Summary
Previously observed effects of rifampicin and chloramphenicol indicate that transcription and translation activity strongly affect the coarse spatial organization of the bacterial cytoplasm. Single-cell, time-resolved, quantitative imaging of chromosome and ribosome spatial distributions and ribosome diffusion in live E. coli provides insight into the underlying mechanisms. Monte Carlo simulations of model DNA-ribosome mixtures support a novel nucleoid-ribosome mixing hypothesis. In normal conditions, 70S-polysomes and the chromosomal DNA segregate, while 30S and 50S ribosomal subunits are able to penetrate the nucleoids. Growth conditions and drug treatments determine the partitioning of ribosomes into 70S-polysomes vs free 30S and 50S subunits. Entropic and excluded volume effects then dictate the resulting chromosome and ribosome spatial distributions. Direct observation of radial contraction of the nucleoids 0-5 min after treatment with either transcription- or translation-halting drugs supports the hypothesis that simultaneous transcription, translation, and insertion of proteins into the membrane (“transertion”) exerts an expanding force on the chromosomal DNA. Breaking of the DNA-RNA polymerase-mRNA-ribosome-membrane chain in either of two ways causes similar nucleoid contraction on a similar timescale. We suggest that chromosomal expansion due to transertion enables co-transcriptional translation throughout the nucleoids.
Keywords: Bacterial organization, Nucleoid, Bacterial chromosome, Ribosome, Transertion, Drug effects, Single-molecule tracking, Live-cell fluorescence microscopy
Introduction
The structure and dynamics of the bacterial chromosome are not well understood. The genome of E. coli consists of 4.6 million base pairs (Blattner et al., 1997), with contour length of 1.5 mm. A rapidly growing E. coli cell is a rod of length ~4 μm and diameter ~1 μm. The cytoplasm contains multiple genome equivalents of chromosomal DNA that occupy an irregular sub-region called the nucleoids (Robinow & Kellenberger, 1994). In spite of the large amount of DNA, the nucleoids do not fill the entire volume of the cytoplasm (Robinow & Kellenberger, 1994, Pettijohn, 1982). The detailed shape of the nucleoids may be governed by the ring topology of DNA and spatial confinement effects on the ring polymer (Jung et al., 2012, Youngren et al., 2014). The overall spatial extent of the nucleoids has been suggested to arise from a balance of nucleoid-expanding and nucleoid-compacting forces (Zimmerman, 2006, Woldringh et al., 1995).
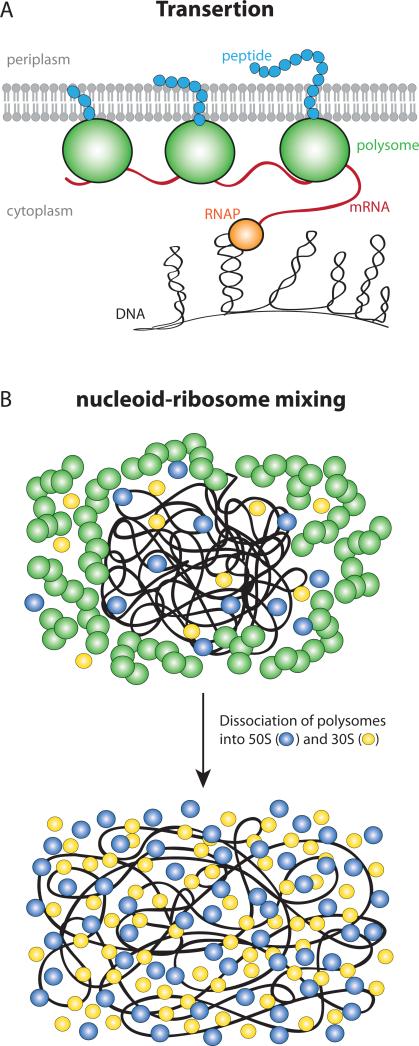
The primary hypothesized expanding force is “transertion”, the simultaneous transcription, translation, and insertion of proteins into the cytoplasmic membrane (Woldringh, 2002, Woldringh et al., 1995). Transertion is viewed as a dynamic process in which numerous membrane-DNA tethers are constantly forming and breaking during normal transcription and translation events (Woldringh, 2002, Norris & Madsen, 1995). Compacting forces on the chromosomal DNA may include depletion-attraction of DNA arising from macromolecular crowding by myriad small proteins (Zimmerman & Murphy, 1996); configurational entropy of the confined DNA polymer, causing the polymer to avoid walls (Mondal et al., 2011); inter-strand coupling by DNA binding proteins such as H-NS (Dame, 2005, Wang et al., 2011); bending of DNA by IHF (Dame, 2005); and net supercoiling of the DNA by Gyrase and Topoisomerase I (Woldringh et al., 1995).
The effects of drugs that inhibit transcription or translation on nucleoid morphology has led to the suggestion that these biochemical processes also play important roles in determining overall nucleoid morphology (van Helvoort et al., 1996, Zimmerman, 2002, Jin et al., 2013, Cabrera et al., 2009, Chai et al., 2014, Sanamrad et al., 2014). Chloramphenicol (Cam) treatment, which halts translation elongation by blocking access of charged tRNA to the ribosomal A site (Wolfe & Hahn, 1965, Sohmen et al., 2009, Hansen et al., 2003), leads to compaction of the chromosomal DNA (van Helvoort et al., 1996, Zimmerman, 2002). This effect plus observation of net migration of membrane protein genes towards the cell membrane after expression is turned on (Libby et al., 2012) provide the primary evidence in support of the transertion hypothesis. However, under the transertion hypothesis the nucleoids should also contract after treatment with rifampicin (Rif), which blocks transcription initiation. Instead, several labs have observed nucleoid expansion after rifampicin treatment, typically ~30 min after addition of the drug (Fishov & Woldringh, 1999, Cabrera et al., 2009).
Here we use quantitative single-cell fluorescence microscopy to examine drug effects on chromosome and ribosome spatial distributions and on ribosomal diffusion in living E. coli cells, in real time with sub-minute time resolution. We are able to discern subtle effects on nucleoid spatial extent on both short (0-5 min) and long (5-20 min) timescales. Some key effects are lost in static images of cells sampled, fixed, and imaged long after drug addition.
One significant finding is that both Rif and Cam cause nucleoid contraction on the 0-5 min timescale. This better supports the transertion hypothesis. In addition, the new data motivate a novel nucleoid-ribosome mixing hypothesis, which views the chromosome and ribosomes as a coupled physical-biochemical system. The physiology of normally growing cells or drug-treated cells determines the partitioning of ribosome components between 70S-polysomes and free 30S and 50S ribosomal subunits. That partitioning combined with the presence or absence of transertion then dictates the spatial distributions of the chromosomal DNA and ribosomes, governed by the physical effects of excluded volume, configurational entropy, and translational entropy (Mondal et al., 2011).
In normal growth conditions, transertion expands the nucleoids to a degree that permits 30S and 50S to penetrate and initiate co-transcriptional translation. However, 70S-polysomes are excluded, causing strong overall segregation of ribosomes from chromosomal DNA. Most protein production occurs in the ribosome-rich regions (Bakshi, et al., 2012). Cam treatment inhibits translation elongation, which terminates transertion, converts almost all 30S subunits to 70S-polysomes, and causes essentially complete segregation of ribosomes from the highly compacted nucleoids. Rif treatment inhibits transcription initiation. On long timescales, existing mRNA is degraded and almost all 70S-polysomes are converted to 30S and 50S subunits. Chromosomal DNA and ribosomal subunits mix in order to maximize total entropy, and the nucleoids expand to fill most of the cytoplasm. This nucleoid-ribosome mixing hypothesis is supported by observations of DNA and 30S-YFP spatial distributions in normal and drug-treated cells, by measurements of 30S diffusion vs time after drug treatment, and by a simple statistical model of DNA and ribosome components confined in a spherocylindrical volume (Mondal et al., 2011).
In this picture, transertion enables ribosomal subunits to penetrate the nucleoids and initiate co-transcriptional translation. That in turn provides important protection of nascent mRNA (Deana & Belaska, 2005, Deneke, et al., 2013) and prevents backtracking of RNA polymerase (McGary & Nudler, 2013). Thus transertion is essential not only for insertion of membrane proteins, but also for optimal use of ribosomes and RNAP.
Results
Chromosome spatial distribution in normally growing cells
All the strains in this study use the same VH1000 background (MG1655: lacI- lacZ- pyrE+) (Jasiecki & Wegrzyn, 2003). Cells are grown in EZ rich, defined medium (EZRDM) at 30°C and observed in a simple microfluidics chamber at 30°C. The bulk doubling time is 43 min. Instead of DAPI, we use the DNA stain SYTOX Orange to image nucleoid morphology. SYTOX Orange enables long-term, non-perturbative imaging of chromosomal DNA in live bacteria cells without affecting growth (Bakshi et al., 2014). Alternating phase contrast and SYTOX Orange fluorescence images were obtained at 6-s intervals (12-s cycle time) to monitor the nucleoid morphology and cell growth over time. The exposure time is 50 ms during each frame.
In the absence of antibiotics the cells grow and divide normally (Bakshi et al., 2014). In Fig. 1A and B, phase contrast images and SYTOX Orange fluorescence images are shown over 25 min for one fairly long cell. In our moderately fast growth conditions, most cells exhibit two major lobes of DNA. The phase contrast images show gradual cell growth and development of a septum. The fluorescence images show gradually increasing separation between major nucleoid lobes and evolution in time of more detailed structure within each major lobe. The phase contrast images are analyzed to obtain the relative tip-to-tip cell length vs time: Lcell (t)/Lcell (t = 0) (Sec. S1 in SI). Without staining or laser illumination, relative cell length increases in exponential fashion with a mean doubling time of 48 ± 3 min (Fig. 1D). We showed earlier (Bakshi et al., 2014) that neither Sytox Orange staining nor this level of laser illumination affects the mean doubling time or the plots of Lcell (t).
