 expression in our expanded analysis of the glycolysis pathway and also contain c-Myc peaks (Supplementary Fig. 6). In conjunction with previous studies15,17, these data suggest that c-Myc and HIF-1α play a role in directly activating a subset of genes important in glycolysis and associated pathways and that Bcl-6 plays a repressive role in direct opposition to this gene program.
T-bet is required for enhanced glycolysis in TH1 cells
Bcl-6 has emerged as one of the critical factors that functionally regulates memory potential, and the activity of Bcl-6 must be precisely controlled to initiate either the memory or TFH cell gene programs5,7,8,22. In TH1 cells, T-bet–Bcl-6 complex formation masks the Bcl-6 DNA binding domain because the DNA-binding zinc fingers are also required for the interaction with T-bet9,29. Thus, T-bet can hold Bcl-6 activity in check in effector TH1 cells because a high ratio of T-bet to Bcl-6 promotes complex formation, which dampens the potential of Bcl-6 to regulate its own target gene program. Previous studies have shown that the relative balance between T-bet and Bcl-6 varies substantially between effector and memory T cells, and that high T-bet expression is important for the development of short-lived effector cells4,36–38. Collectively, these observations led us to hypothesize that the relative balance between T-bet and Bcl-6 may play a role in defining the state of the glycolysis pathway gene program.
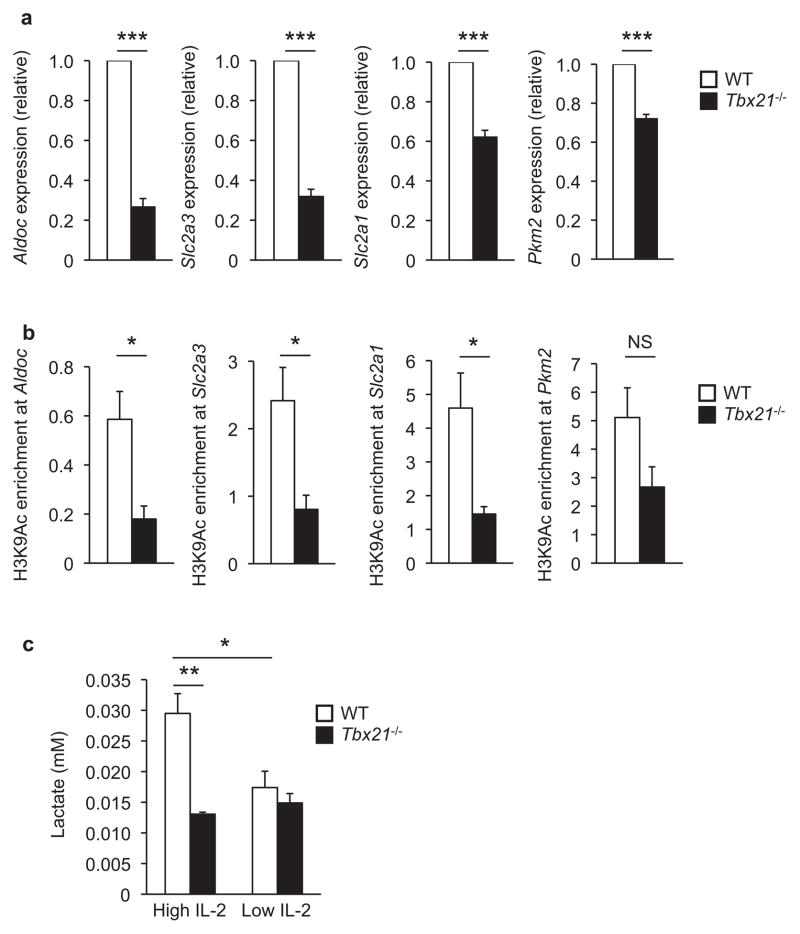
To address this possibility, we first examined the expression pattern of genes in the glycolytic pathway in wild-type versus T-bet-deficient primary TH1 cells. If T-bet inhibits Bcl-6 from repressing genes involved in glycolysis in effector TH1 cells, then the expression of glycolytic target genes would be inhibited in T-bet-deficient cells because Bcl-6 is no longer held in check by T-bet in this setting9. Consistent with this prediction, the expression of numerous genes in the glycolysis pathway were reduced in T-bet-deficient cells in comparison to wild-type effector TH1 cells (Fig. 6a and Supplementary Fig. 7). These included rate-limiting enzymes such as Aldoc and Pkm2 as well as the glucose transporters Slc2a3 and Slc2a1. Consistent with the reduced expression of these genes, there was also a decrease in the permissive histone H3 lysine 9 acetylation (H3K9-Ac) modification at the promoters of Aldoc, Pkm2, Slc2a3, and Slc2a1 in the T-bet-deficient cells in comparison to the primary wild-type effector TH1 cells (Fig. 6b). We observed similar trends in the expression and histone modification patterns for additional target genes identified in the Bcl-6 and HIF-1α microarray comparison (Supplementary Fig. 7).
