Introduction
Grand terms have been used to describe recent progress in biomedical science due to advances in genomics. This is not purely hyperbole. In little more than two decades, we have gone from a having only a rudimentary understanding of a small fraction of the sequence and function of our genome to possessing tens of thousands of complete human genome sequences with a growing instruction manual for their interpretation. Genomics advances relevant to human health and disease not only include information about the human genome but extend to discoveries regarding the genomes of a broad diversity of other organisms. Some genomic advances have had obvious, immediate relevance to the health of the pediatric population, such as the discovery of mutations causing serious single gene disorders and the development of rapid molecular diagnostic tests for pathogens. Others, such as genome sequence data from experimental model organisms have proximate, but not immediate, potential for improving the well-being of children. Less obvious, but potentially with the greatest long term import for human well-being, are genomic discoveries relevant to food production, the environment, and energy resources. Despite such advances, significant challenges must be met to assure the effective translation of scientific discovery to improved health outcomes for the general pediatric population.
This article briefly reviews recent advances in genomic science and several areas in which genomic discoveries are beginning to yield meaningful health advances for pediatric patients. Additionally, we consider some future challenges and opportunities in using genomics to advance “personalized medicine” for the pediatric patient. Personalized medicine has come to be understood by many as the use of individual genome sequence information to ensure the most appropriate selection treatments – often distilled to “the right drug, at the right dose for the right person.” Readers less familiar with genomic concepts may wish to consult recent genomic themed review articles as well as available online glossaries of genomic terms. 1-5
Technological advances driving genomic discovery
At first glance, the human genome is deceptively simple, comprised of 3 billion base pairs of only four repeating chemical bases: adenine (A), guanine (G), cytosine(C), and thymidine (T). Each somatic cell has two copies of the genome, for a total of 6 billion base pairs. However, on deeper consideration, it is mind-numbingly difficult to grasp the amount of data this code encompasses, and the challenges extracting it posed only two decades ago. The Human Genome Project (HGP) developed methods for rapidly, inexpensively, and accurately determining the entire linear sequence of the genome to replace the slow, expensive, and labor-intensive approaches available at its launch in October 1990.6 This led to the completion of the draft sequence of the human genome, as announced at the White House in June 2000 to much fanfare in the scientific and lay press.7 The HGP ended in April 2003, with a more finished sequence, fifty years to the month after Watson and Crick’s seminal description of the DNA double helix.8
At the conclusion of the HGP, the National Human Genome Research Institute of the National Institutes of Health published a vision for the future of genomics research. This vision included technology development as a centerpiece and articulated the goal of achieving the goal of sequencing an entire genome at very low cost, which has come to be referred to as the “$1000 genome.”9 The article recognized that rapidly and inexpensively measuring individual variations in the genome (genotyping) would be of great value. The push to develop inexpensive means for measuring human genetic variation came from awareness that to understand genome function in health and disease would require genotyping or sequencing tens, if not hundreds of thousands, of human genomes, along with the genomes of numerous other organisms. Also appreciated was that the ultimate clinical use of genomics would require highly accurate, rapid, and inexpensive ways to measure variation without reliance on teams of trained scientists and highly specialized equipment.
Gene chips
Fluidics miniaturization (similar to that used in inkjet printers), fluorescent detection methods, and the evolution of powerful informatics approaches have led to mass-produced “gene chips.” A single assay on a gene chip platform, costing tens to hundreds of dollars, can highly accurately analyze a DNA sample for the presence or absence of millions of variations in hours.10 Systems commercially available for research and clinical applications can measure single base pair changes, small deletions, or duplications in genomes, as well as whole-scale genome rearrangements (Table 1). Possibly the most evident biomedical research use of gene chips that detect single base pair variations has been the ground-breaking approach for learning about the genomic underpinning of common complex conditions known as genome-wide association studies (GWAS).3 These studies are predicated on inexpensively measuring variations known as single nucleotide polymorphisms (SNPs) at known points across the genome in large numbers of individuals (hundreds to tens of thousands) with and without a particular common condition. These genotypes are then queried for associations between genome variants and disease status. The first major GWAS, published in 2005, revealed a previously unknown relationship between variations in the complement factor H gene and age-related macular degeneration.11 This publication heralded an avalanche of subsequent studies of common conditions – including such pediatric concerns as asthma, type 1 diabetes, inflammatory bowel disease, and autism.12 Gene chips can be configured for specific clinical diagnostic and prognostic purposes, and can inexpensively measure large numbers of point mutations in a sample. The chips’ accuracy depends on the specific platform, but is generally quite high, especially for single base pair changes measured in a DNA sample, where sensitivity and specificity can exceed 99%.
