Abstract
The principal mosquito innate immune response to virus infections, RNA interference (RNAi), differs substantially from the immune response to bacterial and fungal infections. The exo-siRNA pathway constitutes the major anti-arboviral RNAi response and its essential genetic components have been identified. Recent research has also implicated the Piwi-interacting RNA pathway in mosquito anti-arboviral immunity, but Piwi gene-family components involved are not well-defined. Arboviruses must evade or suppress RNAi without causing pathogenesis in the vector to maintain their transmission cycle, but little is known about mechanisms of arbovirus modulation of RNAi. Genetic manipulation of mosquitoes to enhance their RNAi response can limit arbovirus infection and replication and could be used in novel strategies for interruption of arbovirus transmission and greatly reduce disease.
Keywords: RNA interference, RNAi, innate immunity, arbovirus, transgenic mosquito
1. Introduction
Insects, as do all metazoans, mount an innate immune response upon exposure to infectious agents. Innate immunity is the cellular-level first line of defense against infection and is initiated by detection of a pathogen-associated molecular pattern (PAMP) by a host pattern-recognition receptor (PRR). Insect genomes do not encode elements of protein-based adaptive immune responses, and thus must rely totally on innate immunity for protection. The fruit fly Drosophila melanogaster is a model organism for the study of anti-arboviral defense in mosquitoes; both are members of the order Diptera and can be infected with arboviruses, although Drosophila are not arbovirus vectors. In addition, experimental infections of Drosophila by intrathoracic injection with high viral doses frequently result in pathogenesis. Arbovirus infections of mosquitoes naturally occur by infectious blood-meal ingestion and are generally non-pathogenic and persistent, possibly due to the balance that has evolved between the mosquito innate anti-viral response to control pathogenesis and the arboviral evasion without complete suppression of this response. Availability of the Drosophila genome sequence [1] and use of facile genetic techniques have provided a framework for genetic comparisons with Anopheles gambiae [2,3], Aedes aegypti [4,5] and Culex quinquefasciatus [6,7] as these mosquito genome sequences have been published.
2. Drosophila innate immunity
In the canonical Drosophila innate immune response following bacterial or fungal injection, transcriptional signaling cascades induced by detection of PAMPs by host PRRs result in activation of transcription factors from the NF-κB family (e.g.,Dif, Relish and dorsal) and, ultimately, release of antimicrobial peptides (AMPs) into the insects’ hemolymph. Two major signaling pathways with well-defined microbial PAMPs and insect PRRs are Toll, which responds to fungal and Gram-positive bacterial infections and Imd (immune deficiency), which is triggered by Gram-negative bacterial infections [8]. Several studies of the Drosophila transcriptional response to viral infection have implicated elements of Toll and Imd pathways in antiviral immunity, depending upon the virus used [9-13]. Viral PAMPs and identities and functions of host effectors for these pathways have not been characterized for the most part. Injection with Drosophila C virus (DCV) induces the JAK/STAT pathway, which also can be activated by septic injury [14], resulting in transcription of several genes with STAT-binding elements in their promoters [15]. One such STAT-regulated, DCV-responsive gene encodes a small protein called Vago that was shown to control DCV load in the fat body after infection. Intriguingly, induction of vago transcription was dependent on the DExD/H-box domain of Dicer 2, the initiator of the anti-viral RNAi response [16].
Recently, Kemp et al. [17] explored the role of the JAK/STAT pathway in Drosophila innate immunity to a diverse set of viruses. Each virus infection resulted in a unique pattern of gene induction; however, flies with a mutation in hopscotch, their sole Janus kinase (JAK) gene, were susceptible to significantly increased mortality only after infection with members of the Dicistroviridae insect virus family, and not with the arbovirus Sindbis (SINV, Alphavirus) or any of the other four viruses tested. In contrast, Drosophila with a mutation in the dcr2 gene, encoding the PRR for the exo-siRNA pathway of the RNAi response, exhibited increased mortality after infection with insect viruses of all families, including a DNA-containing iridovirus. Their study confirmed previous findings that RNAi, which is the Drosophila innate immune response unique to viral infections, is the most effective and wide-ranging antiviral response in insects [16,18].
3. RNAi in mosquitoes
3.1 RNAi is an antiviral defense mechanism in invertebrates and plants
Silencing of gene expression by introduction of long double-stranded (ds)RNA with cognate sequence to the silenced gene was described in both the nematode Caenorhabditis elegans [19] and Drosophila [20] in 1998. It was soon recognized that a similar gene silencing phenomenon had been previously observed in plants, and ultimately that RNA-mediated gene silencing was an antiviral defense mechanism in both plants and invertebrates [21-23]. Modeling plant virus studies, we described inhibition of arbovirus replication in mosquitoes and mosquito cells resulting from expression of both virus genome-derived positive-sense and negative-sense RNA from alphavirus transduction vectors (presumably expressed as dsRNA during alphavirus replication) and from expression of virus genome-derived inverted repeat RNA from a plasmid, which formed dsRNA in the mosquito cell nucleus [24-28]. Following the description of dsRNA-mediated gene silencing (RNAi) in C. elegans and D. melanogaster, we proceeded to characterize the mechanism and machinery of mosquito RNAi in An. gambiae [29] and Ae. aegypti [30,31].