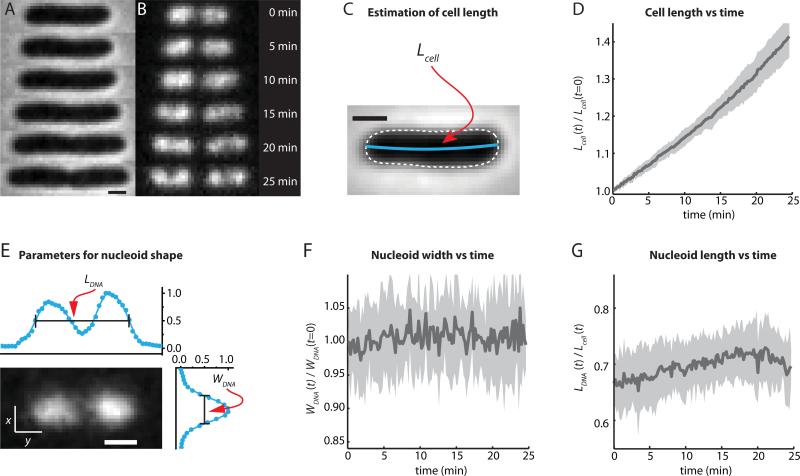
Figure 1.
E. coli nucleoid morphology vs time from SYTOX Orange staining of a single cell. (A) Time course of phase contrast images. (B) Time course of SYTOX Orange-stained DNA images. (C) Length of the cell (Lcell) is determined from contour length of the central spine of the phase contrast image. (D) Normalized cell length vs time for 12 cells from the same field of view. The mean is plotted as a thick black line. The grey swath shows the extremes of cell-to-cell variability. The culture is not synchronized. Part of the cell-to-cell variability is due to the time-independent contributions of the two endcaps to total cell length, a larger fraction of Lcell for short cells than for long cells. (E) Fluorescence image of a cell stained with SYTOX Orange and imaged with the 561 nm laser, defining the nucleoid length (LDNA) and width (WDNA). (F) Width WDNA (t) of the nucleoids vs time for the same 12 cells as in (D), normalized to WDNA (t = 0). (G) Length of the nucleoids LDNA (t) relative to the cell length Lcell (t) for the same 12 cells. Scale bars in (A), (C), and (E) are 1 μm. The mean behavior shown in panels D, F, and G reproduced across five separate experiments with at least 10 cells each.
The orientation of the cell is determined by fitting an ellipse to the binary mask generated from the phase contrast image. The two principal axes of the cell are used as the coordinate system. The long axis is referred to as x (axial dimension) and the short axis is referred to as y (transverse dimension). In order to quantitatively describe the overall spatial distribution of the chromosomal DNA vs time, we define two parameters measured from the SYTOX Orange fluorescence intensity distributions projected along x and y (Bakshi et al., 2014). The axial distribution was characterized by the overall length LDNA, measured as the “outside” full-width at half-maximum height (FWHM) of the intensity distribution along x (Fig. 1E). Cells that septate during the observation period are excluded from the analysis of Fig. 1D, F, and G. The included cells are short at t = 0 and lengthen without evidence of septation over 25 min. The width WDNA, a rough measure of the mean nucleoid diameter, is defined as the FWHM of the projection of intensity along the transverse coordinate y (Fig. 1E). For normal cell growth, the measured width of the nucleoid is constant over time to within ±2% (Fig. 1F). The nucleoid length increases roughly proportionally to cell length (Fig. 2D in (Bakshi et al., 2014)). To partially compensate for overall cell and nucleoid growth over 25 min, we find it useful to present the nucleoid length relative to the tip-to-tip cell length, LDNA (t) /Lcell (t), as in Fig. 1G. Because we omit cells that septate within 25 min and the nucleoids never occupy the endcap regions (which contribute a constant amount to Lcell), the ratio LDNA (t) /Lcell (t) gradually increases by about 8% over 20 min.
Figure 2.
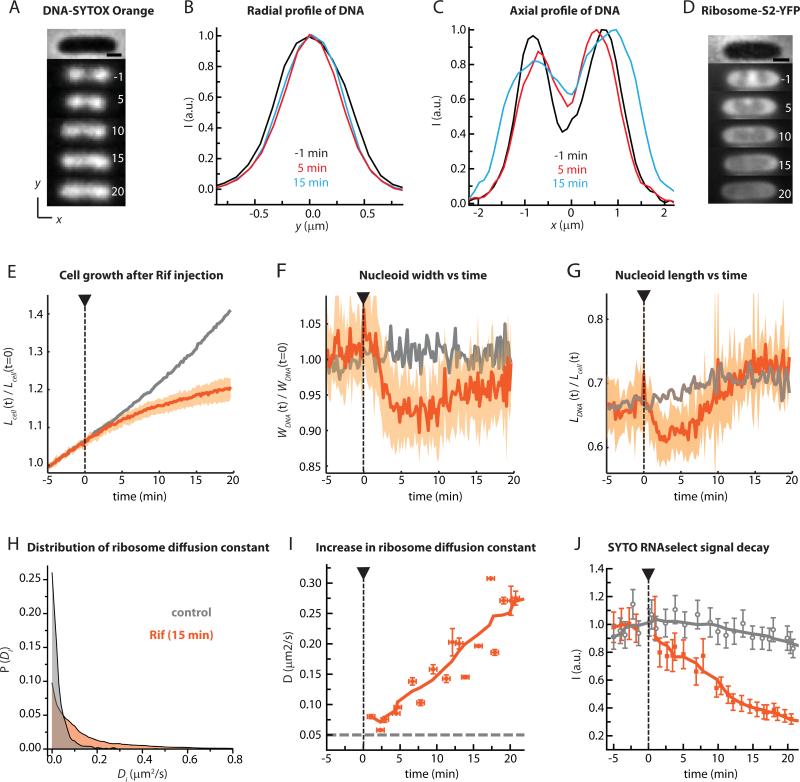
Effect of rifampicin on nucleoid morphology and ribosome spatial distribution. (A) Snapshots of a SYTOX Orange-stained cell before and after injection of Rif (300 μg/mL) at t = 0. Times in minutes. The phase contrast image of the cell is shown at top. Scale bar =1 μm. (B) SYTOX Orange radial fluorescence intensity profile (along the short, y-axis) for three different times. (C) SYTOX Orange axial fluorescence intensity profile (along the x-axis) for three different times. (D) Snapshots of ribosomal 30S subunits labeled as S2-YFP from a different Rif-treated cell of similar length to that in (A). Scale bar =1 μm. (E) Mean normalized cell length vs time (thick orange line) and range of lengths (orange swath) for 36 Rif-treated cells. Grey line is mean for untreated cells. (F) Mean and range of normalized nucleoid width vs time for 11 Rif-treated cells (orange) from the same experiment. Grey is mean for untreated cells. The experiment was repeated five times and consistently yielded the same result. (G) Mean and range of normalized nucleoid length vs time for 11 Rif-treated cells (orange). Grey is mean for untreated cells. (H) Distribution of single-molecule diffusion constants Di from 30S-mEos2 trajectories of untreated cells (grey) and Rif-treated cells (orange). Di values are estimated from 6-step, 60 ms/frame trajectories with lag time τ = 180 ms. (I) Mean diffusion constant of the 30S-mEos2 label at different times after injection of Rif. Horizontal bars represent the duration of data acquisition; vertical bars represent estimated errors in initial slope of MSD plots. (J) Orange: Mean total intensity of SYTO RNAselect fluorescence from cells at different time lags after Rif injection. Grey: Control cells growing without Rif treatment. The mean behavior shown in panels E, F, and G reproduced across five separate experiments with at least 10 cells each.
Time-dependent effects of rifampicin (Rif) on the chromosome spatial distribution
Rifampicin (Rif) is a well-known transcription inhibitor that prevents bacterial RNA polymerase (RNAP) from entering the mRNA elongation phase (Campbell et al., 2001). RNAP stalls at the initiation stage. Copies already engaged in elongation continue transcribing until the message is complete. Over a timescale of ~30 s, Rif should gradually prevent synthesis of new mRNA transcripts (Proshkin et al., 2010). The average half-life of complete mRNA transcripts in E. coli is ~5 min (Bernstein et al., 2002). Therefore, on a timescale of ~15 min, ~90% of the cellular mRNA will have been partially degraded by ribonucleases. Ribosomes that were translating mRNA when Rif is added should complete protein synthesis in ~30 s on average and dissociate to 30S and 50S subunits (Forschhamer & Lindahl, 1971, Proshkin et al., 2010). As mRNA is degraded, free 30S subunits of ribosomes will no longer be able to find mRNA to translate. The 30S subunit population should gradually be converted from 70S-polysomes to free 30S and 50S subunits.
We stained cells with SYTOX Orange and plated them in the microfluidics chamber. At t = 0, we injected 300 μg/mL of Rif (20 times the MIC, Fig. S2 in SI). The observations were similar at 100 μg/mL or 600 μg/mL Rif. Supporting Movie 1 shows the effects of Rif on several cells. Snapshots of the SYTOX Orange-stained nucleoids from one representative short cell at different time points before and after Rif injection are shown in Fig. 2A; compare the normally growing cell in Fig. 1A. Intensity profiles of the SYTOX Orange fluorescence along the y-axis (Fig. 2B) and the x-axis (Fig. 2C) are shown for three time points during the experiment. Within 5 min of the injection of Rif, the nucleoid spatial distribution has changed significantly. The nucleoid width WDNA has decreased from 810 nm to 745 nm (a decrease of ~8%, Fig. 2B). On the same 5-min timescale, the two axial peaks move closer together as the dip between major nucleoid lobes fills in substantially (Fig. 2C). Total nucleoid length LDNA decreases slightly. During the interval 5-20 min, LDNA reverses, eventually expanding beyond the normal length as the two axial intensity peaks broaden and separate again. At all times after 2 min the nucleoid width remains significantly smaller than in normal cells (Fig. 2F).
These short-time size effects are subtle, but they are corroborated by plots of the mean behavior across cells. Each field of view contains some 50 cells. To avoid complications of septation and multi-peaked nucleoids, we typically give results only for the 10-15 cells that are short at t = 0 (Lcell < 4 μm) and do not show visible evidence of septation during the 25-min observation period. Throughout this paper (including Figs. 1, 2, and 4), we display data for 10-15 cells from a single field of view in a single experiment in order to ensure the same drug injection timing for all cells. In all cases, the results were highly reproducible across at least five experiments including at least 10 cells each.
