Abstract
Novel scaffolds are of uttermost importance for the discovery of functional material. Three different heterocyclic scaffolds easily accessible from isocyanoacetaldehyde dimethylacetal 1 by multicomponent reaction (MCR) are described. They can be efficiently synthesized by a Ugi tetrazole multicomponent reaction of 1. We discuss the synthesis, 3D structures, and other physicochemical properties.
Novel scaffolds form the basis for success in the discovery of bioactive compounds, which eventually can be developed to drugs for the treatment of unmet medical needs. A decade ago the NIH started an initiative (Molecular Libraries Program (MLP)) to assemble a large chemical library to be screened by academic institutions to yield, after optimization, in vitro tool compounds (molecular probes) for novel targets showing activity and selectivity in cell based systems.1 These tool compounds can be accessed by interested researchers and are of importance to elucidate the interplay of novel targets in biology and disease.2 The European Lead Factory (ELF), a public private partnership, is a complementary European initiative with similar targets aiming for a library of 500 000 novel compounds by 2017.3 The availability of molecular probes (small molecule or antibody) has been recently and impressively demonstrated to be a key determinant of progress in basic biology and disease areas.4
From a practical point of view the synthesis of medium sized high quality libraries is demanding. The use of a “universal building block” in the synthesis of different scaffolds has great advantages in the parallel synthesis of larger libraries. For example, unprotected α-amino acids have been used recently in different multicomponent reaction chemistry to stereoselectively afford a diversity of novel cyclic and acyclic scaffolds, including amido-aminophosphonates,5 boneratamide analogues,6 iminodicarboxamides,7 iminobenzazocineacetamide,8 boropeptides,9 thiolactone and thiomorpholino, diketopiperazines,10 seleno amino acid,11 imidazol,12 or indol derivatives.13
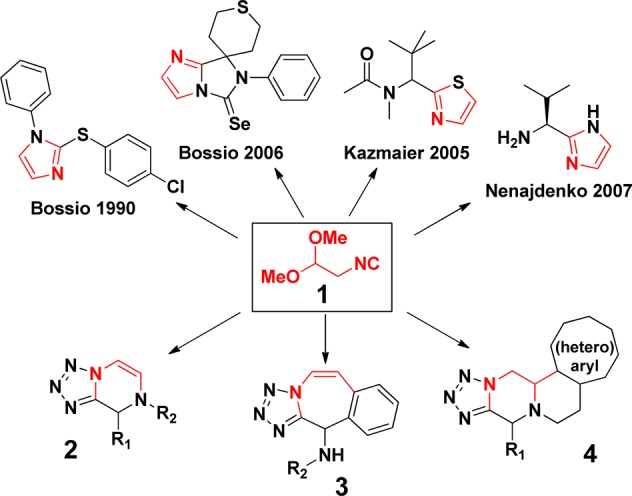
Isocyanide 1 as its diethyl acetal was first described by Hardtke in 1966 and now robust large scale syntheses exist.14 Competitive analysis shows few applications of the building block 1 in heterocycle synthesis toward 1-aryl-2-arylthio-1H-imidazoles and imidazo[1,5-a]imidazoles (Scheme 1).15 Other scaffolds reported include thiazoles and chiral imidazoles both by a thio Ugi reaction.16 Here we describe the easy synthesis of different scaffolds 2–4 using the same easily accessible building block 1, while not compromising diversity aspects.
Scheme 1. Previously Elaborated Heterocyclic Scaffold Using Isocyanide 1 and Our Work toward Three Novel Heterocycles.
