Abstract
The Esx/WXG-100 (ESAT-6/Wss) exporters are multiprotein complexes that promote protein translocation across the cytoplasmic membrane in a diverse range of pathogenic and nonpathogenic bacterial species. The Esx-1 (ESAT-6 System-1) system mediates virulence factor translocation in mycobacterial pathogens, including the human pathogen Mycobacterium tuberculosis. Although several genes have been associated with Esx-1-mediated transport and virulence, the contribution of individual Esx-1 genes to export is largely undefined. A unique aspect of Esx-1 export is that several substrates require each other for export/stability. We exploited substrate “codependency” to identify Esx-1 substrates. We simultaneously quantified changes in the levels of 13 Esx-1 proteins from both secreted and cytosolic protein fractions generated from 16 Esx-1-deficient Mycobacterium marinum strains in a single experiment using MRM/SRM targeted mass spectrometry. This expansion of measurable Esx-1 proteins allowed us to define statistical rules for assigning novel substrates using phenotypic profiles of known Esx-1 substrates. Using this approach, we identified three additional Esx-1 substrates encoded by the esx-1 region. Our studies begin to address how disruption of specific genes affects several proteins in the Esx-1 complex. Overall, our findings illuminate relationships between Esx-1 proteins and create a framework for the identification of secreted substrates applicable to other protein exporters and pathways.
Keywords: targeted proteomics, secretion, Esx-1, Mycobacterium marinum, EsxA, RD1, MRM/SRM, substrate identification
Introduction
Mycobacterium tuberculosis is the etiological agent of human tuberculosis (TB). Despite efforts aimed at the treatment and prevention of TB, the disease remains a global health problem. TB is particularly associated with individuals with compromised immune function, including people co-infected with HIV and those with diabetes. As the diabetes epidemic expands in the coming decades, M. tuberculosis poses an ongoing threat.1−3
The Esx/WXG-100 (Wss) protein exporters directly promote virulence of both mycobacterial and Gram-positive pathogens.4−8 The genomes of pathogenic mycobacteria can encode up to four or five independent Esx exporters (Esx-1–5).9,10 The Esx/Wss exporters are also conserved and functional in several nonpathogenic bacteria.9,11−16 In mycobacterial pathogens, the Esx-1 system promotes the export of several small protein substrates to both the cell surface and the bacteriological media in vitro.6,8,17−26In vivo, the Esx-1 system permeabilizes the macrophage phagosomal membrane and engages the human immune response to promote both mycobacterial infection and host resistance.27−31
The Esx-1 system is of particular interest because it is lacking from the genome of the M. bovis Bacillus Calmette-Guérin (BCG) vaccine strain, which is the only TB vaccine currently in use.7 The BCG strain is not efficacious at preventing adult pulmonary TB.32,33 Understanding how the Esx-1 system functions at the molecular level and defining secreted virulence factors that are poised to interface with the human immune response may provide insight into why the vaccine is not protective.
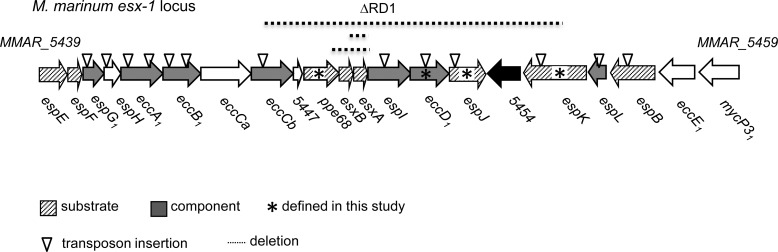
In M. tuberculosis, three genomic regions are required for Esx-1-mediated export. These include the esx-1 locus (including the RD1, Region of Difference-1, deletion missing from the BCG strain), the espACD (Esxsecretion-associated protein-A etc.)34 locus, and the espR gene.6−8,18,22,35 Orthologous loci are conserved in the genome of the mycobacterial pathogen Mycobacterium marinum, which is an established model for mycobacterial Esx-1 export.36−40 In M. marinum, the major esx-1 locus includes the espE (MMAR_5439) through mycP3_1 (MMAR_5459) genes (Figure 1).36 Five protein substrates are encoded within the esx-1 locus, including EsxA (ESAT-6; Early Secreted Antigenic Target, 6 kDa), EsxB (CFP-10; Culture Filtrate Protein, 10 kDa), EspE, EspF, and EspB. Proteins are characterized as substrates if disruption of the Esx-1-associated genes results in retention in the mycobacterial cell in vitro.34 Accordingly, all five substrates are exported across the M. marinum cytoplasmic membrane either to the cell wall or cell surface or into the bacteriological media in vitro in an Esx-1-dependent manner (Figure 1).17,19,21,25,37,41−43 Two additional substrates, PE35 and PPE68_1 (MMAR_0186), are encoded at another genomic locus but are not known to be required for Esx-1 substrate export.43 In M. tuberculosis, EspAmt and EspCmt (subscript “mt” indicates the proteins are from M. tuberculosis, and “mm” indicates M. marinum) are also exported by Esx-1, but this has not been confirmed for the M. marinum EspAmm and EspCmm proteins.17,22,35 However, when the EspAmt and EspCmt proteins were expressed in M. marinum, they were exported in an Esx-1-dependent manner demonstrating that the mechanisms of substrate recognition are conserved between the two species.17
Figure 1.
The esx-1 region in M. marinum. The ∼10 Kb genomic region including the Esx-1 secretion system from M. marinum is shown. Genes are named according to Bitter et al.34 Briefly, Esp = Esx secretion-associated proteins; Ecc = Esx conserved components. Subscript “1” indicates the gene is found at the esx-1 locus. Genes include letters based on the order of the genes at the esx-1 locus. Transposon (Tn) insertion strains used in this study are represented with an inverted open triangle. Strains used in this study are in Supplemental Table S1. Deletions are indicated by a dotted line above the genes within the deletion. MMAR_5454 is not conserved in M. tuberculosis. * indicates substrates or components defined in this study.
Additional proteins encoded by genes within the esx-1 locus likely form part of the Esx-1 apparatus based on conservation across Esx systems and predicted cellular localization.43−45 The “components” include two predicted chaperones, EspG1 and EccA1 (Esx conserved components34); two FtsK-SpoIIIE-like AAA ATPases, EccCa and EccCb; and two predicted membrane-associated proteins, EccB1 and EccD1.21,37,38,43,46,47
Several additional proteins encoded by the esx-1 locus (EspK, EspH, EspL, EspJ, and EspI) are required for substrate export. It is unclear if these proteins are Esx-1 components or substrates (or both).19,25,37,42,48 We have recently identified a second genomic region (MMAR_1663–1668) in M. marinum that is required for Esx-1 export and virulence.49
In both mycobacterial species, disruption of genes within Esx-1-associated loci generally results in the loss of Esx-1 substrate export in vitro and attenuation of virulence in cell culture and animal models of infection.6−8,18,22,25,35,37,40,41,48−52 However, recent studies have identified both attenuated M. tuberculosis strains that still secrete Esx-1 substrates and virulent M. tuberculosis strains that do not secrete Esx-1 substrates into the bacteriological media in vitro.53−56 We found that in M. marinum substrates localized to the cell surface, and not necessarily the culture supernatant, correlate with virulence.49 Therefore, the complex relationship between Esx-1 export and virulence requires further investigation.
Further confounding our understanding of this relationship are limitations with how we measure the export of Esx substrates; in vitro Esx-1 export and secretion assays measuring the presence of substrates in the bacteriological media are generally performed using Western blot analysis. It has been difficult to precisely quantitate change in Esx protein secretion using this approach in part because of antibodies that are of poor quality or lacking altogether for most proteins associated with the mycobacterial Esx-1 system.47,57 Multiple reaction monitoring (MRM) has emerged as the proteomics tool to target and precisely determine protein levels directly from complex biological mixtures.58 In practical terms MRM functions as a replacement for Western blot analysis, allowing highly parallel stacking of targets (proteins). For microbes in particular, MRM data is also largely portable, and online databases to collect and curate these targets are available.59,60
The Esx-1 export apparatus is a multiprotein complex, spanning at least the mycobacterial cytoplasmic membrane. It is not yet known how or if the Esx-1 system promotes translocation of substrates across the mycolate outer membrane (MOM) and out of the cell.61,62 It is also not well-defined how or if individual Esx-1-associated proteins affect the levels and stability of other Esx-1 proteins in the export complex. A lack of understanding of how each Esx-1-associated protein contributes to protein export makes it difficult to establish how the Esx-1 system works at the molecular level. Elucidating Esx-1 protein translocation at the molecular level will advance our understanding of mycobacterial virulence mechanisms and potential courses of treatment and prevention of human TB.
It has been reported several times that Esx-1 substrates require each other for export.17,21,22,25,35 This phenomenon is referred to as “codependent secretion”.22 The basis for this codependency is not understood. Direct interaction between substrates or shared genetic requirements for export could underlie this observation.17,44 We hypothesized that the quantification of substrate codependency could define rules for identifying novel Esx-1 substrates. In a single experiment, we simultaneously examined the changes in the levels of 13 Esx-1-associated proteins. We employed targeted mass spectrometry to differentially quantify changes in the levels and export of these Esx-1-associated proteins from both secreted and cytosolic protein fractions across the 16 Esx-1-deficient M. marinum strains as compared to the wild-type (WT) M. marinum M strain.