Figure 6. The relative balance between T-bet and Bcl-6 regulates the expression of genes involved in the glycolysis pathway.
(a) Primary wild-type (white bars) or Tbx21−/− (black bars) CD4+ T cells were cultured in TH1 polarizing conditions and transcript abundance for the indicated genes were determined by quantitative RT-PCR and normalized as described in Fig. 1. (b) Primary CD4+ T cells purified from either wild-type (white bars) or Tbx21−/− (black bars) mice were treated as described in (a). Chromatin samples were immunoprecipitated with an antibody specific to either H3K9Ac or a nonspecific IgG antibody control. The indicated promoter regions were monitored by qPCR and the samples were normalized and represented as in Fig. 3c. (c) Primary wild-type (white bars) or Tbx21−/− CD4+ T cells were cultured in TH1 polarizing conditions and either high or low IL-2 as indicated. Lactate production was monitored from equal cell numbers within each treatment condition. (a, b) Three or (c) four independent experiments were performed and error bars represent SEM. NS, not significant; *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
We next monitored lactate production in the setting of wild-type versus T-bet-deficient CD4+ T cells to determine whether the observed gene expression changes had a functional consequence on glycolysis activity. Similar to previous findings in CD8+ T cells17, lactate production was induced in wild-type TH1 cells in the presence of high environmental IL-2 conditions in comparison to low IL-2 (Fig. 6c). Lactate production was significantly reduced in T-bet-deficient cells cultured in high environmental IL-2 conditions in comparison to the wild-type TH1 cells in the same IL-2 conditions. Notably, the lactate produced by T-bet-deficient cells exposed to high environmental IL-2 conditions was similar to the lactate concentrations found in wild-type cells exposed to low IL-2 (Fig. 6c). These data suggest that in TH1 cells, T-bet expression is required for the IL-2-dependent increase in glycolysis activity. Consistent with the observation that the robust induction of Bcl-6 expression in low environmental IL-2 conditions is sufficient to bypass T-bet-mediated control9, lactate production in wild-type and T-bet-deficient cells exposed to low environmental IL-2 conditions was similar (Fig. 6c). Together, these data support the hypothesis that T-bet is required to inhibit the modest amounts of Bcl-6 expressed in effector TH1 cells to prevent Bcl-6 from dominantly repressing the expression of glycolysis pathway genes and that this activity is functionally important for promoting glycolysis in effector TH1 cells. In contrast, low environmental IL-2 conditions substantially enhance Bcl-6 expression, which overcomes T-bet–mediated control in TH1 cells, allowing Bcl-6 to downregulate glycolysis and create a metabolic state more compatible with memory cell formation.