3.2 Components and mechanisms of mosquito antiviral RNAi
RNAi in mosquitoes is now known to comprise three major pathways, named for the effector RNAs that are their end products: small interfering (si)RNA, micro (mi)RNA, and Piwi-interacting (pi)RNA pathways. Each has a distinct role in either antiviral defense, regulation of development and gene expression, or defense of the genome against transposon mobilization and expression, although in Drosophila some interconnections have been noted [32]. The mosquito genes that encode major participants in each pathway have been identified by homology to Drosophila genes [33]. The exogenous (exo)-siRNA pathway represents the major antiviral innate immune response in mosquitoes [34] and this antiviral response will be the focus of this review. The potential role of piRNAs in antiviral defense is less clear and will be discussed in section 3.2.2.
3.2.1. The exo-siRNA pathway
The exo-siRNA response in arbovirus-infected mosquitoes can be triggered by dsRNA >150 bp in length [35] (Figure 1). In virus-infected cells, the source of dsRNA is thought to be genome replication intermediates, intra-strand RNA secondary structures [30,36-38], or convergent transcription of DNA virus genomes [17]. Other virus-specific RNA structures might also serve as PAMPs, since it has been shown that viruses with negative-sense RNA genomes generate little dsRNA during replication [39], yet infection of insect cells with vesicular stomatitis virus (VSV, Vesiculovirus), with a non-segmented negative-sense genome, and La Crosse and Rift Valley bunyaviruses, with three negative sense and/or ambisense genome segments, generates typical virus-specific small RNAs [40-43].
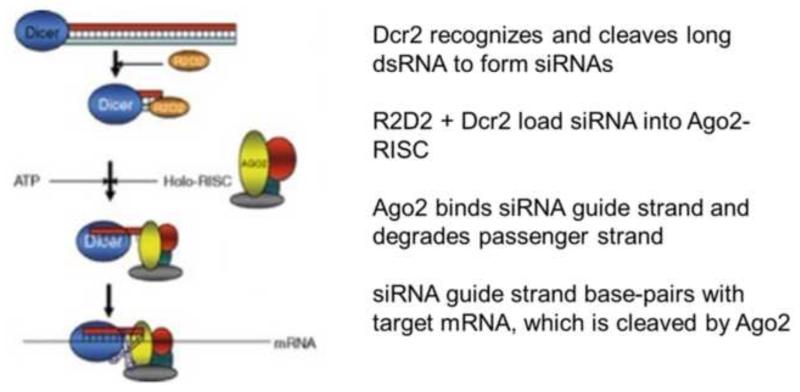
Figure 1.
Diagram showing the essential components of the mosquito exo-siRNA pathway that is the major anti-viral immune response: long virus-specific dsRNA, which serves as the PAMP; Dicer 2 (Dicer), which serves as the PRR; R2D2, the dsRNA-binding protein; Argonaute 2 (Ago2), which acts as effector in cleaving target viral mRNA.
Many of the components and activities of the insect siRNA pathway have been elucidated with in vitro reconstitution assays using Drosophila or mosquito cell lysates or purified proteins (Figure 1). The dsRNA PAMP is recognized by the mosquito ortholog of Drosophila Dicer 2 (Dcr2), an RNase III-family dsRNA endonuclease that serves as a PRR to initiate the exo-siRNA pathway [33,44]. Dcr2 is a large protein (1658 amino acids in Ae. aegypti) with a DExD/H helicase domain at the N terminus, followed by a second helicase domain, a dsRNA-binding domain, a PAZ domain required for recognition and alignment of dsRNA ends, and two RNase III domains near the C-terminus [33]. Dcr2 binds and cleaves dsRNA into 19-22 bp siRNA duplexes with 2-nt overhangs at the 3′-OH-ends. In arbovirus-infected mosquitoes, the siRNAs are derived from both genome-sense and antisense viral RNA from sites distributed throughout their lengths [37,38,45]. The dsRNA-binding protein R2D2 associates with the siRNA-Dcr2 complex to facilitate loading into the multi-protein RNA-induced silencing complex (RISC), which is the effector for antiviral RNAi [46]. The essential component of the RISC is Argonaute 2 (Ago2), which contains an endonuclease V-homologous Piwi domain with “slicer” activity [47] and a PAZ domain, which binds and unwinds the siRNA, allowing endonucleolytic degradation of the “passenger” strand [48]. The RISC-bound siRNA “guide” strand forms a perfectly base-paired duplex with the complementary sequence on target viral mRNA [48] and the target is cleaved at the center of the duplex by Ago2 endonuclease activity [47,49,50]. The non-capped, 5′-phosphorylated product of Ago2-cleaved viral mRNA [51,52] can serve as a substrate for further degradation by the cellular 5′-3′ exonuclease XRN1 [53,54]. The essential roles of Dcr2, Ago2 and the exo-siRNA pathway in mosquito antiviral defense were shown by injection of dsRNAs derived from their respective ago2 and dcr2 gene sequences into An. gambiae and Ae. aegypti, followed by arbovirus infection. This RNAi-mediated knock-down of gene expression resulted in ~10-fold higher infectious o’nyong-nyong virus (ONNV, Alphavirus) production in An. gambiae and dengue virus type 2 (DENV2, Flavivirus) in Ae. aegypti [29,31]. In addition, we discovered that the C6/36 cell culture line (Ae. albopictus) has a single nucleotide deletion in the dcr2 gene, resulting in a non-functional Dcr2 protein and 10-to-100-fold enhanced arbovirus replication compared to other mosquito cell lines such as Aag2 (Ae. aegypti) [38,42]
3.2.2. The piRNA pathway
