Summary
Bacterial pathogens are exposed to toxic molecules inside the host and require efficient systems to form and maintain correct disulfide bonds for protein stability and function. The intracellular pathogen Francisella tularensis encodes a disulfide bond formation protein ortholog, DsbA, which previously was reported to be required for infection of macrophages and mice. However, the molecular mechanisms by which F. tularensis DsbA contributes to virulence are unknown. Here, we demonstrate that F. tularensis DsbA is a bifunctional protein that oxidizes and, more importantly, isomerizes complex disulfide connectivity in substrates. A single amino acid in the conserved cis-proline loop of the DsbA thioredoxin domain was shown to modulate both isomerase activity and F. tularensis virulence. Trapping experiments in F. tularensis identified over 50 F. tularensis DsbA substrates, including outer membrane proteins, virulence factors, and many hypothetical proteins. Six of these hypothetical proteins were randomly selected and deleted, revealing two novel proteins, FTL_1548 and FTL_1709, which are required for F. tularensis virulence. We propose that the extreme virulence of F. tularensis is partially due to the bifunctional nature of DsbA, that many of the newly-identified substrates are required for virulence, and that the development of future DsbA inhibitors could have broad anti-bacterial implications.
Keywords: Francisella tularensis, tularemia, oxidoreductase, isomerase
Introduction
The constant struggle between bacterial pathogens and their hosts ultimately determines health or disease. Host cells use a variety of strategies to thwart intracellular bacteria, including generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS) (Ray et al., 2009, Thi et al., 2012). Conversely, successful bacterial pathogens encode an array of defensive proteins, including superoxide dismutase (SOD) and catalase-peroxidase (Imlay, 2003). Despite these defensive mechanisms, the bacterial envelope, including surface-exposed outer membrane proteins (OMPs), is vulnerable to ROS- and RNS-induced damage (Imlay, 2003, Graves, 2012). Whereas misoxidized OMPs can be removed by mechanisms such as outer membrane vesiculation (Manning & Kuehn, 2013), bacterial oxidoreductase pathways repair damaged proteins in a much more efficient manner. Because many bacterial virulence factors require disulfide bonds for proper folding and function, oxidoreductase studies are important for understanding bacterial pathogenesis and developing novel therapeutics (Heras et al., 2009).
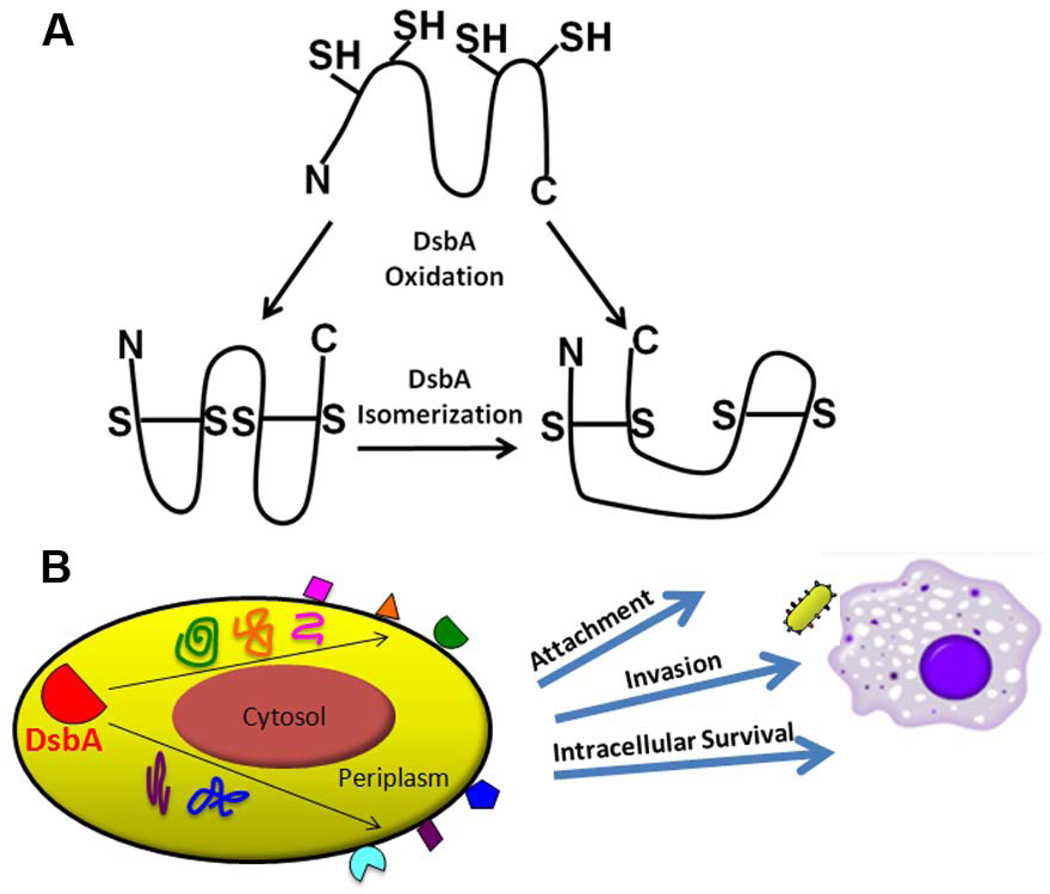
The oxidoreductase pathway has been well-studied in E. coli. Disulfide bond formation protein A, DsbA, forms disulfide bonds between consecutive cysteine residues in envelope proteins as they pass through the inner membrane translocation system (Bardwell et al., 1991, Kadokura & Beckwith, 2009, Berkmen et al., 2005). The E. coli DsbA (EcDsbA) structure includes a thioredoxin domain (Martin et al., 1993), with a pair of redox-active cysteines in a cysteine – any amino acid – any amino acid – cysteine (CXXC) motif and a cis-proline (cisPro) loop that are separated in the primary amino acid sequence but adjacent in the three-dimensional structure (Ren et al., 2009). DsbB is an inner membrane-bound quinone reductase that reoxidizes reduced DsbA via a disulfide bond exchange reaction (Bader et al., 1999). Because DsbA tends to misoxidize substrates that require non-consecutive disulfide bonds (Berkmen et al., 2005), the DsbC-DsbD pathway is responsible for editing mismatched disulfide bonds. Cytosolic electrons are transferred to inner membrane-bound DsbD, which then reduces DsbC (Katzen & Beckwith, 2000). DsbC functions as the major isomerase to correct mismatched disulfides either from DsbA misoxidation or external oxidative stress (Zapun et al., 1995).
Francisella tularensis is a Gram-negative bacterium that is the causative agent of the zoonotic disease tularemia. Due to its low infectious dose, multiple routes of infection, and high morbidity and mortality rates, F. tularensis is one of the most dangerous pathogens known (Dennis et al., 2001, Keim et al., 2007). F. tularensis recently was designated a Tier 1 Select Agent, highlighting that it is a severe threat to human health and has the potential to be used as a bioterrorism agent. Two F. tularensis subspecies are clinically significant: subsp. tularensis (Type A) and subsp. holarctica (Type B). Whereas Type A and B strains share 99% genomic sequence identity, they have distinct geographic distributions and virulence (Keim et al., 2007). Despite recent advances in F. tularensis research, little is understood about detailed mechanisms of F. tularensis virulence (Celli & Zahrt, 2013). Previous studies demonstrated that F. tularensis DsbA (FtDsbA; FTT_1103 in Type A strain SchuS4; FTL_1096 in Type B Live Vaccine Strain [LVS]) is an OMP required for intracellular replication and animal death (Huntley et al., 2007, Qin et al., 2009). FtDsbA, also referred to as FipB (Francisella infectivity potentiator protein B), is distinct from other bacterial DsbA orthologs in that it contains two putative domains: an amino-terminal Forskolin-binding protein-N (FKBP-N) dimerization domain found in macrophage infectivity potentiator (Mip) proteins and a carboxy-terminal DsbA-like domain (Qin et al., 2011). A very recent study reported that the FtDsbA/FipB amino-terminal Mip domain possesses isomerase activity but only in the presence of another F. tularensis Mip ortholog, FipA (FTT_1102) (Qin et al., 2014). In that same study, the FtDsbA/FipB amino-terminal Mip domain was found to be dispensable for intracellular replication and virulence in mice. Conversely, other studies have shown that the thioredoxin-like CXXC motif in the carboxy-terminal DsbA-like domain was required for virulence in macrophages and mice (Qin et al., 2011, Schmidt et al., 2013). Whereas many of those previous studies referred to FtDsbA as an essential virulence factor, the true role of FtDsbA in virulence remains obscure for a number of reasons. First, the well-characterized function of EcDsbA and other bacterial DsbA proteins in substrate disulfide bond formation (McMahon et al., 2014) suggests that FtDsbA performs a similar function and is not a direct virulence factor. Although a generally-accepted definition of ‘virulence factor’ does not exist (Casadevall & Pirofski, 2009), we presumed that FtDsbA does not directly interact with or damage the host and, thus, is not a true virulence factor. However, given that FtDsbA knockouts were deficient for virulence in mice and macrophages, we predicted that FtDsbA substrate identification would reveal any number of new virulence factors. Second, the difference between the periplasmic localization of EcDsbA (Bardwell et al., 1991) and outer membrane localization of FtDsbA (Huntley et al., 2007) indicated that FtDsbA might have unique functions or characteristics that confer its extreme virulence.
Here, we sought to fully characterize the molecular function(s) of FtDsbA, to assess if FtDsbA contributes to differences in Type A and Type B virulence, and to identify FtDsbA substrates. By comparing FtDsbA sequences from 26 Type A and Type B strains, we discovered a naturally-occurring amino acid polymorphism in the cisPro loop that modulates FtDsbA isomerase activity and F. tularensis virulence. More importantly, we generated a FtDsbA CXXS active site mutant to trap DsbA-substrate complexes in F. tularensis and identified over 50 FtDsbA substrates, including OMPs, virulence factors, and 25 hypothetical proteins. Using biochemical and genetic approaches, we verified that DsbA oxidizes disulfide bonds in these substrates and isomerizes mismatched disulfide bonds to promote fully-functional proteins. The results from this study clarify the role of FtDsbA in virulence, reveal that the cisPro motif modulates FtDsbA disulfide isomerase activity, and offer a new subset of F. tularensis virulence factors for future investigations.
Results
Amino acid polymorphism in FtDsbA cisPro-1 residue modulates virulence
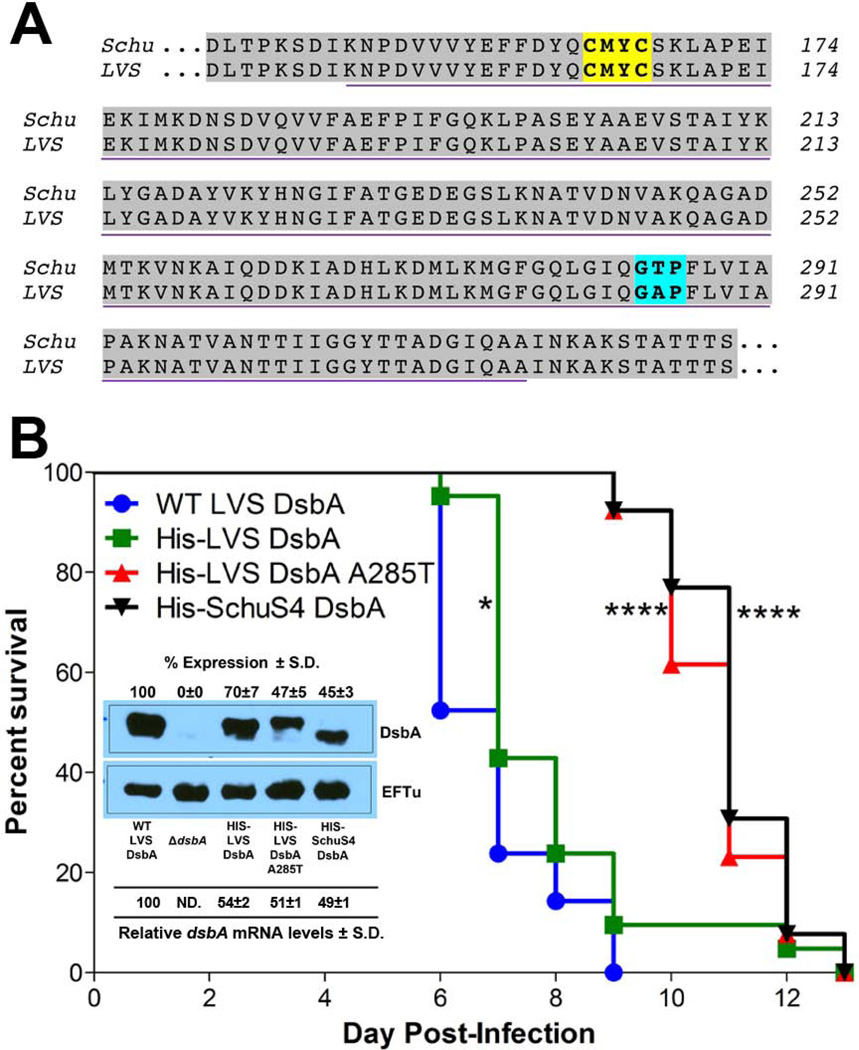
Previous studies have demonstrated that FtDsbA is required for F. tularensis virulence in mice and host cells, that the FtDsbA CXXC active site is required for virulence, and that FtDsbA possesses oxidoreductase activity (Qin et al., 2011, Straskova et al., 2009, Qin et al., 2009, Schmidt et al., 2013). However, major questions still remain about the detailed molecular mechanisms by which FtDsbA promotes virulence and if differences between Type A and Type B FtDsbA contribute to known differences in subsp. virulence. To begin to address these questions, Type B LVS DsbA (FTL_1096) and EcDsbA (YP_491589.1) sequences were compared to confirm that FtDsbA is a DsbA ortholog and to reveal thioredoxin-fold molecular features that may indicate function. Despite only 10% sequence identity, the carboxy-terminal portion of LVS DsbA encodes a DsbA-like domain, including a conserved CXXC active site motif and a putative cisPro motif (Gly-Ala-Pro; Figs. S1A and S1B). The thioredoxin cisPro motif and the amino acid immediately amino-terminal (cisPro-1) to the conserved proline were first described in E. coli and subsequent evidence has confirmed the importance of the cisPro-1 residue side chain in various thioredoxin-fold proteins, including DsbA and DsbC, affecting both redox properties and substrate interactions (Ren et al., 2009). Next, SchuS4 (FTT_1103) and LVS DsbA amino acid sequences were compared, demonstrating over 99% sequence identity between the C-terminal DsbA-like domains, including identical CMYC active site motifs (Fig. 1A). Interestingly, only one amino acid difference exists between SchuS4 and LVS DsbA-like domains and is located in the cisPro motif: Gly-Thr-Pro (GTP) in SchuS4 and Gly-Ala-Pro (GAP) in LVS (Fig. 1A). This cisPro-1 amino acid polymorphism suggested that SchuS4 and LVS DsbA may possess different redox properties and that FtDsbA may have different redox properties from EcDsbA, which encodes Gly-Val-Pro (GVP; Fig. S1A) (Quan et al., 2007). Importantly, the FtDsbA cisPro-1 amino acid polymorphism was not isolate-specific, as DsbA sequences for all available Type A and Type B F. tularensis genomes in the GenBank database were compared, demonstrating that all 15 sequenced Type A strains encoded GTP and all 9 sequenced Type B strains encoded GAP in the cisPro motif (Fig. S1C).