T-bet restrains Bcl-6 activity at glycolysis target genes
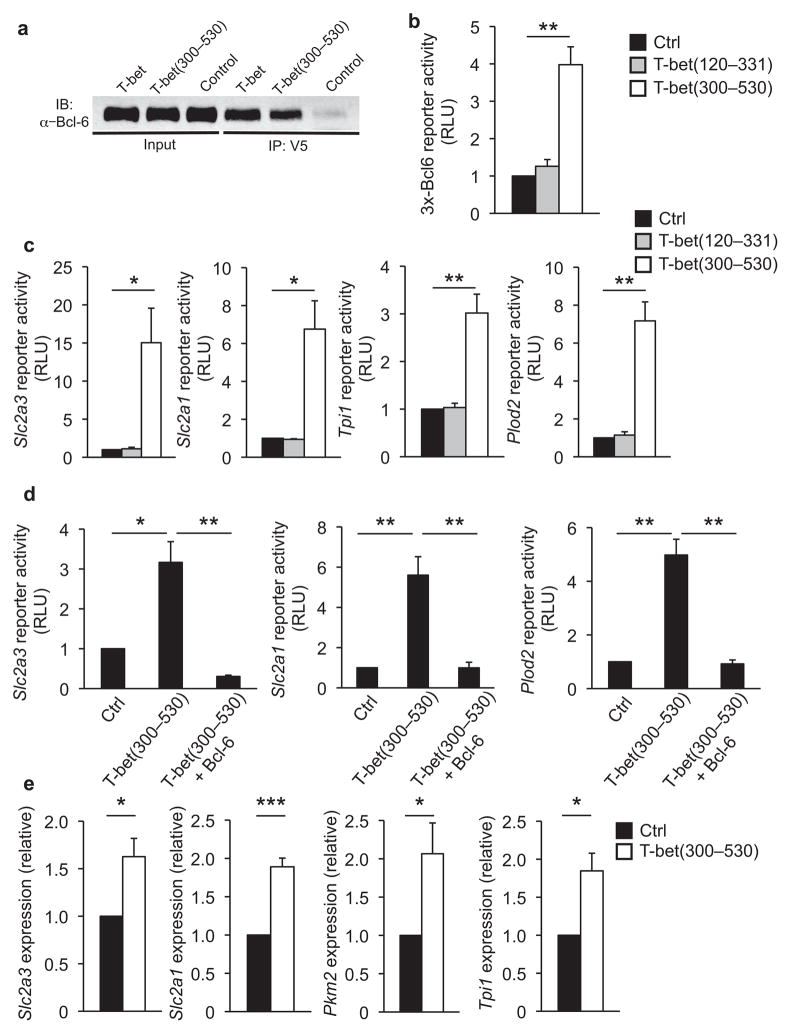
We next wanted to more directly address whether the role for T-bet in promoting the expression of glycolysis pathway genes might be mediated through its ability to functionally regulate Bcl-6 activity. To address this question, we needed to separate the mechanistic activity that T-bet utilizes to modulate Bcl-6 activity from all other aspects of the transcriptional regulatory potential of T-bet. To accomplish this goal, we created a construct that contained only the C-terminal domain of T-bet (T-bet amino acids 300–530), which was the domain that we have previously shown to be required for the physical interaction with Bcl-6 (ref. 9). Importantly, this truncated protein lacks the centrally located T-box DNA binding domain as well as an N-terminal domain that is needed for the transactivation potential of T-bet.
We first performed a co-immunoprecipitation analysis to demonstrate that the C-terminal domain of T-bet alone was sufficient to mediate the interaction with Bcl-6 (Fig. 7a). We next examined whether the expression of T-bet(300–530) was sufficient to inhibit Bcl-6 from repressing its direct DNA binding elements. To address this question, we first performed a series of experiments with a promoter-reporter construct that contains three Bcl-6 DNA binding elements upstream of a minimal promoter (3x-Bcl6)9. The repression of the 3x-Bcl6-promoter-reporter construct is completely dependent upon the Bcl-6 DNA binding sites9. We co-transfected the 3x-Bcl6-promoter-reporter construct into A20 B cells with either a control vector or one expressing T-bet(300–530). Notably, there was an enhancement in 3x-Bcl6 promoter-reporter activity in the presence of T-bet(300–530) (Fig. 7b). In contrast, the expression of another domain of T-bet that does not interact with Bcl-6 (T-bet(120–331)) had no effect on the activity of the 3x-Bcl6-promoter-reporter construct (Fig. 7b). As an additional specificity control, T-bet(300–530) had no activity when Bcl-6 expression was diminished by stimulation (Supplementary Figs. 8b, c). Together, these data support the interpretation that the physical interaction between T-bet and Bcl-6 inhibits Bcl-6 from repressing its own DNA binding sites and that this experimental system recapitulates the principles of the T-bet-dependent control of Bcl-6 without introducing the DNA-binding-dependent transcriptional activity of T-bet into the cell.
Figure 7. The C-terminal domain of T-bet inhibits the Bcl-6-dependent repression of a subset of glycolysis pathway genes.
