Abstract
Glioblastoma multiforme (GBM) is the most aggressive malignant primary brain tumor, with a dismal mean survival even with the current standard of care. Although in vitro cell systems can provide mechanistic insight into the regulatory networks governing GBM cell proliferation and migration, clinical samples provide a more physiologically relevant view of oncogenic signaling networks. However, clinical samples are not widely available and may be embedded for histopathologic analysis. With the goal of accurately identifying activated signaling networks in GBM tumor samples, we investigated the impact of embedding in optimal cutting temperature (OCT) compound followed by flash freezing in LN2 vs immediate flash freezing (iFF) in LN2 on protein expression and phosphorylation-mediated signaling networks. Quantitative proteomic and phosphoproteomic analysis of 8 pairs of tumor specimens revealed minimal impact of the different sample processing strategies and highlighted the large interpatient heterogeneity present in these tumors. Correlation analyses of the differentially processed tumor sections identified activated signaling networks present in selected tumors and revealed the differential expression of transcription, translation, and degradation associated proteins. This study demonstrates the capability of quantitative mass spectrometry for identification of in vivo oncogenic signaling networks from human tumor specimens that were either OCT-embedded or immediately flash-frozen.
Keywords: glioblastoma multiforme, signaling, iTRAQ, mass spectrometry, tyrosine phosphorylation, signal transduction
Introduction
Glioblastoma multiforme (GBM) accounts for 12–15% of all intracranial tumors and 50–60% of all astrocytic tumors, making it the most common primary brain tumor. The current standard of care for GBM consists of surgical removal, radiotherapy, and chemotherapy treatment with the alkylating agent Temozolomide.1,2 Despite these interventions the median survival times remain at 9–14 months following diagnosis.3 Over the past decade, genetic and transcriptional characterization of GBMs has led to the identification of multiple genetic aberrations across a diverse array of genes.4 Specifically, the overexpression of RTKs in >80% of all GBMs indicates a significant involvement of RTKs in the tumor remodelling of cells in GBM.4 The epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor alpha (PDGFRα),5,6 platelet-derived growth factor receptor beta (PDGFRβ),5 and the hepatocyte growth factor receptor (Met/HGFR)7 have each been shown to play critical roles in GBM pathology and resistance to RTK targeted therapeutics. The combination of genotyping and gene expression profiling has led to the identification of four subclasses of GBM tumors classical, mesenchymal, neural, and proneural, with each subtype driven by mutation/deregulated expression of EGFR, NF1, FBXO3, and PDGFRα/IDH1 respectively.6,8,9 Thus, the significant alteration of RTKs across human GBMs indicates a need for an increased understanding of activated phosphotyrosine signaling pathways at the molecular level.
Stratification of patients based on molecular tumor characteristics to enable more effective treatment strategies (i.e., personalized medicine) relies on the identification of molecular markers indicative of survival.10,11 Identification of regulatory signaling networks in GBM would be of significant importance for stratifying patients in clinical trials. It has recently become possible to quantify phosphorylation events across patient samples with high sensitivity and throughput. For instance, panels of antibodies to phosphorylation sites can be used to probe tumor lysates on reverse phase protein arrays.12,13 Furthermore, immunoprecipitation of phosphotyrosine peptides followed by mass spectrometric analysis has been used to identify activated signaling pathways within clinical lung carcinoma and prostate tumors.14,15 Additionally, we have previously quantified phosphotyrosine signaling differences across a panel of eight human GBM patient derived xenograft (PDX) tumors with differing expression of EGFRvIII.16 Identification and quantification of phosphotyrosine signaling and protein expression profiles emphasized the significant heterogeneity of the disease across the eight PDX tumors and allowed the identification of proteins that were correlated with poor survival in EGFRvIII driven GBM.16
Although the identification of signaling in in vivo systems is critically important to understand the effects of altered RTK activation in glioblastoma, differences in extracellular growth environments have a significant impact on molecular profiles of cells, as tumor cells grown intracranially, subcutaneously and on tissue culture plastic were have been shown to induce the expression of distinct sets of genes.17 Accurate identification of physiological tumor signaling therefore necessitates a move toward quantitative analysis of protein phosphorylation in clinical samples. The limited availability of clinical samples for proteomic analysis remains an issue that is further compounded by the differential processing of tumor samples prior to pathological analysis and long-term storage. Embedding tissues for frozen sectioning is an important practice in histopathologic analysis and as such formalin-fixed paraffin embedded (FFPE) and optimal cutting temperature (OCT) compound embedding is routine in pathology laboratories.18 To exploit differentially preserved tumor material it is paramount to evaluate the ability to quantify activated signaling networks and protein expression profiles across these tumors.19,20
With limited availability of clinical samples it is important to utilize human GBM tumor sections that have been prepared and stored in different ways. To investigate the effect of these alternate storage methods on protein stability or protein post-translational modifications, we have quantified activated phosphotyrosine networks and profiled global protein expression across eight pairs of human GBM tumor sections that have been either embedded in OCT compound followed by flash freezing in LN2 or immediately flash frozen in LN2. Samples were labeled with isobaric tags and subsequent enrichment of phosphotyrosine peptides was carried out. Once phosphotyrosine profiling was completed, peptides were fractionated and protein expression profiling was carried out across the panel of human GBM tumors.16 Quantitative proteomic analysis of these clinical samples has allowed us to identify effects of sample storage on the signaling and protein expression profiles and enabled the identification of oncogenic signaling processes. Correlation analysis and functional analysis of the quantitative proteomic data indicate groups of related proteins that are coexpressed in GBM tumors and led to the identification of activated phosphotyrosine networks related to GBM biology in vivo. Ultimately, these data demonstrate the utility of quantitative phosphotyrosine analyses to identify the activation of kinases and downstream signaling pathways in vivo in the context of the inter- and intratumoral heterogeneity present in GBM.
Materials and Methods
Tissue Homogenization
Tumors were sourced from the brain tumor tissue bank of Canada (www.Braintumourbank.com). The GBM tumors included in this study have not been genetically subtyped. Tumors were resected and immediately flash frozen in LN2 or embedded in OCT compound and flash frozen in LN2 within 5 min (Table 1). OCT compound embedded tumors were rinsed in ice cold PBS to remove the OCT compound around the tissue prior to homogenization. Tumor sections were homogenized (Polytron) in ice-cold 8 M urea for mass spectrometric analyses or modified ice-cold radioimmunoprecipitation assay (RIPA) buffer for immunoblotting. Lysis buffers were supplemented with 1 mM sodium orthovanadate, 0.1% NP-40, and protease and phosphatase inhibitor cocktail tablets (Roche). Samples were homogenized on ice. Protein concentrations were quantified using a bicinchoninic acid (BCA) assay (Pierce), and the total homogenate was stored at −80 °C.