Experimental Procedures
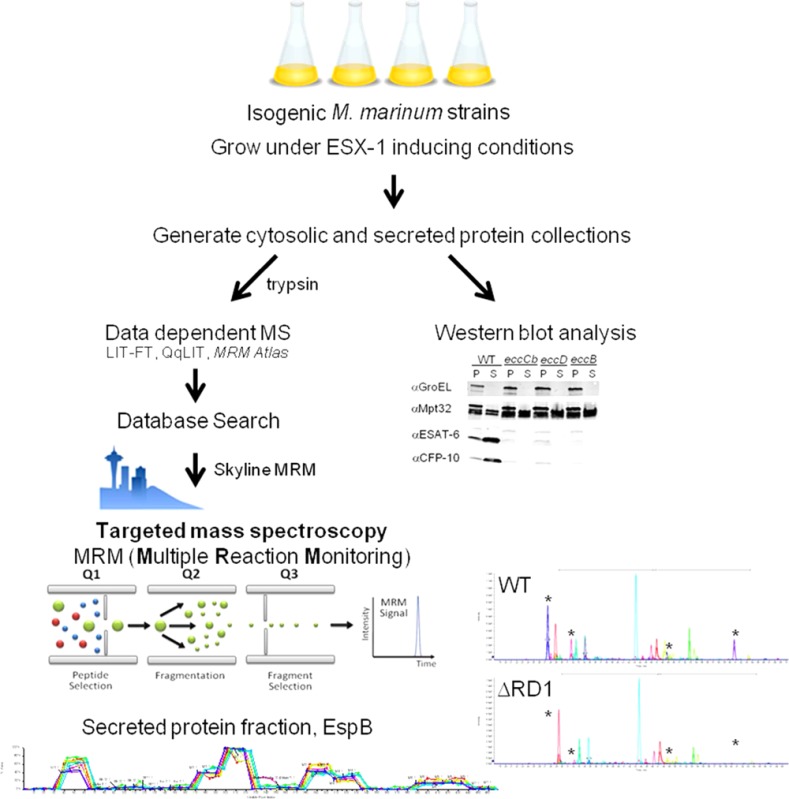
Growth of M. marinum Strains
M. marinum strains were grown at 30 °C and maintained on Middlebrook 7H11 agar (Accumedia, Lansing, MI) or in 7H9 broth (Accumedia, Lansing, MI) supplemented with 0.1% Tween-80 and kanamycin (20 μg/mL; IBI, Poesta, IL) when appropriate. M. marinum strains used in this study are in Supplemental Table S1. All strains were derived from the parent M. marinum M strain. The majority of the strains in this study were obtained from the laboratory of Eric J. Brown; these strains are Esx-1-deficient and attenuated for virulence.21,37 Strains bearing transposon (Tn) insertions in the eccB1 (F11), eccD1 (17), and espL (2) genes were identified from a nonsaturating M. marinum Tn-insertion library.47 The Tn-insertions were mapped by extracting and digesting genomic DNA from each strain and generating a plasmid library in pBluescript as previously explained.47,49 The plasmid bearing the Tn-insertion was selected for kanamycin resistance in E. coli, and the precise site of each insertion was mapped by sequencing analysis using the 821A and 822A primers.47,49,63 The loss of Esx-1 secretion in vitro was confirmed using Western blot analysis and nLC–MRM (see Supplemental Figure S2 for Western blot analysis).
Esx-1-mediated Secretion Assays
Esx-1-mediated secretion assays were performed essentially as described.17 Briefly, M. marinum strains were grown in 7H9 broth to saturation and diluted to an OD600 of 0.8 in 50 mL of Sauton’s defined broth with 0.05% Tween-80. After 48 h of growth, the cells were harvested by centrifugation, and the supernatant was filtered to generate culture filtrates (CF). The cell lysate (CL) fractions were generated using a Mini-Bead-Beater-24 (BioSpec, Bartlesville, OK). Culture filtrates were concentrated ∼100-fold using Amicon Ultra-4 Centrifugal Filter Units (EMD Millipore, Temecula, CA). Protein concentrations were determined using the MicroBCA Protein Assay according to the manufacturer’s instructions (Thermo Scientific Pierce). Western blot analysis was performed as described previously with the following changes.47 A 10 μg portion of cell lysates and cell filtrates was separated using 4–20% Mini PROTEAN TGX Tris-Glycine Polyacrylamide Gels (Bio-Rad). Nitrocellulose membranes were incubated with antibodies against ESAT-6 (EsxA) (1:3000, Abcam ab26246), CFP-10 (EsxB) (1:5000, Pierce PA1-19445), MPT32 (1:5000, NR-13807; BEI Resources), or GroEL (1:5000, Assay Designs SPS-875) as described previously.17,47 Immunoblots for EsxA, EsxB, and MPT32 were incubated with goat anti-mouse IgG (1:5000, HRP-conjugate) or goat anti-rabbit IgG (1:5000; HRP-conjugate, Millipore). The immunoblots were imaged using the LumiGLO Chemiluminescent Substrate Kit (KPL, Gaithersburg, MD). The immunoblot for GroEL was incubated with DyLight anti-rabbit IgG (1:10000, 680 conjugate) (Cell Signaling Technologies, Danvers, MA). The immunoblot was imaged using a LI-COR Odyssey Imager and analyzed with LI-COR Odyssey Software.47 Bands resulting from the Western blot analysis were quantified using ImageJ as described here: http://www.di.uq.edu.au/sparqimagejblots.
Proteomics
Protein Preparation and Digestion
A 50 μg portion of protein as determined by Micro BCA (Pierce) from either culture filtrate or whole cell lysate was prepared and digested with trypsin as previously described17,47 with the exception that samples were resuspended in 0.2% trifluoroacetic acid (TFA) to a final concentration of 500 ng/μL, and 5 μg aliquots were desalted using a C18 zip tip (Millipore) according to manufacturer’s instructions and then dried. Samples were resuspended for LC–MS/MS as described below.
MRM/SRM Generation
We utilized two empirical data sources to determine and develop our MRM/SRM transitions: existing LC–MRM results and LC–MS/MS identifications similar to that described.17,60,64−66 We maintain an in-house database of identified M. marinum proteins from culture filtrates and whole cell lysates and an in-house atlas of MRM transitions derived from these protein identifications, many of which have been previously used in other publications and validated with synthetic stable-isotope analogues of the peptides.17,47,67 In general, our available proteome was collected by triplicate analysis of M. marinum cell lysates and culture filtrates by LC–MS/MS as described in Li et al., using nano UHPLC–MS/MS.67 mgf (Matrix Science) peak lists from the LTQ-Velos Orbitrap data were searched against the current M. marinum M strain FASTA database using the Paragon algorithm in ‘Rapid search mode’. FDR rates were determined using the target-decoy method of Elias et al.,68,69 and FDR rates were <0.001% for all proteins used in this study. MRM transitions were determined from these data using Skyline software from the MacCoss lab (University of Washington) and MRM Pilot (AB Sciex, Framingham, MA). Peptide and protein validation was performed using a procedure nearly identical to that described for empirical LC–MS/MS data from the Aebersold group.60,70 Cross validation was made using independent Western blot analysis (Supplemental Figure S2) as well as the unique aspect here of the availability of mutant strains for most of the monitored proteins. Two independent mutant EccA1 and EccB1 strains were analyzed for verification of phenotypes, clustering, and statistical reproducibility. Individual MRM transitions were tested to ensure peptide profiles degenerated in the presence of strains with genes disrupted for the respective protein product. These bioinformatics approaches yielded a final MRM-transition list containing 166 transitions. Stable heavy-isotope analogues of several Esx-1 proteins, to validate the normalization and lysis controls, were used as in Kennedy et al. for further correction.49 The list of endogenous transitions used in the relative method are included as Supplemental Figure S1.
nLC–MRM
Nano-UHPLC–MS/MS was performed essentially identically to our previous publications.17,49,71 Protein samples (5 μg) were resuspended in 0.3% TFA (20 μL). Then 2 μL (500 ng) was injected onto a 100 mm × 100 μm C18BEH column (Waters, Milford, MA) and separated over a 90 min gradient from 2% to 40% B (A = 0.1% formic acid in water) (B = acetonitrile, 0.1% FA) on an Nano Ultra2D UHPLC (Eksigent, Dublin, CA) running at 650 nL/min. HPLC solvents were of the highest grade available (Burdick and Jackson). MRM-based MS/MS acquisition was performed on a QTRAP 5500 (AB Sciex) running in triple-quadrupole mode. Each sample was analyzed in-triplicate with biological duplicates. In total >200 LC–MS/MS runs were collected, including blanks and standards.
Data Processing
MRM data were integrated, normalized, and corrected for lysis as described.17,49,72,73 Peaks were integrated using MultiQuant 2.0 (AB Sciex) using Intelliquan integration with a 3 point Gaussian smooth with no baseline correction. The apex transition was used for quantification (Quantifier), and the remaining transitions were used as qualifiers to determine relative peptide levels and ensure transition ratios. Multiple peptides (1–4) were averaged together to arrive at the average peak area for the protein.65 In several cases, the low-molecular weight substrates yielded one well-integrated peptide, which passed all other criteria. Statistical analysis for protein quantitation was done using propagation of errors. [(CV % 1)2 + (CV % 2)2 + (CV % n)2)0.5].
The peak area of the proteotypic peptides from each protein was converted to an area response/ratio against the GroES or GroEL2 cytosolic protein standards to control for lysis. Area ratios of each protein were averaged and normalized to the peptide response from the WT strain to allow direct comparison among proteins between strains. Direct comparisons in protein levels between CL (cell lysate) and CF (culture filtrate) were largely not performed because the sample complexity and dynamic range vastly differ.60 This approach is sensitive to low levels of autolysis and can detect proteins in the secreted fraction that are not necessarily secreted.73 We exploited coincidental detection to normalize sample amounts and correct for cytosolic contamination of the CF.17,47,73 Further validation of the stability of differential area-ratio analysis was performed by selected nUHPLC–MS injections using stable heavy-isotope (AQUA) versions of peptides corresponding to EsxA and GroES49 (New England Peptide, Gardner, MA). Data from these selected absolute quantitative standards are in excellent agreement with the ratios observed by the differential analysis described here.74
Statistical Analysis and Hierarchical Clustering
Pearson’s correlation coefficients of the proteins were calculated independently from normalized area ratios of each protein compared to the peptide response found in wild-type M. marinum for the cell pellet and cell lysate data sets, and their p values were computed by fitting a linear model using R, an open source statistical analysis environment.91 The R code and the raw data files used to calculate the correlation and its significance are included in the Supporting Information (Supplemental Figure S6 and Supplemental files). Hierarchical clustering was performed using HCE 3.5 software (Schneiderman Lab) using normalized peak area MRM data as cell input with peptide (column) and mutant/strain (row) for consistency. Data were normalized by Row (Protein) across strains using (X – m)/σ to normalize the response ratio between high and low abundance proteins relative to the internal standards. Hierarchical aggregate clustering was employed to cluster the data for both M. marinum strains and Esx-1-associated proteins as described in Seo et al.75 Thresholds were set at approximately 1SD for color graphing.
Our results were further statistically analyzed at each level of experimental reporting. First, we determined calculated protein ratios from the MRM-peptide measurements, their respective % CV, and then the propagated error to determine the analytical robustness. A paired two-tailed Student’s t test was used to compare the protein ratios for each protein between the WT M. marinum and Esx-1-deficient strains (Microsoft Excel). p values ≤0.05 were considered significant. This was performed for both cell lysate and culture filtrate fractions separately.