Fig. 1. FtDsbA cisPro-1 residue amino acid polymorphism modulates virulence.
A. Amino acid alignment of FtDsbA from SchuS4 (Type A strain) and LVS (Type B strain). The purple underline indicates the predicted DsbA domain. The yellow box indicates the predicted DsbA CXXC active site. The blue box indicates the predicted cisPro motif.
B. Groups of C3H/HeN female mice were intranasally-infected with 104 CFU of isogenic LVS strains containing either WT LVS DsbA (n=21), His-LVS DsbA (n=21), His-LVS DsbA A285T (n=13), or His-SchuS4 DsbA (n=13). Mean time-to-death differences were calculated by log-rank Mantel-Cox test (* P < 0.05 versus WT LVS DsbA; **** P < 0.0001 versus WT LVS DsbA). Inset top, analysis of WT and isogenic knock-in DsbA protein expression. Bacterial lysates from WT LVS, ΔdsbA strain, or ΔdsbA strains with isogenic knock-ins of either His-LVS DsbA, His-LVS A285T DsbA, or His-SchuS4 DsbA were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotting was performed using antiserum specific for either DsbA or EFTU (loading control; antibodies indicated to the right of each blot). Densitometry was used to quantitate DsbA expression levels in each isogenic knock-in strain, relative to WT (set to 100%), with % expression ± standard deviation (S.D.) noted on the top of the DsbA blot. Inset bottom, analysis of WT and isogenic knock-in dsbA mRNA levels by qRT-PCR. RNA was purified from overnight cultures grown on CDM agar. The relative % dsbA mRNA levels ± S.D. were normalized to RNA polymerase subunit α for each isogenic knock-in strain.
Previous studies demonstrated that both SchuS4 and LVS dsbA knockouts were avirulent in mice (Qin et al., 2009, Straskova et al., 2009). For the purposes of this study, we generated our own LVS ΔdsbA and confirmed that it was completely avirulent in mice (LD50 > 107), whereas wild-type (WT) LVS quickly killed all mice (0% survivors by day 5 post-infection; LD50 ~ 104; Fig. S2). To test whether the FtDsbA cisPro-1 polymorphism contributed to differences in F. tularensis virulence, we replaced the entire LVS dsbA (including amino-terminal Mip domain and carboxy-terminal DsbA domain) either with a C-terminal histidine-tagged (His-) LVS DsbA, His-LVS DsbA A285T (Type A cisPro-1 residue), or His-SchuS4 DsbA (all knock-in constructs included both the amino-terminal Mip domain and carboxy-terminal DsbA domain). Mouse infection experiments demonstrated that WT LVS DsbA and His-LVS DsbA had identical median time-to-death (7 days; Fig. 1B), despite two His-LVS DsbA-infected mice surviving through days 12 and 13 post-infection. Interestingly, infection of mice with either His-LVS DsbA A285T or His-SchuS4 DsbA resulted in identical median time-to-death (11 days; Fig 1B), which were both significantly delayed when compared with either WT LVS or His-LVS DsbA. These results were unexpected, given that Type A F. tularensis is generally-accepted to be more virulent than Type B F. tularensis. To assess possible replication defects in the LVS isogenic mutants due to genetic manipulation, we monitored the in vitro growth of each isogenic knock-in strain in liquid media but did not observe obvious growth defects (Fig. S3). Next, we examined DsbA protein expression of each isogenic knock-in strain by western blot, with elongation factor (EF)-Tu (FTL_1751) serving as a loading control. When compared with WT LVS DsbA protein expression (set to 100%), His-LVS DsbA (70±7% of WT), His-LVS A285T DsbA (47±5% of WT), and His-SchuS4 DsbA (45±3% of WT) exhibited significantly reduced levels of DsbA expression (Fig. 1B inset). To more accurately quantitate DsbA expression levels, we compared mRNA transcript levels from WT dsbA and each of the three dsbA knock-in variants by qRT-PCR, with RNA polymerase subunit α (FTL_0261) serving as the reference gene. qRT-PCR results indicated that dsbA transcripts in each of the knock-in strains (His-LVS DsbA, His-LVS A285T DsbA, and His-SchuS4 DsbA) was approximately half of WT dsbA (Fig. 1B inset). At this time, we are unable to completely explain why dsbA knock-in variants express DsbA at lower levels than WT, but suspect that: (1) the C-terminal histidine tag may affect RNA or protein stability; (2) nucleotide scars created both upstream and downstream of dsbA during knock-in generation may affect transcription; (3) the cisPro-1 residue may affect DsbA protein stability in F. tularensis, as indicated by decreased levels of DsbA in His-LVS A285T DsbA and His-SchuS4 DsbA versus WT and His-LVS DsbA. Regardless, differences in DsbA expression levels did not completely explain identical median time-to-death of WT DsbA and His-LVS DsbA (Fig. 1B). To test whether observed virulence differences might be recapitulated in a different F. tularensis strain background, we performed similar FtDsbA knock-in experiments in SchuS4, replacing SchuS4 dsbA with either LVS dsbA or SchuS4 dsbA T285A (Type B-like cisPro-1 residue). However, we did not observe any SchuS4 virulence differences in mice when comparing WT and dsbA knock-in strains (Fig. S4), likely due to the highly-infectious nature of SchuS4 (LD100 approx. 40 CFU). Although we cannot completely rule out that differences in DsbA expression contributed to differences in LVS virulence in mice (Fig. 1B), our results demonstrated that the DsbA cisPro-1 residue differs between Type A and Type B F. tularensis strains and that this single amino acid polymorphism significantly alters F. tularensis virulence in the host. Based on these findings, we speculated that inherent DsbA characteristics, such as enzymatic activity or redox properties, likely contributed to observed differences in mouse virulence.
FtDsbA oxidizes protein disulfide bonds
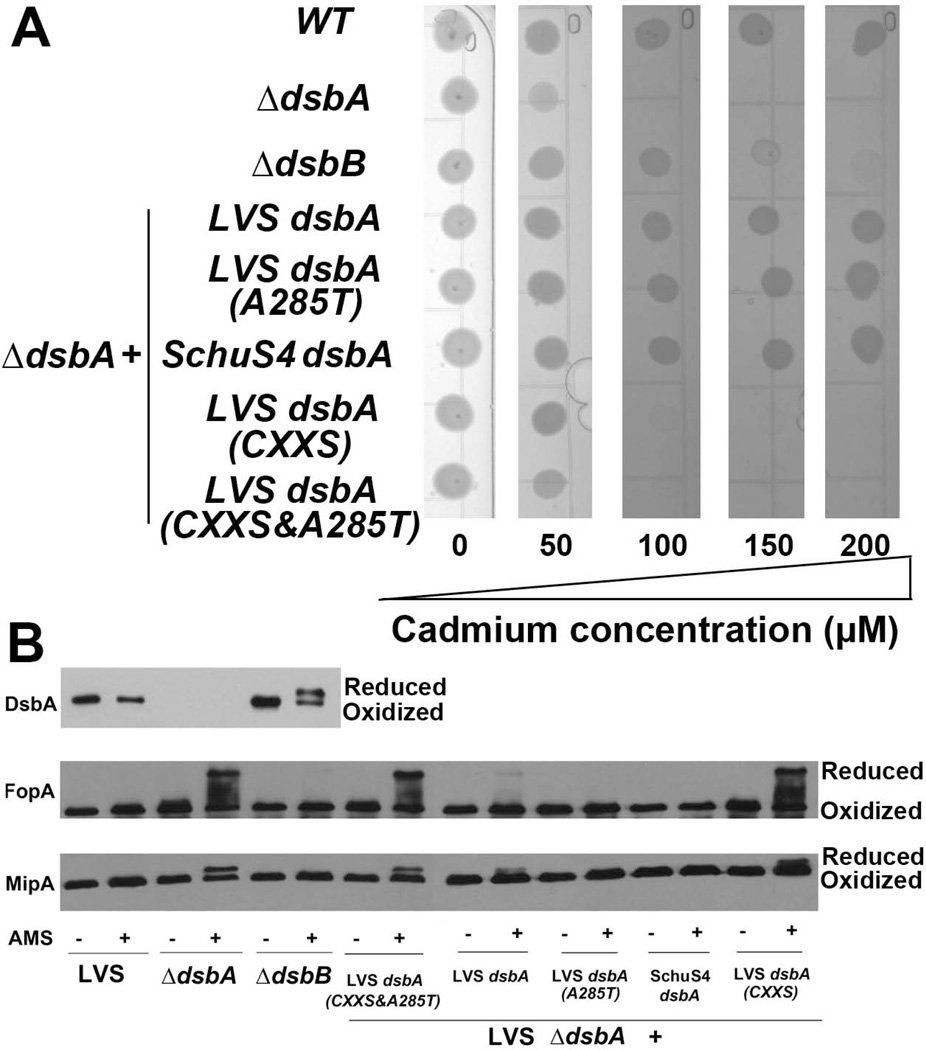
Previous studies reported oxidoreductase activity of FtDsbA (Straskova et al., 2009, Schmidt et al., 2013). In addition, although a F. tularensis DsbB ortholog was identified, its function was not assessed and its association with DsbA is still unknown (Qin et al., 2008). Analogous to the E. coli DsbA-DsbB pathway, we speculated that the F. tularensis DsbB ortholog functions as the upstream oxidizing partner of DsbA. Here, cadmium sensitivity was tested as an indicator of oxidase capacity in F. tularensis, as E. coli ΔdsbA previously was reported to be cadmium sensitive due to the high affinity of Cd2+ for protein free thiols (Vallee & Ulmer, 1972). WT LVS, LVS ΔdsbA, LVS ΔdsbB, or various LVS dsbA point mutants were grown in the presence of increasing concentrations of cadmium (0 – 200 µM). WT LVS was resistant to concentrations up to 200 µM cadmium, but both ΔdsbA and ΔdsbB were cadmium sensitive (Fig. 2A). Notably, ΔdsbA was three times more cadmium sensitive than ΔdsbB, similar to the cadmium phenotype previously reported for E. coli (Stafford et al., 1999). Complementation of ΔdsbA with LVS dsbA, LVS dsbA A285T, or SchuS4 dsbA equally restored cadmium resistance (Fig. 2A). However, complementing with LVS dsbA CXXS (active site mutant) or LVS dsbA CXXS&A285T (contains both a CXXS mutation and the Type A cisPro-1 residue) failed to restore cadmium resistance to WT levels (Fig. 2A). These results indicate that the CXXC active site, but not cisPro-1 residue, plays a major role in FtDsbA oxidase activity.
Fig. 2. FtDsbA oxidizes protein disulfide bonds.
F. tularensis WT LVS, ΔdsbA, ΔdsbB, or ΔdsbA complemented with either LVS dsbA, LVS dsbA A285T, SchuS4 dsbA, LVS dsbA CXXS, or LVS dsbA CXXS&A285T were tested in cadmium sensitivity assays (A) by spotting 106 CFU of each bacterial strain onto CDM containing increasing concentrations of cadmium (0 to 200 µM) and AMS trapping assays (B) to examine the redox status of DsbA and predicted DsbA OMP substrates FopA and MipA, following growth in liquid CDM. Bacteria were lysed and precipitated in TCA, incubated without (−) or with (+) 10 mg ml−1 AMS, separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotting was performed using antisera specific either for DsbA, FopA, or MipA, as noted in the left margin of each blot. Reduced and oxidized forms of each protein are noted in the right margin of each blot.