In Drosophila, Piwi-interacting RNAs (piRNAs) are 24-30 nt small RNAs that regulate transposable element (TE) transcription and transposition and thus protect the genome. piRNA biogenesis in Drosophila involves three proteins from the Piwi clade of Ago family proteins, Piwi, Aubergine (Aub) and Ago3, but is Dicer-independent. Drosophila piRNAs are generated by “ping-pong” amplification, resulting in biases for 5′ U (U1) on antisense strands and position 10 A (A10) on sense strands [55,56]. Several recent studies have implicated piRNAs in mosquito antiviral defense [38,42,43,45,57]. We found that in DENV2 (Flavivirus)-infected, Dcr2-defective C6/36 cells (see section 3.2.1), the predominant virus RNA-derived small RNAs were 27 nt long, whereas viral small RNAs in DENV2-infected, Dcr2-competent Aag2 cells and Ae. aegypti mosquitoes were predominantly 21 nt siRNAs. The vast majority (96%) of 27-nt viral small RNAs in C6/36 cells had sense polarity and a strong preference for A10, unlike piRNAs generated by ping-pong amplification [38]. Morazzani et al. [45] found that chikungunya virus (CHIKV, Alphavirus)-infected Ae. albopictus mosquitoes generated a large proportion of piRNA-like virus-specific small RNAs in addition to siRNAs, and although the 27-nt RNAs largely had positive polarity, they displayed the A10-positive strand, U1-negative strand signature suggesting ping pong amplification. Similarly, Vodovar et al. [58] showed that exo-siRNA-competent cultured Ae albopictus and Ae aegypti cells produced both siRNA and piRNA-like virus-specific small RNAs with a ping-pong signature after SINV infection. Léger et al. [43] also observed Rift Valley fever virus (RVFV, Phlebovirus)-specific small RNAs with both Dcr2 and Piwi signatures from infected Dcr2-competent Aag2 and U4.4 (Ae. albopictus) cell cultures. Interestingly, the 21-nt siRNAs were predominant early in infection whereas 24-27-nt piRNAs were more prominent after persistent infection was established. Overall, the piRNA pathway appears to play a role in mosquito antiviral defense that varies depending upon the mosquito species and possibly tissue, the arbovirus family and the magnitude of the exo-siRNA response. By comparison to Drosophila, the Piwi-subfamily genes have expanded greatly in culicine mosquitoes. Ae. aegypti have ago3 and piwi1-7 genes from the Piwi subfamily and Cx. quinquefasciatus have ago3 and piwi1-6 [33]. The potential role of each in mosquito antiviral pathways has not been clearly defined, although ago3 was implicated in ONNV-infected An. gambiae [29] and piwi4 in Semliki Forest virus (Alphavirus)-infected Aedes cell cultures [59].
4. Other potential antiviral immune mechanisms in mosquitoes
4.1. Transcriptional induction
A number of studies have examined transcriptional activation of known immunity genes after arbovirus infection by phylogenomic comparisons with Drosophila and/or between mosquitoes [7,60-62]. In a meta-analysis, Bartholomay et al. [7] determined the response of Cx. quinquefasciatus to infection by West Nile virus (WNV) and compared previous studies of Ae. aegypti, Ae. albopictus and An. gambiae mosquitoes and cultured cells infected by SINV, WNV, DENVs and ONNV. Although large numbers of infection-response genes were induced in both midgut and carcass of Cx. quinquefasciatus by 14 days after a WNV-infectious blood-meal, reflecting spread of infection throughout the midgut and dissemination to fat body, salivary glands and other tissues, relatively few of these were canonical immunity genes. Activated genes included orthologs of a few Toll, Imd and JAK/STAT pathway components; activation of Toll and JAK/STAT genes had previously been observed to play a role in control of DENV infection by Ae. aegypti [61-63]. Arboviral PAMPs and effector molecules were not identified in most studies; however, Souza-Neto et al. [62] identified two DENV restriction factors that contain putative STAT-binding elements in their promoter regions and Luplertlop et al. [64] demonstrated that the anti-bacterial peptide cecropin, which was induced in DENV-infected Ae aegypti salivary glands, had anti-DENV and anti-CHIKV activity as well as anti-bacterial activity. In addition, Paradkar et al. [65,66] showed that the Culex pipiens molestus ortholog of Drosophila vago (See Section 2) was induced in a Dcr2-dependent manner following WNV infection and restricted viral infection in cultured mosquito cells by activating the JAK/STAT pathway.
It is important to note that although Cx. quinquefasciatus, like Ae. aegypti, encodes an expanded repertoire of RNAi pathway orthologs, none of these had significantly modulated expression levels during infection by WNV, SINV, or DENV2 [7,30,57]. This might be related to the necessity for a balance between the potentially potent RNAi response of the host and arbovirus evasion of this immune response without pathogenesis in order for virus persistence and transmission to occur [34].