Table 1. Comparison of selected mutation detection technologies currently in clinical and research use and their characteristics.
| Technology | Variant types detected? |
Novel variants detected? |
Number of variants detected in an assay? |
Speed of assay per variant detected? |
Cost per variation detected? |
Accuracy? |
|---|---|---|---|---|---|---|
| Allele-specific oligonucleotide hybridization |
Single nucleotide polymorphisms (SNPs), duplications, deletions, epigenetic changes |
No | Low | Low | High | High |
| Polymerase chain reaction (PCR) based assays |
SNPs, small insertions/duplications, deletions, epigenetic changes |
Yes (depending on platform) |
Medium | Low | Medium | High |
| Fluorescent in situ hybridization |
Large insertions/duplications, deletions, chromosome rearrangements |
Yes | Medium | Low | High | Variable |
| DNA microarrays | SNPs, insertions/duplications, deletions, chromosome rearrangements, epigenetic changes |
Yes (depending on platform) |
High | High | Low | Medium |
| Sanger sequencing |
SNPs, small insertions/duplications, deletions, epigenetic changes |
Yes | Medium | Medium | Medium | High |
| Next-generation sequencing |
SNPs, small insertions/duplications, deletions, epigenetic changes |
Yes | High | High | Low | Variable |
The ability of chips to detect other types of variations in the genome, such as copy number variations and structural rearrangements, is also quite good. Chips designed to detect structural changes are rapidly supplanting traditional karyotyping techniques as the first line laboratory investigation for children with developmental and congenital abnormalities.13 Another clinical use of gene chips is for expression profiling.14 In this application, RNA is harvested from a sample (a tumor, for example), processed, and incubated with the chip. The location of binding in the microarray provides an idea of the type of RNA expressed and the signal intensity at the binding site provides a measure of the expression level of that particular RNA species. This type of analysis can give insights into gene expression patterns in healthy or diseased tissues, as well as the genome’s response to environmental influences including medications. Patterns of gene expression have proven to be useful guides for prognostication and treatment selection; examples include the FDA-approved MammaPrint® and Oncotype DX™ assays for prognosis and treatment selection in breast cancer.15
High throughput sequencing
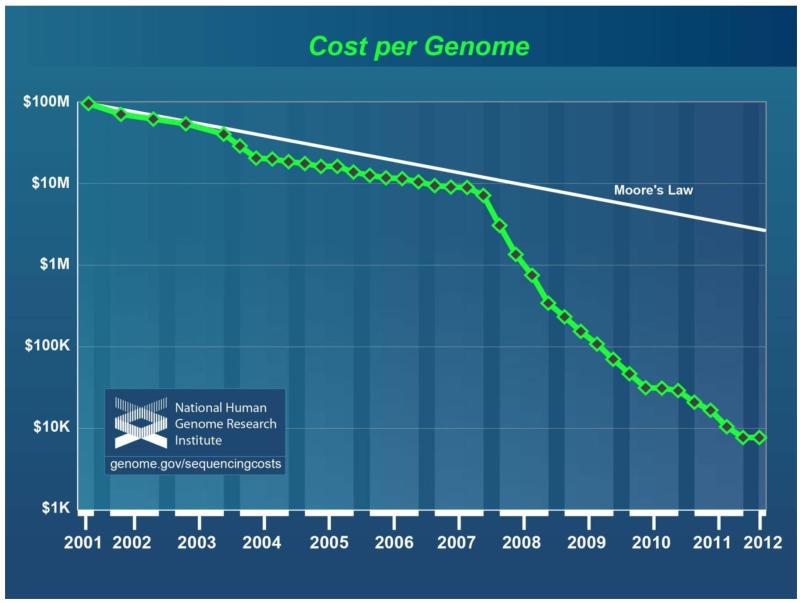
While gene chips efficiently detect known single nucleotide variations at specific points in the genome, detecting previously unsuspected small variations in the genome often requires DNA sequencing. Sequencing technology advances in recent years have been remarkable; costs of high quality DNA sequencing are now approximately 1/100,000 of what they were five years ago.16 (Figure 1) Consequently, both biomedical research and clinical medicine have begun to take advantage of sequencing the protein coding regions of the genome (the exome), and even entire genomes. New sequencing machines can fit on a desktop and generate high quality sequence data in hours that took teams of individuals, with rooms of equipment, years to complete a decade ago.
Figure 1.
Decrease in sequencing cost per genome plotted on a logarithmic scale in U.S. dollars. The white line represents the cost that would have resulted if sequencing cost decreases had followed the mathematics of Moore’s Law of semiconductors. It is important to note that these cost figures do not include genome interpretation. Adapted from the website of the National Human Genome Research Institute, National Institutes of Health (http://www.genome.gov/sequencingcosts/).
