Abstract
Gender-related differences in colorectal cancer (CRC) are not fully understood. Recent studies have shown that CRC arising in females are significantly associated with CpG island methylator phenotype (CIMP-high). Using array comparative genomic hybridization, we analyzed a cohort of 116 CRCs (57 males, 59 females) for chromosomal copy number aberrations (CNA) and found that CRC in females had significantly higher numbers of gains involving chromosome arms 1q21.2–q21.3, 4q13.2, 6p21.1 and 16p11.2 and copy number losses of chromosome arm 11q25 compared to males. Interestingly, a subset of male CRCs (46%) exhibited a “feminization” phenomenon in the form of gains of X chromosomes (or an arm of X) and/or losses of the Y chromosome. Feminization of cancer cells was significantly associated with microsatellite-stable CRCs (p-value 0.003) and wild-type BRAF gene status (p-value 0.009). No significant association with other clinicopathological parameters was identified including disease-free survival. In summary, our data show that some CNAs in CRC may be gender specific and that male cancers characterized by feminization may constitute a specific subset of CRCs that warrants further investigation.
Keywords: X chromosome, comparative genomic hybridization, colorectal cancer, copy number aberration, gender
1. Introduction
Genomic instability is an important molecular event in the development of colorectal cancer (CRC) [1], encompassing chromosomal instability, microsatellite instability and aberrant DNA methylation. Chromosomal instability is the most common type of genomic instability and is an early event in colorectal carcinogenesis [2] causing chromosomal copy number aberrations (CNA). CNAs in CRC have been extensively studied using cytogenetic techniques and comparative genomic hybridization (CGH) [3,4,5,6,7,8,9]. Yet, despite recent advances in CRC genomics, gender-specific CNAs have only been sporadically documented by CRC genomic studies. Excluding gender-specific cancers (e.g., prostate, breast), males and females in general differ in their susceptibility to different cancers and differ in clinical outcome, therefore, understanding gender differences is important in order to gain insight into disease biology and underlying pathogenic mechanisms [10]. Studying gender-related genomics can also help optimize therapeutic strategies particularly in the era of personalized medicine.
With respect to CRC, males and females show many epidemiological, clinical and pathological differences. It is plausible to hypothesize that at least some of these gender differences are related to genes encoded in the sex chromosomes. It has been previously demonstrated that X chromosome CNAs are frequent in CRC cells in both male and female patients [11,12]. However, a significantly higher frequency of these X chromosomal aberrations has been demonstrated in CRCs from male patients [6,13,14]. The clinical significance of this phenomenon is yet to be elucidated.
Are copy number aberrations in CRC gender-specific? To address this question, a cohort of 116 male and female patients with CRC was explored for chromosomal CNA by high-resolution array CGH (aCGH), with special attention to sex chromosomal aberrations. The aim was to specifically shed more light on the differences in CNAs between male and female colorectal cancer cells, and to correlate the aCGH findings with clinicopathological parameters, an aim that, to the best of our knowledge, has not been previously well-addressed.
2. Results
2.1. Demographic/Pathologic Data
Table 1 shows that of 116 patients included in this study, 59 (50.9%) were females and 57 (49.1%) were males. Mean age was 64.2 years (range from 35 to 94). Tumors were from different colonic sites with 28 tumors (24.2%) being right-sided (cecum to transverse colon), 47 tumors (40.5%) being left-sided (descending colon and sigmoid), 23 (19.8%) rectal, and 18 (15.5%) of unknown site. Histological differentiation was as follows: 13 tumors (11.2%) were well-differentiated, 84 (72.4%) were moderately-differentiated, 11 (9.5%) were poorly-differentiated, and in eight cases (6.9%) differentiation could not be determined. Most tumors (82.8%) in both male and female patients were Dukes’ stage B (84.2% in males and 81.4% in females). The majority of patients (75%) had microsatellite-stable cancers with wild-type BRAF gene (72% in males and 77.9% in females). Outcome data were available for 87 (75%) patients and the mean period of clinical follow-up was 7.5 years. In total, 63 patients (54.3%) were alive and disease-free 10 years after surgery, while 24 patients (20.7%) had disease relapse in the form of local recurrence or distant metastasis (Table 1). Female patients showed more recurrences than male patients (28.8% and 12.3%, respectively), but the disease-free survival did not significantly differ between the two genders (data not shown).
Table 1.
Clinicopathological characteristics of the colorectal cancers classified based on gender.
| Characteristics | Males No. (%) | Females No. (%) | Total No. (%) |
|---|---|---|---|
| No. Of Patients | 57 (49.1) | 59 (50.9) | 116 (100) |
| Mean Age in Years | 63 | 65.4 | – |
| Age | |||
| 31–40 years | 1 (1.8) | 4 (6.8) | 5 (4.3) |
| 41–50 years | 8 (14.0) | 8 (13.6) | 16 (13.8) |
| 51–60 years | 15 (26.3) | 11 (18.6) | 26 (22.4) |
| 61–70 years | 14 (24.5) | 9 (15.2) | 23 (19.8) |
| 71–80 years | 18 (31.6) | 17 (28.8) | 35 (30.2) |
| >81 years | 1 (1.8) | 10 (17.0) | 11 (9.5) |
| Tumor Site | |||
| Right side of colon | 16 (28.1) | 12 (20.3) | 28 (24.2) |
| Left side of colon | 18 (31.6) | 29 (49.2) * | 47 (40.5) |
| Rectum | 11 (19.3) | 12 (20.3) | 23 (19.8) |
| Unspecified | 12 (21.0) | 6 (10.2) | 18 (15.5) |
| Histological Grade | |||
| Well-differentiated | 4 (7.0) | 9 (15.2) | 13 (11.2) |
| Moderately-differentiated | 44 (77.2) | 40 (67.8) | 84 (72.4) |
| Poorly-differentiated | 6 (10.5) | 5 (8.5) | 11 (9.5) |
| Unspecified | 3 (5.3) | 5 (8.5) | 8 (6.9) |
| Dukes’ Stage | |||
| Stage B | 48 (84.2) | 48 (81.4) | 96 (82.8) |
| Stage C | 8 (14.0) | 10 (16.9) | 18 (15.5) |
| Stage D | 1 (1.8) | 1 (1.7) | 2 (1.7) |
| Microsatellite Status—BRAF | |||
| MSS—wild type BRAF | 41 (72.0) | 46 (77.9) | 87 (75.0) |
| MSS—mutated BRAF | 2 (3.5) | 1 (1.7) | 3 (2.6) |
| MSI—wild type BRAF | 6 (10.5) | 4 (6.8) | 10 (8.6) |
| MSI—mutated BRAF | 4 (7.0) | 4 (6.8) | 8 (6.9) |
| Unspecified | 4 (7.0) | 4 (6.8) | 8 (6.9) |
| Follow-Up | |||
| Relapse, local or metastatic | 7 (12.3) | 17 (28.8) ** | 24 (20.7) |
| Relapse-free | 35 (61.4) | 28 (47.5) | 63 (54.3) |
| Unknown | 15 (26.3) | 14 (23.7) | 29 (25.0) |
* p-value = 0.040; ** p-value = 0.023.