4.2. Apoptosis
Apoptosis is a highly-regulated process of programmed cell death required for development and homeostasis in metazoans through removal of unneeded or damaged cells, and may also serve in innate antiviral immunity in insects [67]. Apoptosis correlated with WNV infection in midguts of a refractory strain of Cx. pipiens pipiens and was proposed as the basis for limited midgut infection and inhibition of disseminated infection [68]. Large accumulations of apoptotic cells were also observed in Cx. p. quinquefasciatus salivary glands at 28 days after WNV infection and appearance of this pathology was associated with a lower proportion of virus-transmitting mosquitoes [69]. To answer the question whether apoptosis is an early innate immune response that can prevent/limit viral infection or simply one of the cellular outcomes associated with viral infection, Liu et al. [70] provided DENV-infectious blood-meals to both refractory and susceptible strains of Ae aegypti. They observed that the pro-apoptotic gene michelob_x (an ortholog of Drosophila reaper) was rapidly induced only within the refractory mosquito strain. On the other hand, some arboviruses may exploit the apoptotic pathway to enhance disseminated infection in mosquitoes. Induction of apoptosis in Ae. aegypti by injection of dsRNA to silence the inhibitor of apoptosis gene (Aeiap) followed by oral infection with SINV resulted in increased midgut infection and dissemination to other tissues, and inhibition of apoptosis by silencing the initiator caspase Aedronc had the opposite effect [71]. Viral PAMPs, potential virus-encoded regulators of apoptosis, and detailed mechanisms for apoptotic effects on arboviral replication have not been determined.
4.3. Autophagy
Autophagy is also a normal cellular process in which damaged or unwanted proteins and organelles are sequestered in double-membrane structures for degradation in response to nutrient deprivation or stress such as virus infection [72]. A potential protective role for autophagy was shown in VSV infection of Drosophila [73]; however, little is known about autophagy in arbovirus-mosquito interactions. Studies in mammalian cell cultures have shown that arboviruses such as DENV subvert the autophagic mechanism for use in formation of the cytoplasmic double-membrane vesicles required to sequester viral RNA replication [74] and to regulate lipid metabolism [75], suggesting that the autophagy machinery might also facilitate DENV replication in mosquito cells. Thus, autophagy, as well as several other cellular processes normally assumed to participate in innate antiviral immunity, might play diverse anti-or pro-viral roles depending on the arbovirus-vector combination.
5. Genetic manipulation of mosquitoes to enhance innate immunity
The powerful anti-viral RNAi response of mosquitoes suggests novel ways of controlling arbovirus replication and transmission. As early as 2000, plant geneticists developed transgenic Arabidopsis thaliana that transcribed inverted repeat (IR) RNA that mediated silencing of expression of targeted plant genes [76]. The RNA transcript contained tandem gene-specific sequences in inverted sense and antisense orientations that formed dsRNAs to trigger an RNAi response. In 2003, Drosophila geneticists developed IR transgenes that expressed as heritable phenotypes with RNAi-targeted gene silencing. The IRs were separated by a functional intron such that the transgene transcript formed a loopless hairpin RNA following splicing [77]. Inclusion of a functional intron was necessary because IR gene sequences are difficult to clone in bacterial plasmids and the intron spacer greatly enhanced their stability, facilitating cloning. Using a similar strategy to target DENV2 instead of a host gene, we generated a transposable element (mariner, Mos1)-transformed Ae. aegypti line, Carb77 [78] that was DENV2-resistant. The transgenic mosquitoes expressed 587 nt IR RNA from the DENV2 prM gene in sense and antisense orientations separated by an active Ae. aegypti sialokinin intron sequence. Expression of the transgene was placed under control of the carboxypeptidase A promoter so that IR RNA transcription was induced soon after the mosquito acquired a blood-meal. In a more recent study, we developed new transgenic lines using the identical transgene as Carb77. One of these lines, Carb109M, has been genetically stable and refractory to DENV2 infection for >33 generations [79]. Genetic analyses and physical chromosome mapping identified two closely linked transgene integration sites in Carb109M mosquitoes associated with chromosome 3. Northern blot analysis detected abundant, transient expression of the IR-RNA 24h after a blood-meal. NexGen sequencing of midgut small RNAs from blood-fed, but uninfected, Carb109M revealed that the IR-RNA was rapidly processed into 21 nt small RNAs with sequences corresponding to the 587-base target region of the DENV2 RNA genome. Carb109M mosquitoes were refractory to infection with different DENV2 genotypes, but not to other DENV serotypes due to the sequence specificity of RNAi. Expression of a DENV2 sequence-derived IR-RNA in the mosquito midgut initiated the antiviral intracellular exo-siRNA response early in the initial site of infection, efficiently blocked DENV2 infection and profoundly impaired vector competence for DENV2. The two transgene integration sites were stable after multiple generations and following introgression into a genetically-diverse laboratory strain (GDLS) Ae. aegypti population from Mexico. Introgression of the transgene into GDLS, with a different genetic background from Carb109M, changed the GDLS population from a highly DENV2-permissive phenotype to a DENV2-refractory phenotype. Significantly, the DENV2-refractory homozygous line, Carb109M/GDLS.BC5.HZ, exhibited (relative to GDLS) minimal fitness loss associated with the transgene [79].