Figure 4.
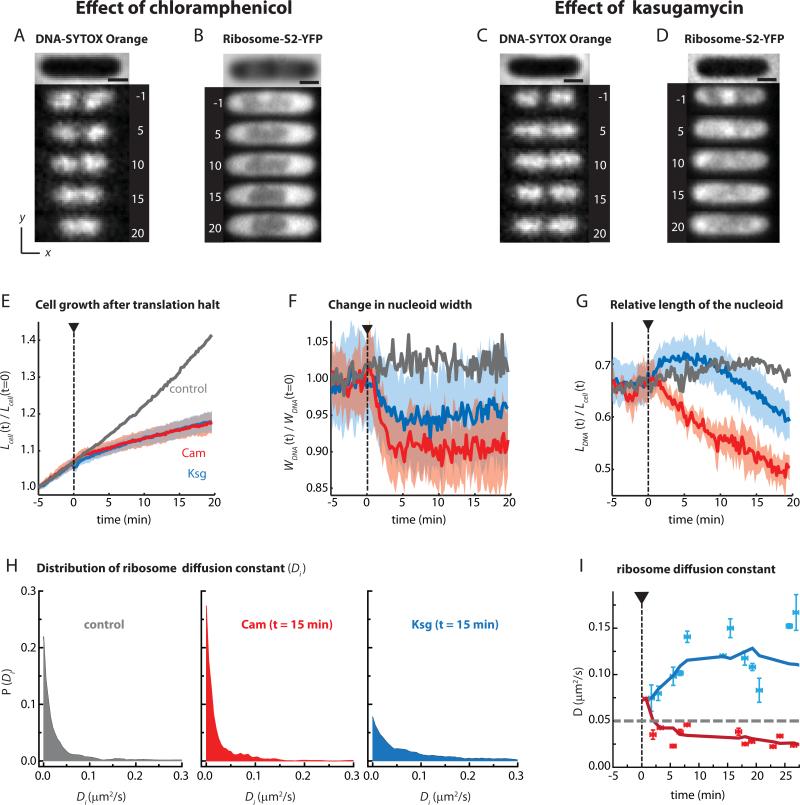
Effect of translation inhibitors on nucleoid morphology and ribosome distribution. (A) Snapshots of SYTOX Orange-stained cell before and after injection of Cam (300 μg/mL) at t = 0. Times in minutes. The phase contrast image of the cell is shown at top. (B) Snapshots of ribosome (30S-YFP) distribution from another Cam-treated cell of comparable size. (C and D) Snapshots of one SYTOX Orange-stained cell and a different 30S-YFP labeled cells before and after injection of Ksg (5 mg/mL) at t = 0. Times in minutes. Normalized cell length (E), normalized nucleoid width (F) and relative nucleoid length (G) are plotted vs time for cells treated with Cam (300 μg/mL) and Ksg (5 mg/mL). The mean trend is the thick line. The colored swath shows the cell-to-cell variability. Ncell = 13 for Cam and 18 for Ksg. Mean of data from untreated cells is heavy grey line. Cells treated with Cam are Ksg are shown in red and blue, respectively. (H) Distribution of ribosome (30S-YFP) diffusion constants Di for untreated cells (left, gray), cells treated with Cam for 15 min (middle, red), and cells treated with Ksg for 15 min (right, blue). Di values are estimated from 6-step, 100 ms/frame trajectories with lag time τ = 300 ms. (I) Mean ribosome (30S-YFP) diffusion constant from MSD plots at different time intervals after injection of Cam (300 μg/mL) and Ksg (5 mg/mL). Heavy lines are from 5-point smoothing of data. Scale bars in parts (A) to (D) are 1 μm. The mean behavior shown in panels E, F, and G reproduced across five separate experiments with at least 10 cells each.
In Fig. 2E-G, we display the mean trend (heavy line) and range (shaded swaths) of normalized cell length Lcell(t)/ Lcell(t = 0), normalized nucleoid width WDNA(t)/WDNA(t = 0), and relative nucleoid length LDNA(t)/ Lcell(t) for 11 cells from a single field of view following Rif treatment. Individual traces for all the cells are provided in Fig. S3. Rif effects on longer cells are similar (data not shown). Prior to injection of Rif, cells were growing normally as monitored by relative cell length (Fig. 2E, orange). The mean relative cell length shows discernible deviation from normal growth within 2–3 min of drug injection. The mean relative nucleoid width and length both begin to decrease within 1-2 min of drug injection, suggesting the drug substantially penetrates both cell membranes on a timescale of ~1 min. On average, the nucleoids continue to contract in both length and width for 3-4 minutes after Rif injection. Relative length expands over the following 15 min, exceeding that of control cells 20 min after Rif injection. The width expands slightly over the interval t = 5-20 min, but remains significantly smaller than in untreated cells.
Although Rif is known to bind specifically to bacterial RNAP, it is possible that non-specific interactions of the drug might obscure the true effect of transcription halting on nucleoid morphology and 30S diffusion. As a control, we repeated the SYTOX Orange experiments using the RifR mutant strain SB7. RifR cells continue to grow normally after rifampicin injection (Fig. S4B) and the morphology of the nucleoids remains normal (Fig. S4A and D). The nucleoids in RifR cells undergoes radial contraction upon Cam treatment (Fig. S4B and D), as observed with VH1000 cells (described below). Evidently the nucleoid radial contraction at t = 0-5 min after Rif treatment of the normal VH1000 strain is induced by the halting of transcription elongation, not by some other non-specific effect of the drug.
Time-dependent effects of rifampicin (Rif) on ribosome spatial distribution and diffusion
The short-time contraction of the nucleoid width supports the transertion hypothesis. The nucleoid-ribosome mixing hypothesis predicts that the initially segregated chromosomal DNA will expand and mix with 30S and 50S monomeric subunits on the timescale of overall mRNA degradation. We therefore tested whether or not the 5-20 minute timescale of axial nucleoid expansion, the timescale of nucleoid-ribosome desegregation, and the timescale of mRNA degradation are comparable. Furthermore, if 70S-polysomes are converting to free 30S and 50S subunits, then the diffusion of 30S subunits should be enhanced on the same timescale.
The changes in nucleoid morphology (Fig. 2F, G) indeed correlate in time with changes in the distribution of 30S-YFP ribosomal subunits (example in Fig. 2D; details in Fig. S5). The labeling is due to expression of the ribosomal protein S2-YFP from the chromosome (Bakshi et al., 2012). Because the 10-min maturation time of YFP (Nagai et al., 2002) is much longer than the ~2-min assembly time of 30S subunits (Lindahl, 1975), we expect most observable S2-YFP copies to be incorporated into 30S subunits. However, a 30S-YFP subunit could be free or bound as a full 70S ribosome translating mRNA, usually a member of a 70S-polysome chain.
As described in our previous work (Bakshi et al., 2012), in normal growth in EZRDM medium at 30°C (45-min doubling time), ribosomes are highly segregated from the E. coli chromosomal DNA. Peaks in the ribosome distribution faithfully correspond to dips in the DNA density distribution. Before Rif injection, the axial 30S distribution in Figs. 2D and S5 exhibits three “ribosome-rich regions”, two in the endcaps and one at the cell center (between the two major nucleoid lobes). Over 5 min, the period during which the gap between major nucleoid lobes narrows, the central ribosome peak diminishes in relative intensity; the axial DNA contraction seems to displace ribosomes. The two ribosome-rich endcap regions persist and expand axially, seemingly to accommodate the “extra” ribosomes (Fig. S5E). Over the same timescale, the transverse 30S profile (along y) develops modest peaks at y = ±400 nm, as if the radial DNA contraction has opened up additional space for ribosomes near the cylindrical cell membrane (Fig. S5F). Up to t = 5 min, DNA-ribosome spatial segregation remains strong. Over the longer interval t = 5-20 min, extensive mixing occurs. As the nucleoids gradually expand axially, the spatial distribution of 30S subunits gradually becomes more uniform, seemingly occupying the entire cytoplasm (Fig. S5D). All three axial 30S peaks are diminished and the overall distribution becomes much more uniform. However, even at t = 25 min, the nucleoid-30S mixing remains incomplete. For two-color imaging of ribosomes and DNA in the same cell 30 min after Rif treatment, both radial and axial profiles of the 30S subunits stay wider than those of the DNA (Fig. S6). The chromosomal DNA continues to avoid the cytoplasmic membrane, enabling the 30S subunits to concentrate near the membrane.
We expect free 30S subunits to diffuse more rapidly than 30S subunits incorporated into 70S-polysomes. Thus we also studied the time dependence of single-molecule diffusion of 30S subunits labeled by an S2-mEos2 construct expressed from the chromosome (Bakshi et al., 2012). As before, images were collected with 10 ms exposure time and 30 ms between frames. As shown in Fig. 2H, the distribution of estimated single-molecule diffusion constants P(Di) at t = 15 min after Rif treatment exhibits a long tail compared with that of untreated cells. The mean 30S diffusion constant <Di> was extracted from the initial slope of MSDr(τ) plots (Bakshiet al., 2012, Bakshi et al., 2011); see Sec. S7 of SI for details. Its value increases monotonically by a factor of 4 over 20 min following Rif treatment, from 0.07 μm2-s−1 to 0.3 μm2-s−1 (Fig. 2I). The enhanced diffusion is consistent with conversion of 70S-polysomes to free 30S subunits.
To examine whether free 30S subunits that dissociate from 70S-polysomes after Rif treatment are able to sample the entire length of the cytoplasm over minutes, we used a photoactivation-chase method to monitor longer term dynamics of 30S-mEos2 (SI Sec. S8). After 30 min of Rif treatment, 30S diffusion fits a model of free diffusion with D30S ~ 0.5 μm2/s. This is consistent with the value of 0.45 μm2/s obtained earlier from single-molecule tracking data (Bakshi et al., 2012).