2.2. Array Comparative Genomic Hybridization (aCGH)
Chromosomal CNAs were noted in male and female CRCs involving both autosomes and sex chromosomes. CRCs from female patients showed significantly more gains in chromosomal arms 1q21.2–q21.3, 4q13.2, 6p21.1, and 16p11.2, as well as copy number losses in chromosome arm 11q25 compared to male patients (Figure 1A–D and Table 2). This analysis was based on a global frequency statistical approach called Significance Testing for Aberrant Copy number (STAC) [15]. STAC-based algorithm is a robust method, which identifies a set of aberrations that are stacked on top of each other from different patients or microarrays such that it would not occur by chance. We further enhanced the analysis by using a second algorithm termed “GISTIC” for Genomic Identification of Significant Targets in Cancer, which identifies functionally significant CNAs by giving more weight to high copy gains and homozygous losses (amplitudes) that may be functionally relevant to the successful evolution of the cancer genome [16]. The GISTIC analysis confirmed the STAC determined CNAs (Figure 1E). The details of the GISTIC determined CNAs are presented in Supplementary File 1.
Figure 1.
aCGH analysis of 116 colorectal cancers showing chromosomal copy number aberrations in male and female patients. Gains are depicted in blue and deletions in red. (A) 1q21.2–q21.3 locus; (B) 4q13.2 locus; (C) 6p21.1 locus; (D) 11q25 locus; (E) aCGH analysis using GISTIC algorithm showing copy number profiles of the autosomes and X-chromosome. The grey bars indicate highly significant aberrations as detected by GISTIC. Asterisks (*) indicate CNAs previously obtained by Significance Testing for Aberrant Copy number (STAC) then confirmed here. Black circles indicate statistically significant CNAs observed in females only, while arrowheads pinpoint CNAs specific to males.
Table 2.
Gender-specific copy number aberrations and associated genes in colorectal cancer.
| Locus | CNA | Candidate Genes | Females % | Males % | p-Value |
|---|---|---|---|---|---|
| 1q21.2 | Gain | LINC00568, AF289612, AK023606, MIR4257, TSRC1, ADAMTSL4 | 50.8 | 17.5 | <0.0001 |
| 1q21.2–1q21.3 | Gain | U78576, PIP5K1A, BC007833, PSMD4, ZNF687, AK023105, PIK4CB, PI4KB, AJ011123, BC040300, RFX5, AK023875, SELENBP1, PSMB4, BX537561, POGZ, CGN, MIR554, TUFT1, AY358610 | 46.7 | 12.3 | <0.0001 |
| 4q13.2 | Gain | AF064819, UGT2B17 | 33.9 | 1.75 | <0.0001 |
| 6p21.31 | Gain | FKBP5, MIR5690 | 37.3 | 5.3 | <0.0001 |
| 6p21.1 | Gain | TBCC, AK096036, KIAA0240, GLTSCR1L, RPL7L1, C6orf226, LOC441150, PTCRA, CNPY3, TNRC5 | 35.6 | 3.5 | <0.0001 |
| 11q25 | Loss | FLJ45436, AY358331, NTM, BC050716, HNT | 47.45 | 14 | 0.0001 |
| 16p11.2 | Gain | QPRT, C16orf54, BC029149, ZG16, KIF22, BC004352, CR590954, AF489858, MAZ, L01420, BC041629, AK074572, PRRT2, LOC112476, AK092265, C16orf53, PAGR1, MVP, AK131349, CDIPT, LOC440356, BC000567, SEZ6L2, AJ245822, ASPHD1, LOC253982, KCTD13, TMEM219, LOC124446, TAOK2 | 54.2 | 20.8 | 0.0003 |
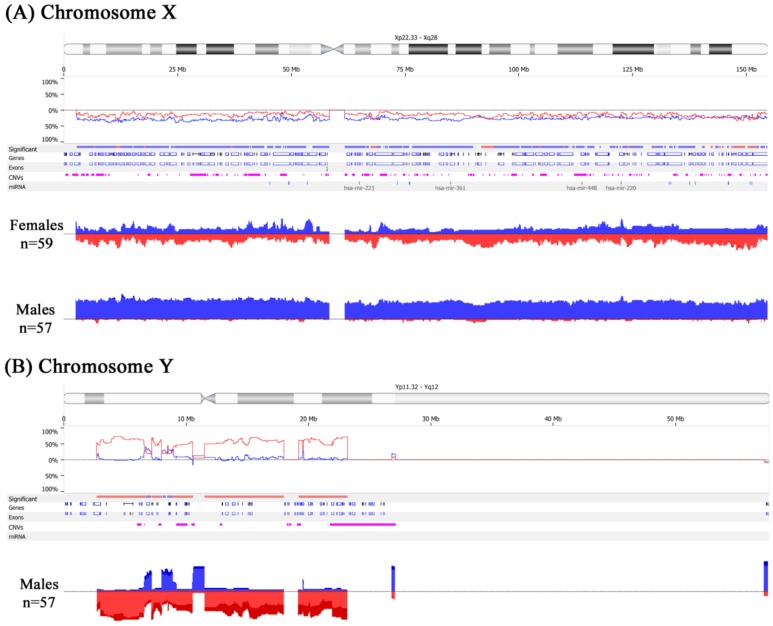
2.3. Feminization of Colorectal Cancer Cells in Males
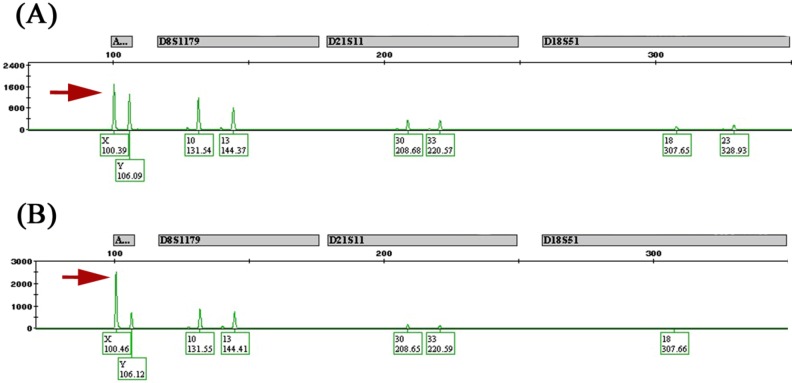
Significant copy number differences of the sex chromosomes were observed in male patients, namely, extensive X chromosome copy number gains associated with Y chromosome losses at multiple loci (Figure 2A,B). We refer to this phenomenon as “feminization” and define it as any CRC arising in male patients showing gains of the X chromosome (or an arm of X chromosome) and/or losses of the Y chromosome. Table 3 shows the loci involved in the X chromosomal gains and Y losses that are of statistical significance along with the candidate genes that may be potentially involved by the CNA at those loci. Cancers arising in females, on the other hand, had equivalent frequencies of gains and losses of the X chromosome (Figure 2A). In total, 26 of 57 (46%) male CRCs showed feminization of cancer cells by aCGH. For confirmation of these results, a number of male and female cases were subjected to reflex testing by FISH and MLPA. Out of 15 male tumors tested by FISH, eight were feminized by aCGH and confirmed by FISH as follows: four showed increased centromeric enumeration probe (CEP) X signals and some loss of CEP Y signals in tumor cells (heterogeneous), three showed increased CEP X signals and complete loss of CEP Y signals in almost all cancer cells analyzed (homogeneous) (Figure 3A,B), and one tumor showed increased CEP X signals with preservation of CEP Y. The remaining seven of the 15 male tumors showed increased CEP X signals by FISH only. Of the 19 male samples tested by MLPA, 14 cancers showed concordant results with that of aCGH. Five feminized cancers by aCGH were not confirmed by MLPA (data not shown). Finally, as a quality control measure, eight tumor samples were subjected to allelotyping and were all confirmed to be from the correct corresponding individuals (i.e., ruling out cross contamination between male and female samples) (Figure 4A,B).