Basic genomic discoveries made possible by technological progress
Currently, much genomics research focuses on genome structure and function. A signature major international research initiative in this area is the ENCODE project (ENCylopedia Of Dna Elements).17 Initiated by the National Institutes of Health (NIH) in 2003, ENCODE’s stated goal was to define the functional elements required for normal genome function. Global collaborative networks of scientists have effectively employed a broad range of approaches, both in silico (computer based) and in bench laboratory investigations, to tackle the dissection of functional elements in the genome. In September 2012 a coordinated release of 40 publications provided a tour de force of the ability of focused ENCODE collaborative effort to advance science. 18 The emerging picture of the genome is far from the old portrait of widely spaced protein coding genes acting in isolation and separated by “junk” DNA; now over 80% of the human genome has an ascribed function, much of which is related to appropriate coordination of gene expression.19 The clinical implications of these very new observations are unclear; however, it seems likely that many variations in these regions are important for disease pathogenesis, perhaps through quantitative and qualitative changes in gene expression.
Gene products: beyond proteins
Expanded understanding of the genome’s normal function has led to recognition that large portions of it are transcriptionally active, and that perhaps a minority of transcribed RNA is translated into protein.20 Most non-translated RNAs fall into a diversity of families that vary by length, stranded-ness (double versus single), and function.21 Developing an understanding of the abundance and diversity of such RNAs remains an area of active investigation, but these RNA species are clearly important contributors to genome regulation. RNA molecules are also increasingly recognized as having roles in a variety of disease processes, including cancers. 22 For example, small non-coding RNA molecules known as microRNA’s have been shown to have an active role in the pathogenesis of Ewing’s sarcoma and may eventually provide both therapeutic targets and prognostic biomarkers for this condition.23
Genome sequence variation
While two randomly selected humans are identical for about 99.5% of their genome sequences, the size of the human genome ensures a remarkable diversity between individuals and population groups. Variations range from common single base pair alterations with no discernible effect on genome structure or function (SNPs) to whole-scale gain or loss of chromosomes (e.g., trisomy 21). Much genome variation is benign; for example, SNPs occur on average about every 1000 base pairs. Separating benign variation from that associated with disease has been a goal of human genetics research since the field’s inception. One approach is to start with groups of affected and “normal” individuals and look for non-random associations between presence of disease and specific variations. Classically, such studies have begun with individuals severely affected by single gene disorders and their families, yielding variations with very high risk of disease (high effect size). These mutations have provided insights into disease pathogenesis, potential treatments, and, sometimes, immediate utility in diagnosis and carrier status detection.
As mentioned previously, GWAS have been quite successful in identifying a portion of the genetic underpinnings of many common conditions and traits; thousands of variations, largely single nucleotide changes, have now been associated with a high degree of certainty with a wide range of conditions.12 GWAS have shed light on the pathogenesis of conditions such as asthma, diabetes, inflammatory bowel disease, and hypertension.24-28 However, it is important to recognize that markers found by GWAS generally confer only a very small increase in disease risk (with odds ratios in the vast majority of SNPs considerably less than 2), and are often not causally related to disease. Since the variations discovered by GWAS studies are often, even in aggregate, only weakly predictive of disease risk, GWAS have not yet had much direct impact on clinical care. This is in contrast to variations (mutations) known to be causal of rare disease. For most common conditions studied to date, family history, clinical characteristics and environmental factors account for a much larger proportion of measurable risk. As more is learned about genetic risk factors for common conditions the situation may change, but for now genotyping has little role in providing risk prediction for common conditions.29 However, GWAS have already proved extremely helpful in better understanding the underlying biology of literally scores of conditions, including many that affect children.27,30,31
Recently, attention has focused on the possibility that substantial portions of the unmeasured heritability of common conditions lie in uncommon variations in the human genome.32 The 1000 Genome Project, initiated by the NIH in 2008, is a systematic effort to catalog the spectrum of genetic variation that occurs with at least a 1% frequency in humans. 33,34,35 The 1000 Genome Project data suggest that substantial disease related variation is present at frequencies below 1%. For example, a study that sequenced a large portion of the protein coding regions of the genome with a high degree of accuracy found more than 500,000 SNPs, of which about 82% were novel and 86% were present in less than 0.5% of the study population.36 Further, of the 2% of variants predicted to change protein function, over 95% were rare. These observations are sobering, since they imply that, if SNPs’ effect sizes are small, reliably detecting associations between such SNPs and health conditions might require enormous populations.