6. Viral counterdefenses to mosquito RNA-mediated innate immunity
Many insect-pathogenic viruses are known to encode proteins that are potent suppressors of RNAi (VSRs) and thus virulence factors; however, none has been identified in arboviruses. Complete suppression of the major mosquito antiviral defense mechanism and resultant viral pathogenesis might be detrimental to establishment of persistent infections [36,80]. Nevertheless, viral evasion of the mosquito immune response may be necessary in order to infect, disseminate and be transmitted. Viruses with RNA genomes (including almost all arboviruses) acquire mutations at every round of replication due to an error-prone RNA-dependent RNA polymerase (RdRP) [81]. Evidence has been presented for selection of genomes with mutations in regions highly targeted by RNAi as a mechanism of evasion [37,82].
A candidate for limited suppression of RNAi is the subgenomic flavivirus RNA (sfRNA) [53]. One proposed suppressive mechanism is inhibition of Dcr proteins; reduced activity of recombinant human Dcr was observed when WNV-sfRNA was added to an in vitro assay. Direct inhibition of mosquito cell Dcr2 was not demonstrated [83]. The sfRNA also was shown to sequester mosquito XRN1, which might be a required downstream step following Ago2 cleavage of viral mRNA to complete degradation [54,84].
7. Future directions
Although evidence has been presented for modulation of arbovirus infections of mosquitoes by canonical innate immune pathways, knowledge that the exo-siRNA pathway is the unique and most potent mechanism of antiviral innate immunity in mosquitoes provides opportunities to develop novel strategies for controlling arbovirus transmission and incentives to more completely understand the mechanism. A few areas in which future research might be pursued are the following: What viral RNA structures in addition to long dsRNA can serve as PAMPs for Dcr2 recognition? What activities are required downstream from Ago2 cleavage of viral RNA to complete the RNAi pathway? What is the role of piRNAs in anti-viral immunity? Which Piwi-clade proteins are involved? What are arboviral strategies for RNAi evasion?
As described in section 5, both genetic manipulation studies and modeling research have suggested the feasibility of transgenic, arbovirus-resistant mosquitoes. Nevertheless, a number of challenges remain to be solved before using RNAi-based genetically modified mosquitoes as an effective strategy for significantly reducing arbovirus transmission and impacting human arboviral disease. In the case of DENVs, IR transgenes need to be developed that target for cleavage RNA genomes of all four DENV serotypes. We are currently designing a tetravalent IR effector gene incorporating the conserved NS5 (RdRP) coding regions of DENV 1-4. In transgenic plants, several examples have been described in which RNAi-based approaches were used to develop resistance to multiple tospoviruses [85,86]. Second, in principle the anti-pathogen gene conferring a DENV-refractory phenotype would require introgression into existing DENV susceptible mosquito populations, in essence replacing DENV-competent mosquito populations with DENV-refractory populations. To accomplish this, the anti-DENV IR effector gene may require linkage with an Ae. aegypti-specific selfish genetic-element or gene drive system to enable fixation of the transgene in the target vector population [87]. Killer-rescue based gene drive systems such as Medea are currently under development in Ae. aegypti [88]; however, gene drive approaches are not likely to be implemented as large-scale public health measures in the near future. Recently, Okamoto et al. [89] used a stochastic, spatially explicit model of Ae. aegypti populations from Iquitos, Peru, to evaluate whether population replacement is feasible absent gene-drive. The modeling indicated that releasing mosquitoes carrying only an anti-pathogen construct can negatively impact vector competence of a natural population at ratios well below those considered necessary for transgenic technologies involving population reduction [89-91]. Moreover, Okamoto and colleagues found that introgression of the effector gene could occur locally in a reasonable timeframe and releasing mosquitoes carrying only an anti-pathogen gene is considerably more robust for immigration into wild-type mosquito populations than other strategies modeled. A third challenge is to determine whether an RNAi-based approach will select for DENV quasispecies populations that escape the heritable RNAi-based strategy. A final challenge is that any genetically modified vector approach will need extensive field testing, encounter regulatory hurdles, and require local and regional consent prior to release of the modified vector [92,93].
Highlights.
RNA interference (RNAi) is the major mosquito immune response against arboviruses
The exo-siRNA pathway plays the key role in anti-arboviral immunity
The piRNA pathway also evidences involvement in anti-arboviral immunity
Arboviruses must evade mosquito RNAi to maintain transmission cycles
Genetic manipulation of RNAi can be used to generate arbovirus-resistant mosquitoes
Acknowledgements
Our research has been funded by NIH grants AI34014 and AI48740 and the Grand Challenges in Global Health through the Foundation for NIH. Funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of this review; and in the decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Address: Arthropod-borne and Infectious Diseases Laboratory, 1692 Campus Delivery, Colorado State University, Fort Collins, CO 80523-1692, USA kenneth.olson@colostate.edu
References
- 1.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. The Genome Sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 2.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, et al. The Genome Sequence of the Malaria Mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 3.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, et al. Immunity-Related Genes and Gene Families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 4.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, et al. Genome Sequence of Aedes aegypti, a Major Arbovirus Vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, et al. Evolutionary Dynamics of Immune-Related Genes and Pathways in Disease-Vector Mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, et al. Sequencing of Culex quinquefasciatus Establishes a Platform for Mosquito Comparative Genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Bartholomay LC, Waterhouse RM, Mayhew GF, Campbell CL, Michel K, et al. Pathogenomics of Culex quinquefasciatus and Meta-Analysis of Infection Responses to Diverse Pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. The publication of the Cx. quincquefasciatus genome sequence enabled this important phylogenomic comparison of immune response genes in 3 genera of arbovirus vectors as well as Drosophila. The paper also presents a meta-transcriptomic analysis of Cx. quincquefasciatus genes responsive to WNV infection.