To examine the role of FtDsbA in OMP oxidation, we used 4-acetoamido-4’-maleimidylstilbene 2,2’-disulfonic acid (AMS) trapping to assess the redox status of FtDsbA and two predicted FtDsbA substrates, FopA and MipA. AMS covalently modifies free thiols, adding 500 Da/thiol to reduced proteins, thus retarding SDS-PAGE migration. FopA, the Francisella outer membrane protein A, and MipA, an ortholog of the Legionella pneumophila macrophage infectivity potentiator, each contain two cysteine residues, are known F. tularensis OMPs (Huntley et al., 2007), and both have been reported to play roles in F. tularensis virulence (Chong et al., 2013, Rasko et al., 2008). In AMS trapping assays, DsbA was fully oxidized in WT LVS but partially reduced in ΔdsbB (Fig. 2B), suggesting that DsbB oxidizes DsbA. In WT LVS and ΔdsbB, FopA and MipA were oxidized whereas in ΔdsbA, both FopA and MipA were partially reduced (Fig. 2B). Complementation of ΔdsbA either with LVS dsbA, LVS dsbA A285T, or SchuS4 dsbA restored oxidation of both FopA and MipA. However, complementation of ΔdsbA with either LVS dsbA CXXS or LVS dsbA CXXS&A285T failed to restore the full oxidation of either FopA or MipA. Together, these results demonstrate that Type A and Type B DsbA possess similar oxidase activities, the CXXC active site is required for FtDsbA oxidase activity, the cisPro-1 residue does not substantially affect FtDsbA oxidase activity, and oxidase activity may not explain the observed in vivo virulence differences between Type A and Type B cisPro-1 residues (Fig. 1B).
cisPro-1 residue controls FtDsbA isomerase activity and correlates with bacterial virulence
In E. coli, the DsbA-DsbB pathway oxidizes protein disulfide bonds, whereas the DsbD-DsbC pathway respectively reduces and isomerizes mismatched disulfide bonds due to DsbA misoxidation or external oxidative stress. Interestingly, many Gram-negative pathogens do not have a DsbC-DsbD pathway but, rather, encode multiple DsbA homologs to maintain high levels of disulfide bond oxidation (Kpadeh et al., 2013, Heras et al., 2009, Jameson-Lee et al., 2011, Tinsley et al., 2004). To better understand the F. tularensis Dsb system, we searched the LVS genome for Dsb orthologs but did not find obvious DsbC or DsbD orthologs. However, in addition to the FtDsbA ortholog (FTL_1096) used throughout this study, we discovered a second DsbA ortholog, FTL_1550, that contains both a CXXC active site motif and a predicted cisPro motif (Ile-Leu-Pro; Fig. S5A). We generated a ΔFTL_1550 strain and found that it was fully-virulent in mice (Fig. S5B). Additionally, we examined the redox status of the two predicted DsbA substrates described above, FopA and MipA, in a ΔFTL_1550 strain. Neither FopA nor MipA redox status was affected by ΔFTL_1550 (Fig. S5C), demonstrating that FTL_1550 does not oxidize FopA or MipA and indicating that FTL_1550 may not be involved in the oxidation of other substrates. In this same assay both FopA and MipA were partially reduced in a ΔdsbA strain (Fig. S5C), indicating that DsbA (FTL_1096) is the major oxidoreductase in F. tularensis. Although it is possible that FTL_1550 has a unique set of substrates (not tested here) or has low levels of intrinsic disulfide bond oxidoreductase activity, our results indicate that either FtDsbA (FTL_1096) compensates for FTL_1550 function or FTL_1550 and/or its substrates are not involved in F. tularensis virulence.
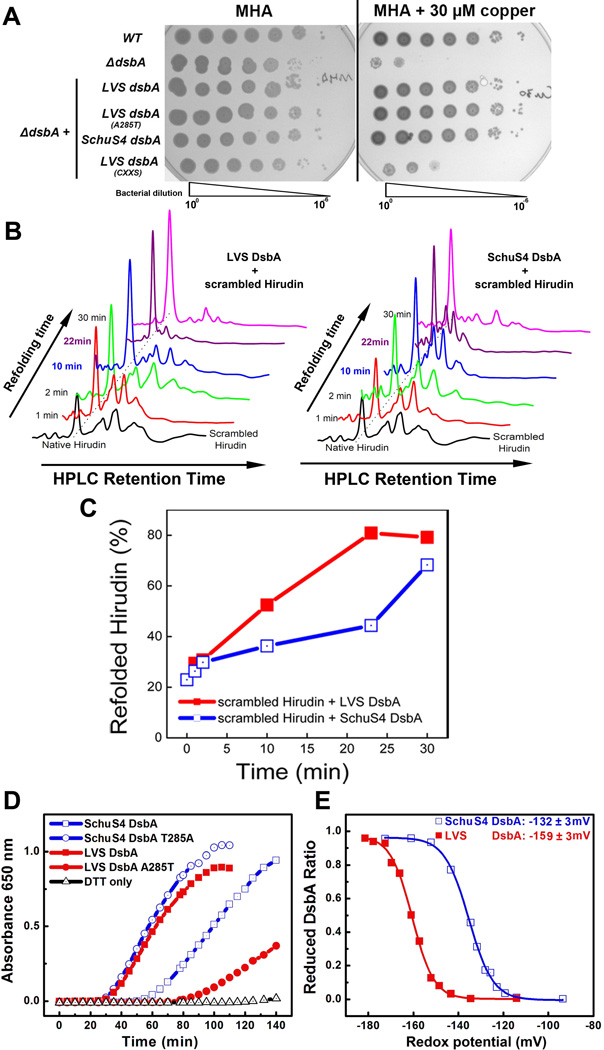
Given that F. tularensis appears to lack a DsbC-DsbD pathway and other functional DsbA orthologs, we hypothesized that FtDsbA could function both as a disulfide oxidoreductase and an isomerase. Another group very recently tested a similar hypothesis but concluded that FtDsbA/FipB isomerase activity was localized to the amino-terminal Mip domain and required another protein, FipA (FTT_1102), for full isomerase activity (Qin et al., 2014). To mimic the oxidative stress encountered during host infection and test if FtDsbA possessed isomerase activity, WT and ΔdsbA were grown in the presence of the redox-active metal copper to compare sensitivities. We decided to use copper for two primary reasons: First, copper rapidly and randomly oxidizes unpaired cysteines through a superoxide mechanism (Kachur et al., 1999, Matsui Lee et al., 2000) and E. coli DsbC isomerase activity previously was shown to be required for copper resistance (Hiniker et al., 2005). In that same study, E. coli ΔdsbA was as copper resistant as WT E. coli, demonstrating that EcDsbA plays a minor role in copper resistance (Hiniker et al., 2005); Second, F. tularensis likely is exposed to copper during mammalian infections as copper has been shown to be specifically released into phagosomes and enhances the bactericidal activity of infected macrophages (White et al., 2009). Here, WT LVS was copper resistant, ΔdsbA was extremely copper sensitive, and complementation with LVS dsbA, LVS dsbA A285T, or SchuS4 dsbA restored copper resistance (Fig. 3A). As expected, complementation with the LVS dsbA CXXS active site mutant did not restore copper resistance (Fig. 3A) because CXXC is required for F. tularensis oxidoreductase activity (Schmidt et al., 2013, Vecerkova et al., 2014). In reference to the second DsbA ortholog, FTL_1550, described above; FTL_1550 was found to be as copper resistant as WT, indicating that FTL_1550 does not affect F. tularensis isomerase activity (Fig. S5D). Whereas these results suggest that FtDsbA can isomerize mismatched disulfide bonds, limitations of the copper sensitivity assay prevented us from more quantitatively comparing isomerase activities between Type A and Type B DsbA or correlating isomerase activity with the cisPro-1 residue.
Fig. 3. FtDsbA cisPro-1 residue modulates disulfide isomerase activity.
A. Copper sensitivity assays were performed by spotting 106 CFU of WT LVS, LVS ΔdsbA, or ΔdsbA strains complemented with either LVS dsbA, LVS dsbA A285T, SchuS4 dsbA, or LVS dsbA CXXS onto MHA with or without 30 µM copper.
B. In vitro refolding of disulfide scrambled hirudin by LVS or SchuS4 DsbA. Equimolar amounts of reduced DsbA and scrambled hirudin were mixed, aliquots were removed at 1, 2, 10, 22, and 30 min (Y-axis), acid-quenched, and evaluated by 220 nm absorbance following reversed phase HPLC (X-axis). Presented data is representative of two independent experiments.
C. HPLC profile analysis of percent hirudin (from panel B) refolded by either LVS or SchuS4 DsbA versus time.
D. Insulin reductase activity was assayed for SchuS4 DsbA, LVS DsbA, SchuS4 T285A DsbA, and LVS A285T DsbA over a period of 140 min. Presented data is representative of three independent experiments.
E. FtDsbA redox potentials (mV) were measured by quantitating the ratios of reduced and oxidized LVS or SchuS4 DsbA following overnight incubation in degassed glutathione redox buffers. The calculated redox potential, ± standard deviation, for LVS DsbA was 159±3, and for SchuS4 DsbA was 132±3. Presented data is representative of three independent experiments.
To more accurately quantitate differences between Type A (SchuS4) and Type B (LVS) FtDsbA isomerase activities, we measured the conversion rate of scrambled hirudin, a 7-kDa protein containing 3 disulfide bonds, into its native disulfide-bonded conformation (Hiniker et al., 2007). Whereas LVS and SchuS4 DsbA isomerized equimolar amounts of hirudin by 30 min (Fig. 3B and C), LVS DsbA was more efficient at isomerizing hirudin at 10 min (Fig. 3C) and had isomerized a majority of the hirudin by 22 min (Fig. 3C). These results suggest that LVS DsbA is a more robust isomerase than SchuS4 DsbA. Given that the initial step of isomerizing mismatched disulfide bonds is disulfide bond reduction, we next compared disulfide reductase activity for LVS DsbA and SchuS4 DsbA using an insulin reduction assay. In agreement with the hirudin refolding assays, LVS DsbA exhibited stronger reductase activity than SchuS4 DsbA (Fig. 3D). Interestingly, FtDsbA reductase activity was directly correlated with the cisPro-1 residue, as LVS DsbA A285T (Type A cisPro-1 residue) exhibited weaker reductase activity than LVS DsbA (Fig. 3D), and SchuS4 DsbA T285A (Type B cisPro-1 residue) exhibited stronger reductase activity than SchuS4 DsbA (Fig. 3D). Because the cisPro-1 residue has been predicted to regulate the redox potential for many thioredoxin fold proteins (Ren et al., 2009), we next measured the redox potential of LVS and SchuS4 DsbAs using a glutathione redox equilibrium assay, demonstrating that LVS DsbA was more reducing (−159±3 mV; Fig. 3E) than SchuS4 DsbA (−132±3 mV; Fig. 3E). Compared to EcDsbA (−118 mV; (Wunderlich et al., 1993)), both Type A DsbA and Type B DsbA are more reducing; however, compared to E. coli thioredoxin (−270 mV; (Krause et al., 1991)) both Type A DsbA and Type B DsbA are much more oxidizing. These results suggest that the redox potential of FtDsbA, mediated by the cisPro-1 residue, carefully balances oxidase and reductase functions for optimal isomerase activity. Similar conclusions have been made for the human protein disulfide isomerase, PDI, which has a redox potential of −160 mV (Chambers et al., 2010). Overall, these results demonstrated that Type A and Type B FtDsbA isomerase activities (Fig. 3B and C) correlated with the Type A and Type B FtDsbA virulence differences observed in our mouse pulmonary infection model (Fig. 1B), suggesting that FtDsbA substrates in the LVS with complex disulfide bond connectivity patterns (i.e., more than three cysteines) heavily rely on efficient isomerase activity for correct folding and function.
Identification of FtDsbA substrates that are virulence factors
Whereas the above results revealed that FtDsbA is a bifunctional protein possessing both oxidase and isomerase activities, the connection between these functions and F. tularensis virulence still was unclear. Presumably, FtDsbA catalyzes disulfide bonds to correctly fold any number of substrates, including envelope proteins and virulence factors. To identify FtDsbA substrates in F. tularensis, we designed a molecular trapping assay using the LVS DsbA CXXS mutant described above to form intermolecular DsbA-substrate disulfide-bonded complexes. Based on previous reports noting that the CXXC active site motif of other thioredoxin superfamily members could be mutated to trap substrates (Depuydt et al., 2009, Motohashi et al., 2001, Denoncin et al., 2010, Sturm et al., 2009), we proposed that the FtDsbA CXXS mutant would be able to form inter-molecular disulfide bonds with substrates, but would lack the ability to release these substrates in the absence of a reducing agent such as 2-mercaptoethanol. In addition, robust Type B FtDsbA isomerase activity in in vitro assays (Fig. 3A, B, and C) suggested that there should be ample FtDsbA-substrate disulfide bond-linked complexes in bacterial lysates. FtDsbA-substrate complexes were affinity purified from LVS lysates, separated by non-reducing SDS-PAGE (Fig. S6), the DsbA-substrate complex lane was excised from the gel, and substrates were identified by mass spectrometry analyses. Two independent analyses were performed, with over 50 putative FtDsbA substrates shared between the two analyses (Table 1), including known OMPs and virulence factors. Roughly 80% (42/53) of the FtDsbA substrates contain at least 3 cysteines in their amino acid sequence (Table 1) and 20 of these substrates contain an odd number of cysteines (Table 1), indicating that they may require FtDsbA isomerase activity to form correct (i.e., non-consecutive) disulfide bond linkages.
Table 1.