Epigenomics
The genome and the environment act in concert to determine all of an individual’s characteristics. A classical pathological example of a gene-environment interaction is phenylketonuria, in which dietary phenylalanine causes severe mental retardation in individuals harboring mutations in the tyrosine hydroxylase gene.37 In this case, elimination of phenylalanine from the diet is preventative. For common conditions such as diabetes and coronary disease, a wealth of epidemiological data show that environmental factors accelerate disease, sometimes with influence spanning generations.38,39 Broadly, external interactions with the genome affecting the expression of traits is known as epigenomics.
The mechanisms through which environmental factors exert lasting effects on genome function are under active study. Classical genetics has long recognized the role of molecular modifications to the primary DNA backbone as a determinant of gene expression; in the era of genomics a wide array of epigenetic changes are known to affect gene expression.40 The classical pediatric example of methylation resulting in differential gene expression is Prader-Willi syndrome, in which the pattern of methylation of cytosine residues at a single genomic locus determines which parental allele is expressed, and thus whether the condition is manifest.41 There is increasing recognition that methylation plays a substantial role in cancer biology, and considerable research is directed at developing drugs that inhibit epigenetic changes.42 There is also great interest in learning how environmental factors interact with the genome to alter the course of common conditions such as obesity, asthma, and diabetes in pediatric populations.43,44
Another particularly active area of epigenomics relates to the toxicology of common environmental agents like bisphenol A that have been demonstrated to have hormone-like properties affecting gene expression in experimental models.45 High throughput methods for measuring epigenetic changes such as methylation should greatly accelerate this area of study.46
Pharmacogenomics
Pharmacogenomics refers to the use of genetic sequence data to help guide the development, selection, and dosing of medications. To date, there are relatively few examples of using genetic testing routinely in the selection or dosing of pediatric drugs; however, one classic example is testing for variations in the TPMT gene to avoid thiopurine toxicity in cancer patients.47 With time, it is likely that an increasing number of drugs will be selected and dosed based on genetic sequence information in pediatric patients. Such advances should dramatically affect pediatric oncology practice, mirroring the situation in adult cancer care. A growing understanding of the molecular basis of both common and rare conditions is having a dramatic affect on the drug development process; in conditions like cystic fibrosis molecularly targeted small molecule therapeutics specific for certain mutations are already a clinical reality.48
Genomics and the pediatric patient
The day when children visit their pediatricians, get genome scans, and leave with personalized plans for their health care over the subsequent 20 years remains distant. However, it is important to recognize that genotyping and low-cost sequencing have created an inflection point in the pace of genomic discovery relevant to clinical care. Microbial genomics, the diagnostic evaluation of undiagnosed disease, and reproductive genetics/newborn screening exemplify the need for the practicing pediatrician to have competency in genomics.
Microbial genomics
The Human Microbiome Project (HMP) has begun to show the diversity of the microbial ecosystems on and in each of us.49 Initiated by the NIH in 2007, this ambitious project uses high throughput sequencing to identify the array of microbes present in human samples. At birth we become dynamic hosts to an eventual population of over a trillion organisms, and until the inception of the HMP, we had only a rudimentary understanding of what organisms were even present, and their relative abundance, since many organisms cannot be cultured outside of the human host.50,51 This complex ecosystem of organisms on and in us is increasingly recognized as a major contributor to many processes in the healthy human and, when either host or microbial factors go astray, disease.
One of the most productive areas of disease-related microbiome research has been in gastrointestinal conditions, since an estimated 80% of gut microbes cannot be cultured using routine methods.52 This has limited the study of a variety of pediatric conditions in which disruption of gut microbial balance is suspected as a key component of pathogenesis, like necrotizing enterocolitis (NEC), obesity, and inflammatory bowel disease. 53, 54 With genomic tools, conditions like NEC are yielding to study; rationally selected treatments including probiotics, although still controversial, may eventually provide real benefits. In inflammatory bowel disease, marked progress in understanding the complex interplay of microbial and host genomic factors that results in aberrant inflammation has helped pave the way for a diversity of new targeted treatment approaches.55
Use of genomic technologies for rapid molecular diagnosis and surveillance of infectious disease has become increasingly important to clinical care and public health practice.56 Specifically, microbial genomic diagnostics have proven useful in a series of high profile disease outbreaks, including the Haitian cholera epidemic, the toxigenic E. coli outbreak in Europe, and a lethal drug resistant Klebsiella outbreak at the NIH Clinical Center.57-60 A recent study retrospectively used microbial genome sequencing to characterize a serious outbreak of MRSA in a neonatal intensive care unit; such techniques may soon become the front line of epidemiologic investigation.61 Numerous commercially available molecular diagnostics tests in clinical care are based on older genetic testing methods for rapid diagnosis of MRSA, gonorrhea, chlamydia, tuberculosis, and a wide variety of viral infections. In many cases, pediatricians ordering these tests are likely unaware that they have ordered “genetic” tests.