(a) V5-epitope tagged wild-type T-bet, T-bet(300–530), or an empty vector control were transfected into A20 lymphoma B cells. Lysates from each transfected sample were immunoprecipitated with a V5 antibody and the co-precipitation of endogenous Bcl-6 was detected in an immunoblot analysis using a Bcl-6-specific antibody. (b) A20 B cells were transfected with an empty vector control (black bars), T-bet(120–331) (grey bars) or T-bet(300–530) (white bars) in combination with the 3x-Bcl6-promoter reporter construct. The luciferase promoter-reporter values were normalized to a co-transfected renilla control and expressed relative to the control sample (set to 1). (c) A20 B cells were transfected with an empty vector control (black bars), T-bet(120–331) (grey bars), or T-bet(300–530) (white bars) in combination with the indicated promoter-reporter constructs. (d) EL4 T cells were transfected with an empty vector control, T-bet(300–530), or T-bet(300–530) and Bcl-6 in combination with the indicated promoter-reporter constructs. (c, d) The luciferase data were normalized and represented as in (b). (e) Ramos human B cells were transfected with either an empty vector control (black bars) or T-bet(300–530) (white bars) and stimulated with anti-CD40. Transcript amounts for the indicated endogenous genes were analyzed by quantitative RT-PCR and normalized to the Rps18 control and the expression was compared relative to the control sample (set to 1). (a–e) Three independent experiments were performed and (b–e) error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
To test the hypothesis that T-bet has the ability to functionally control the activity of Bcl-6 at genes important in glycolysis, we co-transfected promoter-reporter constructs for several different genes in the glycolysis pathway with either an empty vector, a vector expressing T-bet(300–530), or one expressing T-bet(120–331) as a control. The expression of T-bet(300–530), but not the control T-bet(120–331), enhanced promoter-reporter activity for glycolytic target genes including Slc2a3, Slc2a1, Pkm2, Aldoc and Tpi1 (Fig. 7c and Supplementary Fig. 8d). T-bet(300–530) also enhanced the promoter activity of Plod2 and P4ha2 (Fig. 7c and Supplementary Fig. 8d). As a control, overexpression of Bcl-6 in conjunction with T-bet(300–530) restored the Bcl-6-dependent repression of the promoter-reporter constructs (Fig. 7d). Thus, the interaction between T-bet and Bcl-6 functionally inhibits the Bcl-6-dependent repression of the promoter activities for glycolysis pathway target genes.
Finally, we utilized this experimental system to assess whether the molecular balance between T-bet and Bcl-6 could influence the endogenous expression of genes important in glycolysis. Similar to the results with the promoter-reporter constructs, the expression of T-bet(300–530) was sufficient to modestly enhance the endogenous expression of a subset of genes in the glycolysis pathway (Fig. 7e and Supplementary Figs. 8e and 9). Consistent with this mechanism having the potential to be conserved in different settings, these experiments were performed in the human Ramos and murine A20 B cell lines that have constitutive expression of Bcl-6. Collectively, these data indicate that the interaction between T-bet and Bcl-6 functionally controls the ability of Bcl-6 to repress a subset of target genes important for glycolysis, implicating the molecular balance between T-bet and Bcl-6 in regulating the glycolysis pathway gene program.
Discussion
Here we identified a role for Bcl-6 in repressing genes that are important in glycolysis and associated pathways. IL-2-signaling regulates several key transcription factors required for T cell activation and differentiation39,40. IL-2-signaling promotes c-Myc and HIF-1α activity, whereas it inhibits Bcl-6 expression, creating reciprocal expression patterns between these factors in effector versus memory cell populations9,17,27. The close correlation between the expression of Bcl-6 and the repression of genes in the glycolysis pathway in low environmental IL-2 conditions, led us to hypothesize that Bcl-6 may be one of the critical regulatory proteins that inhibits glycolysis in T cells. Indeed, Bcl-6 associated with the loci for numerous glycolysis pathway genes, including rate-limiting enzymes such as Pkm2 and Hk2, and this correlated with their repression in TH1 cells in an IL-2-sensitve manner. These results, in conjunction with the GEO2R Bcl6−/− microarray analysis, the Bcl-6 overexpression studies in primary TH1 cells, and the promoter-reporter experiments indicate that Bcl-6 is an IL-2-sensitve factor that directly represses genes in the glycolysis pathway. Finally, T-bet is required for the IL-2-dependent induction of glycolysis in effector TH1 cells because T-bet can inhibit the ability of Bcl-6 to dominantly repress glycolytic target genes. Therefore, T-bet indirectly serves to promote the expression of genes in the glycolysis pathway in effector TH1 cells by inhibiting Bcl-6 activity.