F. tularensis LVS DsbA substrates identified by mass spectrometry
| Locusa | #Cysb | Protein namec | Accession | Localizationd | Size (kDa) |
Reference |
|---|---|---|---|---|---|---|
| 38 | 4 | HlyD family secretion protein | gi|89255485 | IM | 35 | |
| 93 | 10 | Chitinase | gi|89255533 | Peri | 80 | |
| 125 | 5 | Hypothetical protein | gi|89255562 | Unk | 120 | |
| 174 | 6 | Hypothetical protein | gi|89255608 | Unk | 17 | |
| 181 | 4 | Type IV pili protein | gi|89255615 | Unk | 15 | |
| 187 | 2 | Cyclohexadienyl dehydratase | gi|89255621 | Peri | 27 | |
| 207 | 2 | Pyrrolidone-carboxylate peptidase | gi|89255641 | Peri | 24 | |
| 325 | 4 | OmpA family protein | gi|89255751 | OM/Lipo | 46 | Huntley et al., 2007; Mahawar et al., 2012; Robertson et al., 2014 |
| 336 | 7 | Pal | gi|89255762 | OM/Lipo | 23 | Huntley et al., 2007 |
| 359 | 2 | Type IV pili protein | gi|89255780 | Unk | 33 | |
| 411 | 6 | Hypothetical protein | gi|89255822 | Unk | 32 | |
| 421 | 3 | Tul4-A | gi|89255832 | OM/Lipo | 16 | Huntley et al., 2007 |
| 424 | 3 | Tul4-B | gi|89255834 | OM/Lipo | 16 | Huntley et al., 2007 |
| 466 | 3 | Murein transglycosylase | gi|89255876 | Peri | 72 | |
| 493 | 20 | Hypothetical protein | gi|89255903 | Peri | 30 | |
| 574 | 2 | Hypothetical protein | gi|89255972 | OM | 51 | |
| 701 | 11 | FAD binding family protein | gi|89256090 | Peri | 65 | |
| 823 | 3 | Hypothetical protein | gi|89256186 | Unk | 14 | |
| 834 | 4 | Rhodanese-like protein | gi|89256194 | Unk | 27 | |
| 878 | 3 | DNA/RNA endonuclease family | gi|89256236 | Extracell | 39 | |
| 879 | 2 | Beta-lactamase | gi|89256237 | Peri | 32 | |
| 994 | 7 | Hypothetical protein | gi|89256338 | Unk | 55 | |
| 1029 | 4 | Type IV pili lipoprotein | gi|89256368 | Unk | 33 | |
| 1042 | 4 | MipA | gi|89256380 | OM/Lipo | 30 | Huntley et al., 2007; Rasko et al., 2008 |
| 1060 | 3 | D-alanyl-D-alanine carboxypeptidase | gi|89256395 | IM | 49 | |
| 1064 | 4 | Hypothetical protein | gi|89256399 | Unk | 30 | |
| 1105 | 2 | Hypothetical protein | gi|89256430 | Unk | 16 | |
| 1161 | 4 | PdpE/Hcp | gi|89256483 | Unk | 21 | Barker et al., 2009 |
| 1171 | 5 | PdpB/IcmF | gi|89256493 | Unk | 120 | de Bruin et al., 2011 |
| 1202 | 9 | Hypothetical protein | gi|89256523 | Unk | 36 | |
| 1286 | 3 | Hypothetical protein | gi|89256595 | Unk | 15 | |
| 1306 | 4 | DipA | gi|89256615 | OM | 39 | Chong et al., 2013 |
| 1328 | 2 | FopA | gi|89256632 | OM | 43 | Huntley et al., 2007; Chong et al., 2013 |
| 1363 | 1 | Hypothetical protein | gi|89256662 | Unk | 37 | |
| 1372 | 5 | Hypothetical lipoprotein | gi|89256671 | OM/Lipo | 49 | |
| 1521 | 7 | Chitinase family 18 protein | gi|89256806 | Unk | 84 | |
| 1532 | 3 | Hypothetical protein | gi|89256816 | Unk | 21 | |
| 1548 | 4 | Hypothetical protein | gi|89256832 | Unk | 29 | |
| 1570 | 5 | Phospholipase D | gi|89256847 | Unk | 45 | |
| 1578 | 2 | Hypothetical protein | gi|89256855 | Unk | 13 | |
| 1579 | 4 | Hypothetical protein | gi|89256856 | Unk | 26 | |
| 1581 | 8 | Hypothetical lipoprotein | gi|89256858 | OM/Lipo | 30 | |
| 1582 | 4 | Putrescine-binding protein | gi|89256859 | Peri | 43 | |
| 1629 | 4 | Hypothetical protein | gi|89256906 | Unk | 62 | |
| 1670 | 8 | DsbB | gi|89256943 | Unk | 18 | |
| 1678 | 3 | Hypothetical protein | gi|89256948 | Unk | 37 | |
| 1695 | 1 | Hypothetical protein | gi|89256964 | Unk | 29 | |
| 1709 | 4 | Hypothetical protein | gi|89256978 | Unk | 18 | |
| 1786 | 9 | Succinate dehydrogenase | gi|89257049 | IM | 66 | |
| 1793 | 11 | Hypothetical protein | gi|89257056 | Unk | 104 | |
| 1852 | 2 | Hypothetical protein | gi|89257112 | Unk | 15 | |
| 1896 | 5 | Hypothetical protein | gi|89257144 | Unk | 51 | |
| 1960 | 8 | Peptide methionine sulfoxide reductase | gi|89257196 | Unk | 25 |
F. tularensis LVS gene loci;
Number of cysteines in the predicted coding sequence;
Protein names in genome annotations or previous publications;
Localization predicted by PSORTb version 3.0.2 bacterial subcellular localization prediction program (http://www.psort.org), indicating inner membrane (IM), periplasmic (Peri), outer membrane (OM), or unknown (Unk) localization; lipoproteins (Lipo) predicted by LipoP 1.0 Server (http://www.cbs.dtu.dk/services/LipoP/); confirmed OM proteins are indicated in bold (OM) with relevant references.
Given that the purpose of this trapping analysis was to identify proteins that have a more direct role in virulence, we anticipated that previously-described OMP virulence factors containing cysteines, including FopA (Chong et al., 2013) and MipA (Rasko et al., 2008), would form complexes with DsbA. Indeed, the identification of FopA and MipA as FtDsbA substrates (Table 1) was consistent with our AMS trapping assays (Fig. 2B), confirming that FtDsbA directly oxidizes both proteins. Two additional known virulence factors, FTL_1306 and FTL_0325, were identified as DsbA substrates (Table 1), providing further evidence that FtDsbA is unlikely to be a virulence factor itself, but is required for isomerizing and catalyzing disulfide bonds in virulence factors. FTL_1306, designated as DipA, is a surface-exposed OMP that contains 4 cysteines and recently was shown to be required for in vitro and in vivo virulence of SchuS4 (Chong et al., 2013). The DipA cysteine distribution pattern is conserved between Type A and Type B strains, with the first cysteine (C1) predicted to serve as a lipo-cysteine, and the three remaining cysteines likely require FtDsbA isomerase activity for correct disulfide bond formation (Fig. 4C). FTL_0325, an OmpA-like protein, is an OMP that also contains 4 cysteines and has been reported to be required for in vitro and in vivo virulence in both LVS and SchuS4 (Mahawar et al., 2012, Robertson et al., 2014, Mahawar et al., 2013). FTL_0325 recently was shown to be a lipoprotein (Robertson et al., 2014), indicating that the first cysteine is a lipo-cysteine and the three remaining cysteines likely require FtDsbA isomerase activity for correct disulfide bond formation. Three additional OMPs also were identified as FtDsbA substrates, further validating the utility of our molecular trapping approach: Pal, Tul4-A, and Tul4-B (Table 1) (Huntley et al., 2007). Whereas Tul4-A was reported not to be essential for LVS virulence (Forestal et al., 2008), both Tul4 proteins are well-known to stimulate strong T cell responses that protect from lethal F. tularensis challenge (Ashtekar et al., 2012, Kaur et al., 2012, Valentino et al., 2009). Additional DsbA substrates include type IV pilus components FTL_0181, FTL_0359, and FTL_1029 (Table 1). Type IV pili are important for adherence to host cells by many pathogens (Giltner et al., 2012) and a previous study demonstrated that Type IV pili are a virulence determinant for F. tularensis (Salomonsson et al., 2011). Two proteins from the Francisella pathogenicity island (FPI), PdpE/Hcp (Barker et al., 2009) and PdpB/IcmF (de Bruin et al., 2011), also were identified as FtDsbA substrates (Table 1). The FPI consists of 16–19 ORFs, many of which are required for virulence in macrophages or mice, and is proposed to form a Type VI-like secretion system in Francisella (Broms et al., 2010). Finally, the identification of DsbB (Table 1) validates our above results suggesting that DsbB-DsbA constitute an oxidation pathway in F. tularensis (Fig. 2B).
Fig. 4. Identification of FtDsbA substrates that are virulence factors.
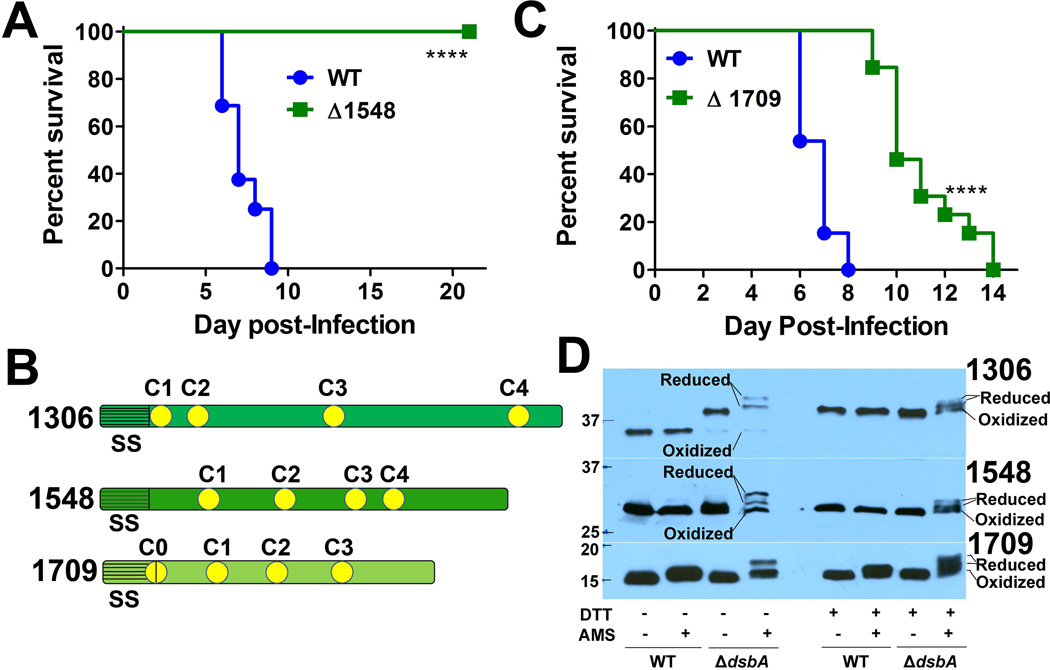
A. Groups of C3H/HeN female mice were intranasally-infected with either 104 CFU of WT LVS (n=16) or LVS ΔFTL_1548 (n=13). Mean time-to-death differences were calculated by log-rank Mantel-Cox test (**** P < 0.0001 versus WT LVS).
B. Groups of C3H/HeN female mice were intranasally-infected with either 104 CFU of WT LVS (n=13) or LVS ΔFTL_1709 (n=13). Mean time-to-death differences were calculated by log-rank Mantel-Cox test (**** P < 0.0001 versus WT LVS).
C. Illustration of DsbA substrates FTL_1306 (DipA), FTL_1548, and FTL_1709. Predicted N-terminal leader sequences (grey) and signal peptidase I or II cleavage sites (SS) are indicated for each protein. Yellow circles indicate relative positions of cysteine residues in each protein. For FTL_1709, C1 (yellow circle with vertical line) indicates the putative lipo-cysteine acylation site.
D. AMS trapping assays to examine the redox status of DsbA substrates FTL_1306 (DipA), FTL_1548, and FTL_1709. Histidine-tagged plasmid-overexpressing strains (noted on the right margin of each blot) were grown in liquid CDM, bacteria were lysed and precipitated in TCA, incubated without (−) or with (+) 10 mg ml−1 AMS, prepared without (−) or with (+) DTT, separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotting was performed using an anti-6× histidine monoclonal antibody. Reduced and oxidized status of each protein are noted in the right margin of each blot. Molecular mass standards with sizes (in kDa) are noted on the left side of each blot.
More importantly, our FtDsbA-substrate trapping assay identified 25 hypothetical proteins in F. tularensis (Table 1), which have not been previously studied and lack homology to any other proteins in the GenBank database. Given the extreme virulence of F. tularensis and our identification of other FtDsbA substrates that are known F. tularensis virulence factors (e.g., FopA, MipA, DipA, FTL_0325), we predicted that many of these hypothetical proteins (Table 1) also would be virulence factors. To test this prediction, we performed a limited genetic screen by randomly selecting and independently deleting six hypothetical proteins, FTL_0424, FTL_0878, FTL_1548, FTL_1581, FTL_1678, and FTL_1709, that contained more than three cysteines. Of the these six hypothetical proteins, two were found to be required for F. tularensis virulence in a mouse pulmonary infection model: FTL_1548 (Fig. 4A) and FTL_1709 (Fig. 4B). Whereas both deletion mutants were significantly attenuated in mice, ΔFTL_1548 was completely avirulent, with 100% of mice surviving through day 20 post-infection. These results demonstrate that many FtDsbA substrates are virulence factors and highlight the importance of our FtDsbA-substrate trapping approach to identify novel F. tularensis virulence factors.