Results and Discussion
Development and Validation of a Targeted MS Esx-1 Secretion Assay
Targeted proteomics (MRM)-based assays are well-suited for the analysis of proteins from microbial systems and particularly well-suited for the analysis of Mycobacterium spp.57,58,60 We previously used targeted proteomics on nano-HPLC-separated peptide digests of bacterial proteins (nLC–MRM-MS) to detect the secretion of individual Esx-1 substrates, including those that were largely undetectable by Western blot analysis.17,49 Moreover, nLC–MRM has recently been used to determine the genetic requirements for export of the EsxH and EsxG substrates of the Esx-3 system in Mycobacterium smegmatis.57 MRM-based analysis is suited to generate relative detection of numerous substrates simultaneously. MRM-based approaches can also be made specific to domains of proteins and facilitate a substantially larger potential set of controls. Contamination, cellular lysis, gene duplications, actual changes in secretion, and protein loading can greatly affect the denominator used to normalize secreted/compartmented proteomics.76,77 Targeted multiplexed MRM allows development of appropriate normalization proteins and schemes simultaneously, including the use of multiple normalization proteins such as MMAR_2929, GroEL2, GroES, MPT64, and MPT32 used here and in numerous mycobacterial studies.
We hypothesized that by using nLC–MRM-MS on a larger scale to measure Esx-1-associated proteins in a collection of isogenic M. marinum strains bearing Tn insertions or deletions in the esx-1 locus, we could both identify Esx-1 substrates and gain a deeper insight into the molecular mechanisms of Esx-1 export. To address this hypothesis, we induced Esx-1 export in M. marinum by growth in Sauton’s defined medium and generated cell lysate (CL) and culture filtrate (CF) protein fractions from each strain (Supplemental Table S1). The protein fractions were digested with trypsin to generate proteotypic peptides for detection and quantitation. We quantified changes in Esx-1 protein levels within and secreted from each Esx-1-deficient M. marinum strain normalized to levels in the WT M. marinum using nLC–MRM in a single experiment (see Supplemental Figure S1 for MRM transitions monitored in this study). The peak area for each peptide was normalized for total quantity and autolysis (false-secretion) to stable cytoplasmic proteins, chiefly GroEL2 and GroES. The area ratio for each protein-derived peptide was then compared directly with the exact same peptide response from the WT M. marinum to allow direct comparison among strains and proteins (see Experimental Procedures).
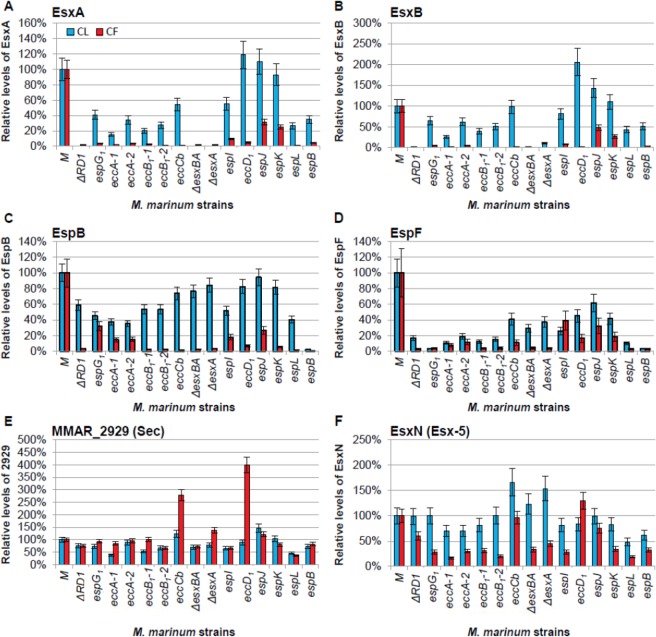
To test our approach, we selected the proteomics data describing the relative levels of the best-characterized and most abundant Esx-1 substrates, EsxA and EsxB. The data from this study are presented as Figure 2A and B (Log2 transformed data in Supplemental Figure S4). In parallel, we also separately conducted Western blot analysis followed by quantitation by densitometry, which is presented as Supplemental Figure S2. For EsxA and EsxB, the M. marinum ΔRD1, ΔesxA, and ΔesxBA strains, which lack the esxA and/or the esxB genes, served as controls for specificity for both nLC–MRM and Western blot analysis. In these strains, EsxA and EsxB were not detected in the CL and CF fractions, indicating that both detection methods were specific for the EsxA and EsxB proteins (Figure 2A and B and Supplemental Figure S2). The Western blot analysis using the same samples (Supplemental Figure S2) contained 10 μg of protein from M. marinum fractions to detect 1–2 protein bands per analysis with virtually no dynamic range or quantification. Here, each proteomic analysis utilized 20-fold less material, with multiplexing of target proteins and controls simultaneously. Further dilution experiments and the inherent linearity of triple-quadrupole instrumentation gave at least 3.5 orders of magnitude in linear dynamic range for all substrates and sensitivities <100 amol on column per protein (confirmed by selected AQUA peptides, data not shown). The dynamic range of Western blots for EsxB and EsxA for which we have reliable antibodies was <2–2.5 orders of magnitude (Supplemental Figure S2). Absolute peak intensities for EsxB often exceed 5 × 107 cps, and we can typically detect these peptides down to ca. 300 cps (s/n > 5:1). This is nearly 5-orders of linear dynamic range and represents a potential reduction in starting material to detect EsxB from less than 0.5 ng of trypsin-digested CF (<0.005 % of what was loaded for Western blot analysis). The reproducibility of the measurements here (CV ≈ 10%) further demonstrates the superior precision of our approach.
Figure 2.
Development and validation of the targeted MS Esx-1 secretion assay. (A) nLC–MRM analysis of EsxA. Relative EsxA levels within the cell lysate (CL, blue bars) or in the culture filtrate (CF, red bars) generated from Esx-1-deficient strains as compared to wild-type M. marinum. (B) nLC–MRM analysis of EsxB. For EsxA and EsxB, the ΔRD1, ΔesxA, and ΔesxBA strains served as negative controls for EsxA and EsxB detection and demonstrated the specificity of the approach. (C) nLC–MRM analysis of the levels of the EspB substrate in the CL and CF relative to the levels of EspB in the wild-type strain. For EspB, the espB::Tn strain served as a negative control for EspB detection and demonstrated the specificity of the approach. (D) nLC–MRM analysis of the levels of the EspF substrate in the CL and CF. (E) nLC–MRM analysis of MMAR_2929. (F) nLC–MRM analysis of EsxN. Error bars represent the average propagated standard error and were calculated as described in the Experimental Procedures. In all graphs changes in the levels of protein in each of the Esx-1-deficient strains as compared to the wild-type strain were considered significantly different from each other if the p value from a two-tailed Student’s t test was ≤0.05. Changes in protein levels between the wild-type and each Esx-1-deficient strain that were not statistically significant (p ≥ 0.05) were as follows: (A) The levels of EsxA in the cell lysates from the espI, eccD1, espJ, and espK strains were not statistically different from the levels in the wild-type strain. (B) The levels of EsxB in the cell lysates from the eccD1, espJ, and espK strains were not statistically different from the levels in the wild-type strain. (C) The levels of EspB in the cell lysates from the eccB1-2, eccCb, esxBA, esxA, eccD1, espK, and espL were not significantly different from the levels in the wild-type strain. (F) The levels of EsxN in the lysates from the ΔRD1, espG1, eccB1-2, espI, eccD1, espJ, espK, espL, and espB strains were not statistically different from the wild-type strain. The levels of EsxN in the culture filtrate generated from the eccCb, eccD1, and espJ strains were not statistically different from the WT strain. The actual p values are listed in the Supporting Information (Table S2). Log2 transformed versions of the CF data are available in Supplemental Figure S4.
Relative to WT M. marinum, the levels of EsxA and EsxB in the CL were significantly reduced in the majority of Esx-1-deficient M. marinum strains. The levels of EsxA were comparable to WT M. marinum in strains bearing transposon insertions in the eccD1, espJ, and espK genes. The decreased level of EsxA in the espI::Tn strain was not significantly different from that in WT M. marinum (p = 0.0777). The levels of EsxB in the eccCb::Tn, espJ::Tn, and espK::Tn strains were not significantly different from that in WT M. marinum. EsxB, but not EsxA, significantly accumulated in the eccD1::Tn strain relative to the wild-type strain (p = 0.0014 and 0.9782 for EsxB and EsxA, respectively, as compared to WT M. marinum). We conclude from this subset of the data that for detecting relative levels of EsxA and EsxB in the CL, the nLC–MRM-MS analysis was more robust at measuring changes in protein levels than Western blot analysis. Moreover, to our knowledge, this is the first report of accumulation of the EsxB substrate in the absence of Esx-1 export.
Esx-1 substrates are defined as proteins that are present in the CF in an Esx-1-dependent manner.34 We expected to measure decreased levels of substrates in the CF fractions generated from the Esx-1-deficient strains. Consistent with previous reports, we found that the levels of EsxA and EsxB from M. marinum were significantly decreased in the CF protein fractions from the Esx-1-deficient strains as compared to those generated from WT M. marinum (Figure 2A and B; EsxB p ≤ 0.0007, EsxA p ≤ 0.0277; Supplemental Table S2). Specific disruption of the eccD1 gene by transposon insertion abrogated the export of both EsxB and EsxA. We conclude from these data that, as in M. tuberculosis, the eccD1 gene in M. marinum is required for Esx-1 secretion.
It was previously reported by Western blot analysis that disruption of espK did not affect the secretion of EsxB.25 The levels of EsxA and EsxB secretion from the espJ and espK Tn-insertion strains were intermediate and significantly different from the levels of EsxA and EsxB in the CFs generated from both WT M. marinum and the eccD1::Tn strains (for EsxB, eccD1 vs espKp = 0.0011; eccD1 vs espJp = 0.0045). Note that transposon insertions in the espK and espJ genes resulted in a loss of EspK and EspJ proteins, respectively (Figure 4). The intermediate secretion phenotype cannot be attributed to intermediate or residual levels of functional EspK or EspJ proteins in the M. marinum strains. To our knowledge this is the first report of an intermediate secretion phenotype for the export of EsxA and EsxB into the bacteriological media during mycobacterial growth in vitro. The biological significance of intermediate secretion is not completely known. However, all of the Esx-1-deficient strains used in this study, including the espK strain,25 are attenuated for virulence. Thus, it is interesting to consider that ∼2-fold reductions in the levels of known Esx-1 substrates in the CF may reflect attenuation of M. marinum strains.25,37
Figure 4.