Table 1. Tumor Sample Information: Sample ID, Diagnosis GBM (Grade IV), Patient Age at Tumor Resection, Patient Gender, the Recurrence Status of the Tumor, Brain Region and Availability of OCT and iFF Tumor Sections.
| tumor # | sample ID | diagnosis | age | gender | recurrent GBM? | brain region | OCT | FF |
|---|---|---|---|---|---|---|---|---|
| 1 | 2539 | GBM | 55 | M | yes | right temporal | x | x |
| 2 | 2568 | GBM | 40 | M | no | right temporal | xx | |
| 3 | 2556 | GBM | 47 | F | yes | right parietal | x | x |
| 4 | 2585 | GBM | 76 | M | yes | left parietal | x | x |
| 5 | 1789 | GBM | 75 | F | no | / | x | x |
| 6 | 2332 | GBM | 43 | F | yes | right posterior thalamus | x | x |
| 7 | 2442 | GBM | 70 | M | yes | right frontal | x | x |
| 8 | 2589 | GBM | 62 | M | no | right frontal | x | x |
Immunoblotting
Cell lysates were separated on a 7.5% polyacrylamide gel and electrophoretically transferred to nitrocellulose (Biorad). Nitrocellulose was blocked with 5% BSA in TBS-T (150 mM NaCl, 0.1% Tween 20, 50 mM Tris, pH 8.0). Antibodies used are as follows: antiphosphotyrosine (4G10, Millipore), anti-EGFR (BD Biosciences), anti-Her3/ErbB3 (CST), anti-PDGFRα (CST), anti-PDGFRβ (CST), anti-Met (CST), anti-AKT (CST), anti-AKT pS473 (CST), anti-p53 (CST), and anti-β-tubulin (CST). Antibodies were diluted in blocking buffer and incubated with nitrocellulose overnight at 4 °C. Secondary antibodies (either goat antirabbit or goat antimouse conjugated to horseradish peroxidase) were diluted 1:10 000 in TBS-T and incubated at room temperature for 1 h. Antibody binding was detected using the enhanced chemiluminescence (ECL) detection kit (Pierce).
Mass Spectrometry Sample Preparation
Proteins were reduced (10 mM DTT, 56 °C for 45 min), alkylated (50 mM iodoacetamide, room temperature in the dark for 1 h), and excess iodoacetamide was quenched with DTT to a final concentration of 25 mM. Proteins were subsequently digested with trypsin (sequencing grade, Promega), at an enzyme/substrate ratio of 1:100, at room temperature overnight in 100 mM ammonium acetate, pH 8.9. Trypsin activity was quenched by adding formic acid to a final concentration of 5%. Urea was removed from the samples by reverse phase desalting using a C18 cartridge (Waters) and peptides were lyophilized and stored at −80 °C.
iTRAQ Labeling
Peptide labeling with iTRAQ 8plex (AB Sciex) was performed as previously described.16 For each analysis, ∼8 mg (wet weight) of tumor (equivalent to 800 μg of peptide before desalting and processing) for each of the tumors was labeled with two tubes of iTRAQ 8plex reagent.
Phosphotyrosine Enrichment
Phosphotyrosine peptides were enriched prior to mass spectrometry analyses using a cocktail of antiphosphotyrosine antibodies followed by immobilized metal affinity chromatography (IMAC) as previously described.16,21
Peptide Isoelectric Focusing and Protein Expression Profiling
iTRAQ labeled peptides were separated into five fractions using the ZOOM isoelectric focusing (IEF) fractionator (Invitrogen) with a set of six ZOOM disks (pH 3.0, pH 4.6, pH 5.4, pH 6.2, pH 7.0, and pH 10) as previously described.14 Each fraction was separated by reverse phase HPLC (Agilent) over a 240 min gradient before nanoelectrospray into a 5600 triple time-of-flight (ToF) instrument (AB Sciex) operated in positive ion mode.
Protein Expression Data Analysis
Relative quantification and protein identification were performed with the ProteinPilot software (version 2.0; AB Sciex) using the Paragon algorithm as the search engine. MS/MS spectra were searched against human protein sequence database (NCBInr, released May 2011, downloaded from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/protein/). The search parameters allowed for carbamidomethylation of cysteines by iodoacetamide and a standard extensive list of biological modifications that were programmed in the algorithm. Identified proteins were grouped by the ProGroup algorithm (AB Sciex) to minimize redundancy. The false discovery rate (FDR) was calculated by searching the spectra against the NCBI nonredundant Homo sapiens decoy database. Before filtering the protein expression data (explained above), the protein level FDR was calculated at 1%, corresponding to 2054, 2304, 2009, and 1842 proteins in analyses 1a, 1b, 2a, and 2b, respectively. After application of the above filter criteria, the estimated FDR value was <1% (at the protein level) for each of the biological replicates analyzed, indicating a high reliability in the proteins identified. Peptide summaries were exported from ProteinPilot and isotope correction and relative quantification was calculated in Excel. The total list of phosphotyrosine peptides and proteins identified and quantified can be found in Supporting Information Tables S1 and S2 respectively.
Functional Data Analysis
All mass spectra corresponding to phosphotyrosine peptides were manually validated using a previously described computer aided manual validation (CAMV) software tool.22 Curated, annotated spectra of the 402 identified and quantified phosphotyrosine peptides can be found in Supporting Information Figure S1. Unsupervised hierarchical clustering of the mean normalized and log2-transformed phosphotyrosine and protein expression quantitative iTRAQ data was generated using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html). One minus Pearson’s correlation was used as a distance metric in all clustering analyses. Pearson’s correlation analysis of the quantitative phosphotyrosine or protein expression profiles of the tumors was carried out using Excel. P values were calculated using t approximation. All heat maps were generated using GENE-E. Gene ontology (GO) annotations were identified by uploading gene lists to the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system (http://www.pantherdb.org/). Interaction maps were generated using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) program version 9.0 5 (http://string-db.org/). The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001038.23
Results
Initial Molecular Characterization of OCT Embedded and Nonembedded Human GBM Tumors
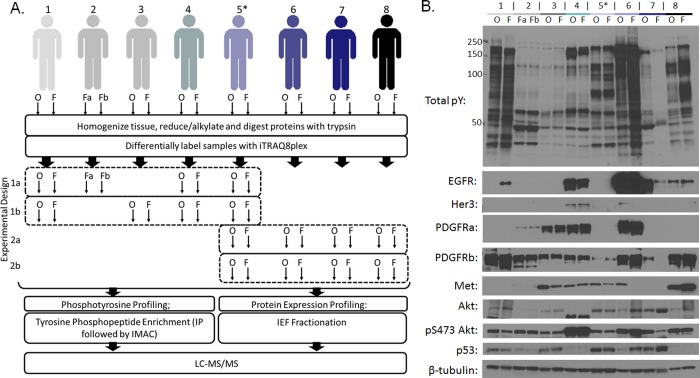
For all analyses, tumor tissue specimens were available from eight patients. Seven of the patients had paired tumor tissue samples, in which one piece of the tumor had been OCT-embedded prior to flash freezing, whereas the other piece of the tumor had been immediately flash-frozen upon resection. For the eighth patient, only two independent immediate flash freezing (iFF) tumor pieces were available (Table 1). These samples were included to assess the physiological variation and potential effects of different preservation and storage methods relative to variation of the analytical process. Initial molecular characteristics of each of the eight patient tumors was carried out using immunoblotting to identify the expression status of EGFR, Her3, PDGFRα, PDGFRβ, Met, Akt pS473, and p53 (Figure 1B). A diverse array of expression levels of these RTKs was detected, with EGFR, Her3, Met, PDGFRα, and PDGFRβ each variably expressed in this panel. To identify the effect of altered expression of these RTKs on signaling in these tumors we also carried out immunoblotting of total phosphotyrosine (Figure 1B). Distinct signaling profiles for each of the eight patient tumors can be seen in this blot, emphasizing the need for quantitative analysis of tyrosine phosphorylation signaling networks with site-specific resolution. Overall, from these blots it was clear that the expression and signaling levels were similar for the OCT compound embedded and the iFF tumor sections (Figure 1B).