Undiagnosed disease
Diagnosing rare diseases can be prolonged and costly, both in emotional distress for families and in medical expenditures. Although not all rare conditions are heritable, a substantial number result from mutations with large effect in one or a few genes. The genetic bases for approximately 40% of the over 7000 suspected Mendelian disorders are now known, though clinical testing is not available for all.62 For conditions with known or suspected causal variants, advances in genotyping and sequencing targeted genomic regions have led to increasingly routine use of these technologies as diagnostic aids. Conversely, the fact that approximately 60% of the suspected 7000 Mendelian disorders lack a known basis leaves significant room for discovery.
Exome and whole genome sequencing may have a role in managing a broad array of conditions, both common and rare. Sequencing forms a centerpiece of the NIH’s Undiagnosed Disease Program, and related efforts elsewhere, which have had some notable successes in diagnosing and treating perplexing clinical problems.63 There is evidence that sequencing might help unravel relatively common conditions relevant to pediatric practice, with a recent report showing a diagnostic yield of 16% from exome sequencing in a series of patient of various ages with unexplained severe intellectual disability.64
Perhaps the most striking foreshadowing of the future that next generation sequencing holds for pediatric patients came in 2010 in the widely publicized case of Nicolas Volker.65 This patient was afflicted by a progressive, inflammatory bowel disease-like condition that resulted in need for repeated abdominal surgery and severe, progressive, debility. Sequencing of his and his parents’ genomes revealed a defect in XIAP (X-linked inhibitor of apoptosis protein) in Nicolas. Subsequent to the discovery, a bone marrow stem cell transplant resulted in cure.
Genomics and newborn health
Identification of the genetic basis of a large number of single-gene disorders, combined with the ability to detect genetic variations rapidly and inexpensively in small samples, has opened a range of possible genomics applications relevant to newborn health. However, scientific and ethical controversy surrounds a variety of these applications.
Preconception tests are commercially available that detect mutations, associated with over 150 rare single gene disorders, in a single assay for less than $1000.66 Although evidence suggests that the mutation detection accuracy is high, interpreting the consequences of harboring a mutation can be challenging. In the prenatal period, technological advances have facilitated non-invasive aneuploidy testing using cell-free fetal nucleic acid present in maternal blood.67 More sophisticated mutation detection approaches applied to cell-free nucleic acids will likely provide non-invasive diagnosis of a broad array of heritable conditions. For couples at increased risk of having an affected child, costly assisted reproductive technologies including pre-implantation genetic diagnosis followed by embryo selection and implantation are options for increasing the likelihood of a healthy pregnancy outcome.68 As these technologies are perfected, and costs come down, the need to address the societal consequences of being able to influence human inheritance will only increase.
For almost everyone born in the U.S., newborn screening provides the first exposure to genomic testing. The “universal” screening panel recommended by the Secretary’s Advisory Committee for Heritable Diseases of Newborns and Children includes over 50 conditions, of which most are heritable.69 Traditionally, newborn screening assays utilize biochemical testing for metabolites, proteins, or enzymatic activity. Recently, incorporating next generation sequencing into newborn screening has been considered. The rationale for integration is that additional information from sequencing can augment biochemical data for conditions already on the universal newborn screening panel by pinpointing the causal genetic variation, while also increasing greatly the number of conditions detected prior to onset of symptoms. Another potential benefit is that sequence data obtained in the newborn period could be available to inform care across the individual’s lifespan. Given the number of newborns involved and the need in this setting for rapid turn-around of highly accurate sequence interpretation, sequence-based newborn screening might have seemed fantastical five years ago, but recent studies suggest that technical feasibility may not be far off.70 The practicalities of implementing sequencing in the context of state public health programs are extremely daunting, especially in the setting of current fiscal constraints facing public health departments. Genome-based newborn screening would also raise a thicket of ethical questions such as what information should be released to whom and when. The National Human Genome Research Institute and the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH have announced a joint program to fund pilot projects that explore technical, medical, and societal issues associated with augmenting traditional newborn screening approaches with sequencing.71
Future challenges
Pediatrics has been at the forefront of genomic technology adoption since the Food and Drug Administration’s approval 30 years ago of human recombinant insulin as the first genetically engineered protein product for human use. For about as long, some have predicted that genomics would precipitate an imminent revolution in the day-to-day practice of medicine. This has yet to happen; in fact, evolution may be a more accurate description of the pace at which clinical changes are occurring. The complexity of genomics, uncertainty regarding the clinical benefits of new and often expensive applications, and a myriad of more pressing health and social issues confronting patients and physicians have limited the uptake of genomic application in many pediatric settings.