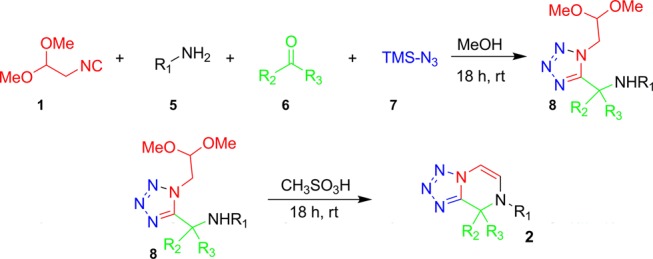
We envisioned that tetrazole annulated piperazine scaffold 2 could be accessed from the Ugi-tetrazole reaction using isocyanide building block 1, primary amines 5, aldehydes or ketones 6, and TMSN37 (Scheme 2) and subsequent cyclization/elimination via the secondary amine. In the optimization campaign the reaction is performed in methanol at ambient temperature. The Ugi adduct 8 was further treated with neat acetic acid at 80 °C but did not yield the cyclized product 2. Similar results were obtained with trifluoroacetic acid, methanesulfonic acid with and without solvents such as CH2Cl2, CH3CN, and toluene at 50–80 °C. However, the Ugi-adduct 8 stirred with neat methanesulfonic acid for 18 h at ambient temperature afforded scaffold 2 in good yields.
Scheme 2. Designed Synthetic Pathway to Tetrazolo Piperazine Scaffold 2.
In general, the Ugi reaction works well with aromatic and aliphatic aldehydes and ketones. In the post-Ugi reaction we observed that aliphatic aldehyde, ketones, and electron-deficient benzaldehydes could be used to give the product 2 in good yield. Electron-rich benzaldehydes also worked, however giving the desired product in low yields (2c). Typical examples are shown in Table 1.
Table 1. Typical Structures and Yields of Scaffold 2.
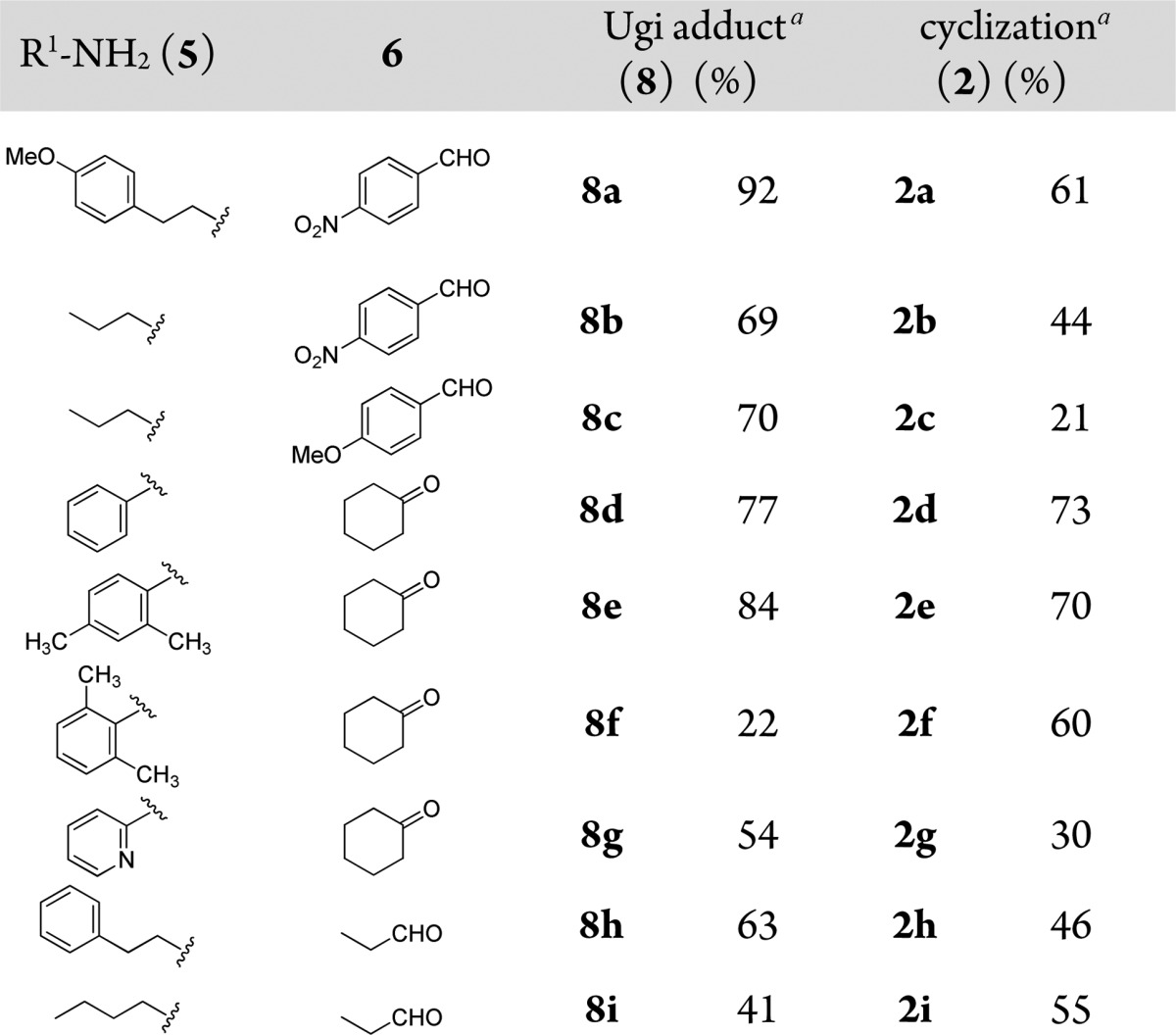
Isolated yields for Ugi reaction and cyclization.