Figure 1.
Quantification of tyrosine phosphorylation signaling and protein expression profiles across eight OCT compound embedded and eight flash frozen GBM human tumors.(A) Experimental mass spectrometric workflow. Human GBM tumors and their processing status either OCT compound embedded (O) or flash frozen (F), are indicated at the top of panel. There were two available sections of flash frozen tumor tissue from patient 2, indicated by Fa and Fb. The * above 5 indicates that tumor tissue 5F was used to normalize the quantitative data across all analyses. Sixteen differentially processed human GBM tumor sections were homogenized, reduced, alkylated, and digested with trypsin, and then peptides were labeled with 8plex- iTRAQ. Phosphotyrosine peptide enrichment was carried out by IP using antiphosphotyrosine antibodies and analyzed by LC–MS/MS. For protein expression profiling, peptides were fractionated by IEF based on their isoelectric point (pI). (B) Total phosphotyrosine levels (pY) were identified across 16 human GBM tumor sections by immunoblotting. Expression levels of EGFR, Her3, PDGFRα, PDGFRβ, Met, Akt, and p53 were identified across the panel of tumors. β-Tubulin was used as a loading control.
Quantification of Protein Expression Profiles and Phosphotyrosine Signaling Across OCT Embedded and Nonembedded Human GBM Tumor Sections
One of the primary concerns in analyzing signaling networks in tumor tissue specimens is that the phosphorylation status may no longer be representative of the physiological state due to alterations associated with the preservation and storage method of the tissue. To assess potential differences arising from OCT-embedding vs immediate flash-freezing of the tumor, we used mass spectrometry, combined with 8-plex iTRAQ isotopic labeling, to perform in-depth quantification of the phosphotyrosine signaling and protein expression profiles across the paired sets of GBM tumor tissue specimens from seven patients, along with replicate samples of the iFF tissue from the eighth patient (Figure 1A). Tyrosine phosphorylation-mediated signaling networks were quantitatively profiled using phosphotyrosine IP followed by liquid chromatography tandem mass spectrometry (LC–MS/MS) as previously described.16 Protein expression profiling was carried out using isoelectric fractionation followed by LC–MS/MS using a 5600 triple ToF instrument.16 Phosphotyrosine and protein expression profiling across the 16 GBM tumor sections resulted in the identification and quantification of 402 phosphotyrosine peptides and 1877 protein groups. The overlap of the identified and quantified phosphotyrosine peptides and proteins are depicted in Venn diagrams (see Supporting Information Figure S2A, B, and C and Supporting Information Figure S2D, E, and F respectively).
Correlation Analysis of Phosphotyrosine Signaling Across Differentially Processed GBM Tumor Sections
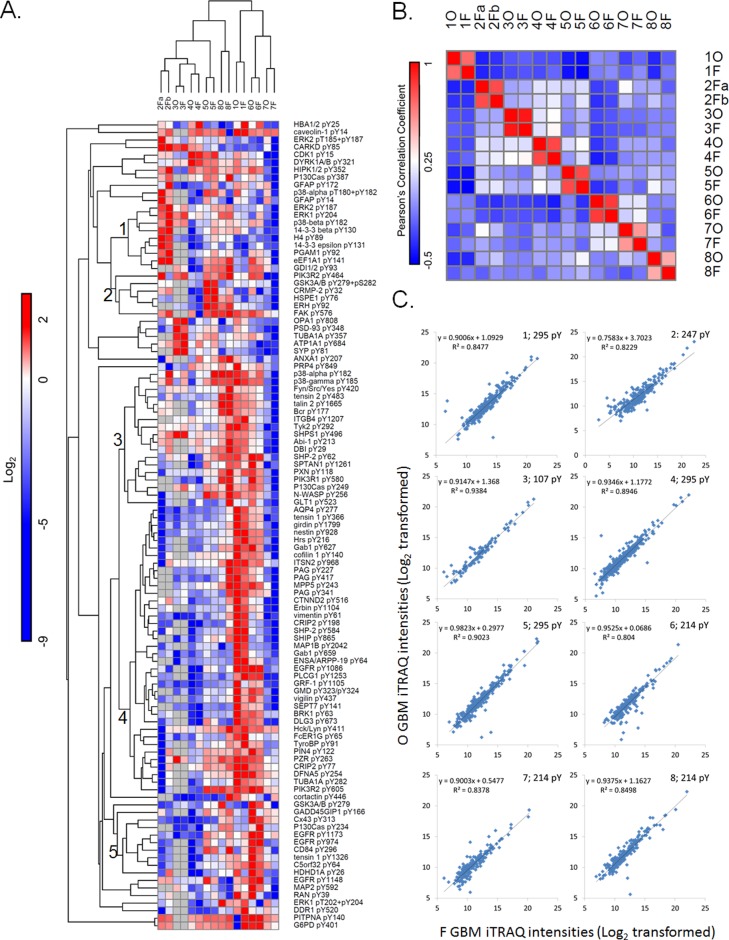
To assess the similarity of the different tumor sections based on their phosphotyrosine profiles, unsupervised hierarchical clustering was performed on the log2-transformed iTRAQ ratios using one minus Pearson’s correlation as a distance metric. Each of the differentially processed human GBM tumor pairs (1, 3, 4, 5, 6, 7, and 8) and the two iFF sections of tumor 2 (2Fa and 2Fb) clustered together (Figure 2A), indicating a high degree of similarity between separate pieces of the same tumor. To generate a more quantitative view of the comparison between OCT and iFF tissue samples from the same patient compared to samples from other patients, we carried out a correlation analysis of the 16 GBM tumor sections based on the 107 phosphotyrosine peptides overlapping in 14 or more of the tumor sections. The correlation matrix for this analysis is shown in Figure 2B. Each of the seven differentially processed tumor pairs can be shown to correlate with an average R2 value of 0.737 ± 0.155. The two iFF sections of tumor 2 correlated with a similar R2 value of 0.792 while the average correlation coefficient for all pairwise analyses was found to be a significantly lower, with an average R2 value of 0.090 ± 0.181. These results indicate that differences resulting from postexcision processing and storage are minimal, especially in the context of the interpatient heterogeneity. These analyses (1) underscore that different pieces of the same tumor are highly similar and thus the signaling networks identified in each piece may be representative of the signaling in the tumor as a whole and (2) provide further proof that tyrosine phosphorylation signaling networks found in each patient’s tumor are distinct, even when those tumors may express similar activated RTKs.
Figure 2.