To validate that the identified FtDsbA substrates (Table 1) were directly oxidized by FtDsbA and were not artifacts of our trapping approach, we assessed the redox status of DipA (FTL_1306) and newly-identified virulence factors FTL_1548 and FTL_1709 in LVS lysates using AMS trapping, as described above. Whereas each of the three proteins contains four cysteines, the protein sizes, cysteine organization patterns, and other molecular features differ among the three proteins (Fig. 4C). As noted above, DipA is a predicted lipoprotein, with the first cysteine (C1) likely serving as the lipoprotein acylation motif (LEQC), and the three remaining cysteines likely require FtDsbA isomerase and oxidoreductase activities to correctly form one disulfide bond (Fig. 4C). Hypothetical protein FTL_1548 has no predicted lipo-cysteine, thus all four cysteines likely require FtDsbA isomerase and oxidoreductase activities to correctly form up to two disulfide bonds (Fig. 4C). For hypothetical protein FTL_1709, the first cysteine (C1) is predicted to serve as a lipoprotein acylation motif (LVTSC), and the three remaining cysteines potentially require FtDsbA isomerase and oxidoreductase activities to correctly form one disulfide bond (Fig. 4C). In AMS trapping assays, DipA, FTL_1548, and FTL_1709 were nearly completely oxidized in the presence of WT FtDsbA (AMS+; Fig. 4D). However, in ΔdsbA, substantial amounts of all three proteins were observed in reduced forms in the presence of AMS (AMS+; Fig. 4D). Considering that this direct redox assessment demonstrated that all three of the examined substrates required FtDsbA for oxidation (Fig. 4D), these results strongly suggest that the majority of FtDsbA substrates identified in this study (Table 1) require FtDsbA for correct disulfide bond formation and protein function.
Interestingly, careful analysis of the DipA redox status in the presence of AMS revealed that DipA may not be a lipoprotein (Fig. 4D). As noted above, if DipA were a genuine lipoprotein, the first cysteine (C1) should be acylated (Fig. 4C), one disulfide bond should be present, and one free cysteine to be available for modification by AMS. Surprisingly, addition of AMS to the WT strain did not result in a molecular weight shift of DipA (Fig. 4D), indicating that no free cysteines were available for AMS modification (i.e., two disulfide bonds were present in DipA; no free thiols; no acylated lipo-cysteine). Further, when AMS was added to ΔdsbA, two reduced forms of DipA were observed (Fig. 4D), suggesting that DipA was present in a fully-reduced form (four free cysteines; upper reduced band) and a partially-reduced form (two free cysteines and one disulfide bond; lower reduced band). AMS trapping results for FTL_1548 were as expected. When AMS was added to WT, no FTL_1548 molecular weight shift was observed (Fig. 4D), indicating that FTL_1548 contains two disulfide bonds. In ΔdsbA, two reduced forms of FTL_1548 were observed in the presence of AMS (Fig. 4D), indicating a fully-reduced form (four free cysteines; upper reduced band) and a partially-reduced form (two free cysteines and one disulfide bond; lower reduced band). Current studies in our laboratory are investigating the mechanisms by which FTL_1548 contributes to F. tularensis virulence, including determining if FTL_1548 is an integral membrane protein, assessing FTL_1548 surface-exposure, studying if FTL_1548 interacts with host cell proteins, and investigating if FTL-1548 is required for SchuS4 virulence. AMS trapping results for FTL_1709 also were as expected, indicating that FTL_1709 is a lipoprotein. When AMS was added to WT, a small (approx. 500 Da) molecular weight shift was observed for FTL_1709 (Fig. 4D), indicating that the first cysteine (C1) is acylated, one disulfide bond is present, and only one free cysteine was available for AMS modification (i.e., FTL_1709 is a lipoprotein). In ΔdsbA, one reduced band and one oxidized band were observed for FTL_1709 when AMS was added (Fig. 4D), indicating that three cysteines were modified by AMS. To confirm that FTL_1709 is a lipoprotein, we grew WT in the presence or absence of the antimicrobial compound globomycin, a selective signal peptidase II inhibitor (Hussain et al., 1980), and analyzed whole-cell lysates by Western blot analysis for increased FTL_1709 molecular weight. In the presence of globomycin (10 or 25 µg ml−1), FTL_1709 was primarily observed at a slightly larger molecular weight, indicating an uncleaved precursor form (Fig. S7). From the same WT lysates, known F. tularensis lipoproteins Pal and Tul4-A (Godlewska et al., 2009, Sjostedt et al., 1991, Huntley et al., 2007) were also observed at increased molecular weights in the presence of globomycin (Fig. S7), indicating similar uncleaved precursor proteins. For FTL_1548 and DipA, globomycin did not result in increased molecular weights (Fig. S7), providing further evidence that neither protein is a lipoprotein. As controls, globomycin did not result in increased molecular weights for either integral OMP FopA or cytoplasmic protein EFTu (Fig. S7). Whereas both AMS trapping and globomycin inhibition studies suggest that FTL_1709 is a lipoprotein, additional analyses such as Triton X-114 partitioning (Radolf et al., 1988) or [3H] palmitate labeling (Belisle et al., 1994) may be needed to confirm that FTL_1709 is a lipoprotein. Our laboratory currently is performing detailed characterizations of FTL_1709, including assessments of FTL_1709 outer membrane localization and surface exposure, studying the role of FTL_1709 in SchuS4 virulence, and investigating why FTL_1709 only partially attenuates (6 day delay in time-to-death) F. tularensis.
Discussion
As an intracellular pathogen, F. tularensis encounters at least temporary phagosomal ROS or RNS exposure before it escapes into the cytosol (Chong & Celli, 2010). Previous studies demonstrated that although F. tularensis encodes both superoxide dismutase and catalase to counteract oxidative stresses, mutants of either gene exhibited varying levels of attenuation in macrophages and mice, suggesting that F. tularensis possess additional mechanisms to counteract oxidative stress (Melillo et al., 2009, Melillo et al., 2010, Lindgren et al., 2007). Indeed, F. tularensis has been shown to inhibit neutrophil activation by two mechanisms, one involving a series of five to six bacterial-encoded acid phosphatases which suppress or neutralize the neutrophil ROS burst and a second involving inhibition of neutrophil NADPH oxidase activity by an unknown bacterial factor (McCaffrey et al., 2010, Mohapatra et al., 2013). Thus, although it appears that F. tularensis can inhibit or neutralize a majority of the host ROS burst, any residual ROS could damage the bacteria, its virulence factors, and restrict bacterial growth. The results of this study suggest that the bifunctional nature of FtDsbA adds another layer of ROS protection, by reducing and isomerizing misoxidized disulfide bonds in envelope proteins, including OMPs and virulence factors, and re-oxidizing disulfide bonds to restore protein folding and function.
Disulfide bond oxidation is known to be required for bacterial virulence (Heras et al., 2009) but this is the first study that correlates disulfide bond isomerase activity with bacterial virulence. During the review of this manuscript, we became aware of a very recently-accepted manuscript demonstrated similar oxidase and isomerase activities for FtDsbA (also referred to as FipB; (Qin et al., 2014)). However, that study found that both isomerase and oxidase activities of FtDsbA/FipB required another F. tularensis accessory protein, FipA (FTT_1102; immediately upstream of FtDsbA/FipB). Additionally, that study noted that although the amino-terminal Mip domain of FtDsbA/FipB was required for isomerase activity, the Mip domain, and thus isomerase activity, was not essential for intracellular growth or mouse virulence (Qin et al., 2014). To put those findings in context with the findings from our study: First, we focused solely on one protein, FtDsbA, and found that FtDsbA alone possessed both oxidoreductase and isomerase activities; Second, we focused our molecular analyses on the carboxy-terminal portion of FtDsbA, given that this contains the DsbA-like domain; Third, we found that a single amino acid polymorphism in the cisPro-1 residue of the DsbA-like domain modulated both isomerase activity and virulence; Fourth, we trapped and identified over 50 FtDsbA substrates, including known and newly-identified virulence factors. Taken together, our studies clarify the role of FtDsbA in virulence, provide evidence that FtDsbA plays an indirect role in virulence, and propose a new set of F. tularensis virulence factors (Table 1) for future studies.
As reviewed earlier, many groups have published studies on FtDsbA. Whereas most of those studies aimed to characterize molecular aspects of various FtDsbA mutants that resulted in defects in intracellular replication and/or virulence in mice, our primary interest was to trap and identify FtDsbA substrates in order to potentially reveal new virulence factors. One other group had a similar goal, but used proteomic analysis to compare LVS and ΔdsbA protein profiles, speculating that proteins that depend on FtDsbA for folding would either increase or decrease (due to instability) in abundance in the absence of FtDsbA (Straskova et al., 2009). That group only identified 10 total proteins with significantly altered expression in ΔdsbA, five of which, FTL_1060, FTL_1306 (DipA), FTL_1521, FTL_1532, and FTL_1579, also were identified here. Whereas the overlap between that study and our trapping approach is encouraging, we identified substantially more FtDsbA substrates (53 total) and the vast majority of our FtDsbA substrates contain more than two cysteines, indicating that they require FtDsbA for complex disulfide bond connectivity. As compared with the previous study, our thiol trapping method was much more stringent, requiring covalent linkages between FtDsbA and substrates. As proof of stringency differences between the two studies, one of their proposed FtDsbA-interacting proteins, FTL_0694, does not contain any cysteines and was not identified in our analysis.
The lack of DsbC and DsbD orthologs in F. tularensis originally led us to speculate that, in addition to oxidoreductase activity, FtDsbA also may possess isomerase activity. Our prediction and demonstration of the bifunctional nature of FtDsbA is not unfounded, as Legionella pneumophila DsbA2 recently was reported to exhibit both oxidoreductase and isomerase activities (Kpadeh et al., 2013). Here, we demonstrated FtDsbA oxidase and isomerase activities in vitro and in F. tularensis, and showed a positive correlation between isomerase activity and F. tularensis virulence. We propose that the bifunctional activity of FtDsbA is not serendipitous, but is an efficiency mechanism used to promote intracellular pathogenesis. Compared with the E. coli DsbC/DsbD isomerase/reductase pathway which consumes cytosolic NADPH/ATP to correct mismatched disulfide bonds (Katzen & Beckwith, 2000), bifunctional FtDsbA is much more efficient by directly cycling electrons between itself and protein substrates to form correct disulfide bonds. As noted above, F. tularensis encounters at least temporary ROS or RNS exposure before it escapes into the cytosol and FtDsbA may gain additional efficiency by using phagocyte oxidative stress as a disulfide bond oxidative source. Intrinsic chaperone activity (Schmidt et al., 2013), which recognizes misfolded proteins, may further enhance FtDsbA efficiency. However, these efficiencies appear to come with a tradeoff. Whereas the E. coli DsbC/DsbD pathway tolerates ≥ 10 mM copper by consuming extra NADPH (Ren et al., 2009), F. tularensis is completely inhibited by > 100 µM copper (Materials and Methods), indicating that FtDsbA cannot counteract strong oxidative stress. These observations indicate that FtDsbA isomerase and oxidoreductase activities are largely dependent on ROS inactivation mechanisms, including SOD, catalase-peroxidase, and acid phosphatase (Chong & Celli, 2010). Although speculative, our FtDsbA results and the work of others (Kpadeh et al., 2013) suggest that any bacterium with merged isomerase and oxidoreductase activities, and the ability to inactivate phagocyte ROS, could be a successful intracellular pathogen.
One surprising finding was that Type B DsbA conferred increased virulence when compared with Type A DsbA in the F. tularensis LVS (Type B) background (Fig. 1B). These results need to be interpreted with caution, as they do not suggest that Type B F. tularensis strains are more virulent than Type A F. tularensis strains. At this time, we cannot definitively explain these paradoxical results but we offer the following possibilities: First, as noted in the Results, we did observe decreased FtDsbA expression among isogenic knock-in strains, including approximately 50% decreased expression of His-LVS A285T (Type A-like) and His-SchuS4 DsbA. Whereas it remains possible that reduced expression of Type A or Type A-like FtDsbA contributed to the observed differences in virulence between DsbA variants, the reduced expression of His-LVS DsbA does not fully-explain these virulence differences. Second, although our virulence comparison of isogenic FtDsbA knock-in strains suggests that the increased virulence of Type B DsbA may be directly attributed to the cisPro-1 residue Ala (WT LVS DsbA and His-LVS DsbA) versus the Type A DsbA cisPro-1 residue Thr (His-LVS A285T and His-SchuS4 DsbA), we cannot rule out the possibility that differences in the cisPro-1 residue, and associated isomerase activities, may have conferred different substrate specificities. As previously noted, detailed studies in E. coli have demonstrated that variations in the cisPro-1 residue affect substrate specificity (Ren et al., 2009). Indeed, by placing the Type A cisPro-1 residue (His-LVS A285T) or Type A DsbA (His-SchuS4 DsbA) in the Type B background (LVS), we may have limited the pool of virulence substrates for Type A FtDsbA and, thus, the full virulence potential of Type A DsbA may have been masked in LVS. Potential substrate differences are highlighted by the fact that although LVS and SchuS4 share 99.3% genetic identity, there are at least 35 genes with sequence variations (nonsense mutations, frameshifts, single nucleotide deletions resulting in protein fusions, and large nucleotide deletions) between LVS and SchuS4 (Rohmer et al., 2006). Taken together, it remains possible that our observed differences between Type A and Type B FtDsbA virulence were due to differences in FtDsbA substrate specificity and/or available substrates.