Figure 2.
Sex-chromosome copy number aberrations in colorectal cancer cells by aCGH. Gains are depicted in blue and deletions in red. (A) Male cancers show extensive X chromosome gains while female cancers show equivalent frequencies of gains and losses; (B) Male cancers also show Y chromosome losses.
Table 3.
Copy number aberrations involving sex chromosomes in male and female colorectal cancers by array comparative genomic hybridization.
| Locus | CNA | Candidate Genes | Females % | Males % | p-Value |
|---|---|---|---|---|---|
| Xp22.32–p22.31 | Gain | NLGN4X, AK125309, MIR4770, VCX3A, HDHD1A, HDHD1, MIR4767, STS, VCX, PNPLA4, MIR651 | 9.3 | 42.3 | <0.01 |
| Xp22.31–p22.2 | Gain | VCX-C, VCX3B, KAL1, FAM9A, AK097501, FAM9B, AY459291, TBL1X, GPR143, X83543, SHROOM2, APXL, CR749271, LOC100288814, KIAA1280, WWC3, BC035601, CLCN4, MID1, HCCS, AMELX, ARHGAP6, MSL3L1, MSL3, PDZK10, FRMPD4, PRPS2, TLR7, TLR8, AY358296, TLR8-AS1, TMSB4X, FAM9C, LOC100093698, ATXN3L, LOC100133123 | 10.7 | 45.8 | <0.01 |
| Xp22.12–p22.11 | Gain | SH3KBP1, AL833278, CXorf23, AK094661, LOC729609, MIR23C, AK098768, MAP7D2, BC089400, FLJ14503, EIF1AX, SCARNA9L, RPS6KA3, CNKSR2, RP11-450P7.3, KLHL34, SMPX, BC036465, YY2, MBTPS2, SMS, PHEX, PHEX-AS1, ZNF645, LOC100873065, DDX53 | 12.5 | 46.2 | <0.01 |
| Xp22.11–p21.2 | Gain | PDK3, BC045634, PCYT1B, SCARNA23, POLA, POLA1, ARX, MAGEB18, MGC33889, MAGEB6, MAGEB5, VENTXP1, SMEK3P, AK131475, AK057304, DCAF8L2, MAGEB10, DCAF8L1, IL1RAPL1, MAGEB2, MAGEB3, MAGEB4, MAGEB1, NR0B1 | 9.4 | 44.8 | <0.01 |
| Xp21.1–p11.4 | Gain | DMD, MIR3915, BC036103, FAM47A, TMEM47, FAM47B, MAGEB16, CXorf22, RP13-11B7.1, CHDC2, CXorf30, FAM47C, LOC442444, AK125992, FTH1P18, PRRG1, AK130368, LANCL3, XK, CYBB, TCTE1L, DYNLT3, CXorf27, SYTL5, SRPX, RPGR, OTC, TSPAN7, TM4SF2, MID1IP1, LOC286442, AY316592, BCOR, ATP6AP2, BC010395, MPC1L, CXorf38, BC025334, AL832829, CRSP2, MED14 | 11.4 | 45.1 | <0.01 |
| Xq21.1–q21.31 | Gain | APOOL, SATL1, ZNF711, ZNF6, BC067294, AK128541, AK026445, AK025039, POF1B, AF309774, BC017500, FLJ38564, CHM, BC032237, DACH2, AK022715, KLHL4, AB051474, LAMR1P15, CPXCR1 | 9.1 | 42.6 | <0.01 |
| Xq22.3 | Gain | MID2, TEX13B, VSIG1, PSMD10, ATG4A, APG4A, AK054927, COL4A6 | 11.9 | 42.6 | 0.001 |
| Xq13.1 | Gain | EDA, MIR676, DGAT2L4, AWAT2, OTUD6A, HSHIN6, IGBP1 | 4.2 | 35.1 | 0.001 |
| Xq22.1–q22.2 | Gain | BEX4, BEXL1, TCEAL8, TCEAL5, BEX2, TCEAL7, WBP5 | 13.5 | 43.8 | 0.002 |
| Xp11.22 | Gain | LOC401589, SNORA11D, SNORA11E, AF329733, MAGED4, MAGED4B, XAGE2B, XAGE1A, XAGE1C, XAGE1D, XAGE1E, XAGE1, BC009538, XAGE1A, XAGE1B, XAGE2, XAGE1D, XAGE1B, SSX8, SSX7, SSX2, SSX2B, SPANXN5, SPANX-N5, XAGE5 | 8.5 | 39.6 | 0.004 |
| Xq21.1 | Gain | BX649166, HDX, CXorf43 | 11.2 | 41.6 | 0.008 |
| Xp22.12 | Gain | AK131412, MAP3K15 | 16.95 | 47.4 | 0.010 |
| Xq22.1 | Gain | RAB40AL, BEX1 | 10.2 | 40.4 | 0.010 |
| Xq13.1 | Gain | KIF4A, GDPD2, DLG3, AB033058 | 14.7 | 45.6 | 0.016 |
| Xq22.3 | Gain | MUM1L1, AK056478 | 8.5 | 38.6 | 0.019 |
| Xq23 | Gain | AMOT | 10.2 | 40.4 | 0.020 |
| Xq26.2 | Gain | AL832725, PHF6, HPRT1 | 17.3 | 48.2 | 0.022 |
| Xq22.3 | Gain | AK001040, CXorf57, BC070110, FLJ10178, AK024253, RNF128 | 11.9 | 42.1 | 0.023 |
| Xq23 | Gain | SLC6A14 | 12.3 | 44.3 | 0.028 |
| Xp11.23 | Gain | WDR45, GPKOW, MAGIX, FLJ21687, PLP2, PRICKLE3, LMO6, SYP, SSX1 | 18.97 | 49.7 | 0.030 |
| Xq22.2 | Gain | RAB40A | 11.4 | 42.3 | 0.030 |
| Xq27.1 | Gain | SRD5A1P1, F9 | 11.9 | 42.1 | 0.040 |
| Xq28 | Gain | AFF2, FMR2 | 11.9 | 42.1 | 0.042 |
| Yp11.31–p11.2 | Loss | SRY, RPS4Y1, ZFY, LINC00278, TGIF2LY, PCDH11Y, TTTY23, TTTY23B, TSPY1, TSPY2, TTTY1B, TTTY1, TTTY2B, TTTY2, TTTY21, TTTY7B, TTTY8B, TTTY8 | – | 67.89 | <0.01 |
| Yp11.2 | Loss | PRKY, TTTY16, TTTY12 | – | 54.6 | <0.01 |
| Yq11.1–q11.221 | Loss | GYG2P1, TTTY15, USP9Y, DDX3Y, UTY, BC071744, TMSB4Y, VCY, VCY1B, BC032567, NLGN4Y, NLGN4Y-AS1 | – | 57.4 | <0.01 |
| Yq11.222–q11.223 | Loss | TTTY9A, HSFY2, NCRNA00185, CYorf14, CD24, TTTY14, BCORL2, BCORP1, CYorf15A, TXLNG2P, BC035312, CYorf15B, D87072, KDM5D, SMCY, TTTY10, EIF1AY, RPS4Y2, U88898, RBMY2EP, AK026367, RBMY1A1, BC070298, RBMY1F, RBMY1B, TTTY13, RBMY1D, RBMY1E, RBMY1A1, TTTY6, PRY, PRY2, AY597808, TTTY6B, RBMY1F, RBMY1J, RBMY1A1, BC047768, TTTY5, RBMY2FP, TTTY6 | – | 60.01 | <0.01 |
Figure 3.