Phosphotyrosine signaling is distinctly different across different GBM patients but similar between OCT and flash frozen processed tumors. (A) The 107 overlapping phosphotyrosine sites quantified across at least 14 OCT compound embedded and flash frozen GBM tumor sections are visualized in the heat map. iTRAQ ratios were normalized to tumor section 5F, normalized to the mean, and log2 transformed. Tumor sections and tyrosine phosphorylation sites were hierarchically clustered using one minus Pearson’s correlation distance metric. Missing values in tumors 2Fa, 2Fb, 3O, and 3F are shaded gray. (B) A correlation matrix of all 16 tumors sections based on the quantitative phosphotyrosine data. The Pearson’s coefficients between every pair of tumor sections are displayed in each box. The color bar indicates the correlation coefficients, where red indicates positive correlation and blue indicates negative correlation. (C) Log2-transformed iTRAQ intensities from the OCT (O) compound embedded tumor sections (y axis) were plotted against the corresponding flash frozen (F) tumor sections (x axis). A simple linear regression line was drawn through the data points and the R2 values were determined for all eight GBM tumor pairs.
To gain additional insight into the potential differential effects of OCT-embedding compared to immediate flash-freezing, we directly compared the mass spectrometry signal intensity for each phosphopeptide in each matched pair of tumor samples by plotting the log2 iTRAQ intensity for the OCT section of the tumor vs the iFF section of the tumor derived from the same patient and carried out simple linear regression (Figure 2C). The average R2 value for the correlation of each matched pair of tumor samples was 0.878 ± 0.046 for the seven tumor pairs, whereas the two iFF sections of tumor 2 correlated with an indiscriminate R2 value of 0.823. By comparison, the average R2 correlation for the OCT and iFF sections from non-matched patients was 0.599 ± 0.074 (Supporting Information Figure S3). These correlation values for matched pairs were found to be significantly greater (p = 9.878 × 10–5) than correlation values for non-matched pairs, as determined by a paired TTEST.
Correlation Analysis of Protein Expression Profiles Across Differentially Processed GBM Tumor Sections
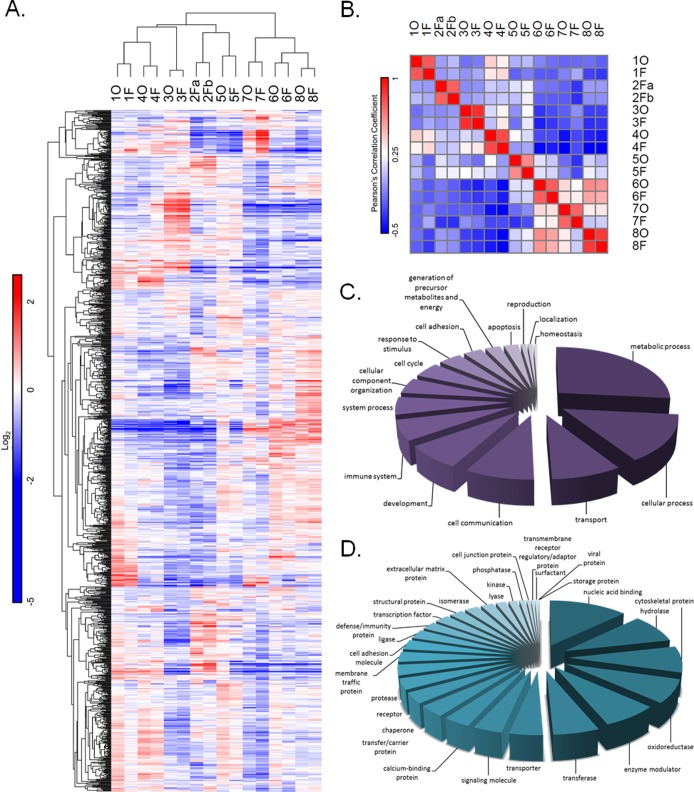
Protein expression analysis resulted in the identification and quantification of 1877 protein groups across human GBM tumors, with 1037 protein groups quantified across all 16 tumor sections. To identify the similarity of the different tumor sections based on their protein expression profiles we carried out hierarchical clustering on these overlapping proteins. As with the phosphotyrosine data, all eight pairs of GBM tumors clustered together in this analysis (Figure 3A). Correlation analysis of the 16 tumor sections based on the 1037 overlapping proteins (Figure 3B) further emphasized the similarity between 2 separate pieces of the same tumor. The seven tumor pairs that were differentially processed correlate with an average R2 value of 0.767 ± 0.089, while the two iFF sections of tumor 2 correlate with a similar R2 value of 0.735. In contrast, the average correlation coefficient for all pairwise analyses was found to be a significantly lower, with an R2 value of −0.046 ± 0.258. These results stress the significant differences in protein expression profiles between each of the eight tumor samples, indicating a large degree of interpatient heterogeneity at the signaling level and at the protein expression level. This analysis indicates that the intertumor heterogeneity is significantly greater than the combination of intratumor heterogeneity and any alterations introduced through the OCT embedding procedure.
Figure 3.
Protein expression profiling highlights significantly different expression patterns across GBM patients but a high degree of similarity between OCT and flash frozen processed tumors. (A) The 1037 overlapping protein groups quantified across all 16 OCT compound embedded and flash frozen GBM tumor sections are visualized in the heat map. iTRAQ ratios were normalized to tumor section 5F, normalized to the mean and log2 transformed. Tumor sections and protein groups were hierarchically clustered using one minus Pearson’s correlation distance metric. (B) A correlation matrix of all 16 tumors sections based on the quantitative protein expression data. The Pearson’s coefficients between every pair of tumor sections are displayed in each box. The color bar indicates the correlation coefficients, where red indicates positive correlation and blue indicates negative correlation. (C) A pie chart diagram displaying the PANTHER GO biological processes annotation analyses carried out for the 1037 overlapping protein groups. (D) A pie chart diagram displaying the PANTHER GO protein classes’ annotation analyses carried out for the 1037 overlapping protein groups.
To identify coverage of the protein expression analysis we carried out GO annotation analysis. The protein classes and the biological functions of the 1037 overlapping proteins are displayed in pie charts in Figure 3C and D respectively. We attempted to identify proteins that were differentially affected by the OCT vs iFF preservation techniques within each of these classes and biological functions; however, we were unable to identify changes that were statistically significant across multiple tumors. Although we could not identify any statistically significant changes with OCT embedding, we did identify subtle but significant changes in a group of phosphorylation sites that were quantified across all experiments (Supporting Information Figure S4). These subtle changes are likely indicative of either intratumor heterogeneity or patient-specific effects of OCT-embedding. In either case, the variance in paired pieces from a given tumor was muted when compared to intertumor heterogeneity.