When more electron-rich benzaldehydes (10) are used the intermediate Ugi tetrazole product undergoes a different cyclization pathway and can be reacted further to yield tetrazolo-phenyl-azepine scaffold 3 involving a o-phenol addition and elimination (Scheme 3). 3,5-Dimethoxy benzaldehyde gives better yields then 3,4,5-trimethoxy and 3,4-dimethoxy benzaldehydes. Generally both primary and secondary amines used gave moderate to good yields while the use of anilines caused the yield of our desired product to drop (3h). Typical examples and yields are shown in Table 2.
Scheme 3. Designed Synthetic Pathway to Benzotetrazolo Azepinamine Scaffold 3.
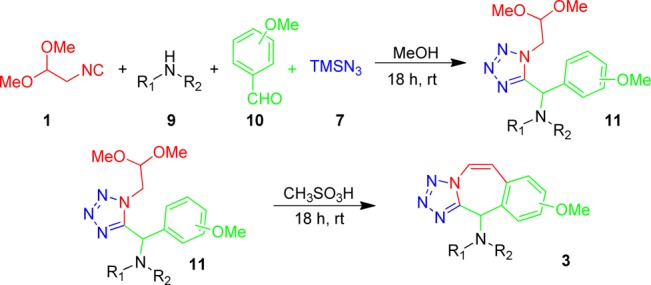
Table 2. Typical Structures and Yields of Scaffold 3.
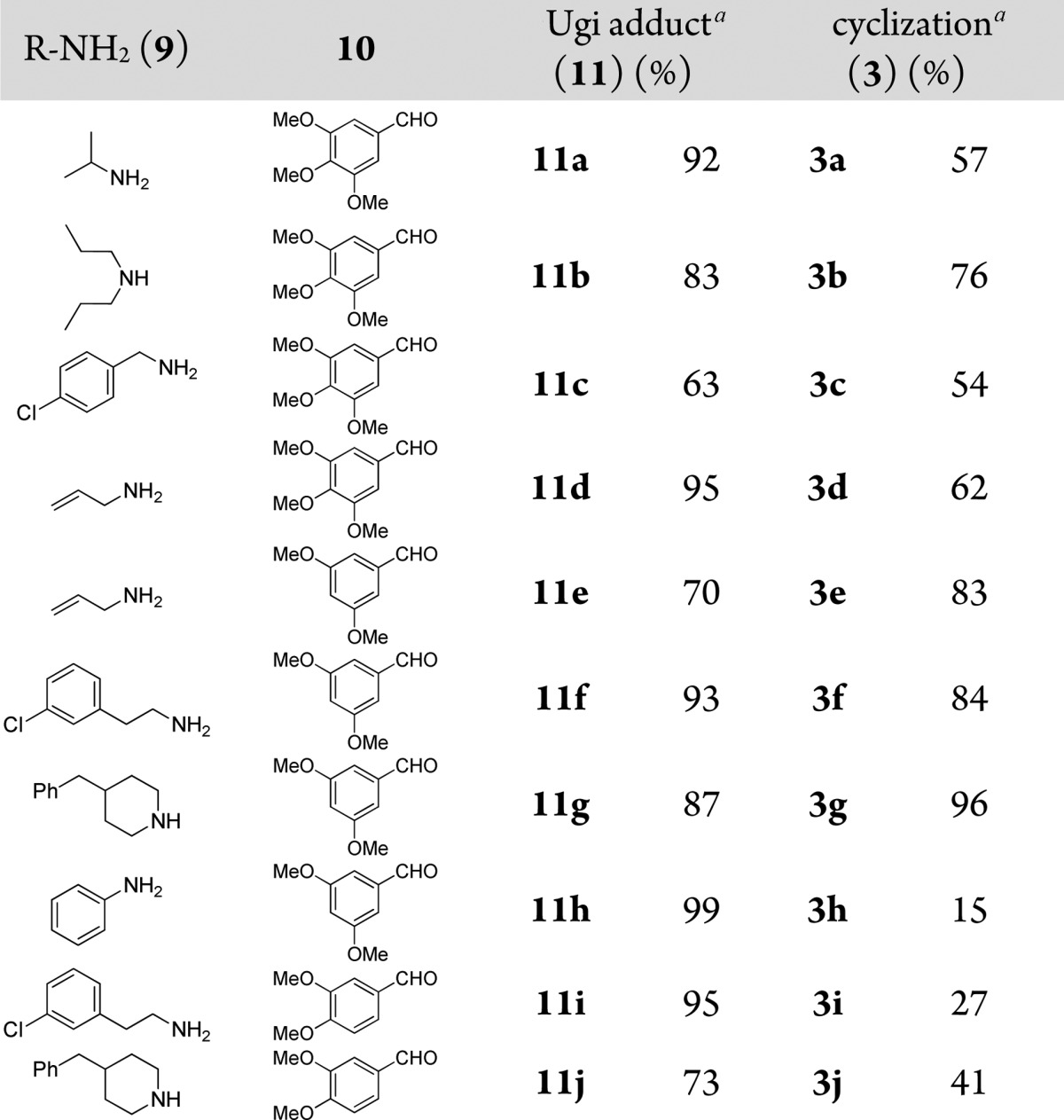
Isolated yields for Ugi reaction and cyclization.