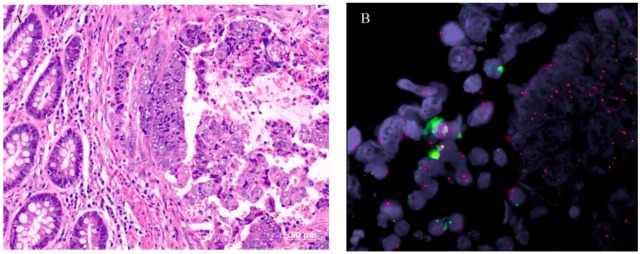
Morphology of a male colorectal cancer with feminization. (A) A hematoxylin and eosin stained histological section showing tumor on the right-hand side and normal colonic mucosa on the left; (B) Fluorescence in-situ hybridization of tumor cells (right-hand side) show multiple red centromeric enumeration probe (CEP) X signals indicating X chromosome gains and absence of green CEP Y signals indicating Y chromosome losses, while both red and green signals are present in non-neoplastic interstitial cells (left-hand side).
Figure 4.
Allelotyping. (A) Normal DNA from a male patient showing X and Y peaks of equal height (arrow); (B) Tumor DNA from same patient showing X peak more than twice the height of the Y peak (arrow) indicating X gains.
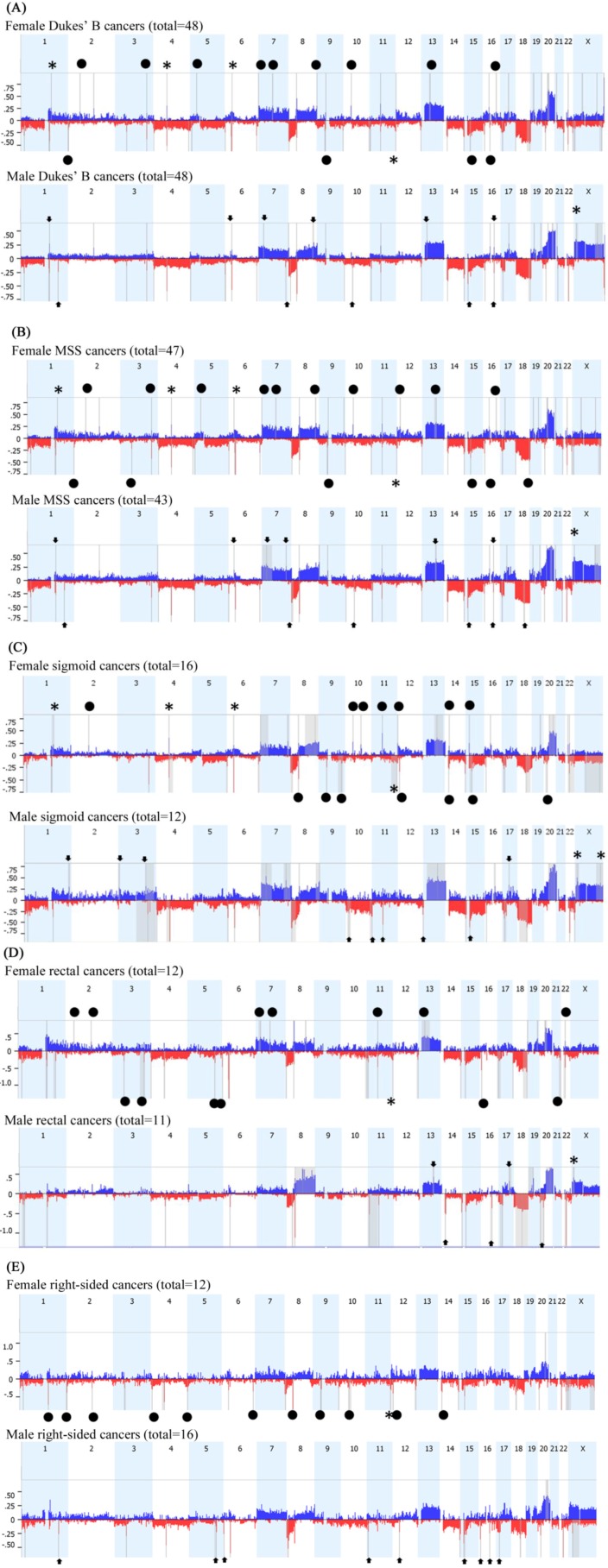
In order to minimize the potential of confounding factors, we have stratified the data by Dukes’ stage B (Figure 5A), microsatellite stable (MSS) status (Figure 5B), and cancer subsite for sigmoid colon (Figure 5C), rectum (Figure 5D) and right side of colon (Figure 5E). The resultant data were closely related to the unstratified GISTIC analysis presented in Figure 1E (Supplementary Files 2 and 3). Interestingly, CRC site appears to have a dramatic influence on gender-specific CNAs. While sigmoid colon cancers appear to display the same gender-specific CNAs seen in all CRCs overall, albeit with additional sigmoid-specific CNAs (Figure 5C and Supplementary File 4), rectal cancers appear to have only maintained the 11q25 loss in females and Xp22.32 gain in males (Figure 5D and Supplementary File 5). In right-sided colon cancer, gain of chromosome X in males was not significantly observed, and all other gender-specific CNAs were not detected except for loss of 11q25 in females, but other copy number losses were obvious between females and males (Figure 5E and Supplementary File 6).
Figure 5.
aCGH analysis of the autosomes and the X-chromosome using GISTIC algorithm limited to: (A) 96 cases of Dukes’ B cancer; (B) 90 cases of MSS cancer; (C) 28 cases with sigmoid colon cancer; (D) 23 cases with rectal cancer; (E) 28 cases with right-sided colon cancer. Gains are depicted in blue and deletions in red. The grey bars indicate highly significant aberrations as detected by GISTIC. Asterisks (*) indicate CNAs previously obtained by STAC then confirmed here. Black circles indicate statistically significant (p < 0.05) CNA specific to females, while arrowheads pinpoint CNA specific to males.
2.4. Clinicopathological Significance
Statistically significant parameters in male “feminized” CRCs, were microsatellite stability (MSS) status and BRAF gene mutation status (Table 4). Of the 26 feminized male cancers, 24 (92%) tumors showed MSS/wild-type BRAF (p-value 0.003). This constitutes 58.5% of all MSS/wild-type BRAF in the male group. For BRAF status alone, all of the 26 feminized tumors showed a wild-type codon 600 of the BRAF gene (p-value 0.009), constituting 53% of all wild-type BRAF tumors in the male group. No significant differences were detected between feminized and non-feminized cases with respect to age, histological differentiation, Dukes’ stage, KRAS mutation status, metastatic relapse or disease-free survival.
Table 4.
Clinicopathological characteristics of feminized colorectal cancers in male patients.
| Characteristics | Feminized Male CRC (Total = 26) No. (%) | Non-Feminized Male CRC (Total = 31) No. (%) | Total Males (Total = 57) No. (%) | p-Value |
|---|---|---|---|---|
| MSS—wild type BRAF | 24 (92.3) | 17 (56.7) | 41 (72.0) | 0.0026 * |
| Wild-type BRAF | 26 (100) | 23 (74.2) | 49 (86.0) | 0.0088 * |
| MMR proficient | 24 (92.3) | 19 (61.2) | 43 (75.4) | 0.0107 * |
* Statistically significant.