We also monitored the concentration of mRNA vs time using SYTO RNASelect, a permeable dye that becomes fluorescent on binding to RNA and is five-fold less sensitive to DNA. Over 20 min following Rif treatment, the mean intensity of SYTO RNASelect fluorescence gradually decreases to 30% of its initial value (Fig. 2J, Fig. S9). The control curve for normal cells shows that the decay after Rif treatment is not due to photobleaching. The timescale of mRNA decay semi-quantitatively matches the timescales of enhancement of 30S diffusion and of strong DNA-30S mixing. The timescale of SYTO RNAselect signal loss is significantly longer than the ~5-min mean half-life of full-length mRNA transcripts in E. coli (Bernstein et al., 2002). It seems likely that SYTO RNAselect would bind to the shorter, cleaved intermediate products of mRNA degradation by ribonucleases. The total population of such intermediates should persist longer than the parent full-length transcripts, since many successive degradation steps are required to abrogate SYTO RNAselect binding. As a control, we monitored SYTO RNAselect intensity in RifR (SB7) cells after Rif injection (data not shown). No significant decay in the SYTO RNASelect signal was observed over 30 min. This eliminates the possibility of a direct effect of Rif on the SYTO RNAselect signal.
In summary, the Rif-induced short-time contraction of the nucleoids in the radial dimension (t = 0-5 min, Fig. 2F) is qualitatively consistent with the transertion hypothesis. If transertion indeed provides a significant expanding force on the nucleoids in untreated cells, the timescale of the Rif-induced contraction would be determined by the penetration time of the drug (apparently ~1-2 min; see Fig. 2), the time to completion of transcription of genes for typical membrane proteins (estimated to be ~30 s, Bremer, Chung et al., 2001), and the relaxation time of the released DNA polymer configuration (timescale unknown). Short-time radial contraction is consistent with loss of transertion. The short-time axial contraction may also arise from loss of transertion; see Discussion below. Later, the nucleoids undergo expansion on the same 5-20 min timescale as mRNA degradation, as observed previously. The mean diffusion coefficient of 30S subunits increases significantly as 70S-polysomes dissociate and the chromosomal DNA mixes with free 30S subunits. These results are consistent with the nucleoidribosome mixing hypothesis, which asserts that after Rif treatment free 30S and 50S subunits can mix with the chromosomal DNA. Next we test that concept against a physical model of DNA-ribosome mixing.
Monte Carlo simulation of rifampicin effects
In earlier work we developed a simple physical model incorporating realistic densities of plectonemic DNA and of 70S-polysomes, and including only excluded volume and entropic effects (Mondal et al., 2011). That model was designed to mimic normal growth conditions, in which some 80% of 30S and 50S subunits are present as complete 70S ribosomes, usually in the form of polysomes. The model does not include effects of transertion, although this could be incorporated into future work. The model placed one chromosome equivalent of model DNA plus 20,000 70S particles organized as polysome 13-mers into a spherocylinder. We found strong segregation of polysomes from DNA arising purely from excluded volume effects and entropy maximization (Fig. 3B). Our interpretation was that the DNA polymer avoids the wall to maximize its configurational entropy. The polysomes occupy the regions near the wall and the endcaps to maximize their translational entropy (motion in space) and configurational entropy. The mutual interaction of chromosomal DNA and polysomes results in compression of the DNA, which preferentially distributes near the center of the cell and away from the endcaps and the cylindrical wall.
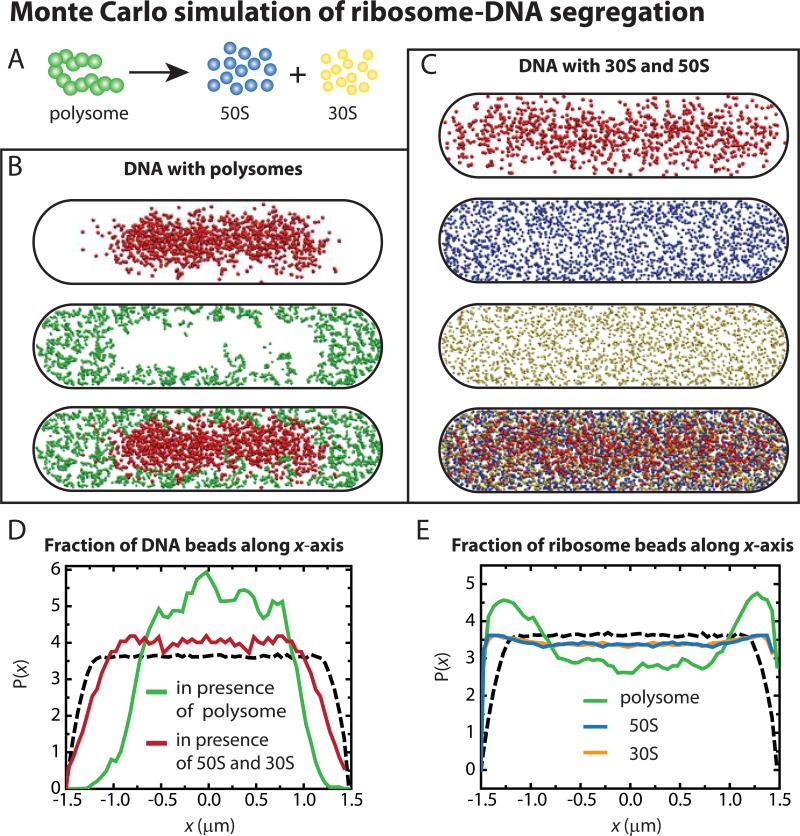
Figure 3.
(A) Model polysome of thirteen 70S beads (20-nm diameter) dissociates into 50S beads (17 nm diameter) and 30S beads (14 nm diameter). (B) Equilibrated distribution of DNA and polysomes within a 50-nm thick central slice showing DNA (top) and polysomes (middle) and the composite (bottom). (C) 50-nm thick central slice through a snapshot of the equilibrated distributions of DNA polymer (first image) in presence of 50S subunits (second) and 30S subunits (third), along with the composite (fourth). (D) Relative axial density of DNA beads (along the x-axis) in DNA-polysome simulation (green) and in DNA-30S-50S simulation (red). Dashed line shows distribution for a uniformly filled spherocylinder. (E) Relative axial density (along the x-axis) of polysome beads (green) in DNA-polysome simulation and of 50S (blue) and 30S (yellow) subunits in DNA-30S-50S simulation. Dashed line shows distribution for a uniformly filled spherocylinder.
To examine whether dissociation of polysomes into 30S and 50S subunits upon Rif treatment might lead to mixing of free ribosomal subunits with the DNA and expansion of the nucleoids, we have performed a new set of Monte Carlo simulations based on the same model of DNA. One chromosome equivalent of DNA, 20,000 30S spheres and 20,000 50S spheres were introduced into the same size spherocylinder as before. Details are provided in Methods and Sec. S10 of SI. For two different initial configurations (Fig. S10), we find that 30S and 50S particles mix fairly thoroughly with the DNA polymer (Fig. 3C), in strong contrast to the DNA-polysome results of Fig. 3B. Comparing the axial dimensions of the nucleoid in these two situations, we see that the nucleoid is more expanded in the model with free 50s and 30S particles (Fig. 3D). The nucleoid width also expands (radial profiles in Fig. S10). This is reminiscent of the observed expansion of the nucleoid length and width from 5-20 min after Rif injection (Fig. 2F and G). The 50S and 30S particles appear to be well mixed with the DNA (Fig. 3E). However, comparison with the distribution for a uniformly filled spherocylinder (dashed lines in Fig. 3D and E) shows that the DNA in fact continues to avoid the endcaps and the ribosomal subunits concentrate near the endcaps. Similar residual segregation occurs at the straight cylindrical walls (Fig. S10). This is also reminiscent of the experimental distributions from two-color imaging on the same cell (Fig. S6) after Rif treatment. The likely underlying causes of enhanced mixing on dissociation to 30S + 50S are discussed below.
Expectations for the translation inhibitors chloramphenicol (Cam) and kasugamycin (Ksg)
According to the transertion hypothesis, inhibition of translation initiation or of translation elongation should cause nucleoid contraction similar to that caused by Rif on a comparable timescale. Cam is a translation elongation inhibitor that prevents peptide bond formation by binding to the 23S rRNA of the 50S subunit (Wolfe & Hahn, 1965, Sohmen et al., 2009, Hansenet al., 2003). The 70S ribosomes within polysomes should stall on mRNA and remain as polysomes, maintaining segregation from the chromosomal DNA over a long period. The nucleoids should contract radially on the timescale of drug penetration and completion of transcription events (the short, 0-5 min timescale). Like Cam, Kan inhibits translation elongation, but it interacts with 30S subunit proteins (Misumi & Tanaka, 1980, Sohmen et al., 2009, Pestka, 1974). Cam and Kan serve as a kind of mutual control for possible non-specific binding effects unrelated to translation inhibition. The effects of Kan are quite similar to those of Cam. These are presented in Sec. S11 of SI.
Ksg specifically inhibits translation initiation at canonical mRNA (i.e., mRNA having a Shine-Dalgarno leader sequence (Schluenzen et al., 2006, Tai et al., 1973). The most recent crystallographic and competitive binding data suggest that Ksg interferes with proper binding of the initiator tRNAfMet to the AUG sequence at the P-site of the 30S subunit (Schluenzen et al., 2006). Ksg evidently does not affect translation initiation of leaderless mRNA (Schluenzen et al., 2006). We expect Ksg to allow translating 70S ribosomes to complete protein synthesis and dissociate into 30S and 50S subunits. If a significant fraction of translation occurs on leaderless mRNA, then only part of these free subunits would continue to carry out translation cycles. If so, then net partial conversion of 70S-polysomes to free 30S and 50S subunits should occur on the short timescale. The transertion hypothesis predicts that the nucleoids will contract radially on the 0-5 min timescale. The nucleoid-ribosome mixing hypothesis predicts nucleoid expansion and mixing with 30S and 50S subunits on the same timescale. This provides a useful contrast to the effects of Rif (which should convert 70S-polysomes to 30S + 50S on the 20-min timescale of mRNA degradation) and of Cam and Kan (which should freeze 70S ribosomes as polysomes).