The use of electron-rich 2-(heteroaromatic)ethylamines 13 allows for access to polycyclic scaffold 4. The intermediate Ugi tetrazole 14 yet undergoes a different reaction pathway and can be further reacted in a Pictet–Spengler transformation according to Scheme 4. Various aldehydes and ketones were tested, and all generated the product in satisfactory yields (Table 3). In most cases the products were obtained as a single major diastereomer; however, in the case of tryptophan methyl ester the Ugi products were obtained as a diastereomeric mixture of 14i (3:2) and 14j (2:1) and corresponding cyclized products 4i (9:1) and 4j (2:1).
Scheme 4. Designed Synthetic Pathway to Pyridotetrazolo Piperazine Scaffold 4.
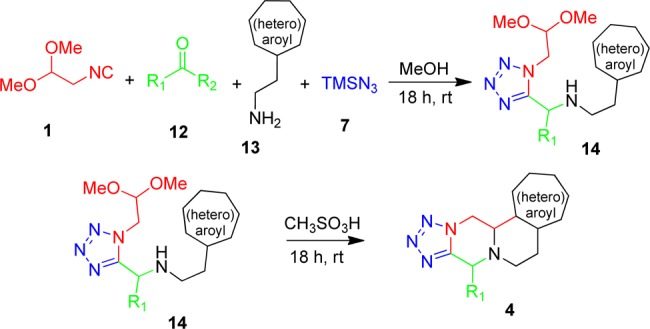
Table 3. Typical Structures and Yields of Scaffold 4.