3. Discussion
Male and female CRCs demonstrate fundamental differences in several epidemiological, pathological, and clinical aspects. For example, men have a higher incidence of CRC than women overall [17,18]. With respect to cancer localization, left-sided CRC tends to be more frequent in men while right-sided CRC more frequent in women [19,20,21]. In our current study, however, left-sided tumors were more frequently seen in female patients (Table 1). CRCs demonstrating microsatellite instability (MSI) and CpG island methylator phenotype (CIMP-high) are significantly associated with the female gender, BRAF mutations and wild-type KRAS [22,23,24,25], whereas microsatellite-stable CRCs with less extensive promoter methylation (CIMP-low) are associated with the male gender and KRAS mutations [25,26]. Additionally, women with CRC seem to have an age-dependent survival advantage over men [18,27,28,29], with better survival seen in young to middle-aged women with localized disease [30,31] and worse survival in older women after menopause [32,33]. Some studies did not show a difference in survival between the two genders [34,35]. Sex hormonal influences and the protective effect of estrogen in women have long been proposed as a factor in this gender bias in the incidence and behavior of CRC [30,36], as opposed to testosterone, which is generally thought to contribute to increased aggressiveness of cancer in males [37,38]. Of note, the hormonal differences typical of the specific gender are unlikely to be seen in female patients older than 55 and male patients above 62 years of age. Nevertheless, the exact mechanisms underlying gender-related differences in CRC remain largely unexplained. Our study showed more cancer recurrences in female patients, however, there were no statistically significant differences in disease-free survival between men and women. This may be partly due to a selection bias of postmenopausal women in this study, since the majority of female patients were over the age of 50 years, or may be due to topographic differences in this region of the world. A recent study in Kuwait showed a mean recurrence-free survival time in CRC patients of 72.8 months and 65.2 months in males and females, respectively (p-value 0.025) [39].
We used high-resolution aCGH (244 K), a technique that screens the entire tumor genome for genetic gains and losses, in order to study chromosomal CNAs in CRC cells in relation to gender. Significant differences in CNAs between females and males were noted at several loci but the differences in the X and Y chromosome imbalances were particularly interesting. Twenty-six male CRCs, comprising 46% of all male cancers in this study, showed “feminization” in the form of extensive X chromosome gains with or without Y chromosome losses. Our aCGH results were further confirmed by multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridization (FISH). In cases subjected to MLPA, 73% showed strong correlation with aCGH data, while those subjected to FISH 57% showed strong correlation with aCGH data. Five of the feminized male cancers identified by aCGH were not confirmed by MLPA. This discrepancy between aCGH and MLPA is expected and can be explained by the fact that the MLPA technique detects a small gene locus on the X chromosome that may not be involved in the CNA, and therefore MLPA offers a more limited view of the X chromosome.
The gender-specific wide-genome CNAs reported here might have been influenced by several confounding factors, although in this cohort female and male CRCs did not differ significantly in terms of clinicopathological characteristics (Table 1). For this reason, we have stratified our CNA analysis to take into account the potential influences of Dukes’ stage, microsatellite status and cancer subsite. After stratification, the gender-specific aberrations were confirmed, but interestingly our data indicate that these gender-specific CNAs may be site-specific. For example, while sigmoid colon cancers in males appear to display significant feminization, right-sided colon cancer did not demonstrate significant gains of the X chromosome. This intriguing observation deserves attention and confirmation in larger cohorts.
X chromosome gains have previously been observed in a variety of tumors including hepatoblastoma [40], prostate cancer [41], and testicular germ cell tumors [42], amongst others. With respect to CRC, Dutrillaux et al. [11,12] first described the gain of early-replicating (active) X chromosomes in male and female CRCs, demonstrating that the X gains were associated with a frequent loss of either the late-replicating (inactive) X chromosome in females or the Y chromosome in males. De Angelis et al. [6] demonstrated a high level of X chromosome gains by CGH in a series of 45 CRCs with a higher percentage of Xq gain in male patients and X losses in females. Brim et al. [14] also demonstrated the preferential gain of chromosome X in male CRC cells in a study of 30 CRCs from African American male patients, a high-risk group for CRC. The hypothesis postulated is that increased “dosage” of expressed X-linked genes may play a key role in oncogenesis [6,11,43] and that the higher incidence of CRC in males may be related to the lack of X-chromosomal inactivation [13]. In normal female cells, random inactivation of one of the X chromosomes occurs early in life making female somatic tissues mosaic with respect to whether the maternal or paternal X chromosome is active [43]. Therefore, gain of X chromosomes in male cancer cells may increase the expression of oncogenes carried on the X chromosome that are related to CRC [13].
Similar to the results of Unotoro et al. [13], the current study shows significant gains of the Xp21 locus in male CRC cells (p-value < 0.01), which is the location of the MAGE-B gene family (melanoma-associated antigen gene) [44]. Another significantly gained locus was Xp11.22 (p-value 0.004), which hosts the XAGE genes as well as the SSX genes (human synovial sarcoma on X chromosome) that are rearranged in synovial sarcoma. MAGE, SSX, and XAGE genes all belong to the cancer-testis antigen family of genes that are normally expressed in the testis and aberrantly expressed in a wide array of human cancers [45,46]. MAGE and SSX co-expression has been recently correlated with liver metastasis in colorectal cancer [47]. At the present time, however, it is difficult to know, which genes are of significance in feminized CRCs due to the large genomic areas affected by the X chromosomal aberrations. With respect to the Y chromosome losses in male CRC observed here, other studies also support our findings [11,12,48].
Other than sex chromosome CNAs, tumors from females showed significantly more gains at loci 1q, 4q, 6p and 16p compared to males and significantly more losses at 11q. Gains of 1q and 4q have previously been demonstrated in CRC [49] but were not correlated to gender. Several other chromosomal gains and losses have also been described in CRC, for example gains of 3q, 7p, 8q, 13q, 17p, 18q, 20q [4,6,7,9] and losses of 1p, 4, 5q, 18q and 17p [5,7,8,9]. However, the relationship between these CNAs and gender has rarely been studied. What kind of influence these gender-associated CNAs have on CRC needs further exploration in larger cohorts.
In the present study, feminized CRCs correlated with MSS and wild-type BRAF status. Conversely, tumors that lacked feminization were mostly MSI tumors, BRAF-mutated, and right-sided. MSS is an expected finding in feminized CRCs, as MSI and chromosomal instability (feminization is a form of chromosomal instability) are less likely to coexist in the same tumor [50,51]. However, more recent evidence suggests that MSI and chromosomal instability may not be mutually exclusive [52,53,54]. Since MSS CRCs may have a worse prognosis than MSI cancers [55,56,57,58], we have restricted our disease-free survival analysis to the “MSS group” of patients and found that males with and without feminized tumors did not differ in disease-free survival rate (data not shown). No significant correlation was found between feminization and age, histological differentiation, Dukes’ stage, KRAS mutation status or disease-free survival. Larger studies are required to confirm these results.
In summary, differences in chromosomal CNAs between genders may be an important aspect in the development of CRC, and feminization may constitute a unique pathway in colorectal carcinogenesis in a subset of male patients, which warrants further investigation using larger cohorts.