Time-dependent effects of Cam and Ksg on chromosome and ribosome spatial distributions
We repeated the imaging experiments with chloramphenicol (Cam, 300 μg/mL, 20x the MIC), kanamycin (Kan, 300 μg/mL, 200x the MIC) and kasugamycin (Ksg, 5 mg/mL, 30x the MIC; Sec. S2 in SI). The concentrations were chosen to enable direct comparisons with previous work. We briefly tested other concentrations, including 100 μg/mL Cam, 100 μg/mL Kan, and 2 mg/mL Ksg, and the results were quite similar.
Time-dependent effects of Cam and Ksg on the nucleoids are shown in Supporting Movies 2 and 3. The qualitative effects of Cam and Ksg on chromosome and ribosome spatial distributions are shown in the DNA and ribosome images of Fig. 4A-D. In each case, the DNA and ribosome images are of different cells of similar initial length. At the concentrations used, both Cam and Ksg decelerate cell growth within 1-2 min of injection (Fig. 4E). Again, short cells were chosen for quantitative analysis due to their simpler nucleoid and cell wall morphology. We provide the individual traces of all the cells in SI (Fig. S12).
Careful measurements of nucleoid width vs time (Fig. 4F) show that on average both Cam and Ksg cause radial contraction at t = 0-5 min, qualitatively like the short-time effect of Rif (Fig. 2F). The mean fractional width contraction is about 10% for Cam, 8% for Rif, and 5% for Ksg. In all three cases, the mean width remains significantly below that of normally growing cells throughout the 20-min observation period. The short-time radial contraction induced by all three drugs is consistent with the transertion hypothesis. The smaller degree of radial contraction induced by Ksg is qualitatively consistent with a short-time competition between nucleoid contraction due to loss of transertion and nucleoid expansion due to mixing with free 30S and 50S subunits. A similar competition should occur after Rif treatment, but on the longer 20-min timescale of mRNA degradation. Accordingly, the short-time fractional contraction of nucleoid width for Ksg is comparable to the long-time contraction for Rif, about 5% in each case.
On the longer time scale of 5-20 min, Cam induces continuous axial nucleoid contraction while strong DNA-ribosome segregation is maintained. The normalized quantity LDNA(t)/Lcell(t) shrinks by about 25% over 20 min (Fig. 4G). On the same timescale, the cells lengthen, but only by 10% (Fig. 4E). Thus most of the decrease in the ratio LDNA(t)/Lcell(t) is due to actual nucleoid contraction. B ~25 min after Cam injection, the axial contraction of the nucleoids is typically complete (example in Fig. S13). Because Cam treatment should freeze 70S ribosomes as polysomes, this axial contraction is consistent with the hypothesis that DNA and 70S-polysomes do not mix. Similar effects on the DNA spatial distribution were observed following treatment with Kan (Fig. S11).
In sharp contrast, shortly after Ksg injection the nucleoids expand axially on the same 0-5 min timescale of the modest width contraction (Figs. 4F, G). This corroborates the presumed Ksg mechanism of partial conversion of polysomes into free 30S and 50S subunits on a short timescale. Recall that for Rif the axial expansion and DNA-30S mixing occurred on the 5-20 min timescale (Fig. 2G). These results are again consistent with the nucleoid-ribosome mixing hypothesis. For Rif, conversion of polysomes to 30S + 50S must wait for degradation of the mRNA population. For Ksg, the conversion begins on the ~30-s timescale of completion of single translation events.
The long-term Ksg-induced effects on nucleoid morphology seem intermediate between the behaviors induced by Cam and by Rif. After injection of Ksg, the relative nucleoid length peaks at t = 5 min and then gradually decreases over the 5-20 min interval. On this timescale, some chromosome-30S mixing has occurred, but the two major nucleoid lobes remain well segregated and the ribosomes remain fairly concentrated in the endcaps. This suggests that many polysomes still exist (presumably bound to leaderless mRNA), but the enhancement of the populations of free 30S and 50S subunits enables some degree of chromosome-ribosome mixing.
After treatment with Ksg the mean diffusion constant of 30S subunits immediately begins to increase (Fig. 4I), but the behavior is again intermediate between that induced by Cam (Fig. 4I) and by Rif (Fig. 2I). Fifteen min after Cam treatment, the distribution of single-molecule 30S diffusion coefficients Di is very similar to that in control cells; in fact, the peak near t = 0 (attributed to 70S-polysomes) is slightly enhanced (Fig. 2H). Fifteen min after Ksg treatment, the distribution of Di values has grown a tail towards faster diffusion, but not as broad a tail as for Rif (Fig. 2H). This suggests a mixture of free 30S subunits and 70S-polysomes. These results are consistent with a significant fraction of translation occurring on leaderless mRNA and the recent suggestion of differential Ksg effects on ribosome interactions with canonical vs leaderless mRNA (Schluenzen et al., 2006).
The Cam data described thus far include only cells that are short at t = 0. In these shorter cells Cam induces merger of the two major nucleoid lobes with each other on the 20-min timescale (Fig. 2A). Longer cells typically exhibit two nucleoid sub-lobes within each major lobe. They also exhibit a greater degree of separation of the two major lobes from each other; note the progress of segregation over time in Fig. 1A. For such longer cells, Cam usually causes each the major lobes to contract independently on a timescale of ~10 min (Fig. S14A and B). The contracted major lobes typically do not coalesce with each other even at times as long as 30 min. The three ribosome-rich regions (cell center and two endcaps) remain intact and expand in volume (Fig. S14C).
Remarkably, the longer cells (Lcell > 4.5 μm) consistently initiate septation into two daughter cells about 10 min after Cam addition, as evidenced by pinching of the phase contrast image or actual septation (left panel in Fig. S14A). This is similar to the time at which the self-contraction of the major nucleoid lobes is essentially complete (right panel in Fig. S14A). As each of the two major nucleoid lobes contracts on itself independently, DNA density is removed from the mid-cell region. Septation occurs shortly thereafter, often prematurely compared with the normal cell cycle. These observations support the nucleoid occlusion hypothesis (van Helvoort et al., 1996, Zaritsky & Woldringh, 2003), which posits that the presence of DNA at mid-cell prevents initiation of septation. One hour after the Cam treatment, all cells have become very short and fairly uniform in length compared to untreated cells (data not shown).
Discussion
Comparison with earlier imaging studies of nucleoids
Previous studies of drug effects on nucleoid morphology imaged aliquots of cells sampled at lag times of 30-60 min after drug addition (van Helvoort et al., 1996, Zimmerman, 2002, Cabrera et al., 2009). On a 30-min timescale, Cam was observed to severely compact the nucleoids. This effect was ascribed to the alleviation of the expanding force from transertion (Woldringh et al., 1995). On the same 30-min timescale, Rif was observed to expand the nucleoids, in agreement with the present work (Cabrera et al., 2009, Binenbaum et al., 1999). The long-term expansion of the nucleoids by Rif treatment stood in apparent conflict with the transertion hypothesis.
The Jin lab used two-color, widefield imaging of RNAP and DNA spatial distributions within the same cell to study the possible role transcription might play in organizing the nucleoid (Cabrera et al., 2009, Jin et al., 2013). They found that the state of the nucleoids, either “closed” (including both normally compacted nucleoids and nucleoids more strongly compacted by Cam) or “open” (nucleoids expanded by Rif) correlates respectively with the presence or absence of transcription foci. These are small regions of highly concentrated RNAP arising from clustering of multiple, highly transcribed rrn operons (Jin et al., 2013, Lewis et al., 2000). A recent study from the Sanyal lab (Chai et al., 2014) describes two-color widefield imaging of DNA and of ribosomes as a function of time in live cells after treatment with a variety of drugs. The bulk culture was sampled at different lag times after drug addition, but the nucleoids were not imaged over long periods.
The present experiments are unique in providing well-defined, quantitative measurements of cell length and width, nucleoid spatial extent, ribosome spatial distribution, and ribosome diffusion as a function of time after antibiotic addition, with time resolution of 12 s. The cells are never fixed, and the SYTOX Orange nucleoid stain is less perturbing than the usual DAPI staining (Bakshi et al., 2014). By measuring Lcell, LDNA, and WDNA for each cell as a function of time, we are able to discern subtle, time-dependent changes in the scaled quantities WDNA/WDNA (t = 0) and LDNA/Lcell that would be lost by averaging over different sets of cells sampled at different time lags after drug addition. Our long-time (t = 20-30 min) results after Rif and Cam treatment agree with earlier studies, but we report on the short, 0-5 min timescale for the first time.
Transertion
The transertion hypothesis posits direct coupling of the chromosomal DNA to the cytoplasmic membrane mediated by simultaneous co-transcriptional translation and insertion of cytoplasmic membrane proteins (or translocation of periplasmic and outer-membrane proteins) by the Sec translocon or the YidC insertase (Kudva et al., 2013). The resulting expanding force is presumed to stretch the nucleoids to a larger volume than they would occupy if transertion did not occur. The hypothesized chains of DNA—RNAP—mRNA—70S ribosome—membrane linkages (Fig. 5A) should be broken by either Rif or Cam. Rif, which blocks transcription initiation, should break each chain at the DNA-RNAP position on the timescale of completion of transcription of messages for the membrane protein, typically ~30 s (Proshkin et al., 2010). Cam, which halts translation elongation, should maintain the mRNA—70S ribosome—membrane linkage, but again each chain will be broken when transcription of the mRNA is completed (or terminated in the absence of co-transcriptional translation) (Proshkin et al., 2010).
Figure 5.
(A) Schematic of DNA-tethering to the membrane via transertion inside E. coli cell. (B) Schematic of nucleoid-ribosome mixing hypothesis. In normal growth condition 70S-polysomes dominate, and they are immiscible with the nucleoids due to excluded volume and entropic effects. This creates spatial segregation between the polysomes and nucleoid (top). Rif treatment causes dissociation of 70S-polysomes into free 30S and 50S subunits. The 30S and 50S subunits mix readily with the nucleoids and cause expansion of the nucleoid (bottom).