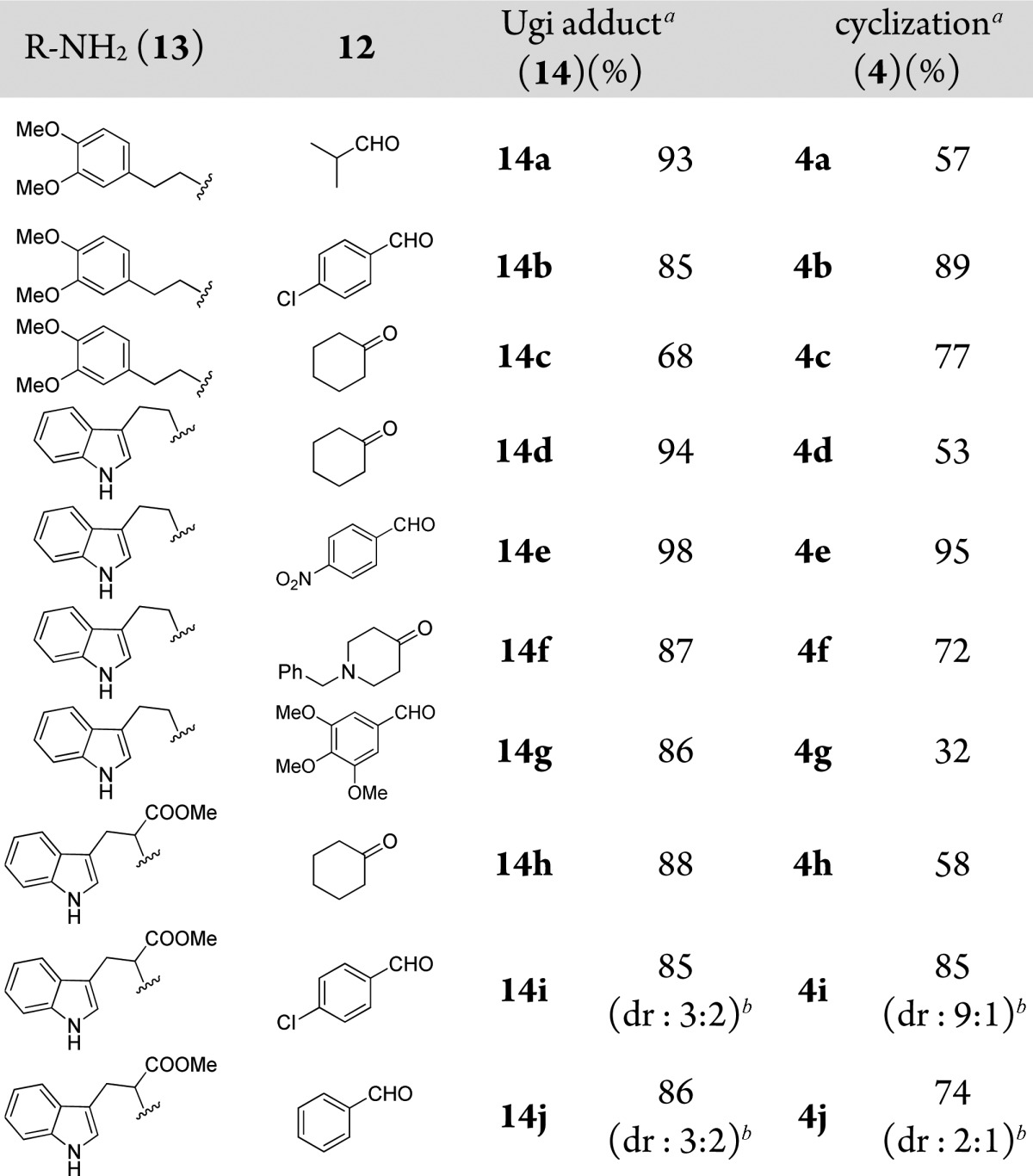
Isolated yields for Ugi reaction and cyclization.
Dr ratio was determined by SFC-MS and 1H NMR.
Next we investigated the structure of exemplary compounds of each scaffold in the solid state (Figure 1). We could grow three crystals of 2e, 3c, and 4f suitable for single crystal structure determination confirming the scaffold design and showing intermolecular interactions.
Figure 1.