- 8.Imler J-L. Overview of Drosophila immunity: A historical perspective. Developmental & Comparative Immunology. 2014;42:3–15. doi: 10.1016/j.dci.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A Novel System for the Launch of Alphavirus RNA Synthesis Reveals a Role for the Imd Pathway in Arthropod Antiviral Response. PLoS Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter J, Hutter S, Baines JF, Roller J, Saminadin-Peter SS, et al. The Transcriptional Response of Drosophila melanogaster to Infection with the Sigma Virus (Rhabdoviridae) PLoS ONE. 2009;4:e6838. doi: 10.1371/journal.pone.0006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa A, Jan E, Sarnow P, Schneider D. The Imd Pathway Is Involved in Antiviral Immune Responses in Drosophila. PLoS ONE. 2009;4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges LM, Johnson KN. Induction of host defence responses by Drosophila C virus. Journal of General Virology. 2008;89:1497–1501. doi: 10.1099/vir.0.83684-0. [DOI] [PubMed] [Google Scholar]
- 13.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agaisse H, Petersen U-M, Boutros M, Mathey-Prevot B, Perrimon N. Signaling Role of Hemocytes in Drosophila JAK/STAT-Dependent Response to Septic Injury. Developmental Cell. 2003;5:441–450. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 15.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- *16.Deddouche S, Matt N, Budd A, Mueller S, Kemp C, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. Demonstration that the DExD/H-box helicase Dcr2, which is the PRR for the Drosophila antiviral RNAi response, is required for induction of the antiviral gene vago. In addition, the DExD/H helicase of Dcr2 is a member of the RIG I-like receptor family, which in mammalian cells are PRRs for viral infection and mediate IFN induction.
- *17.Kemp C, Mueller S, Goto A, Barbier V, Paro S, et al. Broad RNA Interference-Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. The Journal of Immunology. 2013;190:650–658. doi: 10.4049/jimmunol.1102486. A comparison of flies with mutations in either a key RNAi pathway gene or a key JAK/STAT pathway induction gene and their responses to infection with a variety of viruses; RNAi was part of a broad anti-viral response whereas JAK/STAT induction was limited to a single virus family.
- 18.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cellular Microbiology. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 19.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 20.Kennerdell JR, Carthew RW. Use of dsRNA-Mediated Genetic Interference to Demonstrate that frizzled and frizzled 2 Act in the Wingless Pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 21.Lindbo JA, Dougherty WG. Pathogen-Derived Resistance to a Potyvirus: Immune and Resistant Phenotypes in Transgenic Tobacco Expressing Altered Forms of a Potyvirus Coat Protein Nucleotide Sequence. Molecular Plant-Microbe Interactions. 1992;5:144–153. doi: 10.1094/mpmi-5-144. [DOI] [PubMed] [Google Scholar]
- 22.Ratcliff F, Harrison BD, Baulcombe DC. A Similarity Between Viral Defense and Gene Silencing in Plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- *23.Li H, Li WX, Ding SW. Induction and Suppression of RNA Silencing by an Animal Virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. Demonstrates the importance of RNAi as an antiviral defense in Drosophila by showing that an insect-pathogenic virus, Flock House virus, both induces RNAi and encodes a specific protein suppressor of RNAi.
- 24.Powers AM, Kamrud KI, Olson KE, Higgs S, Carlson JO, Beaty BJ. Molecularly engineered resistance to California serogroup virus replication in mosquito cells and mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:4187–4191. doi: 10.1073/pnas.93.9.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *25.Olson KE, Higgs S, Gaines PJ, Powers AM, Davis BS, Kamrud KI, Carlson JO, Glair CD, Beaty BJ. Genetically Engineered Resistance to Dengue-2 Virus Transmission in Mosquitoes. Science. 1996;272:884–886. doi: 10.1126/science.272.5263.884. First demonstration that the virus-induced gene-silencing (VIGS) response of plants, later shown to be due to RNAi, can also be used to engineer arbovirus resistance in a mosquito.
- 26.Gaines PJ, Olson KE, Higgs S, Powers AM, Beaty BJ, Blair CD. Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome. J Virol. 1996;70:2132–2137. doi: 10.1128/jvi.70.4.2132-2137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol Biol. 2001;10:265–273. doi: 10.1046/j.1365-2583.2001.00267.x. [DOI] [PubMed] [Google Scholar]
- 28.Adelman ZN, Sanchez-Vargas I, Travanty EA, Carlson JO, Beaty BJ, Blair CD, Olson KE. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. Publication of the complete Anopheles gambiae genome sequence enabled identification of genes encoding key components of the siRNA pathway and demonstration by RNAi-mediated gene silencing that RNAi controls arbovirus replication in a vector mosquito.
- 30.Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. Publication of the complete Aedes aegypti genome sequence enabled demonstration that RNAi is an important regulator of replication of the most globally-important arbovirus in its natural mosquito vector.
- 32.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Campbell C, Black W, Hess A, Foy B. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. Phylogenomic comparisons of RNAi pathway components in major arbovirus mosquito vectors with Drosophila showed major gene family expansions in mosquitoes.