4. Materials and Methods
4.1. Patients and Tissue Samples
This study was approved by the Ethical Committee (2006-1302-07, 13 April 2008) of the Faculty of Medicine at Kuwait University. A cohort of 116 patients with CRC was used for genetic analysis. Formalin-fixed paraffin-embedded (FFPE) tissue samples were retrieved from pathology archives and clinical information (gender, age, tumor site in the colon, tumor histological grade, Dukes’ stage, and clinical outcome data) was retrieved from patients’ charts.
4.2. Array Comparative Genomic Hybridization (aCGH)
Our standard published protocol was used to process the FFPE tissue samples for aCGH [59]. Briefly, 2 μg of tumor DNA and pooled sex-matched reference DNA (Promega, Fitchburg, WI, USA) were fragmented in a water-bath sonicator (Elmasonic, Singen, Germany), then labeled using the Cy3 and Cy5 universal linkage system dyes (Agilent Technologies, Santa Clara, CA, USA). The labeled DNA was purified using the Agilent-KREApure columns (Agilent Technologies), and hybridized on to Human 244A slides (Agilent Technologies). Hybridization was carried out in SureHyb Chambers (Agilent Technologies) for 40 h at 60 °C. Slides were then washed and scanned at 5 μm resolution using an Agilent microarray scanner. Scanned images were imported, and the background was subtracted and normalized using Feature Extraction Software (Agilent Technologies). The feature extraction software generates a Quality Control Report, which determines the quality of the aCGH. Quality Control metrics such as Derivative of Log Ratio Spread (DLRSpread), Back Ground Noise, Signal Intensity, Reproducibility and Signal to Noise ratio are generated in the QC report.
DLRSpread is defined as the spread of the Log Ratio differences between consecutive probes along all chromosomes. It enhances the ability to measure the noise of the log ratio independently of the number and severity of aberrations found. Samples with DLRSpread higher than 0.5 were excluded from further analysis. The text files representing data ratio points log2 of test/control ratios were imported to Nexus software (Biodiscovery, El Segundo, CA, USA). Quality values ranged between 0.05 and 0.4, which are excellent values given the degraded nature of the samples. To minimize false positive calls and random copy number variations, Fast Adaptive State Segmentation Technique (FASST2) with a stringent significance threshold of 5.0 × 10−6 was used to determine CNAs for each sample. CNA comparisons between the two groups (Males vs. Females) were performed within Nexus using the Fisher’s exact test. The Q-bound value, which corrects for multiple testing by performing False Discovery Rate (FDR) correction, was used to highlight significant CNA differences between the two groups.
4.3. Fluorescence in Situ Hybridization (FISH)
To confirm the aCGH results, sex-chromosome copy number counts by FISH was performed in 30 tumor samples (15 males and 15 females). For that we used the CEP X SpectrumOrange/Y SpectrumGreen DNA Probe Kit (Abbott Molecular, North Chicago, IL, USA), which detects alpha satellite sequences in the centromere region of the X chromosome (Xp11.1–q11.1) and satellite III DNA at the Yq12 region of the Y chromosome. We performed this 2-color FISH on 3-μm-thick tissue sections, which were then counterstained using 4'6-diamidino-2-phenylindole (DAPI). The Capturing and counting of the signals were performed using the “Metafer” system (Metasystems, Newton, MA, USA) on a Carl–Zeiss fluorescent microscope (Zeiss, Oberkochen, Germany).
4.4. Multiplex Ligation-Dependent Probe Amplification (MLPA)
For further verification, 44 tumor samples (19 males, 25 females) were subjected to MLPA to assess the gender of cancer cells. The MLPA assay was performed using the SALSA® MLPA® probemix P294-A1 tumor-loss kit (MRC-Holland; Amsterdam, The Netherlands). The hybridization, ligation, and amplification steps were performed according to the manufacturer’s standard protocol and the data analysis was done using the Coffalyser NET software created by MRC-Holland, Amsterdam, The Netherlands. The data generated from CRC samples were normalized and quality checked against reference controls. MLPA result reports, including descriptive statistics, ratios, 95% confidence intervals, and predictions, were exported to Microsoft Excel software data sheets for further analysis.
4.5. Allelotyping
As a quality control measure, randomly selected eight-paired normal and corresponding tumor tissues were subjected to allelotyping in order to rule-out cross contamination between male and female tissue samples. The identities of tumor tissues and their corresponding normal tissues were validated using PowerPlex-16 HS System (Promega, Fitchburg, WI, USA) according to the manufacturer’s instruction manual. Genomic DNA was extracted using the Qiagen DNA extraction kit (producer, Hilden, Germany). Detection of amplified fragments was performed using Life Technologies 3130xl Genetic Analyzer and GeneMapper software version 4.0 (Foster City, CA, USA).
4.6. Clinicopathological Correlation and Statistical Analysis
Chromosomal CNA data generated from aCGH were correlated with clinicopathological parameters including tumor location within the colon, histopathological differentiation, Dukes’ stage, microsatellite instability (MSI) status, mismatch repair (MMR) gene function, BRAF gene mutation status (p.V600E), and KRAS gene status (pre-determined using the standard Sanger sequencing method). Statistical analysis was calculated using Chi-square or Fisher’s exact tests and p-values of <0.05 were considered statistically significant. Disease-free survivals were quantified by the generation of Kaplan–Meier curves, and detection of cancer recurrence in the form of metastasis or local recurrence was used as an endpoint measurement. Patients who did not relapse (“endpoint”) during the study period (1990–2009) but died of other reasons or were lost to follow-up were censored. The mean period of follow-up was 7.5 years.
5. Conclusions
Feminization of colorectal cancer cells in male patients warrants further investigation using larger cohorts.
Acknowledgments
This work was supported by grant No. 2011-1302-06 and 2006-1302-07 from Kuwait Foundation for the Advancement of Sciences and grant No. MG 02/08 from Kuwait University.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/15/10/17344/s1.