Genomic science
Harnessing the full potential of genomics to improve health is an endeavor that will span decades, if not centuries. In the near term, several key scientific challenges must be met to ensure maximal future benefit. First, research is needed in both in vitro and animal model systems to explore the consequences of multiple human genetic variations acting in concert. Achieving a comprehensive understanding of the functional consequences of genetic variation at the biochemical, cell, organ, and organism level is a monumental undertaking, much exceeding the Human Genome Project in complexity. Second, methods for studying the complicated, longitudinal interactions of environmental factors with human genetic variation are needed. All conditions are affected to some degree by environmental exposures affecting risk, progression, and/or treatment response. To date, only a handful of interactions between genes and environmental factors are well understood. Without a better understanding of such interactions, rational approaches to mitigating or exploiting environmental factors – or genetic ones - will be difficult to achieve. Third, efficient clinical trial methods are needed that leverage genomic information to optimize discovery and patient outcomes. Each child’s genome is unique, even in identical twins, and individual variation may bear meaningfully on responsiveness to health interventions or adverse reactions. Currently, clinical trials rarely integrate genomic information into design and data collection; this represents a major missed opportunity for discovery. Fourth, emerging structures for clinical care and clinical informatics must be adapted to support genomics research. Understanding the health consequences of rare variations in the population will require large numbers of individuals, and prospective research cohorts are extremely costly to assemble and follow over time. Effective “learning health care systems” that leverage clinical information stored in electronic health records for research, while respecting individual privacy and confidentiality, would dramatically accelerate clinical genomics research.72 Such settings would provide rapid correlations between genotype and clinical observations such as new diagnosis, adverse reactions to medications, treatment outcomes, and disease related complications at considerable less expense than would multiple independent clinical studies.
Translating genomics to practice
Effectively integrating genomics into routine care will require studies that go beyond demonstration of the efficacy of a new molecular diagnostic or targeted therapeutic in highly controlled settings. Translational research, including studies evaluating the effectiveness, comparative effectiveness, and public health outcomes related to emerging genomic applications, is needed. Traditional public funding sources in the U.S. have typically not funded such genomics research at levels comparable to genomic discovery science, although the establishment of both the NIH National Center for Advanced Translational Sciences (NCATS) and the Patient Centered Outcomes Research Institute (PCORI) signal an increasing emphasis on translational research.73
Genomics and society
Genomics will continue to present a diverse array of societal issues that may prove even more challenging to address than technology or biology.74 These include topics as profound as the ethics of allowing embryo selection to increase chances of having a “normal” child to subjects as prosaic, but crucial, as the effect genomics will have on health care costs. Given the rapidity with which the field is evolving, pediatricians and their patients will have to grapple with myriad topics, often without much guidance. The utility of genomics in clinical care will depend partly on developing more effective and inclusive national and international frameworks for grappling with these important issues.
Future opportunities
Genomics is irrevocably changing the face of biomedical research and, more slowly, clinical medicine. Pediatricians have the unique and exhilarating responsibility to help ensure that young patients derive maximal benefit from genomics during this time of transition in biomedicine. For the foreseeable future, genomics will provide pediatricians new and often unexpected insights into the biological basis of health and disease. Likewise, genomic research will afford new health care options requiring informed and sometimes challenging choices of physicians and patients.
One can imagine a not far-off world in which knowledge of patients’ genomes improves diagnosis and, through informed prediction of individual drug metabolism and responsiveness, the selection of therapeutics. Even that hallmark of pediatric practice, anticipatory guidance, will likely be affected. With the advent of sequenced-based newborn screening, pediatricians would have access to a huge volume of information with import for not just the current and imminent health of their patients, but also their more distant future. The increasing availability of genomics is a major driver of recent intensified interest in the “developmental origins of adult health and disease.”75 One can foresee a day in which anticipatory guidance is not only about the next six months or even five years of a patient’s life, but about the next eight decades.
Education of the provider will be key to the clinical use of genomics; organizations such as the American Academy of Pediatrics and the National Coalition for Health Professional Education in Genetics have worked to ramp up the genomic literacy of the profession.76 Developing pediatrician competency in genomics, especially for those well away from training, is a daunting task, but one that the specialty can and must accomplish in the near future. Achieving such competency should provide substantial rewards; effectively integrating genomics into practice will improve pediatricians’ effectiveness in caring for patients current health concerns and make pediatricians the guides to lifelong health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Korf BR, Rehm HL. New approaches to molecular diagnosis. JAMA: the journal of the American Medical Association. 2013 Apr 10;309(14):1511–1521. doi: 10.1001/jama.2013.3239. [DOI] [PubMed] [Google Scholar]
- 2.Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. The New England journal of medicine. 2010 May 27;362(21):2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 3.Manolio TA. Genomewide association studies and assessment of the risk of disease. The New England journal of medicine. 2010 Jul 8;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 4. [Accessed May 6, 2013];Genetics Home Reference. http://ghr.nlm.nih.gov/
- 5. [Accessed May 6, 2013];Talking Glossary of Genetic Terms. http://www.genome.gov/glossary/index.cfm?