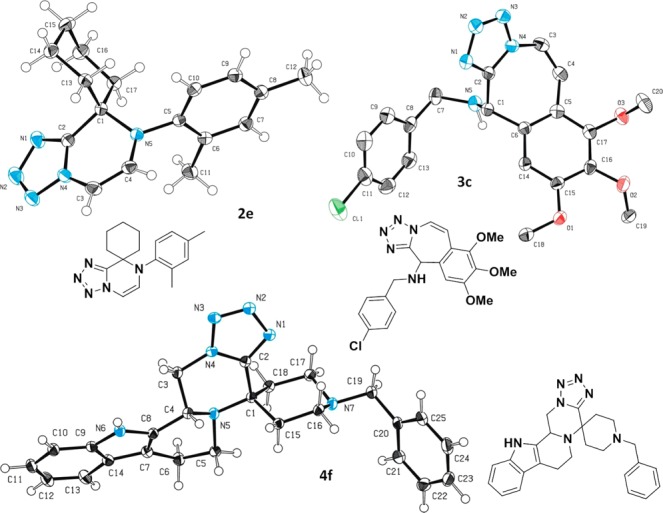
3D structure of examples of scaffolds 2–4 in the solid state.
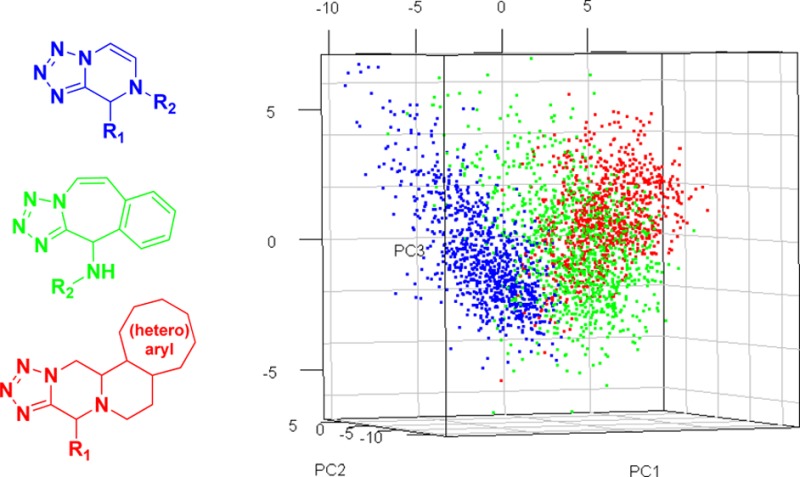
With the general synthesis of these scaffolds we set forth to create a virtual library of all three tetrazole reactions to analyze some of their general physicochemical features. We randomly generated 1000 examples of each scaffold. To visualize the distribution in a 3D chemical space unbiased molecular descriptors were analyzed by principal component analysis (PCA) (Figure 2B and Supplemental Figure 2). Interestingly, even though these scaffolds are all derived from the same first Ugi tetrazole multicomponent reaction they possess very different characteristics in terms of their chemical space due to their connectivity, substitution pattern, and ring sizes. To visualize this difference, 3D generation and alignment of the tetrazole ring on all three scaffolds was done (Figure 2A). As can be seen, due to the different rings that stem from the tetrazole base the three scaffolds each occupy a different space.
Figure 2.
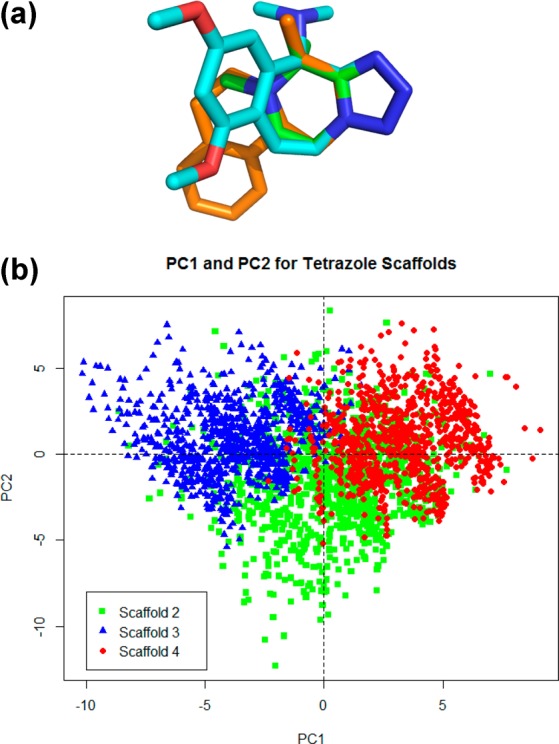
(a) Overlay of energy-minimized conformers of the backbones of the three scaffolds described in this paper (green = scaffold 2, blue = scaffold 3, orange = scaffold 4). (b) 2D principal component analysis of 1000 randomly generated compounds of each of the three scaffolds described in this paper (green = scaffold 2, blue = scaffold 3, red = scaffold 4).