The cisPro motif of thioredoxin proteins is of great interest because of its role in substrate specificity. Mutation of the conserved proline residue in the EcDsbA cisPro motif resulted in delayed substrate release during co-translocational oxidation, allowing for substrate trapping and identification (Kadokura et al., 2004). Importantly, more recent evidence suggests that the cisPro-1 residue of thioredoxin-fold proteins may modulate substrate specificity (Ren et al., 2009). The EcDsbA cisPro-1 residue side chain is spatially located within 4 Å of the CXXC active site and amino acid substitutions of the cisPro-1 residue directly affected EcDsbA substrate interactions and redox properties, primarily due to side chain hydrophobicity (Ren et al., 2009). That study suggested that the cisPro-1 side chain has steric effects that dominate the binding interface between thioredoxin fold proteins and substrates. Here, we discovered inherent differences in the cisPro-1 residue of Type A (Thr) and Type B (Ala) F. tularensis strains. These residues differ in their side chain hydrophobicity and, thus, it is not surprising that we observed differences in Type A and Type B FtDsbA reductase and isomerase activities. Importantly, the novel finding from this study was the discovery that differences in isomerase activity directly correlated with differences in mammalian virulence (i.e., enhanced isomerase activity of Type B FtDsbA correlated with the virulence of the LVS in mice).
This study demonstrates the utility of carefully analyzing subtle genetic changes to understand protein function and bacterial virulence. We discovered that, as compared to EcDsbA, FtDsbA is a bifunctional protein encoding both isomerase and oxidase activities. Further, a single amino acid polymorphism in the FtDsbA cisPro motif affects both isomerase activity and virulence in animals. Together, these activities are necessary for the structure and function of proteins that are directly involved in virulence (Fig. 5). Although not directly tested here, those functions likely include processes involved in host cell attachment, invasion, and intracellular survival (Fig. 5). Our identification of FtDsbA substrates, including known OMPs and virulence factors, the discovery of new virulence factors such as FTL_1548 and FTL_1709, and a large number of yet uncharacterized FtDsbA substrates, highlights the utility of our DsbA-substrate trapping approach. This study provides a detailed molecular mechanism to explain why FtDsbA mutants are attenuated and may aid in the development of inhibitors against intracellular pathogens that require DsbA for virulence, including F. tularensis, L. pneumophila, Mycobacterium tuberculosis, and Burkholderia pseudomallei (Kpadeh et al., 2013, Ireland et al., 2013, Chim et al., 2013).
Fig. 5. FtDsbA protein disulfide isomerase activity and its correlation with bacterial virulence.
A. Unlike EcDsbA, FtDsbA is a bifunctional protein that oxidizes and isomerizes disulfide bonds. FtDsbA directly oxidizes correct disulfide bonds or oxidizes into mismatched intermediates, then subsequently reduces and isomerizes the mismatched disulfides to form correct disulfide bonds.
B. FtDsbA is not a direct virulence factor but, rather, is responsible for folding many envelope proteins, including OMPs and virulence factors, into their correct, functional conformation. The collective effects of these substrates explain the essential role of DsbA in F. tularensis virulence.
Experimental procedures
Bacterial strains, plasmids, and culture conditions
A complete list of bacterial strains and plasmids used in this study are listed in Table S1. Following all federal and institutional select agent and biosafety regulations, F. tularensis Type A strain SchuS4 and Type B strain LVS were obtained from BEI Resources. All work with SchuS4 was conducted under strict BSL3 containment conditions at the University of Toledo Health Science Campus BSL3 laboratory, including the use of liquid-impervious personal protective equipment (PPE) and powered air purifying respirators (PAPRs). All work with LVS was performed using BSL2 containment conditions. F. tularensis stock cultures were grown at 37 °C with 5% CO2. Routine F. tularensis growth was performed on modified Mueller-Hinton agar (MHA): Mueller-Hinton broth (Becton Dickinson) was mixed with 1.6% (wt vol−1) Bacto Agar (Becton Dickinson), autoclave sterilized, cooled to 55 °C, and further supplemented with 0.1% (wt vol−1) glucose, 0.025% (wt vol−1) iron pyrophosphate, 2.5% (vol vol−1) donor calf serum (Mediatech), and 2% (vol vol−1) IsoVitaleX (Becton Dickinson). F. tularensis for mouse infection experiments were grown on Brain Heart Infusion (BHI) agar, prepared as previously described (Hazlett et al., 2008). Following conjugation, F. tularensis was grown on modified chocolate agar (CHOC): Mueller-Hinton medium was mixed with 1% (wt vol−1) tryptone, 0.5% (wt vol−1) NaCl, and 1.6% (wt vol−1) agar, autoclave sterilized, cooled to 55 °C, and further supplemented with 1% (wt vol−1) bovine hemoglobin powder (Neogen), 0.1% (wt vol−1) glucose, and 2% (vol vol−1) IsoVitaleX (Becton Dickinson). For AMS trapping and DsbA-substrate trapping experiments, F. tularensis was grown on chemically-defined medium (CDM) agar, prepared as previously described (Chamberlain, 1965). For cadmium sensitivity assays, CDM contained only 0.02% cysteine to facilitate F. tularensis growth but minimize spontaneous protein disulfide bonds oxidation. When needed, F. tularensis growth media were supplemented with 200 mg L−1 hygromycin (Hyg), 10 mg L−1 kanamycin (Kan), or 100 mg L−1 polymyxin B (PxB). When needed, sucrose was added to MHA or CHOC at a concentration of 8% (wt vol−1) prior to autoclave sterilization. To test for potential growth defects of F. tularensis isogenic mutants, bacteria were initially grown on MHA, then inoculated into liquid Mueller-Hinton medium supplemented with 1.23 mM calcium chloride dihydrate, 1.03 mM magnesium chloride hexahydrate, 0.1% (wt vol−1) glucose, 0.025% (wt vol−1) iron pyrophosphate, and 2% (vol vol−1) IsoVitaleX. F. tularensis were adjusted to an optical density at 600 nm (OD600) of 0.005 to 0.01 and growth was monitored over a period of 15 h based on OD600 measurements of the liquid cultures. E. coli XL-10 gold was routinely used for plasmid manipulation and recombinant protein expression. E. coli S17-1 was used as the donor for F. tularensis conjugation. E. coli were grown using Luria-Bertani (LB) broth or agar at 37 °C, supplemented as needed with 200 mg L−1 Hyg, 50 mg L−1 Kan, or 100 mg L−1 ampicillin (Amp).
Sequence alignments and bioinformatics predictions
Amino acid sequence alignments were performed using either BlastP analysis (http://blast.ncbi.nlm.nih.gov) or ClustalW alignment (MacVector, version 12.6) with E. coli DsbA (YP_491589.1), LVS DsbA (FTL_1096), and SchuS4 DsbA (FTT_1103) sequences. Conserved domains were predicted using the conserved domain database search within BlastP. Conserved residues in the F. tularensis DsbA cisPro motif were identified by comparing either 15 Type A DsbA sequences available from GenBank or 9 Type B DsbA sequences available from GenBank. Type A and Type B cisPro motif sequence logos were generated by WebLogo, version 2.8.2 (http://weblogo.berkeley.edu/logo.cgi). Signal peptidase cleavage sites were predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Lipoprotein acylation motifs were predicted by LipoP 1.0 Server (http://www.cbs.dtu.dk/services/LipoP/).
DsbA cloning, site-directed mutagenesis, and overexpression constructs
F. tularensis LVS and SchuS4 genomic DNA were extracted using TRIzol reagent (Invitrogen). LVS and SchuS4 DsbA coding sequences (complete genes, including both the amino-terminal Mip domain and carboxy-terminal DsbA domain) were PCR-amplified using High Fidelity Platinum Taq Polymerase (Life Technologies), digested with Nco I and Bgl II (New England Biolabs), and ligated into similarly-digested pQE-60 (Qiagen) using T4 DNA Ligase (New England Biolabs). The resulting C-terminal 6× histidine fusion-tagged LVS and SchuS4 DsbA coding regions served as templates for subsequent amplifications, mutant generation, or recombinant protein expression. DsbA mutants, including LVS dsbA A285T, LVS dsbA CXXS, LVS dsbA CXXS & A285T, and SchuS4 dsbA T286A, were generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and primers encoding the indicated DsbA mutations. Briefly, PCR amplification was performed using pQE-LVS DsbA or pQE-SchuS4 DsbA and one of the designated mutation primers, products were digested with Dpn I, precipitated using Pellet Paint (Novagen), suspended in ddH2O, transformed into XL-10 gold chemically-competent cells (Agilent), and incubated on LB-Amp plates at 37 °C overnight to select ampicillin-resistant colonies. Clones were randomly selected, plasmid DNA was purified, and DNA sequencing was performed to confirm the intended point mutation and verify integrity of the remainder of the dsbA coding sequence. For AMS trapping and redox assessments of FTL_1306, FTL_1548, and FTL_1709, full-length coding regions of each gene were PCR-amplified as described above, digested with Nco I and BamH I (New England Biolabs), and ligated into similarly-digested pQE-60 as described above. The resulting C-terminal 6× histidine fusion-tagged constructs (FTL_1306-His, FTL_1548-His, and FTL_1709-His) were confirmed by DNA sequencing before each histidine-tagged coding region was further sub-cloned into pFNLTP6-gro-GFP (Maier et al., 2004) using restriction enzymes EcoR I (New England Biolabs) and BamH I. Overexpression constructs were electroporated into electrocompetent LVS using 0.1 cm gap electroporation cuvettes and a GenePulser Xcell electroporation unit (2500 V, 25 µF capacitance, 600 Ω resistance; BioRad). Electrocompetent LVS were prepared by washing the bacteria 3µ with a solution containing 0.5 M sucrose and 1 mM EDTA, pH 7.5.
F. tularensis gene knock-outs and knock-ins by homologous recombination
Splicing-overlap extension (SOE) PCR was used to generate three-component gene knock-out or knock-in constructs. For gene knock-out constructs, the full coding region of the targeted gene was replaced with the FLP recombination target (FRT)-flanked Pfn-kanamycin resistance cassette (FRT-Pfn-kan-FRT) from pLG66a (Gallagher et al., 2008). For knock-in constructs, the full coding region of the targeted gene was replaced with either LVS dsbA, SchuS4 dsbA, or the one of site-directed dsbA mutants described above. For both gene knock-out and knock-in constructs, approx. 500 bp upstream and downstream regions immediately adjacent to the targeted gene were PCR amplified, SOE PCR was used to fuse the three DNA components (e.g., upstream region, FRT-Pfn-kan-FRT, downstream region), the resulting amplicon was digested with Apa I, and the amplicon was ligated into Apa I-digested pTP163 (Robertson et al., 2013). Constructs were transformed into E. coli S17.1 and conjugation was performed with F. tularensis on CHOC plates without antibiotics. For gene knock-outs, transconjugants were initially recovered on CHOC supplemented with Kan and PxB, passaged once on MHA supplemented with Kan to allow secondary recombination, and finally passaged on MHA supplemented with Kan and 8% (wt vol−1) sucrose to select for clones that had undergone deletion of both the wild-type gene and the sacB sucrose sensitivity marker. For dsbA knock-ins, transconjugants were initially recovered on CHOC supplemented with PxB, passaged once on MHA to allow for secondary recombination, and finally passaged on MHA supplemented with 8% (wt vol−1) sucrose to select for clones that had undergone deletion of both the Kan cassette and the sacB sucrose sensitivity marker. DsbA expression was quantitated (Fig. 1B inset) from WT, ΔdsbA, and isogenic knock-in strains (His-LVS DsbA, His-LVS DsbA A285T, His-SchuS4 DsbA) by diluting each strain to equivalent OD600 0.55 measurements and preparing lysates in SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE and immunoblot analyses were performed as previously described (Huntley et al., 2007). DsbA and EF-Tu were detected in immunoblots using rat polyclonal antisera generated in previous studies (Huntley et al., 2007). EF-Tu was used as the loading control for each bacterial strain and DsbA expression levels were quantitated by densitometry analysis using ImageJ (http://rsb.info.nih.gov/ij/index.html). DsbA expression levels for each isogenic strain are presented as percent expression relative to WT LVS DsbA (set to 100%).
RNA Isolation and quantitative RT-PCR
Total bacterial RNA was isolated using TRIZOL (Invitrogen) following the manufacturer's protocol and was treated with DNase I to remove any residual genomic DNA. RNA was further purified using RNeasy RNA Mini Kit (Qiagen). SuperScript Vilo (Life Technologies) was then used to reverse transcribe 1 µg of RNA. Quantitative real-time PCR (qRT-PCR) reactions included 10-fold dilutions of cDNA, HotStarTaq Plus DNA Polymerase (Qiagen), SYBR Green (Life Technologies), 1x PCR buffer, and 0.2 µM of each primer. qRT-PCR primers were designed using PrimerQuest (Integrated DNA Technologies). dsbA primers were 5’-DsbA 5’-GCTGGCGCAGATATGACTAAAG-3’ and 3’-DsbA 5’-GCAGGAGCTATTACTAGGAATGG-3’. RNA polymerase subunit α (FTL_0261) served as the internal control. FTL_0261 primers were 5’-rpoA1 5’-AGATCAGCCAATAGCTACTTTGACA-3’ and 3’-rpoA1 5’-TCGGTTGGTATCGCAGAAAGTATTC-3’. All qRT-PCR reactions were performed in triplicate and samples without reverse transcriptase were used as negative controls to assess genomic DNA contamination. qRT-PCR reactions were performed and analyzed using a CFX96 Real-Time PCR Detection System instrument (Bio-Rad). Relative dsbA mRNA levels (Fig. 1B inset) were calculated based on FTL_0261 mRNA expression and are presented as percent values relative to WT LVS (set to 100%).