Defining Pathways by Coclustering of Quantitatively Similar Phosphorylation Sites
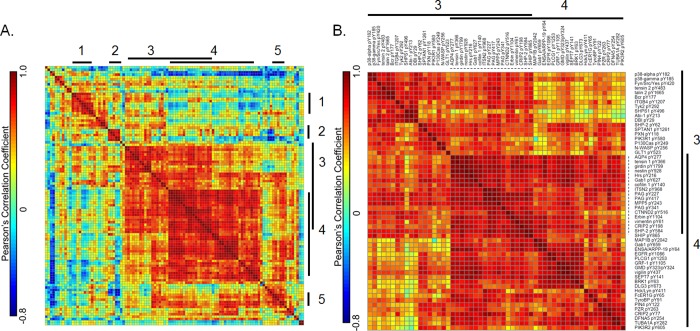
Because the tyrosine phosphorylation data was highly similar between two sections of a given tumor, we reasoned that this phosphorylation data might be an accurate representation of the signaling networks in the overall tumor. Extracting activated signaling networks from relative quantification data for hundreds of phosphorylation sites can be challenging, especially given the high interpatient heterogeneity and the limited number of samples. Here, to highlight the phosphorylation sites that were most commonly coregulated and may therefore define networks, we combined hierarchical clustering and correlation analysis of the 107 tyrosine phosphorylation sites quantified in all 16 tumor tissue specimens. The correlation matrix resulting from this approach (Figure 4A) revealed five distinct clusters containing five or more phosphorylation sites. Correlation coefficient values and corresponding p-values for these five clusters can be found in Supporting Information Table S3. Clusters 3 and 4, the two largest clusters, were composed of 56 tyrosine phosphorylation sites; these clusters are depicted in Figure 4B. Although these clusters of sites represent highly correlated phosphorylation sites, coregulation does not necessarily define a network. To extract networks from these coregulated clusters, we queried the STRING database to identify known and predicted protein–protein interactions (see Supporting Information Figure S5A and B).
Figure 4.
Correlation analyses across the 107 overlapping phosphotyrosine sites reveal clusters of phosphorylation sites with similar quantitative profiles across the 16 tumor sections. (A) Phosphotyrosine peptides were hierarchically clustered using one minus Pearson’s correlation distance metric prior to correlation analysis. The cluster numbers are indicated on the top and right-hand side of the correlation matrix. (B) Zoomed in correlation matrix showing cluster 3 and 4, which includes 56 phosphotyrosine sites. Protein and phosphotyrosine site is labeled at the top and right-hand side of the correlation matrix. The color bars in (A) and (B) indicate the correlation coefficients, where red indicates positive correlation and blue indicates negative correlation. The 18 phosphotyrosine sites that are overlapping cluster 3 and 4 are highlighted by a dashed line.
Activated Signaling Networks Defined by Quantitative Tyrosine Phosphorylation Data
Clusters 3 and 4 were both found to be highly interactive. Cluster 3 includes many proteins and phosphorylation sites associated with the Integrin-Src-FAK signaling pathway which regulates focal adhesions, migration, and cell invasion, and is most strongly phosphorylated in GBM tumor sections 8O, 8F, 1O, and 1F (see cluster 3, Figure 2A). Many of the phosphotyrosine sites in this cluster are ≥2 fold higher in tumors 1 and 8 relative to the remaining tumors; this change is statistically significant relative to the average coefficient of variation (CV) across the technical replicate analyses (15.69 ± 4.090%). Furthermore, the average CV for biological replication between the two differentially processed tumor sections (23.37 ± 7.870%) was found to be slightly, but significantly, greater than the technical variation (p = 0.03). Phosphorylation sites on proteins in this cluster known to be involved in focal adhesions and the integrin-Src-FAK pathway include the Src family kinases (SFK) Src/Fyn/Yes (pY420), p130Cas (pY249; also known as BCAR1), SHP-2 (pY62), Paxillin (pY118), N-WASP (pY256), ITGB4 (pY1207), SHP-2 (pY62), SHPS1 (pY496), and PIK3R1 (pY580). This pathway leads to cytoskeletal remodeling; accordingly, multiple phosphorylation sites on proteins associated with this process were also contained in this cluster: ABI1 (pY213), SPTAN1 (pY1261; also known as spectrin), tensin2 (pY483), and talin 2 (pY1665). Many of the sites on these proteins have been implicated as SFK substrates.
Cluster 4 also features protein phosphorylation sites that have been linked to cell migration and SFK activity. Intriguingly, several sites in this cluster are associated with negative regulatory signaling, including multiple sites (pY227, pY417, and pY341) on phosphoprotein associated with glycosphingolipid microdomains 1 (PAG1), a Src-family kinase substrate whose phosphorylation negatively regulates SFK activity. These sites correlate strongly with the Hck pY411/Lyn pY397 activating tyrosine phosphorylation sites. Cluster 4 also includes sites on Erbin and Hrs, proteins involved in the endocytosis and trafficking of activated RTKs. EGFR 1086, PI3K, and PLC-gamma phosphorylation sites are also included in this cluster; these sites have recently been shown to function coordinately in the macrophage-induced migration and invasion of gastric cancer cells. Sites in cluster 4 were found to be increased in tumor sections 6O, 6F, 1O, and 1F (see cluster 4, Figure 2A). Eighteen phosphorylation sites in cluster 4 significantly correlated with cluster 3 (Figure 4B), potentially due to the association of both clusters with cell migration and SFK activity.
Correlating Signaling Nodes
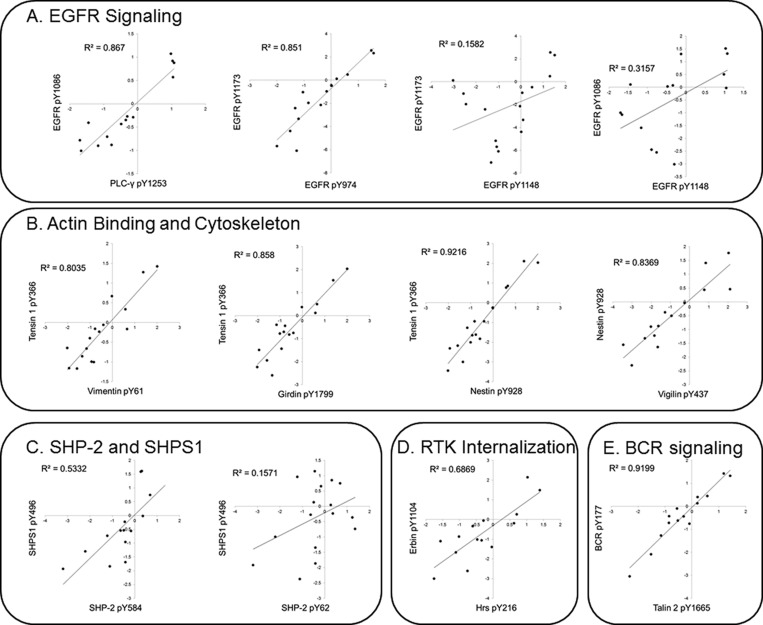
To emphasize signaling nodes that were significantly correlated we selected phosphorylation sites involved in EGFR signaling, actin binding and cytoskeleton, SHP-2 and SHPS1 interactions, RTK internalization and cell adhesion (Figure 5 A–D). Among the EGFR autophosphorylation sites, it is intriguing that EGFR pY1173 is correlated with pY974 (R2 = 0.851) and with pY1086 (R2 = 0.768) but not with EGFR pY1148 (R2 = 0.158). In agreement with a recently published study, EGFR site Y1086 is correlated with PLCG1 pY1253 (R2 = 0.867), although the SH2 domain of PLC-gamma has not been shown to interact with EGFR Y1086 in vitro (Figure 5A). Tyrosine phosphorylation of multiple actin binding and cytoskeletal proteins were found to be highly correlated throughout these analyses, as shown by the strong correlation between tensin 1 pY366 and vimentin pY61 (R2 = 0.8035), tensin 1 pY366 and girdin pY1799 (R2 = 0.858), tensin 1 pY366 and nestin pY928 (R2 = 0.922), and nestin pY928 and vigilin (R2 = 0.8369) (Figure 5B). SHPS1 is a docking protein which induces the translocation of SHP-2 from the cytosol to the plasma membrane, whereas SHPS1 pY496 positively correlates with SHP-2 pY584 (R2 = 0.5332), SHPS1 pY496 does not correlate with SHP-2 pY62 (R2 = 0.157) (Figure 5C), potentially indicating differential kinase or phosphatase regulation of these sites. Tyrosine phosphorylation sites involved in EGFR internalization are coregulated, with Erbin pY1104 positively correlated with Hrs pY216 R2 = 0.687 (Figure 5D).