The identity of the genes that Bcl-6 regulates to perform its role in lineage-commitment decisions has been somewhat enigmatic because Bcl-6 is a transcriptional repressor and it has been difficult to envision the direct pathways that it regulates to promote a specific cellular state. In TFH and B cell commitment, at least one of its direct target genes is Prdm1 (the gene that encodes the transcriptional repressor Blimp-1)2,9,41,42. However, the reciprocal regulation between Bcl-6 and Blimp-1 does not explain many of the altered cellular characteristics that are related to Bcl-6 expression. The role for the Bcl-6-dependent regulation of glycolysis in T cells identifies a new cellular process that Bcl-6 controls which is likely to be important for defining effector versus memory states in T cells. In this regard, the metabolic state of immune cells has a profound impact on their functional capabilities12,13,21,43 and experimentally manipulating the ability of the cell to utilize the glycolysis pathway for energy production can alter the effector versus memory fate decision14,21. Specifically, inhibiting glycolysis promotes memory cell formation21. Therefore, if the glycolysis program in effector cells can be turned off in response to changing environmental cues during the course of the immune response, such as waning IL-2 conditions, this may initiate the transition of an effector cell into a memory cell. Our study demonstrates that Bcl-6 can dominantly repress the glycolysis gene program, even in the presence of HIF-1α and c-Myc. This suggests there is a hierarchy between the competing regulatory pathways that are involved in the differentiation decisions associated with effector and memory potential. This leads to the speculation that if Bcl-6 expression is induced during the course of the immune response in the effector population, the dominant nature of the Bcl-6-dependent repression of the glycolysis pathway could alter the metabolic state of the cell to promote memory formation.
Another intriguing aspect for the finding that Bcl-6 plays a role in the IL-2-sensitive repression of glycolysis pathway genes is that Bcl-6 is also the lineage-specifying factor for TFH differentiation. Recently, it was proposed that the CD8+ memory T cell and CD4+ TFH cell gene expression programs are significantly related22. The current data in the field suggest that a gradient of Bcl-6 expression serves to define whether a memory or TFH program will be initiated. This is consistent with the findings that a similar composition of signaling pathways and regulatory factors contribute to the differentiation potential of both cell populations, but that there may be different thresholds for these events that are necessary for generating each unique cell population4,6,44,45. It will be informative to determine in future experiments whether the abundance of Bcl-6 selects unique subsets of target genes that are more predominant in the memory or TFH phenotypes or rather target specificity is related to co-factor availability. In this context, it will be important to determine how the Bcl-6-dependent regulation of the glycolytic pathway gene program fits into the potential for each of these cellular states.
It is also interesting to speculate how the findings in this study relate to the oncogenic potential of Bcl-6. Highly proliferating cancer cells are prone to express a glycolytic gene program, which is similar to the expression profile of the proliferating effector cells of the immune response12,46. The oncogenic potential of Bcl-6 at first glance appears to be incompatible with both its known role in memory T cell development as well as the newly identified role for Bcl-6 in the inhibition of the glycolysis pathway gene program. However, recent studies examining the survival requirements for leukemia cancer stems cells found that Bcl-6 expression in these cells, a relatively quiescent cell population, was important for their survival and loss of Bcl-6 expression increased the proliferative capacity of the stem cells47. These data suggest that the oncogenic potential of Bcl-6 in cancer stem cells may be similar to its role in long-lived memory cell populations. In the context of these more quiescent cell populations, the inhibition of the glycolysis pathway gene program may be an advantageous activity that Bcl-6 utilizes to promote long-term survival.
Finally, it is interesting to note that creating a gradient of Bcl-6 activity in TH1 cells can also be achieved by modulating the relative molecular balance between T-bet and Bcl-6. This molecular balance is sensitive to environmental signals such as IL-2, and we have previously shown that this allows for flexibility between aspects of the TH1 and TFH-like gene expression patterns9. Here, we extend those findings to demonstrate that the IL-2-dependent induction of glycolysis requires T-bet, most likely because the relative molecular balance between T-bet and Bcl-6 modulates the ability of Bcl-6 to dominantly repress genes involved in glycolysis. This creates a scenario where the high expression of T-bet found in effector TH1 cells serves multiple purposes, which include promoting the expression of TH1-lineage specific genes as well as ensuring that genes in the glycolysis pathway are expressed abundantly. It is worth noting that there is a gradient of T-bet expression between effector and memory T cell populations4,36–38. To date, it has been unclear what aspects of the gene expression program might be impacted by the differences in T-bet expression. The data presented here lead to the hypothesis that the T-bet to Bcl-6 ratio in T cells influences the expression of the glycolysis gene program. This also suggests that there is an essential connection between the lineage-specifying transcription factors that have long been known to define the functional potential of T cells with the regulation of the metabolic state of the cell. Much research still needs to be performed to address these topics, including detailed metabolomic profiling of these cell populations, but our study provides evidence for the involvement of lineage-specifying transcription factors in modulating the expression of the metabolic gene program.