- 34.Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiology. 2011;6:265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blair CD, Adelman ZN, Olson KE. Molecular strategies for interrupting arthropod-borne virus transmission by mosquitoes. Clin Microbiol Rev. 2000;13:651–661. doi: 10.1128/cmr.13.4.651-661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proceedings of the National Academy of Sciences. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. First use of deep sequencing to characterize virus-specific small RNAs produced by Aedes aegypti mosquitoes in the antiviral RNAi response to arbovirus infection.
- *37.Brackney DE, Beane JE, Ebel GD. RNAi Targeting of West Nile Virus in Mosquito Midguts Promotes Virus Diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. Thorough analysis of deep-sequencing data for WNV-specific small RNAs produced in a natural mosquito vector shows that RNAi drives increased genetic diversity of an arbovirus in its mosquito host.
- *38.Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, Olson KE, Blair CD. Comparison of Dengue Virus Type 2-Specific Small RNAs from RNA Interference-Competent and -Incompetent Mosquito Cells. PLoS Negl Trop Dis. 2010;4:e848. doi: 10.1371/journal.pntd.0000848. Shows that production of typical DENV-specific siRNAs is defective in C6/36 mosquito cell cultures and C6/36 cell-lysate assays because of a point mutation in their dicer2 gene, thus allowing 10-100-fold higher virus replication titers.
- 39.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, et al. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proceedings of the National Academy of Sciences. 2010;107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. La Crosse Bunyavirus Nonstructural Protein NSs Serves To Suppress the Type I Interferon System of Mammalian Hosts. J Virol. 2007;81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. C6/36 Aedes albopictus Cells Have a Dysfunctional Antiviral RNA Interference Response. PLoS Negl Trop Dis. 2010;4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Léger P, Lara E, Jagla B, Sismeiro O, Mansuroglu Z, Coppée JY, Bonnefoy E, Bouloy M. Dicer-2- and Piwi-Mediated RNA Interference in Rift Valley Fever Virus-Infected Mosquito Cells. Journal of Virology. 2013;87:1631–1648. doi: 10.1128/JVI.02795-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler J-L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 45.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. Production of Virus-Derived Ping-Pong-Dependent piRNA-like Small RNAs in the Mosquito Soma. PLoS Pathog. 2012;8:e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-Dependent 80S Complex Cleaves Targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 47.Rand TA, Ginalski K, Grishin NV, Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc Natl Acad Sci USA. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 49.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 51.Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarz DS, Tomari Y, Zamore PD. The RNA-Induced Silencing Complex Is a Mg2+− Dependent Endonuclease. Current Biology. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- *53.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, et al. A Highly Structured, Nuclease-Resistant, Noncoding RNA Produced by Flaviviruses Is Required for Pathogenicity. Cell Host & Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. Identification of a possible arboviral suppressor of RNAi, albeit in a genome RNA structural element, not a protein-encoding gene.
- 54.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh A, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 56.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, et al. A Slicer-Mediated Mechanism for Repeat-Associated siRNA 5′ End Formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 57.Hess A, Prasad A, Ptitsyn A, Ebel G, Olson K, Barbacioru C, Monighetti C, Campbell C. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiology. 2011;11:45. doi: 10.1186/1471-2180-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vodovar N, Bronkhorst AW, van Cleef KWR, Miesen P, Blanc H, et al. Arbovirus-Derived piRNAs Exhibit a Ping-Pong Signature in Mosquito Cells. PLoS ONE. 2012;7:e30861. doi: 10.1371/journal.pone.0030861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnettler E, Donald CL, Human S, Watson M, Siu RWC, et al. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. Journal of General Virology. 2013;94:1680–1689. doi: 10.1099/vir.0.053850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders HR, Foy BD, Evans AM, Ross LS, Beaty BJ, Olson KE, Gill SS. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochemistry and Molecular Biology. 2005;35:1293–1307. doi: 10.1016/j.ibmb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Xi Z, Ramirez J, Dimopoulos G. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proceedings of the National Academy of Sciences. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Developmental & Comparative Immunology. 2010;34:625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luplertlop N, Surasombatpattana P, Patramool S, Dumas E, Wasinpiyamongkol L, et al. Induction of a Peptide with Activity against a Broad Spectrum of Pathogens in the Aedes aegypti Salivary Gland, following Infection with Dengue Virus. PLoS Pathog. 2011;7:e1001252. doi: 10.1371/journal.ppat.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paradkar PN, Trinidad L, Voysey R, Duchemin J-B, Walker PJ. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proceedings of the National Academy of Sciences. 2012;109:18915–18920. doi: 10.1073/pnas.1205231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paradkar PN, Duchemin J-B, Voysey R, Walker PJ. Dicer-2-Dependent Activation of Culex Vago Occurs via the TRAF-Rel2 Signaling Pathway. PLoS Negl Trop Dis. 2014;8:e2823. doi: 10.1371/journal.pntd.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clarke TE, Clem RJ. Insect Defenses Against Virus Infection: The Role of Apoptosis. International Reviews of Immunology. 2003;22:401–424. doi: 10.1080/08830180305215. [DOI] [PubMed] [Google Scholar]
- 68.Vaidyanathan R, Scott T. Apoptosis in mosquito midgut epithelia associated with West Nile virus infection. Apoptosis. 2006;11:1643–1651. doi: 10.1007/s10495-006-8783-y. [DOI] [PubMed] [Google Scholar]
- 69.Girard Y, Schneider B, McGee C, Wen J, Han V, et al. Salivary gland morphology and virus transmission during long-term cytopathologic West Nile virus infection in Culex mosquitoes. Am J Trop Med Hyg. 2007;76:118–128. [PubMed] [Google Scholar]
- 70.Liu B, Behura SK, Clem RJ, Schneemann A, Becnel J, Severson DW, Zhou L. P53-Mediated Rapid Induction of Apoptosis Conveys Resistance to Viral Infection in Drosophila melanogaster. PLoS Pathog. 2013;9:e1003137. doi: 10.1371/journal.ppat.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H, Gort T, Boyle DL, Clem RJ. Effects of Manipulating Apoptosis on Sindbis Virus Infection of Aedes aegypti Mosquitoes. Journal of Virology. 2012;86:6546–6554. doi: 10.1128/JVI.00125-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy Is an Essential Component of Drosophila Immunity against Vesicular Stomatitis Virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. Journal of General Virology. 2009;90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- 75.Heaton NS, Randall G. Dengue Virus-Induced Autophagy Regulates Lipid Metabolism. Cell Host & Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chuang C-F, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- *78.Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. Demonstration that the mosquito antiviral RNAi pathway can be exploited for engineering arbovirus resistance in mosquitoes by transgenesis that leads to expression of long dsRNA from the viral genome upon ingestion of an infectious blood-meal.