Figure 5.
Phosphotyrosine sites that cluster together are functionally related. Log2 transformed iTRAQ ratios for all 16 tumor sections are plotted in the graphs, and the associated phosphotyrosine site is labeled on the y and x axis. R2 values associated with pairwise correlation analyses are displayed in the top left-hand corner of each graph. (A) Graphs showing correlations between EGFR autophosphorylation sites. (B) Graphs showing the correlations between phosphorylation sites involved in actin binding and cytoskeleton. (C) Correlation between SHP-2 and the associated SHP-2 docking protein SHPS1. (D) Graphs showing the correlations between RTK internalization phosphorylation sites on Erbin and Hrs are plotted.
Identification of Functionally Related Groups of Proteins in GBM
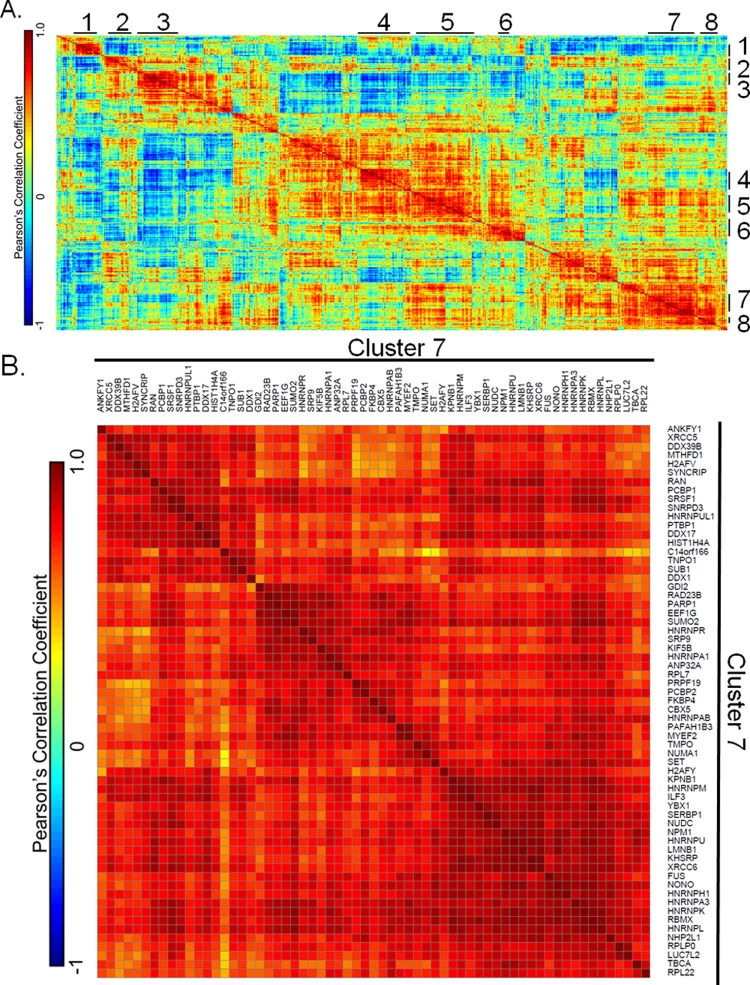
Although activated signaling networks are critical for driving cellular transformation, altered protein expression can also be critically important in regulating cell biology, and may expose additional potential therapeutic intervention points. To identify groups of proteins that are coexpressed, we performed hierarchical clustering followed by correlation analysis to generate a correlation matrix (Figure 6A) of the 1037 overlapping protein expression profiles across 16 GBM tumor specimens. To identify the level of intertumor variation at the protein level, we calculated the average CV across technical replicate analyses (8.900 ± 1.510%) and the CV for biological replicate analyses (11.00 ± 2.483%). Through TTEST analysis, the biological variation was found to be greater than the technical variation between replicate analyses of the same sample (p = 0.05). Correlation coefficient values and corresponding p-values from this analysis are available in Supporting Information Table S4. From these 1037 proteins, we identified eight statistically significant clusters containing 10 or more proteins. To identify previously characterized protein–protein interactions within these clusters we queried the STRING database. Clusters 7 and 8 iterate coregulation of proteins involved in transcription, translation, and protein degradation (Supporting Information Figure 6A and B). For transcription-associated proteins, cluster 7 consists of many members of the heterogeneous ribonucleic complex including HNRNPH1, HNRNPL, HNRNPU, HNRNPR, HNRNPK, HNRNPM, PTBP1, and FUS (Figure 6B), along with the RNA binding proteins RBM8A, THOC4, and RALY. Cluster 8 contains the eukaryotic translation initiation factors EIF4B and EIF4H, both of which are involved in binding mRNA to ribosomes. Translation associated proteins in these clusters include multiple ribosomal proteins: RPL3, RPL4, RPL8, RPL10, RPL13, RPL15, and RPL28 are all present in cluster 7 along with degradation associated proteins PSMC3 and PSMA6 from the 26S proteasome.
Figure 6.
Hierarchical clustering and correlation analysis of protein expression profiles across 1037 protein groups quantified across 16 human GBM sections reveals groups of functionally related proteins. (A) Correlation matrix of 1037 protein groups based on their quantitative profiles. The protein groups were hierarchically clustered prior to the calculation of Pearson’s correlation coefficients. The cluster numbers are indicated on the top and right-hand side of the correlation matrix. (B) Zoomed in correlation matrix showing cluster 7, which includes 63 proteins. Protein names are labeled at the top and right-hand side of the correlation matrix. The color bars in (A) and (B) indicate the correlation coefficients, where red indicates positive correlation and blue indicates negative correlation. The color bars indicate the correlation coefficient, where red indicates positive correlation and blue indicates negative correlation.