Supplementary Files
Author Contributions
Rola H. Ali searched the literature and drafted the manuscript; Makia Marafie and Milad S. Bitar participated in the experiment; Fahad Al-Dousari and Samar Ismael performed the allelotyping; Hussain Bin Haider, Waleed Al-Ali and Sindhu P. Jacob carried out most parts of the experiment; Fahd Al-Mulla designed the study and performed the statistical analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ried T., Knutzen R., Steinbeck R., Blegen H., Schröck E., Heselmeyer K., du Manoir S., Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Bomme L., Bardi G., Pandis N., Fenger C., Kronborg O., Heim S. Clonal karyotypic abnormalities in colorectal adenomas: Clues to the early genetic events in the adenoma-carcinoma sequence. Genes Chromosomes Cancer. 1994;10:190–196. doi: 10.1002/gcc.2870100307. [DOI] [PubMed] [Google Scholar]
- 4.Bardi G., Sukhikh T., Pandis N., Fenger C., Kronborg O., Heim S. Karyotypic characterization of colorectal adenocarcinomas. Genes Chromosomes Cancer. 1995;12:97–109. doi: 10.1002/gcc.2870120204. [DOI] [PubMed] [Google Scholar]
- 5.Al-Mulla F., Keith W.N., Pickford I.R., Going J.J., Birnie G.D. Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer. 1999;24:306–314. doi: 10.1002/(sici)1098-2264(199904)24:4<306::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 6.De Angelis P.M., Clausen O.P., Schjolberg A., Stokke T. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br. J. Cancer. 1999;80:526–535. doi: 10.1038/sj.bjc.6690388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Q.J., Zeng W.F., Sham J.S., Xie D., Yang X.W., Lin H.L., Zhan W.H., Lin F., Zeng S.D., Nie D., et al. Recurrent genetic alterations in 26 colorectal carcinomas and 21 adenomas from Chinese patients. Cancer Genet. Cytogenet. 2003;144:112–118. doi: 10.1016/s0165-4608(02)00959-7. [DOI] [PubMed] [Google Scholar]
- 8.Al-Mulla F., Behbehani A.I., Bitar M.S., Varadharaj G., Going J.J. Genetic profiling of stage I and II colorectal cancer may predict metastatic relapse. Mod. Pathol. 2006;19:648–658. doi: 10.1038/modpathol.3800564. [DOI] [PubMed] [Google Scholar]
- 9.Tsafrir D., Bacolod M., Selvanayagam Z., Tsafrir I., Shia J., Zeng Z., Liu H., Krier C., Stengel R.F., Barany F., et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 10.Dorak M.T., Karpuzoglu E. Gender differences in cancer susceptibility: An inadequately addressed issue. Front. Genet. 2012;3:268. doi: 10.3389/fgene.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutrillaux B., Muleris M., Seureau M.G. Imbalance of sex chromosomes, with gain of early-replicating X, in human solid tumors. Int. J. Cancer. 1986;38:475–479. doi: 10.1002/ijc.2910380404. [DOI] [PubMed] [Google Scholar]
- 12.Muleris M., Dutrillaux A.M., Salmon R.J., Dutrillaux B. Sex chromosomes in a series of 79 colorectal cancers: Replication pattern, numerical, and structural changes. Genes Chromosomes Cancer. 1990;1:221–227. doi: 10.1002/gcc.2870010306. [DOI] [PubMed] [Google Scholar]
- 13.Unotoro J., Kamiyama H., Ishido Y., Yaginuma Y., Kasamaki S., Sakamoto K., Oota A., Ishibashi Y., Kamano T. Analysis of the relationship between sex and chromosomal aberrations in colorectal cancer by comparative genomic hybridization. J. Int. Med. Res. 2006;34:397–405. doi: 10.1177/147323000603400409. [DOI] [PubMed] [Google Scholar]
- 14.Brim H., Lee E., Abu-Asab M.S., Chaouchi M., Razjouyan H., Namin H., Goel A., Schäffer A.A., Ashktorab H. Genomic aberrations in an African American colorectal cancer cohort reveals a MSI-specific profile and chromosome X amplification in male patients. PLoS One. 2012;7:e40392. doi: 10.1371/journal.pone.0040392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diskin S.J., Eck T., Greshock J., Mosse Y.P., Naylor T., Stoeckert C.J., Jr., Weber B.L., Maris J.M., Grant G.R. STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 2006;16:1149–1158. doi: 10.1101/gr.5076506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beroukhim R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J.C., Huang J.H., Alexander S., et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rim S.H., Seeff L., Ahmed F., King J.B., Coughlin S.S. Colorectal cancer incidence in the United States, 1999–2004: An updated analysis of data from the National Program of Cancer Registries and the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1967–1976. doi: 10.1002/cncr.24216. [DOI] [PubMed] [Google Scholar]
- 18.Purim O., Gordon N., Brenner B. Cancer of the colon and rectum: Potential effects of sex–age interactions on incidence and outcome. Med. Sci. Monit. 2013;19:203–209. doi: 10.12659/MSM.883842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeCosse J.J., Ngoi S.S., Jacobson J.S., Cennerazzo W.J. Gender and colorectal cancer. Eur. J. Cancer Prev. 1993;2:105–115. doi: 10.1097/00008469-199303000-00003. [DOI] [PubMed] [Google Scholar]
- 20.McCashland T.M., Brand R., Lyden E., de Garmo P., Project C.R. Gender differences in colorectal polyps and tumors. Am. J. Gastroenterol. 2001;96:882–886. doi: 10.1111/j.1572-0241.2001.3638_a.x. [DOI] [PubMed] [Google Scholar]
- 21.Takada H., Ohsawa T., Iwamoto S., Yoshida R., Nakano M., Imada S., Yoshioka K., Okuno M., Masuya Y., Hasegawa K., et al. Changing site distribution of colorectal cancer in Japan. Dis. Colon Rectum. 2002;45:1249–1254. doi: 10.1007/s10350-004-6400-0. [DOI] [PubMed] [Google Scholar]
- 22.Ward R., Meagher A., Tomlinson I., O’Connor T., Norrie M., Wu R., Hawkins N. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48:821–829. doi: 10.1136/gut.48.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins N., Norrie M., Cheong K., Mokany E., Ku S.L., Meagher A., O’Connor T., Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 24.Ogino S., Cantor M., Kawasaki T., Brahmandam M., Kirkner G.J., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S., Goel A. Molecular classification and correlates in colorectal cancer. J. Mol. Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S., Kawasaki T., Kirkner G.J., Loda M., Fuchs C.S. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: Possible associations with male sex and KRAS mutations. J. Mol. Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsaleh H., Joseph D., Grieu F., Zeps N., Spry N., Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 28.Kotake K., Honjo S., Sugihara K., Kato T., Kodaira S., Takahashi T., Yasutomi M., Muto T., Koyama Y. Changes in colorectal cancer during a 20-year period: An extended report from the multi-institutional registry of large bowel cancer, Japan. Dis. Colon Rectum. 2003;46:S32–43. doi: 10.1097/01.DCR.0000083389.92214.AA. [DOI] [PubMed] [Google Scholar]
- 29.Micheli A., Ciampichini R., Oberaigner W., Ciccolallo L., de Vries E., Izarzugaza I., Zambon P., Gatta G., de Angelis R. EUROCARE Working Group. The advantage of women in cancer survival: An analysis of EUROCARE-4 data. Eur. J. Cancer. 2009;45:1017–1027. doi: 10.1016/j.ejca.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Hendifar A., Yang D., Lenz F., Lurje G., Pohl A., Lenz C., Ning Y., Zhang W., Lenz H.J. Gender disparities in metastatic colorectal cancer survival. Clin. Cancer Res. 2009;15:6391–6397. doi: 10.1158/1078-0432.CCR-09-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majek O., Gondos A., Jansen L., Emrich K., Holleczek B., Katalinic A., Nennecke A., Eberle A., Brenner H. GEKID Cancer Survival Working Group. Sex differences in colorectal cancer survival: Population-based analysis of 164,996 colorectal cancer patients in Germany. PLoS One. 2013;8:e68077. doi: 10.1371/journal.pone.0068077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koo J.H., Jalaludin B., Wong S.K., Kneebone A., Connor S.J., Leong R.W. Improved survival in young women with colorectal cancer. Am. J. Gastroenterol. 2008;103:1488–1495. doi: 10.1111/j.1572-0241.2007.01779.x. [DOI] [PubMed] [Google Scholar]