To study this concept further we performed the principal moment of inertia (PMI)17 analysis to compare the shape distribution of our virtual library of small molecules to that of 1000 randomly selected compounds from the ZINC library18 (Supplemental Figure 1). As expected, based on the previous data, the three scaffolds occupy different 3D spaces and tend to be more 3D in nature to compounds from the ZINC database. The three-dimensionality of lead compounds recently emerged as an important concept to provide “drug-like” properties, e.g. reduced promiscuity or water solubility.19 Interestingly, scaffold 2 occupies a wider array of space compared to scaffolds 3 and 4 which cluster closer together. This is most likely due to the substitution pattern on scaffold 2, using two widely variable starting materials.
Work is ongoing to leverage the new chemical space to discover biologically active compounds and will be reported in due course.
Acknowledgments
The work was financially supported from the NIH (1R01GM097082-01) and by Innovative Medicines Initiative (Grant Agreement No. 115489).
Supporting Information Available
Physical and computational data can be found in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): A.D. is founder, and K.K. is an employee of Carmolex.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Wang Y.; Suzek T.; Zhang J.; Wang J.; He S.; Cheng T.; Shoemaker B. A.; Gindulyte A.; Bryant S. H. Nucleic Acids Res. 2014, 42, D1075–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hong L.; Kenney S. R.; Phillips G. K.; Simpson D.; Schroeder C. E.; Nöth J.; Romero E.; Swanson S.; Waller A.; Strouse J. J.; Carter M.; Chigaev A.; Ursu O.; Oprea T.; Hjelle B.; Golden J. E.; Aubé J.; Hudson L. G.; Buranda T.; Sklar L. A.; Wandinger-Ness A. J. Biol. Chem. 2013, 288, 8531–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Krishnan N.; Koveal D.; Miller D. H.; Xue B.; Akshinthala S. D.; Kragelj J.; Jensen M. R.; Gauss C. M.; Page R.; Blackledge M.; Muthuswamy S. K.; Peti W.; Tonks N. K. Nat. Chem. Biol. 2014, 10, 558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Liang Q.; Dexheimer T. S.; Zhang P.; Rosenthal A. S.; Villamil M. A.; You C.; Zhang Q.; Chen J.; Ott C. A.; Sun H.; Luci D. K.; Yuan B.; Simeonov A.; Jadhav A.; Xiao H.; Wang Y.; Maloney D. J.; Zhuang Z. Nat. Chem. Biol. 2014, 10, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Maddry J. A.; Ananthan S.; Goldman R. C.; Hobrath J. V.; Kwong C. D.; Maddox C.; Rasmussen L.; Reynolds R. C.; Secrist J. A.; Sosa M. I.; White E. L.; Zhang W. Tuberculosis (Edinb). 2009, 89, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Maddry J. A.; Chen X.; Jonsson C. B.; Ananthan S.; Hobrath J.; Smee D. F.; Noah J. W.; Noah D.; Xu X.; Jia F.; Maddox C.; Sosa M. I.; White E. L.; Severson W. E. J. Biomol. Screen 2011, 16, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Sun Q.; Burke J. P.; Phan J.; Burns M. C.; Olejniczak E. T.; Waterson A. G.; Lee T.; Rossanese O. W.; Fesik S. W. Angew. Chem., Int. Ed. 2012, 51, 6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Weïwer