Discussion
Despite the prevalence of genetic alterations in RTKs in GBM tumors, therapeutic agents targeting these putative oncogenic kinases have fared poorly in the clinic. Identification of activated signaling networks downstream of altered kinases may provide novel therapeutic targets while giving insight into the pathways and networks regulating oncogenesis and progression. To assess activated signaling networks, we profiled phosphotyrosine signaling and protein expression in GBM tumors from eight patients, including 16 total tumor specimens: 7 pairs of differentially processed human GBM tumors, and 1 pair of similarly processed tumors (Figure 1). Quantification of profiles across OCT compound embedded and nonembedded flash frozen tissues led to the identification of 402 phosphotyrosine sites and 1037 protein groups across 16 human GBM sections. Hierarchical clustering and correlation analysis of the 16 tumors sections based on the phosphotyrosine and protein expression profiles led to the observation that differentially processed tumor sections clustered together and were significantly correlated at both the protein and phosphotyrosine level. This high degree of similarity indicates that the OCT-embedding procedure does not significantly alter the signaling networks or the protein expression profiles relative to immediate flash-freezing of the resected tumor sample. In fact, more in-depth analysis of the phosphorylation and expression data failed to reveal any consistent, statistically significant changes between the OCT-embedded and the iFF samples. Based on these results, we were confident in using both OCT-embedded and iFF samples for the quantitative analysis of activated signaling networks in vivo (Figure 2 and 3). Quantitative signaling and expression profiling revealed a large amount of interpatient heterogeneity across the 8 GBM tumors at both the phosphotyrosine and protein expression level. Intriguingly, intratumoral heterogeneity, identified across many different cancer types, was not observed within these analyses, as indicated by the high degree of similarity in the protein expression and phosphotyrosine data between two separate specimens of the same tumor sample. We expect that this lack of intratumoral heterogeneity may be due to population averaging across a large number of cells, as each tumor tissue specimen consisted of >107 cells.24,25 Furthermore, the time associated with OCT embedding in this study was minimal; we expect that this has significantly contributed to the maintenance of phosphorylation events in these tissue specimens. We envisage that longer delays between resection and freezing (ischemia time) have the potential to significantly alter the phosphorylation status of signaling networks within the tumor specimens. Thus, when identifying sample preservation methods it is essential to consider the time required, as the ischemia effects may deleteriously affect interpretation of the signaling networks.
Hierarchical clustering and correlation analysis of the quantitative phosphotyrosine profiles uncovered potential network associations that may be important for inducing growth and invasion in these tumors samples. Specifically, the Integrin-Src-FAK signaling pathway was found to have increased phosphorylation in GBM tumors 1 and 8 relative to the other tumor samples (Supporting Information Figure 5A). Because increased phosphorylation of many of the components of the pathway have been associated with increased pathway activation, it is likely that this promigratory pathway is strongly activated in these two tumors.
Phosphorylation of the focal adhesion proteins,PXN (pY118), spectrin 1 (pY1261), talin2 (pY1665), and tensin2 (pY468), are present within cluster 3 and are known to be phosphorylated by Src family kinases.26 Integrin B4 (ITGB4) has also been shown to activate Src signaling and propagate signaling through the Ras-extracellular signal-regulated kinase (ERK) cascade to promote anchorage-independent growth and invasion downstream of Met activation.27,28 These results highlight the activation of the Integrin-Src-FAK pathway in driving migration and invasion in selected GBM tumors. The increased phosphorylation levels on this pathway in GBM tumors 1 and 8 indicate a potential therapeutic role for dasatinib, an SFK-family inhibitor, in these selected tumors.
Further analysis uncovered the identification of phosphorylation events involved in the negative regulation of EGFR/RTK signaling (Supporting Information Figure 5B). Erbin has been shown to inhibit ERK activation by disrupting Raf-1 binding and subsequent activation by active Ras.29 Phosphorylation of CRIP2 was also associated with this group of phosphorylation sites. CRIP2 was identified as a tumor suppression gene in nasopharyngeal carcinoma (NPC) and has been shown to act as a transcription repressor for NF-κB, inhibiting tumor growth and angiogenesis.30,31 In addition, this cluster also includes activating Hck/Lyn tyrosine phosphorylation sites which correlated with PAG1 tyrosine phosphorylation. The interaction between Lyn and PAG has previously been identified to initiate activation of PI3K cascade of signaling events that may play a role in GBM pathogenesis.32 The C-terminal Src kinase binding protein, PAG1 (Cbp), has also been shown to mediate negative feedback to Src family kinases.33 Together, these results assert the importance of transcription regulation, adaptor and scaffolding proteins in the regulation of RTK signaling in GBM.
Interrogation of individual phosphorylation site correlations revealed the significant correlation of SHPS1 (SIRPa) Y496, a docking protein responsible for inducing the translocation of SHP-2 from the cytosol to the plasma membrane, with SHP-2 pY584. The tyrosine phosphorylation of SHPS-1 at both pY449 and pY473 is required for the association of SHPS-1 with SHP-2 that negatively regulates RTK induced cell adhesion.34 Phosphorylation of SHPS1 Y496 was positively correlated to SHP-2 pY584 but not to SHP-2 pY62. This level of detail highlights the site-specific regulation of phosphorylation and indicates potential site-specific interactions that are detectable through correlation analysis of quantitative tyrosine phosphorylation data, as we have shown previously in cell lines.
Protein expression profiling indicated that the eight tumors within this analysis were significantly different at both the phosphotyrosine and protein expression levels, affirming complex interpatient heterogeneity. Correlation analysis of the quantitative protein expression profiles identified functionally related groups of proteins that are significantly differently expressed across the eight GBM tumors. Specifically, transcriptional regulators and heterogeneous ribonucleic complex proteins were differentially expressed, as were multiple splicing factors. Several of these proteins have been previously implicated in tumorigenesis and tumor progression. For instance, the splicing factor PTBP1 (Figure 6B) has previously been identified to play a role in the infiltrative nature of cell in GBM, with PTBP1 involved in cellular proliferation, cell motility and focal adhesion complexes.35 The translation initiator EIF4H1 has been implicated in tumorigenesis;36 deciphering the role of this protein in developed GBM tumors may reveal additional insights about its regulation. Reduced expression of hnRNPH plays a direct role in enhancing tumor aggressiveness by inhibiting apoptosis and promoting invasion in GBM.37 These results highlight multiple potential points of dysregulation in signaling networks, RNA transcriptional processing, and protein translation. Integration of these data sets may reveal novel connections between altered signaling and the detected alterations in protein expression. Whether these results implicate novel points of therapeutic intervention remains to be determined, but through quantitative mass spectrometric analysis of protein expression and phosphorylation, it is possible to detect activated networks in specific tumors. The large amount of interpatient heterogeneity detected here accentuates the complexity of this disease and the difficulty in successfully treating patients.
Results generated in this study highlight the application of quantitative signaling analyses for the identification of activated networks in clinical samples. Moreover, they demonstrate that OCT-embedded samples preserve the integrity of physiological protein expression and protein phosphorylation signaling networks. Finally, clustering and co-occurrence analysis enables the identification of activated signaling networks from complex, heterogeneous data.
Acknowledgments
The authors would like to thank Dr. Abijit Guha, Ms. Takyee Tung, and Ms. Marcela White at the brain tumour bank (www.Braintumourbank.com) for access to patient materials and information. The authors would like to thanks members of the White lab for helpful discussion. This work was supported by in part by a generous gift from the James S. McDonnell Foundation, a grant from the US–Israel Binational Science Foundation, and by NIH grants U54 CA112967 and U24 CA159988.