Our new measurements lend fresh support to the transertion hypothesis. They demonstrate contraction of both the normalized nucleoid width WDNA/ WDNA (t = 0) and the relative nucleoid length LDNA/ Lcell over the first 5 min after Rif, Cam, and Ksg addition (Figs. 2 and 4). During the longer time interval t = 5-20 min, the nucleoids of Cam-treated cells maintain their smaller width and continue to contract in length. The nucleoids of Rif-treated cells maintain about half of the initial decrease in width, but slowly expand in length beyond that of normal cells. The ribosome-DNA mixing hypothesis presented below can explain the long-time behavior of both drugs without recourse to indirect effects of transcription foci (Jin et al., 2013).
The observed contraction of both the relative nucleoid width and length on the same 0-5 min timescale suggests that transertion expands the nucleoids in both the radial and axial dimensions. We recently reported superresolution imaging (~30-nm accuracy) of the spatial distributions of ribosomes (30S-YFP labeling) and RNA polymerase (β’-YFP labeling) under the same growth conditions studied here (Bakshi et al., 2012). We found clear evidence that the distributions of both 30S ribosome subunits and RNAP extend radially outward all the way to the straight, cylindrical part of the cytoplasmic membrane. These observations are also consistent with transertion. However, we found few if any RNAP copies near the endcaps of the cytoplasmic membrane, which are contiguous to the ribosome-rich regions. This argued against transertion as a direct source of axially expanding forces on the nucleoid. What then might explain the short-term axial contraction of the nucleoids after both Rif and Cam treatment? One possibility is that coupling of the DNA to the membrane by transertion causes normal cell wall synthesis to stretch the DNA axially (and perhaps helps maintain chromosomal segregation) (Woldringh, 2002). When the transertion chain is broken, the DNA relaxes axially as well as radially.
Additional support for the transertion hypothesis comes from a study of the spatial distribution of genes coding for two different membrane proteins, with specific genes labeled using the fluorescence operator repressor system (FROS): lacO-LacI-YFP and tetO-TetR-mcherry (Libby et al., 2012). Each distribution shifts outward towards the cytoplasmic membrane within 3 min after these genes are transcriptionally activated. No such shift occurred for two genes coding for cytoplasmic proteins. This provides additional evidence for transertion-mediated membrane tethering of the chromosomal DNA.
Nucleoid-ribosome mixing
The experimental data and the statistical model support a new nucleoid-ribosome mixing hypothesis pictured in Fig. 5. In this view, the chromosomal DNA and ribosomes form a coupled physical-biochemical system. The physiological state of the cell (normal cells or drug-treated cells, and the growth rate) controls the partitioning of ribosome components between 70S-polysomes vs free 30S and 50S ribosomal subunits. That partitioning in turn strongly influences the spatial extent of the nucleoids due to the combined physical effects of DNA configurational entropy, 70S-polysome configurational entropy, free-particle translational entropy, and excluded volume.
Translational entropy is the entropy of movement in space. It increases with the number of free particles and with the volume available to the free particles. The DNA configurational entropy is determined by the number of available conformational states of the DNA polymer. Even without crowding effects of ribosomes, the model polymer avoids walls and does not fill the cytoplasm (SI, Sec. S10). The reason is that placement of a polymer bead near a wall eliminates many potential placements of connected rods and beads, thus decreasing configurational entropy.
Conversion of 70S-polysomes to free 30S and 50S subunits greatly enhances the contribution of ribosomal translational entropy to the total free energy due to formation of a much greater number of free particles. For example, a polysome comprising ten 70S ribosomes counts as one independent particle; if it dissociates after Rif treatment, this creates 20 independent 30S and 50S subunits. The modeling suggests that for a normally expanded DNA polymer, the smaller 30S and 50S subunits are much better able than the 70S-polysomes to mix with the DNA polymer. Free 30S and 50S subunits thus sample a larger volume of cytoplasm, enhancing translation entropy even further. A recent experimental study of the spatial distribution and diffusive properties of labeled 30S and 50S subunits demonstrated this quite directly (Sanamrad, et al., 2014). In normal conditions, the copies that diffuse faster (free 30S and 50S subunits) distributed fairly uniformly throughout the cytoplasm, while the copies that diffuse slowly (70S-polysomes) were strongly excluded from the nucleoid, as shown previously (Bakshi, et al., 2012).
The nucleoid-ribosome states induced by long-time exposure to Rif and to Cam lie at opposite extremes of the spectrum of ribosome component partitioning. Transcription inhibition by Rif converts essentially all 70S-polysomes into 30S + 50S subunits. On the 5-20 min timescale, we directly observe extensive mixing of 30S subunits with the chromosomal DNA and concomitant expansion of the nucleoid length and width (Figs. 2F, G and S6). The mean 30S diffusion coefficient increases gradually on the same timescale (Fig. 2H, I). Importantly, the expanded nucleoids remain smaller in width than normal nucleoids, consistent with the notion that loss of transertion counteracts part of the Rif-induced expansion. The expanded nucleoids still do not fill the cytoplasmic space, likely due to maximization of polymer configurational entropy (Fig. 3).
At the other limit, inhibition of translation elongation by Cam presumably converts most free subunits into 70S-polysomes. Accordingly, the resulting nucleoid-ribosome segregation is even stronger than in normal cells (Fig. 4A and B) and 30S subunits, now completely incorporated into polysomes, diffuse even more slowly than in normal cells (Fig. 4H and I). The Cam-induced nucleoids may well represent the natural degree of compaction that would occur in the absence of transertion. In unpublished work, we found that proteins as small as Kaede tetramers (110 kDa), which are normally distributed homogeneously throughout the cytoplasm, are excluded from the Cam-induced, compact nucleoids.
Conclusion: A comprehensive model
To conclude, we combine the chromosome-expanding effects of transertion with the nucleoid-ribosome mixing hypothesis to paint a comprehensive picture of how transcription and translation work together in live E. coli. For our normal, moderately fast growth conditions (doubling time of 43 min at 30°C), roughly 80% of the ribosomal subunits are contained in 70S-polysomes (Bakshi et al., 2012, Forschhamer & Lindahl, 1971, Bremer). In accord with the mixing hypothesis, super-resolution imaging reveals strong nucleoid-ribosome segregation into ribosome-rich regions and DNA-rich nucleoids (Bakshi et al., 2012). Nevertheless, there are some 30S and 50S subunits within the nucleoids (Sanamrad, et al., 2014), and messages synthesized there by RNAP are presumably being translated during transcription (Proshkin et al., 2010, Miller et al., 1970). When a transcript is completed (in ~20 s), the polysome diffuses fast enough to reach a ribosome-rich region in ~1 s. Once a polysome reaches the ribosome-rich region, it will tend to remain there because polysomes and nucleoids do not mix. Multiple rounds of translation can occur within the 5–10 min lifetime of mRNA relative to degradation (Bakshi, et al., 2012).
This picture implies a circulation of ribosomal components. The diffusive outward flux of polysomes from the nucleoids to ribosome-rich regions must be balanced by a diffusive inward flux that returns free 30S and 50S subunits to the nucleoids, where they can initiate further rounds of co-transcriptional translation. The inward flux is facilitated by the ability of 30S and 50S subunits to penetrate the nucleoids (in accord with the mixing hypothesis). At the same time, the sequestration of most ribosome components within ribosome-rich regions may be important for efficient recycling of 30S and 50S subunits, enabling rapid cell growth (Bakshi et al., 2012).
It has recently been suggested (Sanamrad, et al., 2014) that in exponentially growing cells, active co-transcriptional translation contributes to nucleoid expansion and allows ribosomal subunits to access nascent mRNAs throughout the nucleoid. This results in a positive feedback loop. However, the magnitude of this volume effect must be quite small. As a rough estimate, in normal conditions the nucleoid occupies roughly half the total cytoplasmic volume of ~3 μm3. There are ~50,000 ribosomes per cell, but only 10–15% lie within the nucleoids (Bakshi, et al., 2012). The total volume of 5,000 ribosomes is only ~0.02 μm3. The presence of ribosomes within the nucleoid expands its volume by a few percent at most. Similar arguments show that the ~5000 RNAP copies sequestered within the nucleoids contribute only ~0.003 μm3 of total volume, again a negligible contribution to overall nucleoid volume (Jin, et al., 2013).
Instead, we propose that it is transertion that expands the nucleoids sufficiently to enable 30S and 50S subunits to penetrate and initiate co-transcriptional translation. Co-transcriptional translation is biologically important for at least two reasons. The presence of translating ribosomes on a nascent message provides protection against premature degradation by ribonucleases (Deana & Belaska, 2005, Deneke, et al., 2013). In addition, a growing body of evidence indicates that coupling between the lead ribosome and RNA polymerase prevents RNAP backtracking and enhances the overall transcription rate (McGary & Nudler, 2013). Thus transertion, whose occurrence has long been debated, may be critical to optimal cell growth.
Experimental Procedures
Bacterial strains
Measurements of single-cell growth rate and imaging of the E. coli nucleoids were performed on the strain VH1000 (MG1655: lacI- lacZ- pyrE+) (Jasiecki & Wegrzyn, 2003). All the other strains used in this study use the same VH1000 background. Widefield imaging of ribosome spatial distributions used the strain MSG192 (rpsB::yfp), which contains a translational fusion of the yfp gene to the C-terminus of rpsB, the gene encoding the ribosomal protein S2 (Bakshi et al., 2012). The single-particle tracking study of ribosome diffusion used the strain MSG196 (rpsB::mEos2), which was constructed in the same way as the rpsB::yfp strain (Bakshiet al., 2012). A rifampicin resistant strain RifR (rpoB3595), which has a S522F mutation (codon change TCT → TTT) in the rpoB gene encoding the β subunit of Escherichia coli RNA polymerase (Jin et al., 1988), was used as a control for the effects of rifampicin. The mutation prevents binding of Rif to RNA polymerase. We transduced the mutation into our background strain VH1000 (producing strain SB7) and selected against rifampicin at 50 μg/mL.