Mouse infections
Mouse infections were performed essentially as previously described (Huntley et al., 2008). Groups of 5 to 8 C3H/HeN female mice (6 to 8 weeks old; National Cancer Institute) were anesthetized with a ketamine, xylazine, and acepromazine cocktail and were inoculated intransally (i.n.) with 20 µl of wild-type or isogenic LVS strains (104 CFU in PBS; based on previous OD600 measurements and bacterial enumeration studies). Inocula were grown on CDM (Fig. 1) or BHI (Fig. 4) as described above, serially-diluted, and plated in quadruplet to confirm CFU. Mice were monitored daily through day 20 post-infection or until mice succumbed to infection. Differences in mean time-to-death were calculated for each infection group using the log-rank Mantel-Cox test (GraphPad Prism5 software).
4-acetoamido-4’-maleimidylstilbene 2,2’-disulfonate (AMS) trapping
F. tularensis LVS, isogenic FtDsbA, or overexpression strains were grown overnight on CDM agar plates, bacterial growth was scraped and diluted to OD600 of 0.05 in CDM liquid medium, equal volumes were inoculated into 50 ml of fresh CDM liquid medium, and cultures were grown for 18 h at 37 °C. Following 18 h of growth, bacteria were adjusted to OD600 of 0.5, pelleted by centrifugation at 16,000 µ g for 1 min, lysed in 10% (vol vol−1) trichloroacetic acid (TCA), and precipitated overnight at 4 °C. Precipitated proteins were washed 3µ with cold acetone, suspended in a buffered solution containing 100 mM Tris-HCl pH 7.5, and 1% (wt vol−1) SDS, with or without 10 mg ml−1 AMS, and incubated in the dark at 30 °C for 30 min followed by 37 °C for 10 min. AMS alkylation was stopped by the addition of SDS loading buffer (2% [wt vol−1] SDS, 50 mM Tris, 10% [vol vol−1] Glycerol, 142 mM 2-mercaptoethanol), proteins were separated by SDS-PAGE, and immunoblot analyses were performed as previously described (Huntley et al., 2007). DsbA, FopA, and MipA were detected in immunoblots using rat polyclonal antisera that were generated in previous studies (Huntley et al., 2007). FTL_1306-His, FTL_1548-His, and FTL_1709-His were detected in immunoblots by anti-His antibody (Qiagen).
Spot titers for cadmium and copper sensitivity assays
For cadmium sensitivity (oxidase activity) assays, F. tularensis LVS and isogenic dsbA knock-in strains were grown overnight on CDM agar plates, bacterial growth was scraped and diluted to OD600 of 1.0 (approx. 108 CFU ml−1) in PBS, and 2 µl of each bacterial strain was spotted onto CDM agar plates containing 0, 50, 100, 150, or 200 µM cadmium chloride (Sigma-Aldrich). Following incubation at 37 °C for 48 h, colony growth on each plate was assessed and imaged using an Omega Imager (UltraLum). For copper sensitivity (isomerase activity) assays, F. tularensis LVS and isogenic dsbA knock-in strains were grown overnight on MHA plates, bacterial growth was scraped and diluted to OD600 of 1.0 (approx. 109 CFU ml−1) in PBS, bacteria were 10-fold serially diluted, and 2 µl of each bacterial dilution was spotted onto MHA plates or MHA plates containing 30 µM (Fig. 3) or 40 µM (Fig. S4) copper(II) chloride (Sigma-Aldrich). Preliminary experiments demonstrated that WT LVS was inhibited by copper concentrations > 100 µM. Following incubation at 37 °C for 48 h, colony growth on each plate was assessed and imaged. All spot titer assays were performed in duplicate or triplicate to verify results.
Recombinant protein expression and purification
Non-signal sequence containing regions of F. tularensis LVS dsbA (pQE-LVS nsDsbA), SchuS4 dsbA (pQE-SchuS4 nsDsbA), LVS A285T mutant dsbA (pQE-LVS nsDsbA A285T), and SchuS4 T285A mutant dsbA (pQE-SchuS4 nsDsbA T285A) were generated as described above. Ligations were transformed into E. coli XL-10 chemically-competent cells and sequence verified. Bacteria were grown in LB broth to an OD600 of 0.5, protein expression was induced for 4 h by the addition of isopropyl β-D-thiogalactopyranoside (IPTG) to 50 µM, bacteria were pelleted by centrifugation, and frozen overnight at −20°C to aid in lysis. Cell pellets were suspended in 20 mM Tris and 0.5 M NaCl, sonicated for 30 min at 1 min intervals, insoluble material was removed by centrifugation at 10,000 µ g, and supernatants were collected for affinity purification over pre-equilibrated Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) columns. Eluted recombinant proteins were further purified by anion exchange chromatography using Q Sepharose Fast Flow (GE Healthcare), concentrated in Amicon Ultra-4 centrifugal filter units with 30-kDa cutoff (Millipore), concentrations were determined using the BCA protein assay (Thermo Scientific), and purity (> 90%) was assessed by SDS-PAGE and Imperial protein staining (Thermo Scientific).
Isomerase activity assays with scrambled hirudin
F. tularensis LVS and SchuS4 DsbA isomerase activities were quantitated by measuring the rate of scrambled hirudin refolding, essentially as previously described (Hiniker et al., 2007). LVS and SchuS4 DsbA were reduced by incubation with 20 mM dithiothreitol (DTT) for 2 h at 4 °C, the reactions were desalted on NAP-5 columns (GE Healthcare), and protein fractions were collected. DsbA concentrations were determined by absorbance at 280 nm using extinction coefficients of 17,880 cm−1M−1. Scrambled hirudin (BoTai Bio-Tech, Dalian, Liaoning, China) was diluted to 24 µM in refolding buffer (100 mM sodium phosphate, 1 mM EDTA, pH 7.0) and incubated with or without 24 µM DsbA (LVS or SchuS4) in refolding buffer. Following incubation for 1, 2, 10, 22, or 30 min at room temperature, reactions were quenched by the addition of 10% (vol vol−1) formic acid. Reaction products were separated by reversed-phase HPLC (Waters) on a Vydac 218TP54 C18 column (W.R. Grace) at 55 °C using an acetonitrile gradient (19 to 25%, 30 ml) in 0.1% (vol vol−1) trifluoroacetic acid, eluted proteins were detected by absorbance at 220 nm, and data analyzed by Empower 2 software (Waters).
Reductase activity assays
The abilities of LVS DsbA, SchuS4 DsbA, LVS A285T mutant DsbA, and SchuS4 T285A mutant DsbA to catalyze the reduction of human insulin in the presence of DTT were tested essentially as previously described (Holmgren, 1979). A stock solution of 872 µM insulin was freshly prepared in 100 mM potassium phosphate buffer, pH 7.0, and 2 mM EDTA for each assay. Reaction components were directly added to spectrophotometer cuvettes containing 100 mM potassium phosphate buffer, pH 7.0, 2 mM EDTA, and 0.33 mM DTT. Reactions were started by the simultaneous addition of 131 µM insulin and 5 µM of a DsbA variant, in a final reaction volume of 0.8 ml. After thorough mixing, cuvettes were immediately placed into a spectrophotometer, and 650 nm measurements were recorded once per minute for 140 min. In all experiments, the non-catalyzed (no DsbA variant added) reduction of insulin by DTT was monitored in a control reaction.
Redox potential measurements
The redox potentials of LVS and SchuS4 DsbA were measured by incubating each protein in degassed redox buffers containing various concentrations of L-glutathione reduced (GSH) and L-glutathione oxidized (GSSG), as previously described (Bessette et al., 1999). Briefly, 1 µM of each DsbA protein was incubated for 16 h at room temperature with 1 mM GSSG and 0 to 10 mM GSH, in buffer containing 100 mM sodium phosphate and 1 mM EDTA, pH 7.0. The resulting samples were subjected to AMS trapping as described above, products were separated by SDS-PAGE, stained as described above, the amounts of reduced and oxidized DsbA in each reaction were quantitated by densitometry (ImageJ; National Institutes of Health), and the ratios of reduced DsbA to oxidized DsbA were calculated as reduced DsbA ratios (Fig. 3). To calculate the GSH/GSSG redox buffer potential (mV), the equation of −240 + ln([GSH]2/[GSSG])×8.31×295/(2×9.64×10) was used to convert the GSH and GSSG concentrations in each reaction to mV (Fig. 3).
DsbA-substrate trapping and mass spectrometry protein identification
WT LVS dsbA was replaced in the genome with a LVS dsbA CXXS mutant (His-tagged) through homologous recombination as described above to trap DsbA-substrate complexes. LVS DsbA CXXS mutant and ΔdsbA (negative control) bacteria were grown in 4 L liquid CDM for 18 h at 37 °C, pelleted by centrifugation at 8000 µ g for 30 min, suspended and lysed in 10% TCA, and precipitated overnight at 4 °C. Precipitated proteins were washed 3µ with cold acetone, 100 mM iodoacetamide was added to quench free thiols, DsbA-substrate complexes were purified by Ni-NTA agarose affinity chromatography, and elution fractions were concentrated in Amicon Ultra-4 centrifugal filter units with 50-kDa cutoff (Millipore). DsbA-substrate complexes were treated with or without 142 mM 2-mercaptoethanol, proteins were separated by SDS-PAGE, stained as described above, the non-reduced sample lane was divided into five sections based on molecular weight (< 50 kDa; 50 to 75 kDa; 75 to 100 kDa; 100 to 150 kDa; and > 150 kDa), and sections were excised from the gel. Excised gel sections were subjected to in-gel proteolysis, LC/MS/MS analysis, database searching, and false-discovery-rate assessment, essentially as previously described (Llarrull et al., 2011, Alves et al., 2013). Briefly, extracted trypsinized in-gel digests were extracted and desalted with a C18 ZipTip (Millipore) prior to LC/MS/MS analysis. Samples were re-suspended in 10 µl of 1% (vol vol−1) formic acid. Duplicate injections of approx. 2.5 µl of the material were loaded onto a 100 µm × 100 mm C18BEH column (Waters) running at 750 nl min−1. A 60 min gradient (5 to 45% [vol vol−1] acetonitrile; 0.1% [vol vol−1] formic acid) was used for separation (Waters Acquity) and ESI-LC/MS/MS spectra were acquired on an LTQ Velos Orbitrap (Thermo Scientific) running a data-dependent TOP10 method, as previously described (Li et al., 2012). Peak lists were generated using RAW2MSM (Mann lab) and database searching was performed using Paragon within ProteinPilot (ABSciex) (Shilov et al., 2007) with thorough mode (Orbitrap specifications) and a current version (April 2013) of the F. tularensis FASTA protein database (NCBI). False discovery rates (not needed as FDR for top hits was approx. 0%) were determined using the target-decoy approaches in (Elias & Gygi, 2007, Tang et al., 2008). Two independent trapping and mass spectrometry identification analyses were performed, with duplicated proteins considered positive DsbA substrates. Proteins identified from ΔdsbA lysates were designated as non-specific contaminants and were excluded as potential DsbA substrates.
Globomycin inhibition assays
Globomycin (Sigma) was dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 1 mg/mL. Overnight cultures of F. tularensis were grown in Mueller-Hinton medium to an OD600 of 0.2 as described above, globomycin was added to final concentrations of either 1, 10, or 25 µg/ml, or a final concentration of 2.5% DMSO (vol/vol) was added as a control, and cultures were incubated with shaking at 37°C for an additional 12 h. Bacteria were harvested by centrifugation and lysed for SDS-PAGE and immunoblot analyses as described above. EFTu, FopA, Tul4-A, Pal, FTL_1306, FTL_1548, and FTL_1709 were detected in immunoblots using rat or mouse polycloncal antisera.