- 6.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003 Apr 11;300(5617):286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 7.Marshall E. Human genome. Rival genome sequencers celebrate a milestone together. Science. 2000 Jun 30;288(5475):2294–2295. doi: 10.1126/science.288.5475.2294. [DOI] [PubMed] [Google Scholar]
- 8.International Consortium Completes Human Genome Project [Accessed November 30, 2012];2003 doi: 10.1517/phgs.4.3.241.22688. http://www.genome.gov/11006929. [DOI] [PubMed]
- 9.Collins FS, Green ED, Guttmacher AE, Guyer MS, Institute USNHGR A vision for the future of genomics research. Nature. 2003 Apr 24;422(6934):835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 10.Gresham D, Dunham MJ, Botstein D. Comparing whole genomes using DNA microarrays. Nature reviews. Genetics. 2008 Apr;9(4):291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005 Apr 15;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. [Accessed November 30, 2012];A Catalog of Published Genome-Wide Association Studies. 2012 http://www.genome.gov/gwastudies/
- 13.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. American journal of human genetics. 2010 May 14;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nature reviews. Genetics. 2006 Mar;7(3):200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 15.Marchionni L, Wilson RF, Wolff AC, et al. Systematic review: gene expression profiling assays in early-stage breast cancer. Annals of internal medicine. 2008 Mar 4;148(5):358–369. doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 16. [Accessed November 30, 2012];DNA Sequencing Costs. 2012 http://www.genome.gov/sequencingcosts/
- 17. [Accessed November 30, 2012];Nature ENCODE Explorer. 2012 http://www.nature.com/encode/#/threads.
- 18. [Accessed November 30, 2012];ENCODE data describes function of human genome. 2012 http://www.genome.gov/27549810.
- 19. [Accessed May 6, 2013];ENCODE data describes function of human genome. http://www.genome.gov/27549810.
- 20.Consortium EP, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 Jun 14;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteller M. Non-coding RNAs in human disease. Nature reviews. Genetics. 2011 Dec;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 22.Jansson MD, Lund AH. MicroRNA and cancer. Molecular oncology. 2012 Dec;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dylla L, Moore C, Jedlicka P. MicroRNAs in Ewing Sarcoma. Frontiers in oncology. 2013;3:65. doi: 10.3389/fonc.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. The New England journal of medicine. 2010 Sep 23;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. The New England journal of medicine. 2011 Oct 27;365(17):1612–1623. doi: 10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy MI. Genomics, type 2 diabetes, and obesity. The New England journal of medicine. 2010 Dec 9;363(24):2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler E, Huang N, Bochukova EG, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nature genetics. 2013 Apr 26;45(5):513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wain LV, Verwoert GC, O’Reilly PF, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature genetics. 2011 Oct;43(10):1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009 Oct 8;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feenstra B, Geller F, Krogh C, et al. Common variants near MBNL1 and NKX2-5 are associated with infantile hypertrophic pyloric stenosis. Nature genetics. 2012 Mar;44(3):334–337. doi: 10.1038/ng.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Barcelo MM, Tang CS, Ngan ES, et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2009 Feb 24;106(8):2694–2699. doi: 10.1073/pnas.0809630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schork NJ, Murray SS, Frazer KA, Topol EJ, Common vs. rare allele hypotheses for complex diseases. Current opinion in genetics & development. 2009 Jun;19(3):212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.1000 Genomes A deep catalog of human genetic variation [Accessed November 30, 2012];2012 http://www.1000genomes.org/about.
- 34.Genomes Project C A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genomes Project C. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012 Jul 6;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. [Accessed May 6, 2013];Phenylalanine Hydroxylase Deficiency. http://www.ncbi.nlm.nih.gov/books/NBK1504/
- 38.Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS medicine. 2010 Oct;7(10):e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008 Nov 4;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013 Mar 29;339(6127):1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. [Accessed May 6, 2013];Prader-Willi Syndrome. http://www.ncbi.nlm.nih.gov/books/NBK1330/
- 42.Ho AS, Turcan S, Chan TA. Epigenetic therapy: use of agents targeting deacetylation and methylation in cancer management. OncoTargets and therapy. 2013;6:223–232. doi: 10.2147/OTT.S34680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clinical pharmacology and therapeutics. 2012 Dec;92(6):07–715. doi: 10.1038/clpt.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang IV, Schwartz DA. Epigenetic mechanisms and the development of asthma. The Journal of allergy and clinical immunology. 2012 Dec;130(6):1243–1255. doi: 10.1016/j.jaci.2012.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS genetics. 2013 Apr;9(4):e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki M, Greally JM. Genome-wide DNA methylation analysis using massively parallel sequencing technologies. Seminars in hematology. 2013 Jan;50(1):70–77. doi: 10.1053/j.seminhematol.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 47. [Accessed May 6, 2013];Thiopurine S-methyltransferase. http://www.pharmgkb.org/gene/PA356.