Sample preparation for microscopy
Cell cultures were grown in EZ Rich Defined Medium (EZRDM, Teknova), a chemically defined complete medium suitable for sensitive fluorescence microscopy due to low autofluorescence (Neidhardt. F. C. et al., 1974). Cells were grown overnight with shaking in a 30°C water bath. We subsequently made subcultures by diluting the stationary phase culture at least 1:100 into 2 mL of fresh EZRDM. When cells had grown to midlog phase (OD600 = 0.2-0.4), 500 μL of cell culture was injected into a microfluidics chamber and then rinsed with 1 mL of fresh, warmed, aerated growth medium over 30 s to remove unadhered cells. The microfluidics chamber is made of polydimethylsiloxane (PDMS) adhered to a glass coverslip. It provides a simple rectilinear observation volume of dimensions 11 mm × 6 mm × 50 μm (length × width × height) with inlet and outlet ports to enable flow. Before the experiment, the preassembled chamber was soaked in 0.01% polylysine for 1 hr and then rinsed with water to remove unbound polylysine. The microfluidics chamber was brought into contact with the microscope objective and warmed to 30°C before injecting the cells for imaging. Although there is no flow of medium during imaging, oxygen permeates the PDMS housing and maintains good aerobic growth conditions. Doubling times for single-cell growth in the microfluidics device were estimated by measuring cell length vs time using phase contrast microscopy (Bakshi et al., 2014). The average doubling time of cell length is 48 min in the microfluidics chamber (Sec. S1 in SI). The day-to-day variation in doubling time in the microfluidics device is ±3 min. This is comparable to the 43-min doubling time in the bulk culture estimated from OD measurements.
For imaging nucleoid morphology (Bakshi et al., 2014), a SYTOX Orange solution (50 μM stock concentration, Life Technologies) was added to a growing, midlog phase culture (OD600 = 0.2-0.4) to a final concentration of 500 nM. After 10 min of incubation the cells were twice centrifuged at 8,000 g for 2 min and resuspended in fresh EZRDM, after which 500 μL of cell culture was injected into the microfluidics chamber. Rinsing with 1 mL of fresh EZRDM removed unadhered cells and eliminated background fluorescence from dye molecules adhered to the coverslip. The staining of the nucleoids of adhered cells is not washed away. Time-lapse fluorescence imaging then began. The cells were imaged for ~5 min before injection of fresh medium (EZRDM) containing antibiotic solution. During the remainder of the movie, typically 25 min longer, the effects of the antibiotic on nucleoid morphology were imaged.
To estimate total mRNA content of cells at different time-points after drug injection, we labeled cells with SYTO RNASelect (Life Technologies). In vitro, this dye becomes fluorescent when bound to RNA; it is about five-fold less sensitive to DNA. A 50 μM stock solution of SYTO RNAselect was added to a growing, midlog phase culture (OD600 = 0.2-0.4) to a final concentration of 500 nM. After 10 min of incubation the cells were twice centrifuged at 8,000 g for 2 min and resuspended in fresh EZRDM before injecting into the microfluidics device. Such treatment provided readily detected fluorescence. To avoid signal loss due to photobleaching, we took snapshots of groups of cells in different regions of the coverslip at the different points in time. Mean total fluorescence intensity per cell was used as a measure of total mRNA vs time.
Rifampicin (Rif; Sigma-Aldrich) and chloramphenicol (Cam; Sigma-Aldrich) stock solutions were prepared by dissolving 20 mg and 34 mg of the drugs in 1.0 mL of methanol. Stock solutions of kanamycin (Kan; Sigma-Aldrich) and kasugamycin (Ksg; Sigma-Aldrich) were prepared by dissolving 10 mg of the respective drugs in 1 mL water. These stock solutions were then diluted in fresh EZRDM and injected into the microfluidic chamber during observation. Typically 500 μL of the drug solution was injected over the course of ~5 s, after which there is no flow. The 24-hr MIC value for each drug was estimated as the lowest concentration for which no cell growth could be detected in 96-well plates after 24 h. The details of MIC measurements are provided in the Supporting Information. The drug concentrations used were 300 μg/mL for Rif (20x MIC), 300 μg/mL for Cam (20x MIC), 300 μg/mL for Kan (200x MIC), and 5 mg/mL for Ksg (30x MIC) (Fig. S2). These doses were chosen in accord with typical usage in the literature (Chai et al., 2014, van Helvoort et al., 1996, Cabrera et al., 2009, Zimmerman, 2002). They cause noticeable changes in nucleoid morphology on a convenient timescale that is comparable to previous results.
Time-lapse phase contrast and fluorescence microscopy
E. coli cells were imaged using a Nikon Eclipse Ti inverted microscope equipped with an oil immersion objective (Nikon CFI Plan Apo Lambda, DM 100X Oil, 1.45 NA). The setup includes a Perfect Focus System (PFS, Nikon) to maintain focus over long imaging periods. Alternating fluorescence and phase contrast images were recorded by a back-illuminated EMCCD camera with 512 × 512 pixels of 16 μm × 16 μm each (iXon DV-897, Andor Technology, Connecticut). Each pixel corresponds to 105×105 nm2 at the sample (150X overall magnification). The spatial distribution of SYTOX Orange-stained DNA was monitored with 561 nm laser excitation (CrystaLaser, Reno, Nevada) of intensity 5-10 W/cm2 at the sample. SYTOX Orange fluorescence was collected through a bandpass filter (bright line 617/73, Semrock). The spatial distribution of ribosomal protein S2-YFP in MSG192 was imaged using a 514 nm Ar+ excitation laser (Melles Griot, Carlsbad, CA, USA) of intensity 5-10 W/cm2 at the sample. The yellow emission was collected through a 560/50 emission filter (Chroma). SYTO RNASelect was excited using a 488 nm Ar+ excitation laser (Melles Griot, Carlsbad, CA, USA) of intensity 5-10 W/cm2 at the sample. The emission was collected using a 525/50 emission filter.
Analysis of time-lapse imaging data
The interleaved fluorescence and phase contrast images were separated during image processing. To determine the tip-to-tip cell length Lcell, phase contrast images were analyzed using a MATLAB GUI developed in our lab. First, a binary mask is made from the prior phase contrast image of each cell in each frame. A sub-pixel contour of the cell outline was created from the binary mask, as described in the supporting information (Fig. S1). After the cells are outlined (white dashed boundary in top panel of Fig. 1C), Lcell is calculated as the contour length of the central spine of the outline (blue line in Fig. 1C). From the noise in successive measurements, we estimate the precision of Lcell to be ±30 nm. To compare the growth rates of cells having different initial lengths, we scale each Lcell (t) measurement to Lcell (t = 0) to obtain the relative cell length vs time (Fig. S1). The absolute value of Lcell depends somewhat on the choice of input parameters, but relative changes in cell length are reliable (Barns & Weisshaar, 2013).
Fluorescence images are analyzed as follows. We determine the orientation of the cell by fitting an ellipse to the binary mask generated from the phase contrast image. The two principal axes of the ellipse are used as the coordinate system for the cell. Fluorescence images are projected along x (long axis) or y axis (short axis perpendicular to x-axis) for the analysis of nucleoid morphology (Fig. 1E) and ribosome distribution inside the cell (Bakshi et al., 2014).
Single-molecule tracking of ribosomes and analysis of tracking data
The ribosomal protein S2-mEos2 was used to monitor diffusion of 30S subunits. Because the 130-min maturation time of mEos2 (McKinney et al., 2009) is much longer than the ~2-min assembly time of 30S subunit (Lindahl, 1975), we expect all observable S2-mEos2 copies to be incorporated into 30S subunits. However, a 30S-mEos2 subunit could be free or bound as a full 70S ribosome translating mRNA, typically a member of a polysome chain. The predominant species in normal growth conditions is likely polysomes (Bakshi et al., 2012).
S2-mEos2 was photoactivated with a 405-nm diode laser (CrystaLaser, Reno, Nevada) with a power density of 0.1-1.0 W/cm2 at the sample. Photoactivated S2-mEos2 copies were imaged with a 561 nm laser (Sapphire 561 CW laser, Coherent). Power density of the 561 nm laser was approximately 1.5 kW/cm2. At this power we achieved good signal-to-noise ratio for single molecules with minimal laser damage to the cell, as judged by continuing normal single-cell growth. We collect data for less than 2 min from each field of view to minimize photo-toxicity caused by the laser. Emission is collected through a 617/73 bandpass filter (bright line 617/73-25, Semrock). Analysis of the single-molecule tracking data was performed as described before (Bakshi et al., 2013).
Monte Carlo simulation of ribosome-DNA segregation
Earlier we modeled segregation of polysomes from plectonemic DNA (Mondal et al., 2011). Plectonemic DNA was represented as a hyper-branched, freely jointed chain of hard, spherical beads. Rods of length 200 nm connect the beads, whose functionality (number of rods connecting each bead to other beads) can vary from one to three. Polysomes were represented as freely-jointed chains of 70S ribosomes, each a hard sphere of 20-nm diameter. The ribosome copy number (20,000) and the cell dimensions (350 nm radius, 3.0 μm length) were taken from the best available estimates at the time of the simulation (Bremer, Grossman et al., 1982). We carried out the new simulations using the same parameters. The 19,994 70S particles were organized as 1,538 13-mer polysomes. This copy number is smaller than our more recent estimate in moderate growth condition (~55,000) (Bakshi et al., 2012). We also recently measured accurate cell dimensions in our growth conditions (430 nm radius, 4.9 μm length). The smaller volume and smaller number of ribosomes in the simulations compensate each other to make the number density of DNA and ribosome particles comparable to our estimates from moderate growth condition (Bakshi et al., 2012). There are no attractive interactions between particles, only excluded-volume effects among all components plus translational and configurational entropy.