M.; Bittker J. A.; Lewis T. A.; Shimada K.; Yang W. S.; MacPherson L.; Dandapani S.; Palmer M.; Stockwell B. R.; Schreiber S. L.; Munoz B. Bioorg. Med. chem. Lett. 2012, 22, 1822–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Popovici-Muller J.; Saunders J.; Salituro F.; Travins J.; Yan S.; Zhao F.; Gross S.; Dang L.; Yen K.; Yang H.; Straley K.; Jin S.; Kunii K.; Fantin V.; Zhang S.; Pan Q.; Shi D.; Biller S.; Su S. ACS Med. Chem. Lett. 2012, 3, 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Nat. Rev. Drug Discovery 2013, 12, 173–175. [DOI] [PubMed] [Google Scholar]
- Edwards A. M.; Isserlin R.; Bader G. D.; Frye S. V.; Willson T. M.; Yu F. H. Nature 2011, 470, 163–165. [DOI] [PubMed] [Google Scholar]
- Gargano A. F. G.; Buchinger S.; Kohout M.; Lindner W.; Lämmerhofer M. J. Org. Chem. 2013, 78, 10077–10087. [DOI] [PubMed] [Google Scholar]
- Saito K.; Nishimori A.; Kotsuki H.; Nakano K.; Ichikawa Y. Synlett. 2013, 24, 757–761. [Google Scholar]
- Khoury K.; Sinha M. K.; Nagashima T.; Herdtweck E.; Dömling A. Angew. Chem., Int. Ed. 2012, 51, 10280–10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M. K.; Khoury K.; Herdtweck E.; Dömling A. Chem.—Eur. J. 2013, 19, 8048–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajdlik A.; Wang Z.; Hickey J.; Aman A.; Schimmer A.; Yudin A. Angew. Chem., Int. Ed. 2013, 52, 8411–8415. [DOI] [PubMed] [Google Scholar]
- Borthwick A. D.; Liddle J.; Davies D. E.; Exall A. M.; Hamlett C.; Hickey D. M.; Mason A. M.; Smith I. E. D.; Nerozzi F.; Peace S.; Pollard D.; Sollis S. L.; Allen M. J.; Woollard P. M.; Pullen M. A.; Westfall T. D.; Stanislaus D. J. J. Med. Chem. 2012, 55, 783–796. [DOI] [PubMed] [Google Scholar]
- Haixia L.; Dömling A. Chem. Biol. Drug Des. 2009, 74, 302–308. [DOI] [PubMed] [Google Scholar]
- Sisko J.; Kassick A. J.; Mellinger M.; Filan J. J.; Allen A.; Olsen M. A. J. Org. Chem. 2000, 65, 1516–1524. [DOI] [PubMed] [Google Scholar]
- Bista M.; Wolf S.; Khoury K.; Kowalska K.; Huang Y.; Wrona E.; Arciniega M.; Popowicz G.; Holak T.; Dömling A. Structure 2014, 21, 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartke K. Chem. Ber. 1966, 99, 3163–3172. [Google Scholar]
- a Bossio R.; Marcaccini S.; Pepino R. Liebigs Ann. Chem. 1993, 1229–1231. [Google Scholar]; b Bossio R.; Marcaccini S.; Pepino R.; Torroba T. J. Org. Chem. 1996, 61, 2202–2203. [Google Scholar]
- a Gulevich A. V.; Balenkova E. S.; Nenajdenko V. G. J. Org. Chem. 2007, 72, 7878–7885. [DOI] [PubMed] [Google Scholar]; b Kazmaier U.; Ackermann S. Org. Biomol. Chem. 2005, 3, 3184–3187. [DOI] [PubMed] [Google Scholar]
- Sauer W. H.; Schwarz M. K. J. Chem. Inf. Comput. Sci. 2003, 43, 987–1003. [DOI] [PubMed] [Google Scholar]
- a Irwin J. J.; Shoichet B. K. J. Chem. Inf. Model. 2005, 45, 177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Irwin J. J.; Sterling T.; Mysinger M. M.; Bolstad E. S.; Coleman R. G. J. Chem. Inf. Model. 2012, 52, 1757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering F.; Bikker J.; Humblet C. J. Med. Chem. 2009, 52, 6752–6756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