- 79.Franz AWE, Sanchez-Vargas I, Raban RR, Black WC, IV, James AA, Olson KE. Fitness Impact and Stability of a Transgene Conferring Resistance to Dengue-2 Virus following Introgression into a Genetically Diverse Aedes aegypti Strain. PLoS Negl Trop Dis. 2014;8:e2833. doi: 10.1371/journal.pntd.0002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. Suppression of RNA Interference Increases Alphavirus Replication and Virus-Associated Mortality in Aedes aegypti Mosquitoes. BMC Microbiol. 2009;9:49. doi: 10.1186/1471-2180-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brackney DE, Pesko KN, Brown IK, Deardorff ER, Kawatachi J, Ebel GD. West Nile Virus Genetic Diversity is Maintained during Transmission by Culex pipiens quinquefasciatus Mosquitoes. PLoS ONE. 2011;6:e24466. doi: 10.1371/journal.pone.0024466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siu RWC, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, et al. Antiviral RNA Interference Responses Induced by Semliki Forest Virus Infection of Mosquito Cells: Characterization, Origin, and Frequency-Dependent Functions of Virus-Derived Small Interfering RNAs. J Virol. 2011;85:2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, et al. Noncoding Flavivirus RNA Displays RNA Interference Suppressor Activity in Insect and Mammalian Cells. Journal of Virology. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. The Structural Basis of Pathogenic Subgenomic Flavivirus RNA (sfRNA) Production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bucher E, Lohuis D, van Poppel PMJA, Geerts-Dimitriadou C, Goldbach R, et al. Multiple virus resistance at a high frequency using a single transgene construct. Journal of General Virology. 2006;87:3697–3701. doi: 10.1099/vir.0.82276-0. [DOI] [PubMed] [Google Scholar]
- 86.Peng J-C, Chen T-C, Raja JAJ, Yang C-F, Chien W-C, et al. Broad-Spectrum Transgenic Resistance against Distinct Tospovirus Species at the Genus Level. PLoS ONE. 2014;9:e96073. doi: 10.1371/journal.pone.0096073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.James AA. Gene drive systems in mosquitoes: rules of the road. Trends in Parasitology. 2005;21:64–67. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 88.Chen C-H, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA. A Synthetic Maternal-Effect Selfish Genetic Element Drives Population Replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 89.Okamoto KW, Robert MA, Gould F, Lloyd AL. Feasible Introgression of an Anti-pathogen Transgene into an Urban Mosquito Population without Using Gene-Drive. PLoS Negl Trop Dis. 2014;8:e2827. doi: 10.1371/journal.pntd.0002827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okamoto KW, Robert MA, Lloyd AL, Gould F. A Reduce and Replace Strategy for Suppressing Vector-Borne Diseases: Insights from a Stochastic, Spatial Model. PLoS ONE. 2013;8:e81860. doi: 10.1371/journal.pone.0081860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robert MA, Okamoto K, Lloyd AL, Gould F. A Reduce and Replace Strategy for Suppressing Vector-Borne Diseases: Insights from a Deterministic Model. PLoS ONE. 2013;8:e73233. doi: 10.1371/journal.pone.0073233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lavery JV, Tinadana PO, Scott TW, Harrington LC, Ramsey JM, et al. Towards a framework for community engagement in global health research. Trends in Parasitology. 2010;26:279–283. doi: 10.1016/j.pt.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 93.Ramsey JM, Bond JG, Macotela ME, Facchinelli L, Valerio L, et al. A Regulatory Structure for Working with Genetically Modified Mosquitoes: Lessons from Mexico. PLoS Negl Trop Dis. 2014;8:e2623. doi: 10.1371/journal.pntd.0002623. [DOI] [PMC free article] [PubMed] [Google Scholar]