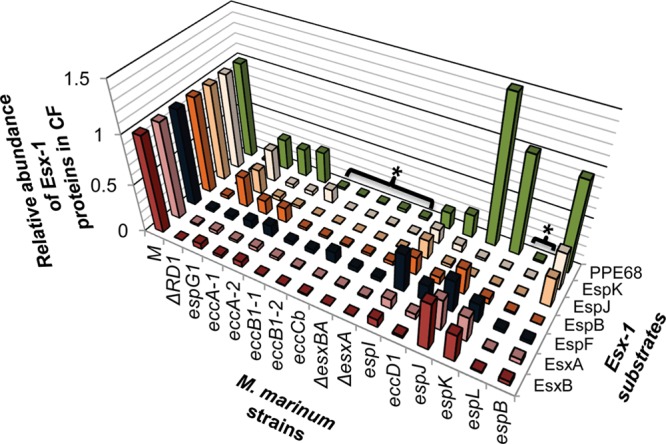
Divergent genetic requirements for substrate export. The graph summarizes the findings presented in Figures 2 and 3. The levels of secreted proteins present in the CF of the Esx-1-deficient strains relative to the WT strain illustrate divergent genetic requirements for substrate export. Brackets with asterisks highlight strains deficient for export of all of the substrates tested, similar to the ΔRD1 strain.
Two additional Esx-1 substrates encoded by the esx-1 locus in M. marinum include EspB and EspF.17,19,25,41 We conducted a similar analysis of the relative levels across Esx-1-deficient strains and WT M. marinum for the EspB and EspF substrates (Figure 2C and D, Supplemental Figure S4). EspB was absent from the CL and CF fractions generated from the espB::Tn strain, strengthening the specificity of our approach. The levels of EspB in the CL generated from the Esx-1-deficient strains were generally comparable to the WT M. marinum, with the exception of the strains with disruptions in espG1, eccA1, and espI, and the ΔRD1 strain (Figure 2C; for these strains, p ≤ 0.0203; Supplemental Table S2). EspB required EccCb and EspK for export into the CF as previously reported.25,41 We found that disruption of eccB1, eccD1, and espL also prevented the export of EspB into the CF, expanding the requirements for EspB export. For the strains with insertions in espG1, espJ, and espI, intermediate levels of EspB were detected in the CF. Interestingly, in previous reports, EspB was not detectable in the CL or CF generated from the eccA1::Tn strains using Western blot analysis.25 Here we observed reduced but detectable levels of EspB in the CL generated from the eccA1::Tn strains, reflecting the sensitivity of this approach. Finally, we found that deletions in the esxA and esxB genes resulted in a loss of EspB export into the CF. These findings agree with those published by Chen et al., which indicate that the export of EsxBA is required for the export of EspB.78
EspF is actively exported from M. marinum.17,19 The levels of EspF were significantly reduced in the CL and CF fractions generated from every Esx-1-deficient strain as compared to the WT M. marinum (Figure 2D, p ≤ 0.0093 for CF and p ≤ 0.0024; Supplemental Table S2). These data indicate that EspF is distinct from the other known substrates in that it is decreased in the absence of a functional Esx-1 system. These results were consistent with our original finding of reduced EspF stability in multiple Esx-1-deficient strains.17
To control the above experiment for changes in general secretion, we next compared the relative levels of MMAR_2929, a secreted protein with a Sec signal sequence, and EsxN, a substrate of the Esx-5 export system, across the WT and Esx-1-deficient M. marinum strains. The Sec secretion system and the Esx-5 system are thought to be independent of the Esx-1 secretion system.44,62,79 The data from MMAR_2929 and EsxN are shown in Figure 2E and F, respectively. We measured significant changes (both increases and decreases) in the levels of MMAR_2929 in the CL and CF fractions generated across the Esx-1-deficient strains as compared to the WT strain (Figure 2E, Supplemental Figure S4). However, compared to EsxA and EsxB (Figure 2A and B), the levels of MMAR_2929 in the CF were decreased only in strains where the levels in the CL were also decreased. Significantly lower levels of EsxN were secreted into the CF when transposon insertion or deletion disrupted the esx-1 in several cases (Figure 2D, Supplemental Figure S4). However, as compared to EsxA and EsxB, none of the disruptions within the esx-1 locus completely abrogated EsxN secretion into the CF. One possible interpretation is that the Esx-1 and Esx-5 systems share genes not included in this study. If the two systems were linked by as of yet-undefined genes, disruption of Esx-1 secretion could lead to decreased EsxN secretion. Alternatively, these results may be statistically significant due to the sensitive nature of our approach, but not biologically relevant. In support of this idea, the export of MMAR_2929 and EsxN do not correlate with the known and additional Esx-1 substrates presented here, as we illustrate below.
From these data, we conclude that we have established a novel assay that accurately and sensitively measures Esx-1-mediated substrate secretion into the bacteriological media in vitro. We found that the nLC–MRM assay recapitulates most of the published observations about Esx-1 substrate stability, as well as the independence of Esx-1 on the general Sec pathway. In doing so, we identified several intermediate Esx-1 secretion defects as a result of disruption of the esx-1 locus. The intermediate secretion defects were widely missed using standard Western blot analysis but commonly observed using the nLC–MRM-MS approach. Importantly, this assay is scalable based on the design of the normalization controls. As new Esx-1-deficient strains are identified, the levels of the proteins of the existing target list can be measured and compared to the data presented here without regenerating the entire data set. Finally, we contribute the first single-data-set quantitative assessment of the changes in both the levels and secretion of the Esx-1 substrates into the bacteriological media in vitro.
Phenotypic Profiling and Correlation Predict Potential Esx-1 Substrates
We postulated that the levels of Esx-1 substrates should statistically correlate across CL and CF fractions generated from the strain collection in this study because they all require the Esx-1 system for export. To test this idea, we calculated the Pearson’s correlation and measured the significance by fitting the data to a linear model to determine if the levels of Esx-1-associated proteins in the CL and CF fractions significantly correlated across the collection of M. marinum strains. The data from this analysis are presented as Table 1. Accordingly, we found that the known substrates, EspF, EspB, EsxA, and EsxB, all significantly correlated with each other in the CF (Table 1, r ≥ 0.9275). Conversely, proteins in the CF as a result of lysis or the active secretion by a non-Esx-1 system did not significantly correlate with Esx-1 substrates. Rather, proteins secreted independently of Esx-1, including MMAR_2929 and EsxN, correlated with the presence of known cytoplasmic or membrane components of the Esx-1 system (for example, EccA1) in the CF, which were detected due to a combination of cell lysis and the high sensitivity of this approach.
Table 1. Levels of ESX-1 Substrates Correlate in the Culture Filtratea.
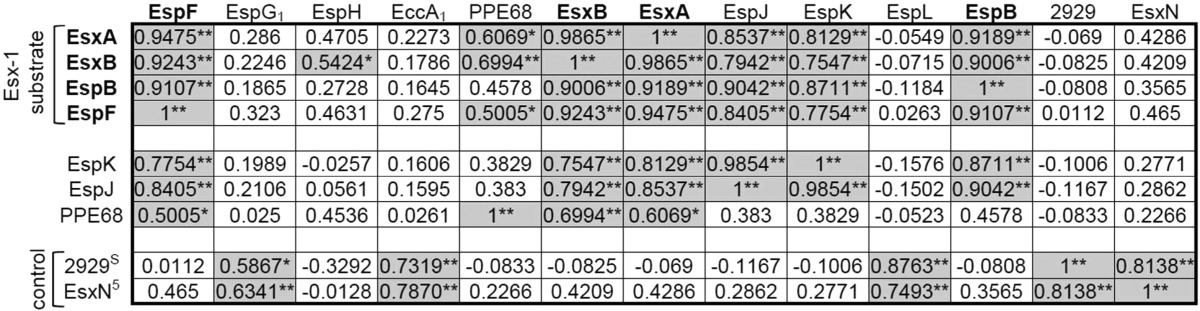
Pearson’s correlation coefficients (r) were calculated using the levels of Esx-1-associated and control (EsxN and MMAR_2929) proteins in the culture filtrate protein fractions. The significance of the correlation coefficients was measured on the basis of p values computed by fitting a linear model. (* p ≤ 0.05; ** p ≤ 0.01, colored grey). All r and p values can be found in Supplemental Figure S3. Bolded proteins are published Esx-1 substrates. 5 and S superscripts refer to proteins secreted by the Esx-5 and Sec secretion systems, respectively. Note that Esx-1 substrates correlated only with other substrates. The control proteins (MMAR_2929 and EsxN) did not significantly correlate with Esx-1 substrates in the CF.
We further reasoned that statistical correlation with known substrates in CF could identify Esx-1 substrates based on correlation-clustering. In the CF, three additional Esx-1-associated proteins, EspJ, EspK and PPE68, significantly correlated with the known Esx-1 substrates but not with the Esx-1-independent controls, MMAR_2929 and EsxN.
To confirm that EspK, EspJ, and PPE68 were Esx-1 substrates, we mined the nLC–MRM-MS experiment to determine the relative levels of EspK, EspJ and PPE68 in and secreted from WT M. marinum as compared to the Esx-1-deficient strains. The results of this analysis are shown in Figure 3. The ΔRD1 strain and the espK::Tn strain should be deficient for EspK and served as controls for specificity. On the basis of the absence of detectable EspK levels in these two strains, we validated the EspK measurements. EspK was also absent from the strain bearing the transposon insertion in espL, the gene transcribed immediately upstream of espK, indicating the two could be operonic (Figure 1). EspK was clearly detected in the CL and CF fractions generated from WT M. marinum (Figure 3A and Supplemental Figure S4). EspK levels were significantly reduced in CL generated from all Esx-1-deficient strains except the espG1::Tn strain (for all strains except espG1::Tn, p ≤ 0.0005 in the CL; for espG1::Tn, p = 0.6519). EspK levels in the CF were significantly reduced in all of the strains tested as compared to WT M. marinum (p ≤ 0.0001). From these results we conclude that the presence of EspK in the CF fraction was dependent upon Esx-1-associated genes, indicating that EspK is indeed an Esx-1 substrate. These findings expand upon Sani et al., who demonstrated that EspK was found in the capsule of M. marinum in an Esx-1-dependent manner.19 It is likely that in the study reported by McLaughlin et al. the addition of N- and C-terminal protein tags to EspK abrogated secretion by Esx-1.25 Our findings support the necessity of analyzing EspK and other Esx components without epitope tags or indirect reporters.
Figure 3.