Methods
Primary cells and cell culture
The Mag Cellect kit (R&D) was used to isolate primary CD4+ or CD8+ T cells from the spleen and lymph nodes of wild-type C57BL/6 or Tbx21−/− mice as previously published9,29. The primary cells were incubated with plate-bound anti-CD3/CD28 (BD Pharmingen) in TH1–TC1 polarizing conditions [anti-IL-4 (5 μg/ml) (NIH) and IL-12 (5 ng/ml) (R & D)]. Cells were split on day three and cultured for an additional two or three days in either high (250 units/ml) or low IL-2 (10 units/ml) (NIH) conditions as previously described9. Experiments with mice were performed with IACUC approval from the University of Washington, Virginia Tech, and the University of Alabama at Birmingham.
Murine EL4 T cells (TIB-39; ATCC), A20 B cells or human Ramos B cells (generously provided by D. Rawlings) were cultured in RPMI supplemented with 10% FBS and pen/strep. The Lonza nucleofection system with program 0–17 and solution V was used for transfection experiments as previously published48. For studies analyzing endogenous gene expression, Ramos B cells were transfected with the T-bet C-terminal domain (T-bet, amino acids 300–530) expression construct or a vector control and the cells were then stimulated with anti-CD40 (553787; BD Pharmingen) 6–8 h post-transfection and harvested 48 h post-transfection for the analysis of endogenous gene expression. For primary CD4+ T cell transfections, the 4D nucleofection system from Lonza was used. Transfections were performed according to the protocol supplied by the manufacturer. In brief, primary CD4+ T cells exposed to high environmental IL-2 conditions were resuspended in solution P3 and nucleofections were carried out with program DN-100. Following nucleofection, primary cells were allowed to recover in high environmental IL-2 conditions for 24 h prior to gene expression analysis.
Co-immunoprecipitation
The co-immunoprecipitation assay was performed as published previously29. The immunoprecipitation was performed with a V5-specific antibody (AB9116; Abcam). For immunoblot analysis to monitor the co-immunoprecipitation of endogenous proteins, a Bcl-6-specific antibody (561520; BD Pharmingen) was used.
qRT-PCR
The First Strand Superscript II Synthesis kit (Invitrogen) was used to prepare cDNA. For qPCR analysis, 20 ng of each cDNA template was amplified with gene specific primers and the qPCR Sybr Green Mix (Biorad). For the experiments in Fig. 3b and Supplementary Figs. 1, 2, 5 and 9 PrimePCR custom plates (Biorad) were used. All samples were first normalized to the Rps18 control and then expressed as a ratio to the indicated comparison condition.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed as previously published9,49,50. Chromatin from primary polarized CD4+ TH1 cells maintained in variable IL-2 conditions was precipitated with antibodies to either Bcl-6 (C-19; Santa Cruz Biotechnology), c-Myc (N-262; Santa Cruz Biotechnology), HIF-1α (NB100-134; Novus Biologicals), or H3K9Ac (AB4441; Abcam). The purified DNA was analyzed by qPCR with the indicated primers. The experimental samples were first normalized to a standardized input DNA control then the IgG antibody control was subtracted to account for the nonspecific background, with this value graphically represented as the percent input of each sample.
Promoter-reporter assay
The 3x-Bcl6 reporter construct has been described previously9. Slc2a3, Slc2a1, P4ha2, Plod2 and Tpi1 reporter constructs were prepared by cloning the promoters of each gene into the pGL3-basic luciferase reporter construct (Promega). EL4 or A20 cells were co-transfected with the indicated expression vectors along with the promoter-reporter constructs as well as a CMV-renilla control plasmid to normalize for transfection efficiency. The samples were harvested 16–24 h post-transfection and then analyzed with the Dual-Luciferase Reporter system (Promega).