The present study uses the same kind of model to explore mixing properties of DNA with 30S and 50S monomers. A single model chromosome of plectonemic DNA plus two different types of monomeric ribosomes (50S and 30S) are placed inside a spherocylinder of the same size as before. The free 50S and 30S ribosomal subunits are represented by hard spheres of appropriate diameter (17 nm and 14 nm, respectively), both present at equal number inside the cell. The subunit diameters are calculated by assuming spherical shape and scaling the volume according to the mass of each subunit. To be consistent with the previous model, 19,994 particles of each kind (50S and 30S) were used to compare with the 1,538 13-mer polysomes used in the previous study. The differential volume exclusion among and between DNA and ribosomal subunits was handled as before. Further details are provided in Sec. S10 of SI.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [NIGMS, R01-GM094510 to J.C.W. and R01-GM093265 to J.C.W. and S. Gellman] and the National Science Foundation [CHE-1213860 to S. Cavagnero and J.C.W]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bakshi S, Bratton BP, Weisshaar JC. Subdiffraction-Limit Study of Kaede Diffusion and Spatial Distribution in Live Escherichia coli. Biophysical Journal. 2011;101:2535–2544. doi: 10.1016/j.bpj.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Choi H, Rangarajan N, Barns KJ, Bratton BP, Weisshaar JC. Non perturbative Imaging of Nucleoid Morphology in Live Bacterial Cells during Antimicrobial Peptide Attack. Applied and Environmental Microbiology. 2014;80:4977–4986. doi: 10.1128/AEM.00989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Dalrymple RM, Li W, Choi H, Weisshaar JC. Partitioning of RNA Polymerase Activity in Live Escherichia coli from Analysis of Single-Molecule Diffusive Trajectories. Biophysical Journal. 2013;105:2676–2686. doi: 10.1016/j.bpj.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Molecular Microbiology. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns KJ, Weisshaar JC. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochimica Et Biophysica Acta-Biomembranes. 2013;1828:1511–1520. doi: 10.1016/j.bbamem.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binenbaum Z, Parola AH, Zaritsky A, Fishov I. Transcription- and translation-dependent changes in membrane dynamics in bacteria: testing the transertion model for domain formation. Molecular Microbiology. 1999;32:1173–1182. doi: 10.1046/j.1365-2958.1999.01426.x. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, ColladoVides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. (2nd ed.) :1559. [Google Scholar]
- Cabrera JE, Cagliero C, Quan S, Squires CL, Jin DJ. Active Transcription of rRNA Operons Condenses the Nucleoid in Escherichia coli: Examining the Effect of Transcription on Nucleoid Structure in the Absence of Transertion. Journal of Bacteriology. 2009;191:4180–4185. doi: 10.1128/JB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Chai Q, Singh B, Peisker K, Metzendorf N, Ge X, Dasgupta S, Sanyal S. Organization of Ribosomes and Nucleoids in Escherichia coli Cells during Growth and in Quiescence. Journal of Biological Chemistry. 2014;289:11342–11352. doi: 10.1074/jbc.M114.557348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, Vese LA. Active contours without edges. IEEE Transactions on Image Processing. 2001;10:266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- Chung YJ, Krueger C, Metzgar D, Saier MH. Size comparisons among integral membrane transport protein homologues in Bacteria, Archaea, and Eucarya. Journal of Bacteriology. 2001;183:1012–1021. doi: 10.1128/JB.183.3.1012-1021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Molecular Microbiology. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–33. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Deneke C, Lipowsky R, Valleriani A. Effect of ribosome shielding on mRNA stability. Phys. Biol. 2013;10:046008, 10. doi: 10.1088/1478-3975/10/4/046008. [DOI] [PubMed] [Google Scholar]
- Fishov I, Woldringh CL. Visualization of membrane domains in Escherichia coli. Molecular Microbiology. 1999;32:1166–1172. doi: 10.1046/j.1365-2958.1999.01425.x. [DOI] [PubMed] [Google Scholar]
- Forschhamer J, Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. Journal of Molecular Biology. 1971;55:563–568. doi: 10.1016/0022-2836(71)90337-8. [DOI] [PubMed] [Google Scholar]
- Grossman N, Ron EZ, Woldringh CL. Changes in Cell Dimensions During Amino-Acid Starvation of Escherichia coli. Journal of Bacteriology. 1982;152:35–41. doi: 10.1128/jb.152.1.35-41.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. Journal of Molecular Biology. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Jasiecki J, Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Reports. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Cagliero C, Zhou YN. Role of RNA Polymerase and Transcription in the Organization of the Bacterial Nucleoid. Chemical Reviews. 2013;113:8662–8682. doi: 10.1021/cr4001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Cashel M, Friedman DI, Nakamura Y, Walter WA, Gross CA. Effects of Rifampicin Resistant rpoB Mutations on Antitermination and Interaction with nusA in Escherichia-coli. Journal of Molecular Biology. 1988;204:247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- Jung Y, Jeon C, Kim J, Jeong H, Jun S, Ha B-Y. Ring polymers as model bacterial chromosomes: confinement, chain topology, single chain statistics, and how they interact. Soft Matter. 2012;8:2095–2102. [Google Scholar]
- Kudva R, Denks K, Kuhn P, Vogt A, Mueller M, Koch H-G. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Research in Microbiology. 2013;164:505–534. doi: 10.1016/j.resmic.2013.03.016. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO Journal. 2000;19:710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Roggiani M, Goulian M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7445–7450. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L. Intermediates and time kinetics of in vivo assembly of Escherichia coli ribosomes. Journal of Molecular Biology. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends in Cell Biology. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madras N, vanRensburg EJJ. Monte Carlo study of the Theta-point for collapsing trees. Journal of Statistical Physics. 1997;86:1–36. [Google Scholar]
- McGary K, Nudler E. RNA polymerase and the ribosome: the close relationship. Curr Opin Micro. 2013;16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Hamkalo BA, Thomas CA. Visualization of bacterial genes in action. Science. 1970;169:392. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Misumi M, Tanaka N. Mechanism of Inhibition of Translocation By Kanamycin and Viomycin - Comparative-Study With Fusidic Acid. Biochemical and Biophysical Research Communications. 1980;92:647–654. doi: 10.1016/0006-291x(80)90382-4. [DOI] [PubMed] [Google Scholar]
- Mondal J, Bratton BP, Li Y, Yethiraj A, Weisshaar JC. Entropy-Based Mechanism of Ribosome-Nucleoid Segregation in E. coli Cells. Biophysical Journal. 2011;100:2605–2613. doi: 10.1016/j.bpj.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture Medium for Enterobacteria. Journal of Bacteriology. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris V, Madsen MS. Autocatalytic Gene Expression OccursviaTransertion and Membrane Domain Formation and Underlies Differentiation in Bacteria: A Model. Journal of Molecular Biology. 1995;253:739–748. doi: 10.1006/jmbi.1995.0587. [DOI] [PubMed] [Google Scholar]
- Otsu N. Threshold Selection Method from Gray-Level Histograms. Ieee Transactions on Systems Man and Cybernetics. 1979;9:62–66. [Google Scholar]
- Pestka S. The use of inhibitors in studies on protein synthesis. Methods in enzymology. 1974;30:261–282. doi: 10.1016/0076-6879(74)30030-4. [DOI] [PubMed] [Google Scholar]
- Pettijohn DE. Structure and Properties of The Bacterial Nucleoid. Cell. 1982;30:667–669. doi: 10.1016/0092-8674(82)90269-0. [DOI] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation Between Translating Ribosomes and RNA Polymerase in Transcription Elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow C, Kellenberger E. The Bacterial Nucleoid Revisited. Microbiological Reviews. 1994;58:211–232. doi: 10.1128/mr.58.2.211-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanamrad A, Persson F, Lundius EG, Fange D, Gynnå AH, Elf J. Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. PNAS. 2014;111:11413–11418. doi: 10.1073/pnas.1411558111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzo M, Nierhaus KH, Yokoyama S, Fucini P. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nature Structural & Molecular Biology. 2006;13:871–878. doi: 10.1038/nsmb1145. [DOI] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Molecular Microbiology. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E77–E81. doi: 10.1073/pnas.1101130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohmen D, Harms JM, Schluenzen F, Wilson DN. SnapShot: Antibiotic Inhibition of Protein Synthesis I. Cell. 2009;138:1248–A1248. doi: 10.1016/j.cell.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Tai P-C, Wallace BJ, Davis BD. Actions of aurintricarboxylate, kasugamycin, and pactamycin on Escherichia coli polysomes. Biochemistry. 1973;12:616–620. doi: 10.1021/bi00728a008. [DOI] [PubMed] [Google Scholar]
- van Helvoort JM, Kool J, Woldringh CL. Chloramphenicol causes fusion of separated nucleoids in Escherichia coli K-12 cells and filaments. J. Bacteriol. 1996;178:4289–4293. doi: 10.1128/jb.178.14.4289-4293.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Li GW, Chen CY, Xie XS, Zhuang XW. Chromosome Organization by a Nucleoid-Associated Protein in Live Bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Molecular Microbiology. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Jensen PR, Westerhoff HV. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiology Letters. 1995;131:235–242. doi: 10.1111/j.1574-6968.1995.tb07782.x. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, Hahn FE. Mode of action of chloramphenicol .IX. Effects of chloramphenicol upon a ribosomal amino acid polymerization system and its binding to bacterial ribosome. Biochimica Et Biophysica Acta. 1965;95:146–155. doi: 10.1016/0005-2787(65)90219-4. [DOI] [PubMed] [Google Scholar]
- Youngren B, Nielsen HJ, Jun S, Austin S. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes & Development. 2014;28:71–84. doi: 10.1101/gad.231050.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A, Woldringh CL. Localizing cell division in spherical Escherichia coli by nucleoid occlusion. FEMS Microbiology Letters. 2003;226:209–214. doi: 10.1016/S0378-1097(03)00580-9. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB. Toroidal nucleoids in Escherichia coli exposed to chloramphenicol. Journal of Structural Biology. 2002;138:199–206. doi: 10.1016/s1047-8477(02)00036-9. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB. Shape and compaction of Escherichia coli nucleoids. Journal of Structural Biology. 2006;156:255–261. doi: 10.1016/j.jsb.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB, Murphy LD. Macromolecular crowding and the mandatory condensation of DNA in bacteria. FEBS Letters. 1996;390:245–248. doi: 10.1016/0014-5793(96)00725-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.