Glossary
Abbreviations
- BCA
bicinchoninic acid
- CAMV
computer aided manual validation
- CV
coefficient of variation
- EGFR
epidermal growth factor receptor
- FFPE
formalin fixed paraffin embedded
- GBM
glioblastoma multiforme
- GO
gene ontology
- IEF
isoelectric focusing
- iFF
immediate flash freezing
- IMAC
immobilized metal affinity chromatography
- IP
immunoprecipitation
- LC–MS/MS
liquid chromatography tandem mass spectrometry
- LN2
liquid nitrogen
- MALDI
matrix assisted laser desorption ionization
- Met/HGFR
hepatocyte growth factor receptor
- OCT
optimal cutting temperature
- PANTHER
Protein Analysis Through Evolutionary Relationships
- PDGFRα
platelet-derived growth factor receptor alpha
- PDX
patient derived xenograft
- PDGFRβ
platelet-derived growth factor receptor beta
- pI
isoelectric point
- PTEN
phosphatase and tensin homologue
- pY
phosphotyrosine
- RIPA
radioimmunoprecipitation assay
- RTKs
receptor tyrosine kinases
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- ToF
time of flight
Supporting Information Available
Curated annotated mass spectra of the 402 quantified phosphotyrosine sites can be found in figure S1. The overlapping phosphotyrosine and proteins present in each independent analyses are depicted in Venn diagrams in figure S2. Graphs showing correlation analyses between unmatched patients tumors are shown in figure S3. Significant changes identified between different sections of the same tumor are depicted in figure S4. STRING data of phosphotyrosine cluster 3 and 4 and protein cluster 7 and 8 can be shown in figures S5 and S6 respectively. The total lists of phosphotyrosine peptides and proteins that were quantified in this study can be found in tables S1 and S2 respectively. Phosphotyrosine and protein expression correlation matrices are shown in tables S3 and S4 respectively. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
§ Signalling Laboratory, The Babraham Institute, Babraham Research Campus, Cambridge, U. K.
Author Contributions
H.J. and F.M.W. designed research; H.J. performed research; H.J. analyzed data; H.J. and F.M.W. wrote the paper.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Stupp R.; et al. Radiotherapy plus concomitant and adjuvant Temozolomide for glioblastoma. N. Engl. J. Med. 2005, 35210987–96. [DOI] [PubMed] [Google Scholar]
- Stupp R.; et al. Effects of radiotherapy with concomitant and adjuvant Temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 105459–66. [DOI] [PubMed] [Google Scholar]
- Furnari F. B.; et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007, 21212683–710. [DOI] [PubMed] [Google Scholar]
- Network T. C. G. A. R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 45572161061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A.; et al. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. Int. J. Cancer 1995, 602168–73. [DOI] [PubMed] [Google Scholar]
- Verhaak R. G.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17198–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. H.; et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. U. S. A. 2007, 1043112867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.; et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One 2009, 411e7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H. S.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 93157–73. [DOI] [PubMed] [Google Scholar]
- Holdhoff M.; et al. Use of personalized molecular biomarkers in the clinical care of adults with glioblastomas. J. Neurooncol. 2012, 1102279–85. [DOI] [PubMed] [Google Scholar]
- Huang P. H.; et al. Uncovering therapeutic targets for glioblastoma: a systems biology approach. Cell Cycle 2007, 6222750–4. [DOI] [PubMed] [Google Scholar]
- Gopal Y. N.; et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010, 70218736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujral T. S.; et al. Profiling phospho-signaling networks in breast cancer using reverse-phase protein arrays. Oncogene 2013, 32293470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. M.; et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc. Natl. Acad. Sci. U. S. A. 2012, 10951643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K.; et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007, 13161190–203. [DOI] [PubMed] [Google Scholar]
- Johnson H.; et al. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol. Cell. Proteomics 2012, 11121724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camphausen K.; et al. Influence of in vivo growth on human glioma cell line gene expression: convergent profiles under orthotopic conditions. Proc. Natl. Acad. Sci. U. S. A 2005, 102238287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steu S.; et al. A procedure for tissue freezing and processing applicable to both intra-operative frozen section diagnosis and tissue banking in surgical pathology. Virchows Arch. 2008, 4523305–12. [DOI] [PubMed] [Google Scholar]
- Loken S. D.; Demetrick D. J. A novel method for freezing and storing research tissue bank specimens. Hum. Pathol. 2005, 369977–80. [DOI] [PubMed] [Google Scholar]
- Chaurand P.; et al. Monitoring mouse prostate development by profiling and imaging mass spectrometry. Mol. Cell. Proteomics 2008, 72411–23. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; et al. Time-resolved Mass Spectrometry of Tyrosine Phosphorylation Sites in the Epidermal Growth Factor Receptor Signaling Network Reveals Dynamic Modules. Mol. Cell. Proteomics 2005, 491240–1250. [DOI] [PubMed] [Google Scholar]
- Curran T. G.; et al. Computer aided manual validation of mass spectrometry-based proteomic data. Methods 2013, 613219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.; et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic acids res. 2006, 34, D659–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 36610883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A.; et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A 2013, 110104009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. K.; Schlaepfer D. D. Integrin-regulated FAK–Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 185516–523. [DOI] [PubMed] [Google Scholar]
- Bertotti A.; et al. Beta4 integrin activates a Shp2-Src signaling pathway that sustains HGF-induced anchorage-independent growth. J. Cell Biol. 2006, 1756993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta U.; Shaw L. M. A key tyrosine (Y1494) in the beta4 integrin regulates multiple signaling pathways important for tumor development and progression. Cancer Res. 2008, 68218779–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Z.; et al. Erbin suppresses the MAP kinase pathway. J. Biol. Chem. 2003, 27821108–14. [DOI] [PubMed] [Google Scholar]
- Cheung A. K.; et al. Cysteine-rich intestinal protein 2 (CRIP2) acts as a repressor of NF-kappaB-mediated proangiogenic cytokine transcription to suppress tumorigenesis and angiogenesis. Proc. Natl. Acad. Sci. U. S. A 2011, 108208390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A. B.; et al. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int. J. Cancer 1999, 824498–503. [DOI] [PubMed] [Google Scholar]
- Sutton P.; et al. Lyn, a Src family kinase, regulates activation of epidermal growth factor receptors in lung adenocarcinoma cells. Mol. Cancer 2013, 12, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi L.; et al. Global Phosphoproteomics Reveals Crosstalk Between Bcr-Abl and Negative Feedback Mechanisms Controlling Src Signaling. Sci. Signaling 2011, 4166ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada T.; et al. Roles of the complex formation of SHPS-1 with SHP-2 in insulin-stimulated mitogen-activated protein kinase activation. J. Biol. Chem. 1998, 273159234–42. [DOI] [PubMed] [Google Scholar]
- Cheung H. C.; et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain 2009, 132Pt 82277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.; et al. An alternative splicing isoform of eukaryotic initiation factor 4H promotes tumorigenesis in vivo and is a potential therapeutic target for human cancer. Int. J. Cancer 2011, 12851018–30. [DOI] [PubMed] [Google Scholar]
- Lefave C. V.; et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011, 30194084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.