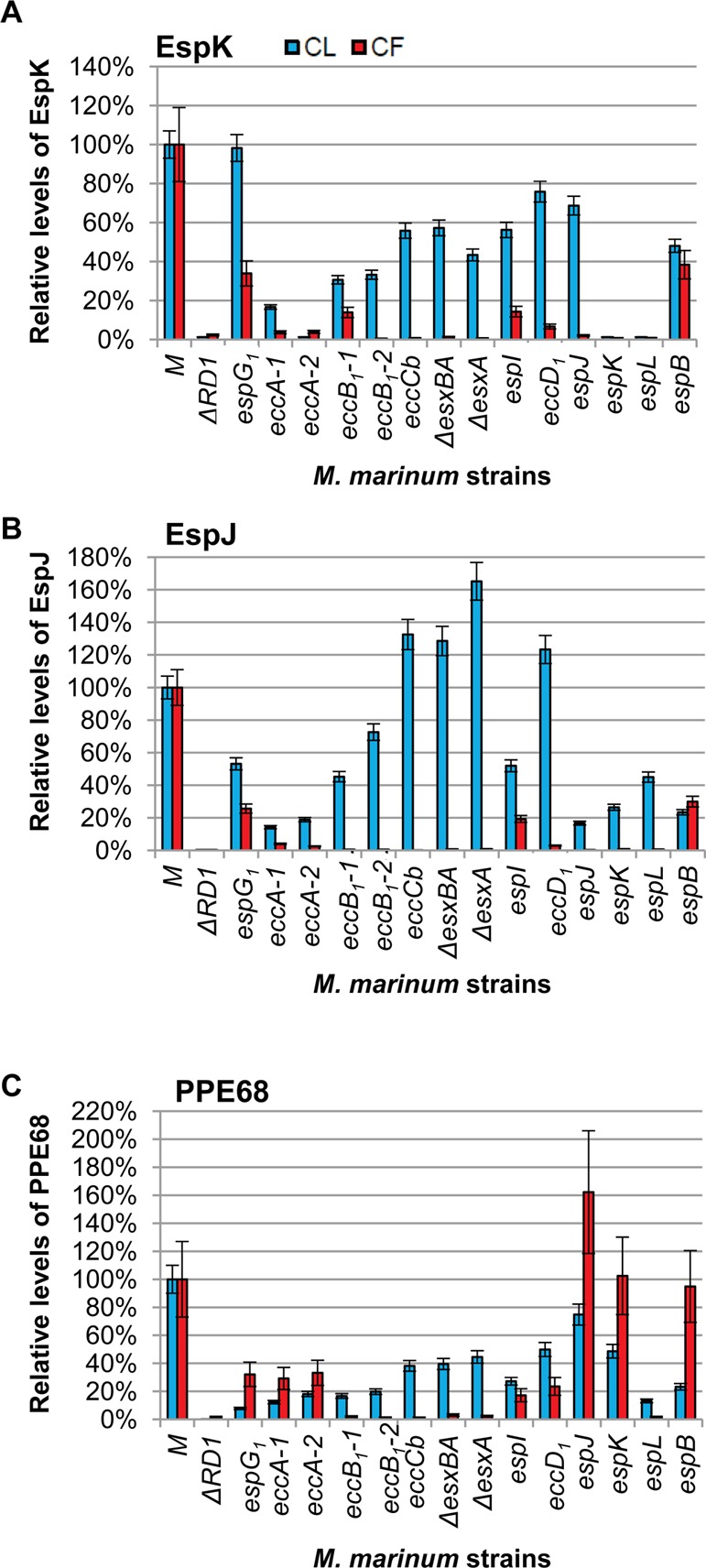
EspK, EspJ, and PPE68 are Esx-1 substrates. (A) nLC–MRM analysis of the levels of EspK in cell lysate (CL, blue bars) and culture filtrate (CF, red bars) relative to the levels of EspK in the WT M strain. The ΔRD1 and espK::Tn strains served as a negative control for EspK detection and demonstrate specificity of the approach. (B) nLC–MRM analysis of the levels of the EspJ substrate in the CL and CF. The ΔRD1 strain and espJ::Tn strain serve as a negative control for EspJ detection. We report a weak false positive signal indicating extremely low levels of EspJ peptides in the espJ::Tn strain. One EspJ peptide was observed at <0.5% of the levels in the espB::Tn strain and <0.2% of the EspJ levels observed in the WT M strain. This low false-positive signal was due to the large EspJ signal observed in the espB::Tn strain, which was the previous nLC injection on the mass spectrometer. For example, the EspJ tryptic peptide TSSMSTAADIYAK was present at ∼1e4cps2 in the M strain and was not detected in ΔRD1 or the espJ::Tn strains. AEPLAVDPAR is a high-intensity proteotypic peptide for EspJ that was measured at >2e6cps2 in the WT strain and 1.3e6cps2 in the espB::Tn strain, which was injected immediately prior to espJ::Tn analysis. Further evidence of this is the gradual reduction in carryover signal with each successive analysis of the espJ::Tn strain. (C) nLC–MRM analysis of the levels of PPE68 in the CL and CF. The ΔRD1 strain served as a negative control for PPE68 detection. Error bars represent the average propagated standard error and were calculated as described in the Experimental Procedures. The differences between the levels of each protein in the WT and each Esx-1-deficient strain were considered statistically significant if p values were less than 0.05 based on a two-tailed Student’s t test. The levels of the indicated protein in each Esx-1-deficient strain were significantly different from the wild-type levels with the following exceptions: The levels of EspK in the cell lysates of the espG strain were not different from the levels of EspK in the wild-type lysate. The levels of PPE68 in the cell lysate from the espJ strain were not significantly different from the wild-type strain. The levels of PPE68 in the culture filtrate generated from the espK strain were not significantly different from the levels of PPE68 in the culture filtrate from the wild-type strain. The actual p values are listed in Supplemental Table S2. Log2 transformed versions of the CF data are available in Supplemental Figure S4.
We readily detected EspJ in both CLs and CFs generated from WT M. marinum (Figure 3B). The espJ gene is absent in the ΔRD1 strain. As such, EspJ was not detectable in the CL from the ΔRD1 strain, indicating that we were specifically detecting EspJ by nLC–MRM. We report here extremely low levels of EspJ peptides in the espJ::Tn strain. The EspJ peptide was observed at <0.5% of the EspJ levels in the espB::Tn and <0.2% of the EspJ levels in the WT strain. This low false-positive signal was inconsistent with the genetics and was found due to the large EspJ signal observed specifically in the espB::Tn strain, which arbitrarily was the prior nLC injection on the mass spectrometer. This was the only case of measurable carryover we observed in the entire experiment (see Figure 3 legend for more detail). The levels of EspJ were otherwise significantly altered in all of the CL fractions for all Esx-1-deficient strains tested (p ≤ 0.0114) even considering this level of background. We interestingly observed a significant increase in the levels of EspJ in several strains indicating accumulation of EspJ in the mycobacterial cell (eccCb::Tn, ΔesxBA, ΔesxA and eccD1::Tn). For all mutant strains tested, the levels of EspJ were significantly decreased in the CF (p ≤ 0.0001). We have experimentally confirmed that EspJ is a substrate of the Esx-1 system, as predicted by Daleke et al. on the basis of the YXXXD/E motif.42
PPE68 was detected in the CL and CF from the M. marinum M strain (Figure 3C). The ppe68 gene is absent in the ΔRD1 strain (Figure 1). Accordingly, we did not detect the PPE68 protein in the CL or CF generated from the ΔRD1 strain, indicating that we were indeed measuring PPE68. PPE68 levels were significantly reduced in the CL derived from all of the Esx-1-deficient strains tested (p ≤ 0.0059) except the espJ::Tn strain (p = 0.3475). Interestingly, the levels of PPE68 in the CF were significantly reduced from all Esx-1-deficient strains (p ≤ 0.0393) as compared to the WT strain, except for the espK::Tn strain (p = 0.7889).
Two reports demonstrated that a protein related to PPE68 (MMAR_0186, PPE68_1) encoded outside of the esx-1 locus was an Esx-1 substrate.19,43 Although PPE68 and PPE68_1 are related proteins, the peptides used in this study to measure PPE68 are not conserved in PPE68_1, allowing us to distinguish between the two proteins. Here we demonstrated for the first time that the PPE68 protein encoded by the esx-1 locus was secreted or shed into the CF in an Esx-1-dependent manner.
Overall, we demonstrate the development of a proteomics-based workflow that successfully identified three additional Esx-1 substrates in M. marinum, whose genes are conserved in the human pathogen M. tuberculosis. We demonstrate that identifying proteins that significantly correlate with known Esx-1 substrates in the culture filtrate can identify novel Esx-1 substrates, which are key virulence factors in disease caused by mycobacterial and Gram-positive pathogens. This approach, while applied here to the Esx-1 system, is broadly applicable to any protein secretion system, many of which play direct roles in bacterial pathogenesis.
Effects of Esx-1 Deficiency on Substrate Export and Protein Levels
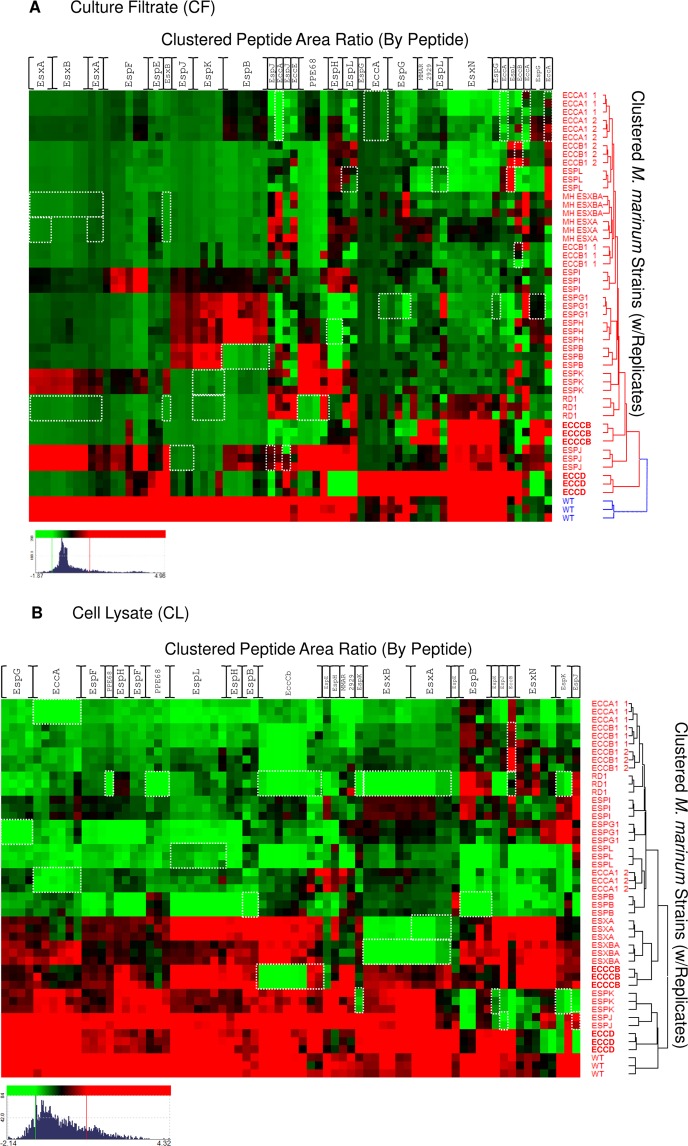
To further interpret the data we generated by this approach and to elucidate relationships between substrates, substrate codependency, and differences between Esx-1-deficient strains, we summarized our findings in two ways. First, we plotted the relative levels of the Esx-1 substrates in the CF from each strain to compare the genetic requirements for Esx-1 substrate export (Figure 4). Second, we performed a hierarchical clustering array of Esx-1-associated proteins across the Esx-1-deficient strains (Figure 5). The clustering of individual peptides (columns) vs strain (rows) as compared to clustering the average for each protein was performed to highlight potential differences in peptide clustering. Most Esx-1-associated peptides cluster together as a protein, such as EsxN, but some such as EspH and EspJ exhibit differential clustering depending on where in the protein (N- or C-terminus) the tryptic peptide was derived. One explanation for this is that the dynamic range of Esx-1 export magnifies the apparent clustering differences; however, there is some evidence of proteoforms of Esx-1 that may be biological or sample-handling-associated.89,90 Preserving the peptide-centric clustering of these substrates presents the possibility of teasing apart what, if any, contribution proteoform fragments play in the underlying biology.