Supplementary Material
Acknowledgments
We thank Dr. Colin Manoil (University of Washington) for providing pLG66a, Dr. Michael Norgard (U.T. Southwestern Medical Center at Dallas) for providing pTP163, Dr. James Bardwell (Howard Hughes Medical Institute, University of Michigan) for HPLC use, and Drs. Akira Takashima and Robert Blumenthal (University of Toledo Health Science Campus) for scientific discussions and critical review of the manuscript. Finally, we thank Dr. Bill Boggess and The Mass Spectrometry and Proteomics Facility (MSPF) at the University of Notre Dame for assistance with protein identification. This work was supported by the National Institute of Allergy and Infectious Disease of the National Institutes of Health grants K22AI083372 and R01AI093351 to J.F.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Alves NJ, Champion MM, Stefanick JF, Handlogten MW, Moustakas DT, Shi Y, et al. Selective photocrosslinking of functional ligands to antibodies via the conserved nucleotide binding site. Biomaterials. 2013;34:5700–5710. doi: 10.1016/j.biomaterials.2013.03.082. [DOI] [PubMed] [Google Scholar]
- Ashtekar AR, Katz J, Xu Q, Michalek SM. A mucosal subunit vaccine protects against lethal respiratory infection with Francisella tularensis LVS. PLoS One. 2012;7:e50460. doi: 10.1371/journal.pone.0050460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo . Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle JT, Brandt ME, Radolf JD, Norgard MV. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J Bacteriol. 1994;176:2151–2157. doi: 10.1128/jb.176.8.2151-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkmen M, Boyd D, Beckwith J. The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J Biol Chem. 2005;280:11387–11394. doi: 10.1074/jbc.M411774200. [DOI] [PubMed] [Google Scholar]
- Bessette PH, Cotto JJ, Gilbert HF, Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- Broms JE, Sjostedt A, Lavander M. The Role of the Francisella tularensis Pathogenicity Island in Type VI Secretion, Intracellular Survival, and Modulation of Host Cell Signaling. Front Microbiol. 2010;1:136. doi: 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. Virulence factors and their mechanisms of action: the view from a damage-response framework. J Water Health. 2009;7(Suppl 1):S2–S18. doi: 10.2166/wh.2009.036. [DOI] [PubMed] [Google Scholar]
- Celli J, Zahrt TC. Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a010314. doi: 10.1101/cshperspect.a010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain RE. Evaluation of Live Tularemia Vaccine Prepared in a Chemically Defined Medium. Appl Microbiol. 1965;13:232–235. doi: 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Tavender TJ, Oka OB, Warwood S, Knight D, Bulleid NJ. The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1alpha. J Biol Chem. 2010;285:29200–29207. doi: 10.1074/jbc.M110.156596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim N, Harmston CA, Guzman DJ, Goulding CW. Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis . BMC Struct Biol. 2013;13:23. doi: 10.1186/1472-6807-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Celli J. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol. 2010;1:138. doi: 10.3389/fmicb.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Child R, Wehrly TD, Rockx-Brouwer D, Qin A, Mann BJ, Celli J. Structure-Function Analysis of DipA, a Virulence Factor Required for Intracellular Replication. PLoS One. 2013;8:e67965. doi: 10.1371/journal.pone.0067965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, et al. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology. 2011;157:3483–3491. doi: 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Denoncin K, Vertommen D, Paek E, Collet JF. The protein-disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential beta-barrel protein LptD. J Biol Chem. 2010;285:29425–29433. doi: 10.1074/jbc.M110.119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, et al. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Forestal CA, Gil H, Monfett M, Noah CE, Platz GJ, Thanassi DG, et al. A conserved and immunodominant lipoprotein of Francisella tularensis is proinflammatory but not essential for virulence. Microb Pathog. 2008;44:512–523. doi: 10.1016/j.micpath.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, McKevitt M, Ramage ER, Manoil C. Genetic dissection of the Francisella novicida restriction barrier. J Bacteriol. 2008;190:7830–7837. doi: 10.1128/JB.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewska R, Wisniewska K, Pietras Z, Jagusztyn-Krynicka EK. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Lett. 2009;298:1–11. doi: 10.1111/j.1574-6968.2009.01659.x. [DOI] [PubMed] [Google Scholar]
- Graves DB. The emerging role of reactive oxygen and nitrogen species in redox biology and some implications for plasma applications to medicine and biology. J Phys D Appl Phys. 2012:45. [Google Scholar]
- Hazlett KR, Caldon SD, McArthur DG, Cirillo KA, Kirimanjeswara GS, Magguilli ML, et al. Adaptation of Francisella tularensis to the mammalian environment is governed by cues which can be mimicked in vitro. Infect Immun. 2008;76:4479–4488. doi: 10.1128/IAI.00610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- Hiniker A, Collet JF, Bardwell JC. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005;280:33785–33791. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- Hiniker A, Ren G, Heras B, Zheng Y, Laurinec S, Jobson RW, et al. Laboratory evolution of one disulfide isomerase to resemble another. Proc Natl Acad Sci U S A. 2007;104:11670–11675. doi: 10.1073/pnas.0704692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- Huntley JF, Conley PG, Hagman KE, Norgard MV. Characterization of Francisella tularensis outer membrane proteins. J Bacteriol. 2007;189:561–574. doi: 10.1128/JB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley JF, Conley PG, Rasko DA, Hagman KE, Apicella MA, Norgard MV. Native outer membrane proteins protect mice against pulmonary challenge with virulent type A Francisella tularensis . Infect Immun. 2008;76:3664–3671. doi: 10.1128/IAI.00374-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Ichihara S, Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980;255:3707–3712. [PubMed] [Google Scholar]
- Imlay JA. Pathways of oxidative damage. Annual review of microbiology. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Ireland PM, McMahon RM, Marshall LE, Halili M, Furlong E, Tay S, et al. Disarming Burkholderia pseudomallei: Structural and Functional Characterization of a Disulfide Oxidoreductase (DsbA) Required for Virulence In Vivo . Antiox Redox Signal. 2013 doi: 10.1089/ars.2013.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson-Lee M, Garduno RA, Hoffman PS. DsbA2 (27 kDa Com1-like protein) of Legionella pneumophila catalyses extracytoplasmic disulphide-bond formation in proteins including the Dot/Icm type IV secretion system. Mol Microbiol. 2011;80:835–852. doi: 10.1111/j.1365-2958.2011.07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur AV, Koch CJ, Biaglow JE. Mechanism of copper-catalyzed autoxidation of cysteine. Free Radic Res. 1999;31:23–34. doi: 10.1080/10715769900300571. [DOI] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- Katzen F, Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/s0092-8674(00)00180-x. [DOI] [PubMed] [Google Scholar]
- Kaur R, Chen S, Arevalo MT, Xu Q, Chen Y, Zeng M. Protective immunity against tularemia provided by an adenovirus-vectored vaccine expressing Tul4 of Francisella tularensis . Clin Vaccine Immunol. 2012;19:359–364. doi: 10.1128/CVI.05384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella . Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- Kpadeh ZZ, Jameson-Lee M, Yeh AJ, Chertihin O, Shumilin IA, Dey R, et al. Disulfide bond oxidoreductase DsbA2 of Legionella pneumophila exhibits protein disulfide isomerase activity. J Bacteriol. 2013;195:1825–1833. doi: 10.1128/JB.01949-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G, Lundstrom J, Barea JL, Pueyo de la Cuesta C, Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- Li Y, Champion MM, Sun L, Champion PA, Wojcik R, Dovichi NJ. Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry as an alternative proteomics platform to ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry for samples of intermediate complexity. Anal Chem. 2012;84:1617–1622. doi: 10.1021/ac202899p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H, Shen H, Zingmark C, Golovliov I, Conlan W, Sjostedt A. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect Immun. 2007;75:1303–1309. doi: 10.1128/IAI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llarrull LI, Toth M, Champion MM, Mobashery S. Activation of BlaR1 protein of methicillin-resistant Staphylococcus aureus, its proteolytic processing, and recovery from induction of resistance. J Biol Chem. 2011;286:38148–38158. doi: 10.1074/jbc.M111.288985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahawar M, Atianand MK, Dotson RJ, Mora V, Rabadi SM, Metzger DW, et al. Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J Biol Chem. 2012;287:25216–25229. doi: 10.1074/jbc.M112.367672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahawar M, Rabadi SM, Banik S, Catlett SV, Metzger DW, Malik M, Bakshi CS. Identification of a live attenuated vaccine candidate for tularemia prophylaxis. PLoS One. 2013;8:e61539. doi: 10.1371/journal.pone.0061539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier TM, Havig A, Casey M, Nano FE, Frank DW, Zahrt TC. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol. 2004;70:7511–7519. doi: 10.1128/AEM.70.12.7511-7519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ. Functional advantages conferred by extracellular prokaryotic membrane vesicles. J Mol Microbiol Biotechnol. 2013;23:131–141. doi: 10.1159/000346548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, Bardwell JC, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- Matsui Lee IS, Suzuki M, Hayashi N, Hu J, Van Eldik LJ, Titani K, Nishikimi M. Copper-dependent formation of disulfide-linked dimer of S100B protein. Arch Biochem Biophys. 2000;374:137–141. doi: 10.1006/abbi.1999.1595. [DOI] [PubMed] [Google Scholar]
- McCaffrey RL, Schwartz JT, Lindemann SR, Moreland JG, Buchan BW, Jones BD, Allen LA. Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis . J Leukoc Biol. 2010;88:791–805. doi: 10.1189/jlb.1209811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon RM, Premkumar L, Martin JL. Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbapap.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Melillo AA, Bakshi CS, Melendez JA. Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J Biol Chem. 2010;285:27553–27560. doi: 10.1074/jbc.M110.144394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo AA, Mahawar M, Sellati TJ, Malik M, Metzger DW, Melendez JA, Bakshi CS. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J Bacteriol. 2009;191:6447–6456. doi: 10.1128/JB.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Rajaram MV, Strandberg KL, Gunn JS. Type A Francisella tularensis acid phosphatases contribute to pathogenesis. PLoS One. 2013;8:e56834. doi: 10.1371/journal.pone.0056834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Mann BJ. Francisella tularensis subsp. tularensis Schu S4 disulfide bond formation protein B, but not an RND-type efflux pump, is required for virulence. Infect Immun. 2008;76:3086–3092. doi: 10.1128/IAI.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Rabideau MM, Moore EA, Mann BJ. Requirement of the CXXC motif of novel Francisella infectivity potentiator protein B FipB, and FipA in virulence of F. tularensis subsp. tularensis . PLoS One. 2011;6:e24611. doi: 10.1371/journal.pone.0024611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Thompson JA, Mann BJ. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun. 2009;77:152–161. doi: 10.1128/IAI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Zhang Y, Clark ME, Rabideau MM, Millan Barea LR, Mann BJ. FipB, an essential virulence factor of Francisella tularensis subspecies tularensis, has dual roles in disulfide bond formation. J Bacteriol. 2014 doi: 10.1128/JB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Schneider I, Pan J, Von Hacht A, Bardwell JC. The CXXC motif is more than a redox rheostat. J Biol Chem. 2007;282:28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- Radolf JD, Chamberlain NR, Clausell A, Norgard MV. Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent triton X-114. Infect Immun. 1988;56:490–498. doi: 10.1128/iai.56.2.490-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- Ren G, Stephan D, Xu Z, Zheng Y, Tang D, Harrison RS, et al. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem. 2009;284:10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GT, Case ED, Dobbs N, Ingle C, Balaban M, Celli J, Norgard MV. FTT0831c/FTL_0325 contributes to Francisella tularensis cell division, maintenance of cell shape, and structural integrity. Infect Immun. 2014 doi: 10.1128/IAI.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GT, Child R, Ingle C, Celli J, Norgard MV. IglE is an outer membrane-associated lipoprotein essential for intracellular survival and murine virulence of type A Francisella tularensis . Infect Immun. 2013;81:4026–4040. doi: 10.1128/IAI.00595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L, Brittnacher M, Svensson K, Buckley D, Haugen E, Zhou Y, et al. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect Immun. 2006;74:6895–6906. doi: 10.1128/IAI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsson EN, Forslund AL, Forsberg A. Type IV Pili in Francisella - A Virulence Trait in an Intracellular Pathogen. Front Microbiol. 2011;2:29. doi: 10.3389/fmicb.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Klimentova J, Rehulka P, Straskova A, Spidlova P, Szotakova B, et al. Francisella tularensis subsp. holarctica DsbA homologue: a thioredoxin-like protein with chaperone function. Microbiology. 2013;159:2364–2374. doi: 10.1099/mic.0.070516-0. [DOI] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics : MCP. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Sjostedt A, Tarnvik A, Sandstrom G. The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect Immun. 1991;59:3163–3168. doi: 10.1128/iai.59.9.3163-3168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford SJ, Humphreys DP, Lund PA. Mutations in dsbA and dsbB, but not dsbC, lead to an enhanced sensitivity of Escherichia coli to Hg2+ and Cd2+ FEMS Microbiol Lett. 1999;174:179–184. doi: 10.1111/j.1574-6968.1999.tb13566.x. [DOI] [PubMed] [Google Scholar]
- Straskova A, Pavkova I, Link M, Forslund AL, Kuoppa K, Noppa L, et al. Proteome analysis of an attenuated Francisella tularensis dsbA mutant: identification of potential DsbA substrate proteins. J Proteome Res. 2009;8:5336–5346. doi: 10.1021/pr900570b. [DOI] [PubMed] [Google Scholar]
- Sturm N, Jortzik E, Mailu BM, Koncarevic S, Deponte M, Forchhammer K, et al. Identification of proteins targeted by the thioredoxin superfamily in Plasmodium falciparum . PLoS Pathog. 2009;5:e1000383. doi: 10.1371/journal.ppat.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7:3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- Thi EP, Lambertz U, Reiner NE. Sleeping with the enemy: how intracellular pathogens cope with a macrophage lifestyle. PLoS Pathog. 2012;8:e1002551. doi: 10.1371/journal.ppat.1002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley CR, Voulhoux R, Beretti JL, Tommassen J, Nassif X. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J Biol Chem. 2004;279:27078–27087. doi: 10.1074/jbc.M313404200. [DOI] [PubMed] [Google Scholar]
- Valentino MD, Hensley LL, Skrombolas D, McPherson PL, Woolard MD, Kawula TH, et al. Identification of a dominant CD4 T cell epitope in the membrane lipoprotein Tul4 from Francisella tularensis LVS. Molecular immunology. 2009;46:1830–1838. doi: 10.1016/j.molimm.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee BL, Ulmer DD. Biochemical effects of mercury, cadmium, and lead. Ann Rev Biochem. 1972;41:91–128. doi: 10.1146/annurev.bi.41.070172.000515. [DOI] [PubMed] [Google Scholar]
- Vecerkova R, Hernychova L, Dobes P, Vrba J, Josypcuk B, Bartosik M, Vacek J. Investigation of protein FTT1103 electroactivity using carbon and mercury electrodes. Surface-inhibition approach for disulfide oxidoreductases using silver amalgam powder. Anal Chim Acta. 2014;830:23–31. doi: 10.1016/j.aca.2014.04.056. [DOI] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Jaenicke R, Glockshuber R. The redox properties of protein disulfide isomerase (DsbA) of Escherichia coli result from a tense conformation of its oxidized form. J Mol Biol. 1993;233:559–566. doi: 10.1006/jmbi.1993.1535. [DOI] [PubMed] [Google Scholar]
- Zapun A, Missiakas D, Raina S, Creighton TE. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli . Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.