- 33.Koo J.H., Leong R.W. Sex differences in epidemiological, clinical and pathological characteristics of colorectal cancer. J. Gastroenterol. Hepatol. 2010;25:33–42. doi: 10.1111/j.1440-1746.2009.05992.x. [DOI] [PubMed] [Google Scholar]
- 34.Ponz de Leon M., Sant M., Micheli A., Sacchetti C., di Gregorio C., Fante R., Zanghieri G., Melotti G., Gatta G. Clinical and pathologic prognostic indicators in colorectal cancer. A population-based study. Cancer. 1992;69:626–635. doi: 10.1002/1097-0142(19920201)69:3<626::aid-cncr2820690305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 35.Manfredi S., Bouvier A.M., Lepage C., Hatem C., Dancourt V., Faivre J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br. J. Surg. 2006;93:1115–1122. doi: 10.1002/bjs.5349. [DOI] [PubMed] [Google Scholar]
- 36.Slattery M.L., Potter J.D., Curtin K., Edwards S., Ma K.N., Anderson K., Schaffer D., Samowitz W.S. Estrogens reduce and withdrawal of estrogens increase risk of microsatellite instability-positive colon cancer. Cancer Res. 2001;61:126–130. [PubMed] [Google Scholar]
- 37.Shahabi S., He S., Kopf M., Mariani M., Petrini J., Scambia G., Ferlini C. Free testosterone drives cancer aggressiveness: Evidence from US population studies. PLoS One. 2013;8:e61955. doi: 10.1371/journal.pone.0061955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orsted D.D., Nordestgaard B.G., Bojesen S.E. Plasma testosterone in the general population, cancer prognosis and cancer risk: A prospective cohort study. Ann. Oncol. 2014;25:712–718. doi: 10.1093/annonc/mdt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Qallaf D.A., Al-Mulla F. Ph.D. Thesis. Kuwait University; Kuwait City, Kuwait: 2012. Colorectal Carcinoma in Kuwait: Apoptosis and Its Relation to Clinicopathological Characteristics, p53 Expression and ki-ras Proto-Oncogene Mutations. [Google Scholar]
- 40.Terracciano L.M., Bernasconi B., Ruck P., Stallmach T., Briner J., Sauter G., Moch H., Vecchione R., Pollice L., Pettinato G., et al. Comparative genomic hybridization analysis of hepatoblastoma reveals high frequency of X-chromosome gains and similarities between epithelial and stromal components. Hum. Pathol. 2003;34:864–871. doi: 10.1016/s0046-8177(03)00351-4. [DOI] [PubMed] [Google Scholar]
- 41.Visakorpi T., Hyytinen E., Kallioniemi A., Isola J., Kallioniemi O.P. Sensitive detection of chromosome copy number aberrations in prostate cancer by fluorescence in situ hybridization. Am. J. Pathol. 1994;145:624–630. [PMC free article] [PubMed] [Google Scholar]
- 42.Looijenga L.H., Oosterhuis J.W. Pathogenesis of testicular germ cell tumours. Rev. Reprod. 1999;4:90–100. doi: 10.1530/ror.0.0040090. [DOI] [PubMed] [Google Scholar]
- 43.Spatz A., Borg C., Feunteun J. X-chromosome genetics and human cancer. Nat. Rev. Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 44.Muscatelli F., Walker A.P., de Plaen E., Stafford A.N., Monaco A.P. Isolation and characterization of a MAGE gene family in the Xp21.3 region. Proc. Natl. Acad. Sci. USA. 1995;92:4987–4991. doi: 10.1073/pnas.92.11.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dos Santos N.R., Torensma R., de Vries T.J., Schreurs M.W., de Bruijn D.R., Kater-Baats E., Ruiter D.J., Adema G.J., van Muijen G.N., van Kessel A.G. Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res. 2000;60:1654–1662. [PubMed] [Google Scholar]
- 46.Simpson A.J., Caballero O.L., Jungbluth A., Chen Y.T., Old L.J. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 47.Choi J., Chang H. The expression of MAGE and SSX, and correlation of COX2, VEGF, and survivin in colorectal cancer. Anticancer Res. 2012;32:559–564. [PubMed] [Google Scholar]
- 48.Bottarelli L., Azzoni C., Necchi F., Lagrasta C., Tamburini E., D’Adda T., Pizzi S., Sarli L., Rindi G., Bordi C. Sex chromosome alterations associate with tumor progression in sporadic colorectal carcinomas. Clin. Cancer Res. 2007;13:4365–4370. doi: 10.1158/1078-0432.CCR-06-2736. [DOI] [PubMed] [Google Scholar]
- 49.Xiao X.Y., Zhou X.Y., Yan G., Sun M.H., Du X. Chromosomal alteration in Chinese sporadic colorectal carcinomas detected by comparative genomic hybridization. Diagn. Mol. Pathol. 2007;16:96–103. doi: 10.1097/PDM.0b013e31803190f2. [DOI] [PubMed] [Google Scholar]
- 50.Lothe R.A., Peltomäki P., Meling G.I., Aaltonen L.A., Nyström-Lahti M., Pylkkänen L., Heimdal K., Andersen T.I., Møller P., Rognum T.O., et al. Genomic instability in colorectal cancer: Relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 51.Muleris M., Dutrillaux A.M., Olschwang S., Salmon R.J., Dutrillaux B. Predominance of normal karyotype in colorectal tumors from hereditary non-polyposis colorectal cancer patients. Genes Chromosomes Cancer. 1995;14:223–226. doi: 10.1002/gcc.2870140312. [DOI] [PubMed] [Google Scholar]
- 52.Goel A., Arnold C.N., Niedzwiecki D., Chang D.K., Ricciardiello L., Carethers J.M., Dowell J.M., Wasserman L., Compton C., Mayer R.J., et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 53.Camps J., Armengol G., del Rey J., Lozano J.J., Vauhkonen H., Prat E., Egozcue J., Sumoy L., Knuutila S., Miró R. Genome-wide differences between microsatellite stable and unstable colorectal tumors. Carcinogenesis. 2006;27:419–428. doi: 10.1093/carcin/bgi244. [DOI] [PubMed] [Google Scholar]
- 54.Trautmann K., Terdiman J.P., French A.J., Roydasgupta R., Sein N., Kakar S., Fridlyand J., Snijders A.M., Albertson D.G., Thibodeau S.N., et al. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin. Cancer Res. 2006;12:6379–6385. doi: 10.1158/1078-0432.CCR-06-1248. [DOI] [PubMed] [Google Scholar]
- 55.Hawkins N.J., Tomlinson I., Meagher A., Ward R.L. Microsatellite-stable diploid carcinoma: A biologically distinct and aggressive subset of sporadic colorectal cancer. Br. J. Cancer. 2001;84:232–236. doi: 10.1054/bjoc.2000.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinicrope F.A., Rego R.L., Halling K.C., Foster N., Sargent D.J., La Plant B., French A.J., Laurie J.A., Goldberg R.M., Thibodeau S.N., et al. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology. 2006;131:729–737. doi: 10.1053/j.gastro.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Kazama Y., Watanabe T., Kanazawa T., Tanaka J., Tanaka T., Nagawa H. Microsatellite instability in poorly differentiated adenocarcinomas of the colon and rectum: Relationship to clinicopathological features. J. Clin. Pathol. 2007;60:701–704. doi: 10.1136/jcp.2006.039081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe T., Kobunai T., Yamamoto Y., Matsuda K., Ishihara S., Nozawa K., Yamada H., Hayama T., Inoue E., Tamura J., et al. Chromosomal instability (CIN) phenotype, CIN high or CIN low, predicts survival for colorectal cancer. J. Clin. Oncol. 2012;30:2256–2264. doi: 10.1200/JCO.2011.38.6490. [DOI] [PubMed] [Google Scholar]
- 59.Al-Mulla F. Microarray-based CGH and copy number analysis of FFPE samples. Methods Mol. Biol. 2011;724:131–145. doi: 10.1007/978-1-61779-055-3_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.