Quantitation of intracellular lactate
Wild-type or Tbx21−/− cells were isolated, cultured, and harvested as described above. Cell pellets (5 × 105 cells/assay) were washed with 1X PBS following harvest to remove residual media, resuspended in 200 μl Lactate Assay Buffer (K607-100; BioVision) and lysed by 20–25 passes through a 22-gauge needle. Lysed samples were centrifuged at 16,100 g for 5 min to remove cell debris before concentration and lactate dehydrogenase (LDH) removal via 10 kDa spin columns (1997–25; BioVision). Intracellular lactate concentrations were measured using the Colorimetric/Fluorometric Lactate Assay Kit (K607-100; BioVision) per the manufacturer’s instructions.
GEO2R and Genomic Analysis
The differential expression of genes between untreated Bcl-6-deficient and wild-type cells was determined by performing a GEO2R51 analysis examining microarray expression data (GSE24813)28 deposited into the GEO database from a previously published study28. The Bcl-6 expression status in the two populations was used to confirm the identity of the Bcl-6-deficient sample. Genes that were functionally repressed by Bcl-6 were identified by analyzing the log2 fold change and P values between the Bcl-6-deficient and wild-type cells. These genes were then compared to the genes that were previously identified to be differentially expressed between wild-type and HIF-1α-deficient cells from a published study17.
ChIP-seq data for Bcl-6 (GSE29282 and GSE46663)30,31, BCOR and SMRT (GSE29282)30 and HIF-1α (GSE39089)35 deposited into the GEO database from previous publications were uploaded onto the UCSC genome browser for visualization (http://genome.ucsc.edu). BED files containing peak calls were used when provided in the GSE file, otherwise the WIG files were uploaded into the UCSC genome browser (human hg18 reference genome). The c-Myc ChIP-seq peaks were derived from the ENCODE consortium ChIP-seq datasets52 generated by the Iyer group at UT-Austin from HUVEC or GM12878 cells. The distribution of the Bcl-6, BCOR, SMRT, HIF-1α, and c-Myc peaks surrounding the loci for select glycolysis and associated pathway genes were then analyzed.
Statistical Analysis
The error bars in all graphs represent the standard error of the mean (SEM). For statistical analysis, an unpaired Student’s t test was performed using GraphPad Prism online software. Experiments and analyses were performed in an unblinded fashion. The number of independent, biological replicate experiments that were performed is indicated in the Figure Legends.
Supplementary Material
Acknowledgments
This research was supported through grants from the NIAID (AI061061) and the American Cancer Society (RSG-09-045-01-DDC) to A.S.W. We are grateful to the NCI preclinical repository for generously providing IL-2 and anti-IL-4. We thank M. Wijaranakula for technical assistance and D. Chisolm for critical reading of the manuscript. We also thank A. Ballesteros-Tato, B. Leon and T. Dadali-Abel for helpful advice and assistance.
Footnotes
AUTHOR CONTRIBUTIONS
K.J.O. and A.S.W conceived of and designed the study, performed experiments, analyzed data and wrote the manuscript. K.A.R., S.E.G., K.P.H., P.W.M., and V.K. performed experiments and analyzed datasets.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 9.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat Immunol. 2013;14:380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 14.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi YS, et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. 2013;190:4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crotty S. Follicular Helper CD4 T Cells (T(FH)) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 24.Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct Waves of BCL6 Expression during T Follicular Helper Cell Development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 25.Ballesteros-Tato A, et al. Interleukin-2 Inhibits Germinal Center Formation by Limiting T Follicular Helper Cell Differentiation. Immunity. 2012 doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurtz C, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208:2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatzi K, et al. A Hybrid Mechanism of Action for BCL6 in B Cells Defined by Formation of Functionally Distinct Complexes at Enhancers and Promoters. Cell reports. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swaminathan S, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao W, et al. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc Natl Acad Sci U S A. 2014;111:3508–3513. doi: 10.1073/pnas.1301138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mimura I, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Intlekofer AM, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007;204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 42.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 43.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi YS, Yang JA, Crotty S. Dynamic regulation of Bcl6 in follicular helper CD4 T (Tfh) cells. Curr Opin Immunol. 2013;25:366–372. doi: 10.1016/j.coi.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayamada S, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 47.Duy C, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beima KM, et al. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 50.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.