Figure 5.
Hierarchical clustering of the Esx-1-associated proteome by peptide. Hierarchical clustering was performed on the quantitative peak area response of Esx-1-derived peptides vs WT and the Esx-1-deficient M. marinum strain library. Red intensity indicates higher expression, and green indicates lower expression vs the mean value for each gene product. Clustering was independently performed on nLC–MRM data from (A) culture filtrate (CF) and (B) cell lysate (CL) peak response. Threshold was set to 1S.D for color response. Displayed dynamic range for CF (Log2 −1.87–4.96 green–red) and CL (Log2 −2.14–4.32 green–red). Novel ESX-1 substrates PPE68, EspJ, and EspK are readily apparent from examination of the CF data, and loss of substrate stability in Esx-1 mutants is also well represented in CL fractions. White boxes indicate peptides that are absent due to interruption or loss of the gene that encodes the protein. For example, in the esxA deletion strains, the peptides corresponding to EsxA are boxed in white. Columns are listed for each protein for simplicity but represent individual peptide responses as in Supplemental Figure S1.
All of the substrates tested had an absolute requirement for EccCb, EccB1, EsxA, EsxB, and EspL for export (Figure 4, marked with an asterisk; see Supplemental Figure S5 for a model). The CF levels of Esx-1 substrates generated from these strains all resembled the CF levels of Esx-1 substrates generated from the ΔRD1 strain, which is deficient for the production of several Esx-1-associated proteins. Cluster analysis performed of the CF fraction at the protein and strain level indicated a relationship among all of these strains with the exception of the eccCb::Tn strain (Figure 5A). In the espL-deficient strain, all of the Esx-1-associated proteins measured in this study were also decreased in the CL as compared to WT M. marinum (Figure 5B). These proteins, including EsxA and EsxB, may form a core part of the Esx-1 apparatus that actively exports the known Esx-1 substrates.80 Alternatively, if there is a temporal order to Esx-1 substrate export, EsxA and EsxB may be required for the export of the other substrates because they are exported prior to other substrates.
Our findings indicate, consistent with previous findings, that EsxA and EsxB share export requirements.6,8,37 The levels of EsxA and EsxB significantly and substantially correlated across all M. marinum strains in the CF (r = 0.9865, p = 2.132 × 10–12). Both proteins were absent from the CF if any gene in the esx-1 region was disrupted or deleted (Figure 4, Figure 5A) with the exceptions of the espK and espJ genes. Likewise, both proteins were exported to intermediate levels in strains with disruptions in the espK or espJ genes (Figure 4, Figure 5A). This highly significant correlation likely reflects the molecular relationship between EsxA and EsxB; the two are encoded in an operon, directly interact and form a 1:1 dimer, and require each other for stability.8,81,82
Interestingly, the EspJ and EspK substrates shared genetic requirements for export into the CF. Both EspJ and EspK were absent from CFs generated from the majority of the strains in this study (Figures 4 and 5A). Disruption of the espG1, espB, and espI genes resulted in intermediate secretion phenotypes for both proteins. Consistent with the shared export requirements, the levels of EspJ in the CF highly and significantly correlated with EspK (r = 0.9854, p = 3.704 × 10–12; Table 1, Supplemental Figure S3). The correlation between EspJ and EspK was comparable to those measured for the EsxA and EsxB pair, indicating that there is likely a molecular relationship between EspJ and EspK that is yet undefined. The export requirements for the EspJ and EspK proteins differed from the EsxA and EsxB proteins, indicating that these substrates are not absolutely dependent upon each other for export. EspJ and EspK are not encoded by genes that can form an operon, so the shared requirements for export cannot simply be due to co-transcription (Figure 1). EspJ and EspK are both proteins of unknown function; EspK is classified as a proline- and alanine-rich protein. Interestingly, bioinformatics analysis revealed that while M. tuberculosis H37Rv has a single gene encoding espK and espJ, the M. marinum genome contains 3 additional espJ paralogs and 5 additional espK paralogs (Supplemental Table S3). The paralogs vary with respect to conservation with each other and with the M. tuberculosis orthologs. Moreover, for all of the espJ paralogs there are espK paralogs either adjacent or nearby. There is one orphan espK paralog. Possible explanations for both of the correlations we observed and for the genomic organization that require further testing include that EspK and EspJ interact or are secreted simultaneously (Supplemental Figure S5).
The EspB, EspF, and PPE68 substrates had distinct genetic requirements for export from the other Esx-1 substrates measured here (Figures 4 and 5A). Based on these comparisons, the PPE68 substrate was clearly divergent as compared to the other monitored substrates. For example, PPE68 did not require the EspK, EspJ, or EspB substrates for export (Figure 4). Instead, PPE68 levels were significantly increased in the CF protein fraction generated from the espJ::Tn strain (p = 0.0123). PPE68 levels correlated significantly with a subset of Esx-1 substrates in the CF, including EsxB, EsxA, and EspF (Table 1). The relative levels of EspF and PPE68 in the Esx-1-deficient strains were very similar in the CL across the strains tested in this study (r = 0.9456, p = 9.48 × 10–9), which likely represents a molecular relationship between the EspF and PPE68 proteins. Consistent with this idea, both EspF and PPE68 directly interact with the component EccA1 (Supplementary Figure S5).17,83
Our findings indicate that the codependency in both secretion and stability of Esx-1 substrates are not absolute. Indeed, several reports have indicated exceptions to codependent export. For example, EspFmt is not required for the secretion of EsxAmt and EsxBmt.53 Likewise, EspCmt is not required for the export of EspBmt.17,78 One possibility is that the different groups of substrates represent components that are not strictly necessary for secretion in vitro or are needed at later points in apparatus assembly or function. As the number of Esx-1 substrates grows, it is likely that the dependency on other substrates will be resolved.
Across all of our analyses, accumulation of Esx-1 substrates in the absence of Esx-1 function was rare. We uncovered only two examples of significant substrate accumulation (EsxB in the CL of the eccD1::Tn strain and EspJ in the CL of the eccCb::Tn, ΔesxBA and eccD1::Tn strains). In a recent report using nLC–MRM to study the Esx-3 substrates, EsxG and EsxH, these proteins failed to accumulate in the CL fractions generated from Esx-3-deficient M. smegmatis strains.57 These findings indicate that the levels of Esx substrates are likely regulated, and stability in the absence of export is rare.
Although the substrate secretion profile generated from some Esx-1-deficient strains mirrored that from the ΔRD1 strain (eccCb::Tn, eccB1::Tn, ΔesxBA, and espL::Tn, discussed above), Tn-insertions in other genes within the deletion in the ΔRD1 strain (espI, eccD1, espJ, and espK) and others in the esx-1 locus resulted in different substrate profiles in the CF (Figures 4 and 5A). These findings demonstrate that all genetic disruptions of the Esx-1 apparatus are not equal; disruption of a single gene or operon is not the same as deleting several Esx-1-associated genes. One possible interpretation of these data is that the Tn-insertions in some Esx-1-deficient strains result in an Esx-1 system that is largely intact, while others result in a lack of stability or assembly of the apparatus. Note that the espJ, espK, and eccD1 strains cluster closest to the WT strain when comparing levels of Esx-1-associated proteins in the cell lysate (Figure 5B). Moreover, the majority of the Esx-1-associated proteins measured from these strains were present at levels comparable to those of WT M. marinum (Figure 5B). In contrast, disruption of the several other Esx-1-associated genes resulted in a decrease in the relative levels in Esx-1 proteins in the cell (Figure 5B). The decreased levels of Esx-1 proteins were linked to disruption of several Esx-1 components (EccA1, EspL, EspG1, EspI, and EccB1) as well as the EspB substrate. EspL and EccB1 have been localized to or associated with the membrane.45,84−86 One possibility is that in the absence of EspL and EccB1, the Esx-1 membrane complex does not form or is destabilized. This has not been reported previously, as there has never before been an approach that could simultaneously quantitate changes to Esx-1 proteins at this scale.
EspG1, EccA1, EspI, and EspB were required for the export of some but not all of the substrates. It was recently reported that EspG1 functions as a chaperone for Esx-1.43 Indeed, disruption of espG1 led to decreased levels of all of the Esx-1 proteins measured in this study. By this analogy, several Esx-1 proteins may function as chaperones (Supplementary Figure S5). For example, EccA1 directly interacts with at least three known Esx-1 substrates, EspC, EspF, and PPE68,17,83 and shares AAA+ domains with chaperones.87,88 EccA1 may function to keep Esx-1 substrates in the properly folded or unfolded state for export. Because EspB is secreted, it is not a true chaperone. However, upon export EspB is digested by the MycP protease.25,52 It may be that in the absence of EspB the apparatus is unstable or that EspB is a secreted part of the apparatus.