- 48. [Accessed May 6, 2013];FDA approves Kalydeco to treat rare form of cystic fibrosis. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm289633.htmFDAapproves Kalydeco to treat rare form of cystic fibrosis.
- 49.Human Microbiome Project C A framework for human microbiome research. Nature. 2012 Jun 14;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012 Jun 14;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012 May;129(5):950–960. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005 Jun 10;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morowitz MJ, Poroyko V, Caplan M, Alverdy J, Liu DC. Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics. 2010 Apr;125(4):777–785. doi: 10.1542/peds.2009-3149. [DOI] [PubMed] [Google Scholar]
- 54.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature reviews. Genetics. 2012 Apr;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malik T, Mannon P. Inflammatory bowel diseases: emerging therapies and promising molecular targets. Frontiers in bioscience. 2012;4:1172–1189. doi: 10.2741/s324. [DOI] [PubMed] [Google Scholar]
- 56.Relman DA. Microbial genomics and infectious diseases. The New England journal of medicine. 2011 Jul 28;365(4):347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin CS, Sorenson J, Harris JB, et al. The origin of the Haitian cholera outbreak strain. The New England journal of medicine. 2011 Jan 6;364(1):33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasko DA, Webster DR, Sahl JW, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. The New England journal of medicine. 2011 Aug 25;365(8):709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snitkin ES, Zelazny AM, Thomas PJ, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Science translational medicine. 2012 Aug 22;4(148):148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loman NJ, Constantinidou C, Christner M, et al. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA: the journal of the American Medical Association. 2013 Apr 10;309(14):1502–1510. doi: 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- 61.Koser CU, Holden MT, Ellington MJ, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. The New England journal of medicine. 2012 Jun 14;366(24):2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011 Feb 10;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 63.Gahl WA, Markello TC, Toro C, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genetics in medicine: official journal of the American College of Medical Genetics. 2012 Jan;14(1):51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. The New England journal of medicine. 2012 Nov 15;367(20):1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 65.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genetics in medicine: official journal of the American College of Medical Genetics. 2011 Mar;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 66. [Accessed November 30, 2012]; https://www.counsyl.com/ 2012 https://www.counsyl.com/
- 67.Committee opinion no. 545: noninvasive prenatal testing for fetal aneuploidy. Obstetrics and gynecology. 2012 Dec;120(6):1532–1534. doi: 10.1097/01.AOG.0000423819.85283.f4. [DOI] [PubMed] [Google Scholar]
- 68.Bodurtha J, Strauss JF., 3rd. Genomics and perinatal care. The New England journal of medicine. 2012 Jan 5;366(1):64–73. doi: 10.1056/NEJMra1105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. [Accessed November 30, 2012];Secretary’s Advisory Committee on Heritable Disorders in Newborns and Children. 2012 http://www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/
- 70.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Science translational medicine. 2012 Oct 3;4(154):154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. [Accessed November 30, 2012];NIH seeks applications to study genomic sequencing in newborn period. 2012 http://www.genome.gov/27549767.
- 72.Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Science translational medicine. 2010 Nov 10;2(57):57cm29. doi: 10.1126/scitranslmed.3001456. [DOI] [PubMed] [Google Scholar]
- 73.Schully SD, Benedicto CB, Gillanders EM, Wang SS, Khoury MJ. Translational research in cancer genetics: the road less traveled. Public health genomics. 2011;14(1):1–8. doi: 10.1159/000272897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hudson KL, Genomics, health care, and society The New England journal of medicine. 2011 Sep 15;365(11):1033–1041. doi: 10.1056/NEJMra1010517. [DOI] [PubMed] [Google Scholar]
- 75. [Accessed November 30, 2012];Scientific Vision Workshop on Developmental Origins of Health and Disease. 2012 http://www.nichd.nih.gov/vision/vision_themes/developmental_origins/Documents/Vision_De vOrig_WP_04212011.pdf.
- 76.Genetics in Primary Care Institute [Accessed November 30, 2012];2012 http://www.medicalhomeinfo.org/gpci.aspx.