Overall, the present study and subsequent analysis allowed us to expand the genetic requirements for export of four known Esx-1 substrates and identified three additional substrates encoded by the esx-1 locus. These findings set the stage for a future work aimed at understanding how these additional substrates promote virulence in M. marinum and M. tuberculosis. We defined rules of assigning new substrates to the Esx-1 system using this approach, which can be expanded as new Esx-1-deficient strains are isolated. Surprisingly, we identified several intermediate phenotypes of Esx-1 secretion that correlated with attenuation of virulence. Importantly, while the application of this approach was directed at Esx-1 export in M. marinum, these findings can directly be tested in the human pathogen M. tuberculosis. This method creates a framework that can be readily applied to studying protein secretion or any complex molecular pathway.
Acknowledgments
Research reported in this study was supported by the National Institute of Allergies and Infectious Diseases of the National Institutes of Health under award numbers R21AI092484 and R01AI106872 to P.A.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. E.A.W. is supported by a graduate student research fellowship funded by the Eck Institute for Global Health at the University of Notre Dame. R.S.P. is supported by the National Institutes of Health Chemistry-Biochemistry-Biology Interface Training Fellowship T32GM075762. We thank the Mass Spectrometry and Proteomics Facility and the Genomics and Bioinformatics Facility at the University of Notre Dame for their assistance. We thank members of the Champion Lab and Dr. Michael Ferdig for helpful discussion on data visualization, presentation, and statistical analysis.
Glossary
Abbreviations
- CL
culture lysate
- CF
culture filtrate
- MRM/SRM
multiple reaction monitoring, selected reaction monitoring
- cps
counts per second
- S/N
signal-to-noise
- nLC, UHPLC
nano/ultra high performance liquid chromatography
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- FDR
false discovery rate
- CV (%)
coefficient of variation (%)
- Esx
ESAT-6-system
- Wss
WXG-100
- Ecc
Esx conserved components
- Esp
Esx secretion-associated protein
- RD1
region of difference 1
- ESAT-6
early secreted antigenic target, 6 kDa
- CFP-10
culture filtrate protein, 10 kDa
- MOM
mycolate outer membrane
- BCG
Bacillus Calmette-Guérin
- TB
tuberculosis
- Tn
transposon
Supporting Information Available
Supplemental tables and figures as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- BCG Vaccine; World Health Organization: Geneva, 2009.
- Tuberculosis & Diabetes: Collaborative framework for care and control of tuberculosis and diabetes; World Health Organization: Geneva, 2011. [PubMed]
- WHO Global Tuberculosis Report 2013; World Health Organization: Geneva, 2013.
- Burts M. L.; Williams W. A.; DeBord K.; Missiakas D. M. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U.S.A. 2005, 10241169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garufi G.; Butler E.; Missiakas D. ESAT-6-like protein secretion in Bacillus anthracis. J. Bacteriol. 2008, 190217004–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn K. M.; Hickey M. J.; Mathur S. K.; Zakel K. L.; Grotzke J. E.; Lewinsohn D. M.; Smith S.; Sherman D. R. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2004, 512359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T.; Hingley-Wilson S. M.; Chen B.; Chen M.; Dai A. Z.; Morin P. M.; Marks C. B.; Padiyar J.; Goulding C.; Gingery M.; Eisenberg D.; Russell R. G.; Derrick S. C.; Collins F. M.; Morris S. L.; King C. H.; Jacobs W. R. Jr. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. U.S.A. 2003, 1002112420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A.; Raghavan S.; Hwang W. W.; Cox J. S. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. U.S.A. 2003, 1002213001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey Van Pittius N. C.; Gamieldien J.; Hide W.; Brown G. D.; Siezen R. J.; Beyers A. D. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001, 210RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P.; Seemann T.; Harrison P. F.; Jenkin G. A.; Davies J. K.; Johnson P. D.; Abdellah Z.; Arrowsmith C.; Chillingworth T.; Churcher C.; Clarke K.; Cronin A.; Davis P.; Goodhead I.; Holroyd N.; Jagels K.; Lord A.; Moule S.; Mungall K.; Norbertczak H.; Quail M. A.; Rabbinowitsch E.; Walker D.; White B.; Whitehead S.; Small P. L.; Brosch R.; Ramakrishnan L.; Fischbach M. A.; Parkhill J.; Cole S. T. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008, 185729–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpe San Roman S.; Facey P. D.; Fernandez-Martinez L.; Rodriguez C.; Vallin C.; Del Sol R.; Dyson P. A heterodimer of EsxA and EsxB is involved in sporulation and is secreted by a type VII secretion system in Streptomyces coelicolor. Microbiology 2010, 156Pt 61719–29. [DOI] [PubMed] [Google Scholar]
- Baptista C.; Barreto H. C.; Sao-Jose C. High levels of DegU-P activate an Esat-6-like secretion system in Bacillus subtilis. PLoS One 2013, 87e67840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse S. E.; Cox J. S. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J. Bacteriol. 2005, 18741238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros A.; Callahan B.; Battaglioli E.; Derbyshire K. M. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 2008, 694794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J. L.; Kowalski J. C.; Karnati P. K.; Derbyshire K. M. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. U.S.A. 2004, 1013412598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. A.; Krywy J. A.; Harold J.; Palumbo M. J.; Derbyshire K. M. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol. 2013, 117e1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion P. A.; Champion M. M.; Manzanillo P.; Cox J. S. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol. Microbiol. 2009, 735950–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S.; Manzanillo P.; Chan K.; Dovey C.; Cox J. S. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 2008, 4547205717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M.; Houben E. N.; Geurtsen J.; Pierson J.; de Punder K.; van Zon M.; Wever B.; Piersma S. R.; Jimenez C. R.; Daffe M.; Appelmelk B. J.; Bitter W.; van der Wel N.; Peters P. J. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010, 63e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet F. X.; Rasmussen P. B.; Rosenkrands I.; Andersen P.; Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 1998, 144Pt 113195–203. [DOI] [PubMed] [Google Scholar]
- Carlsson F.; Joshi S. A.; Rangell L.; Brown E. J. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 2009, 51e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune S. M.; Jaeger A.; Sarracino D. A.; Chase M. R.; Sassetti C. M.; Sherman D. R.; Bloom B. R.; Rubin E. J. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 2005, 1023010676–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinhikar A. G.; Verma I.; Chandra D.; Singh K. K.; Weldingh K.; Andersen P.; Hsu T.; Jacobs W. R. Jr.; Laal S. Potential role for ESAT6 in dissemination of M. tuberculosis via human lung epithelial cells. Mol. Microbiol. 2010, 75192–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majlessi L.; Brodin P.; Brosch R.; Rojas M. J.; Khun H.; Huerre M.; Cole S. T.; Leclerc C. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J. Immunol 2005, 17463570–9. [DOI] [PubMed] [Google Scholar]
- McLaughlin B.; Chon J. S.; MacGurn J. A.; Carlsson F.; Cheng T. L.; Cox J. S.; Brown E. J. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007, 38e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym A. S.; Brodin P.; Majlessi L.; Brosch R.; Demangel C.; Williams A.; Griffiths K. E.; Marchal G.; Leclerc C.; Cole S. T. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 2003, 95533–9. [DOI] [PubMed] [Google Scholar]
- Houben D.; Demangel C.; van Ingen J.; Perez J.; Baldeon L.; Abdallah A. M.; Caleechurn L.; Bottai D.; van Zon M.; de Punder K.; van der Laan T.; Kant A.; Bossers-de Vries R.; Willemsen P.; Bitter W.; van Soolingen D.; Brosch R.; van der Wel N.; Peters P. J. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell. Microbiol. 2012, 1481287–98. [DOI] [PubMed] [Google Scholar]
- Manzanillo P. S.; Shiloh M. U.; Portnoy D. A.; Cox J. S. Mycobacterium Tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 2012, 115469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A.; Cox J. S. Host-Pathogen Interactions During Mycobacterium tuberculosis infections. Curr. Top. Microbiol. Immunol. 2013, 374, 211–41. [DOI] [PubMed] [Google Scholar]
- van der Wel N.; Hava D.; Houben D.; Fluitsma D.; van Zon M.; Pierson J.; Brenner M.; Peters P. J. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 2007, 12971287–98. [DOI] [PubMed] [Google Scholar]
- Watson R. O.; Manzanillo P. S.; Cox J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012, 1504803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishai W.; Sullivan Z.; Bloom B. R.; Andersen P. Bettering BCG: a tough task for a TB vaccine?. Nat. Med. 2013, 194410–1. [DOI] [PubMed] [Google Scholar]
- Lalvani A.; Sridhar S.; Fordham von Reyn C. Tuberculosis vaccines: time to reset the paradigm?. Thorax 2013, 68121092–4. [DOI] [PubMed] [Google Scholar]
- Bitter W.; Houben E. N.; Bottai D.; Brodin P.; Brown E. J.; Cox J. S.; Derbyshire K.; Fortune S. M.; Gao L. Y.; Liu J.; Gey van Pittius N. C.; Pym A. S.; Rubin E. J.; Sherman D. R.; Cole S. T.; Brosch R. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009, 510e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J. A.; Raghavan S.; Stanley S. A.; Cox J. S. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol. Microbiol. 2005, 5761653–63. [DOI] [PubMed] [Google Scholar]
- Kapopoulou A.; Lew J. M.; Cole S. T. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinburgh) 2011, 9118–13. [DOI] [PubMed] [Google Scholar]
- Gao L. Y.; Guo S.; McLaughlin B.; Morisaki H.; Engel J. N.; Brown E. J. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 2004, 5361677–93. [DOI] [PubMed] [Google Scholar]
- Tan T.; Lee W. L.; Alexander D. C.; Grinstein S.; Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell. Microbiol 2006, 891417–29. [DOI] [PubMed] [Google Scholar]
- Tobin D. M.; Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell. Microbiol. 2008, 1051027–39. [DOI] [PubMed] [Google Scholar]
- Volkman H. E.; Clay H.; Beery D.; Chang J. C.; Sherman D. R.; Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004, 211e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Laine O.; Masciocchi M.; Manoranjan J.; Smith J.; Du S. J.; Edwards N.; Zhu X.; Fenselau C.; Gao L. Y. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol. Microbiol. 2007, 663787–800. [DOI] [PubMed] [Google Scholar]
- Daleke M. H.; Ummels R.; Bawono P.; Heringa J.; Vandenbroucke-Grauls C. M.; Luirink J.; Bitter W. General secretion signal for the mycobacterial type VII secretion pathway. Proc. Natl. Acad. Sci. U.S.A. 2012, 1092811342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke M. H.; van der Woude A. D.; Parret A. H.; Ummels R.; de Groot A. M.; Watson D.; Piersma S. R.; Jimenez C. R.; Luirink J.; Bitter W.; Houben E. N. Specific chaperones for the type VII protein secretion pathway. J. Biol. Chem. 2012, 2873831939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah A. M.; Gey van Pittius N. C.; Champion P. A.; Cox J.; Luirink J.; Vandenbroucke-Grauls C. M.; Appelmelk B. J.; Bitter W. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol 2007, 511883–91. [DOI] [PubMed] [Google Scholar]
- Houben E. N.; Bestebroer J.; Ummels R.; Wilson L.; Piersma S. R.; Jimenez C. R.; Ottenhoff T. H.; Luirink J.; Bitter W. Composition of the type VII secretion system membrane complex. Mol. Microbiol. 2012, 862474–84. [DOI] [PubMed] [Google Scholar]
- Joshi S. A.; Ball D. A.; Sun M. G.; Carlsson F.; Watkins B. Y.; Aggarwal N.; McCracken J. M.; Huynh K. K.; Brown E. J. EccA1, a component of the Mycobacterium marinum ESX-1 protein virulence factor secretion pathway, regulates mycolic acid lipid synthesis. Chem. Biol. 2012, 193372–80. [DOI] [PubMed] [Google Scholar]
- Champion M. M.; Williams E. A.; Kennedy G. M.; Champion P. A. Direct detection of bacterial protein secretion using whole colony proteomics. Mol. Cell. Proteomics 2012, 11, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop E. J.; Schipper T.; Huber S. K.; Nezhinsky A. E.; Verbeek F. J.; Gurcha S. S.; Besra G. S.; Vandenbroucke-Grauls C. M.; Bitter W.; van der Sar A. M. Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis. Models & Mech. 2011, 44526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G. M.; Hooley G. C.; Champion M. M.; Medie F. M.; Champion P. A. A novel ESX-1 locus reveals that surface associated ESX-1 substrates mediate virulence in Mycobacterium marinum. J. Bacteriol. 2014, 196101877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P.; Majlessi L.; Marsollier L.; de Jonge M. I.; Bottai D.; Demangel C.; Hinds J.; Neyrolles O.; Butcher P. D.; Leclerc C.; Cole S. T.; Brosch R. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 2006, 74188–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G. M.; Morisaki J. H.; Champion P. A. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba, Acanthamoeba castellanii. Appl. Environ. Microbiol. 2012, 862049–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol Y. M.; Goetz D. H.; Chan K.; Shiloh M. U.; Craik C. S.; Cox J. S. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 2010, 73210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottai D.; Majlessi L.; Simeone R.; Frigui W.; Laurent C.; Lenormand P.; Chen J.; Rosenkrands I.; Huerre M.; Leclerc C.; Cole S. T.; Brosch R. ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J. Infect. Dis. 2011, 20381155–64. [DOI] [PubMed] [Google Scholar]
- Garces A.; Atmakuri K.; Chase M. R.; Woodworth J. S.; Krastins B.; Rothchild A. C.; Ramsdell T. L.; Lopez M. F.; Behar S. M.; Sarracino D. A.; Fortune S. M. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog. 2010, 66e1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P.; de Jonge M. I.; Majlessi L.; Leclerc C.; Nilges M.; Cole S. T.; Brosch R. Functional analysis of ESAT-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex-formation, virulence and immunogenicity. J. Biol. Chem. 2005, 280, 33953–9. [DOI] [PubMed] [Google Scholar]
- Chen J. M.; Zhang M.; Rybniker J.; Basterra L.; Dhar N.; Tischler A. D.; Pojer F.; Cole S. T. Phenotypic Profiling of Mycobacterium tuberculosis EspA point-mutants reveals blockage of ESAT-6 and CFP-10 secretion in vitro does not always correlate with attenuation of virulence. J. Bacteriol. 2013, 195, 5421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M. S.; Steigedal M.; Ahmad R.; Mehra A.; Dragset M. S.; Schuster B. M.; Phillips J. A.; Carr S. A.; Rubin E. J. Mycobacterial ESX-3 requires multiple components for iron acquisition. mBio 2014, 53e1073–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R.; Burlingame A. L.; Bradshaw R. A. Western blots vs. SRM assays: Time to turn the tables?. Mol. Cell. Proteomics 2013, 1292381–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiere F.; Deutsch E. W.; King N. L.; Nesvizhskii A. I.; Mallick P.; Eng J.; Chen S.; Eddes J.; Loevenich S. N.; Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006, 34Database issueD655–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert O. T.; Mouritsen J.; Ludwig C.; Rost H. L.; Rosenberger G.; Arthur P. K.; Claassen M.; Campbell D. S.; Sun Z.; Farrah T.; Gengenbacher M.; Maiolica A.; Kaufmann S. H.; Moritz R. L.; Aebersold R. The M.tb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 2013, 135602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnot C.; Zorgani M. A.; Astruc T.; Desvaux M. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 2013, 4, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion P. A.; Cox J. S. Protein secretion systems in Mycobacteria. Cell. Microbiol. 2007, 961376–84. [DOI] [PubMed] [Google Scholar]
- Gao L. Y.; Groger R.; Cox J. S.; Beverley S. M.; Lawson E. H.; Brown E. J. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 2003, 712922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surinova S.; Huttenhain R.; Chang C. Y.; Espona L.; Vitek O.; Aebersold R. Automated selected reaction monitoring data analysis workflow for large-scale targeted proteomic studies. Nat. Protoc. 2013, 881602–19. [DOI] [PubMed] [Google Scholar]
- Ludwig C.; Claassen M.; Schmidt A.; Aebersold R. Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol. Cell. Proteomics 2012, 113M111 013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llarrull L. I.; Toth M.; Champion M. M.; Mobashery S. Activation of BlaR1 protein of methicillin-resistant Staphylococcus aureus, its proteolytic processing, and recovery from induction of resistance. J. Biol. Chem. 2011, 2864438148–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Champion M. M.; Sun L.; Champion P. A.; Wojcik R.; Dovichi N. J. Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry as an alternative proteomics platform to ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry for samples of intermediate complexity. Anal. Chem. 2012, 8431617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2007, 604, 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 43207–14. [DOI] [PubMed] [Google Scholar]
- Picotti P.; Clement-Ziza M.; Lam H.; Campbell D. S.; Schmidt A.; Deutsch E. W.; Rost H.; Sun Z.; Rinner O.; Reiter L.; Shen Q.; Michaelson J. J.; Frei A.; Alberti S.; Kusebauch U.; Wollscheid B.; Moritz R. L.; Beyer A.; Aebersold R. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 2013, 4947436266–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund S. S.; Bonar N. A.; Champion M. M.; Ghazi J. P.; Houk C. M.; Leming M. T.; Syed Z.; Duffield G. E. Daily rhythms in antennal protein and olfactory sensitivity in the malaria mosquito Anopheles gambiae. Sci. Rep. 2013, 3, 2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Wojcik R.; Dovichi N. J.; Champion M. M. Quantitative multiple reaction monitoring of peptide abundance introduced via a capillary zone electrophoresis-electrospray interface. Anal. Chem. 2012, 84146116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion P. A.; Stanley S. A.; Champion M. M.; Brown E. J.; Cox J. S. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 2006, 31357931632–6. [DOI] [PubMed] [Google Scholar]
- Gerber S. A.; Rush J.; Stemman O.; Kirschner M. W.; Gygi S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 2003, 100126940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.; Gordish-Dressman H.; Hoffman E. P. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 2006, 227808–14. [DOI] [PubMed] [Google Scholar]
- Sancak Y.; Markhard A. L.; Kitami T.; Kovacs-Bogdan E.; Kamer K. J.; Udeshi N. D.; Carr S. A.; Chaudhuri D.; Clapham D. E.; Li A. A.; Calvo S. E.; Goldberger O.; Mootha V. K. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 2013, 34261641379–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette M. A.; Carr S. A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 2013, 10128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. M.; Zhang M.; Rybniker J.; Boy-Rottger S.; Dhar N.; Pojer F.; Cole S. T. Mycobacterium tuberculosis EspB binds phospholipids and mediates EsxA-independent virulence. Mol. Microbiol. 2013, 8961154–66. [DOI] [PubMed] [Google Scholar]
- Ligon L. S.; Hayden J. D.; Braunstein M. The ins and outs of Mycobacterium tuberculosis protein export. Tuberculosis (Edinburgh) 2012, 922121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B.; Palmer T. Microbiology. Mycobacteria’s export strategy. Science 2006, 31357931583–4. [DOI] [PubMed] [Google Scholar]
- Renshaw P. S.; Lightbody K. L.; Veverka V.; Muskett F. W.; Kelly G.; Frenkiel T. A.; Gordon S. V.; Hewinson R. G.; Burke B.; Norman J.; Williamson R. A.; Carr M. D. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 2005, 24142491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw P. S.; Panagiotidou P.; Whelan A.; Gordon S. V.; Hewinson R. G.; Williamson R. A.; Carr M. D. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 2002, 2772421598–603. [DOI] [PubMed] [Google Scholar]
- Teutschbein J.; Schumann G.; Mollmann U.; Grabley S.; Cole S. T.; Munder T. A protein linkage map of the ESAT-6 secretion system 1 (ESX-1) of Mycobacterium tuberculosis. Microbiol. Res. 2009, 1643253–9. [DOI] [PubMed] [Google Scholar]
- de Souza G. A.; Leversen N. A.; Malen H.; Wiker H. G. Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. J. Proteomics 2011, 752502–10. [DOI] [PubMed] [Google Scholar]
- Gu S.; Chen J.; Dobos K. M.; Bradbury E. M.; Belisle J. T.; Chen X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 2003, 2121284–96. [DOI] [PubMed] [Google Scholar]
- Malen H.; Berven F. S.; Fladmark K. E.; Wiker H. G. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 2007, 7101702–18. [DOI] [PubMed] [Google Scholar]
- Neuwald A. F.; Aravind L.; Spouge J. L.; Koonin E. V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999, 9127–43. [PubMed] [Google Scholar]
- Wagner J. M.; Evans T. J.; Korotkov K. V. Crystal structure of the N-terminal domain of EccA ATPase from the ESX-1 secretion system of Mycobacterium tuberculosis. Proteins 2013, 821159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Sun L.; Champion M. M.; Knierman M. D.; Dovichi N. J. Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry for top-down characterization of the Mycobacterium marinum secretome. Anal. Chem. 2014, 86104873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S.; Rosenkrands I.; Stein R.; Andersen P.; Kaufmann S. H.; Jungblut P. R. Analysis of protein species differentiation among mycobacterial low-Mr-secreted proteins by narrow pH range Immobiline gel 2-DE-MALDI-MS. J. Proteomics 2014, 97, 235–44. [DOI] [PubMed] [Google Scholar]
- R Core Team; R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; http://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.