Abstract
Cardiac hypertrophy is a multifactorial disease characterized by multiple molecular alterations. One of these alterations is change in activity of Akt, which plays a central role in regulating a variety of cellular processes ranging from cell survival to aging. Akt activation is mainly achieved by its binding to phosphatidylinositol 3,4,5 triphosphate (PIP3). This results in a conformational change that exposes the kinase domain of Akt for phosphorylation and activation by its upstream kinase PDK1 in the cell membrane. Recent studies have shown that sirtuin isoforms SIRT1, SIRT3 and SIRT6 play an essential role in the regulation of Akt activation. While SIRT1 deacetylates Akt to promote PIP3 binding and activation, SIRT3 controls ROS-mediated Akt activation and SIRT6 transcriptionally represses Akt at the level of chromatin. In the first part of this review, we discuss the mechanisms by which sirtuins regulate Akt activation and how they influence other post-translational modifications of Akt. In the latter part of the review, we summarize the implications of sirtuin-dependent regulation of Akt signaling in the control of major cellular processes like cellular growth, angiogenesis, apoptosis, autophagy and aging; which are involved in the initiation and progression of several diseases.
Keywords: Akt, Sirtuins, cardiac hypertrophy, aging, IGF signaling
Introduction
In the quest to live longer and lead a healthy disease free life, we have invested enormous resources to understand the mechanism of aging process. The only proven approach which is capable of retarding the aging process and aging-associated diseases is calorie-restriction1. One of the most extensively studied signaling pathways associated with nutrition supply is the insulin like growth factor (IGF) signaling2. Disruption of IGF-1 signaling uniformly extends lifespan of animal species ranging from yeast to monkeys3. Accumulating data provide overwhelming evidence that another group of enzymes called sirtuins are also sensitive to calorie restriction, and they too regulate IGF signaling4. One of the key signaling mechanisms that is activated following insulin like growth factor receptor (IGF-1R) stimulation is the PI3K/AKT pathway, which plays a central role in cellular signaling and regulation of variety of cellular processes. Recent advancements in cell biology have identified sirtuins as major regulators of Akt activation. Here, in this review we will discuss the mechanism of activation of Akt signaling by sirtuins, and its implications in the development of cardiac diseases and the aging process.
Sirtuin deacetylases
Lysine acetylation is a reversible post translational modification process where histone acetyltransferases (HATs) transfer the acetyl moiety from acetyl coenzyme A to the -amino groups of lysine (K) within a protein, resulting in its charge neutralization. The opposite reaction is cairred out by another group of enzymes called histyome deacetylases (HDACs), which remove the acetyl moiety from target proteins. Sirtuins belong to class-III HDACs, which need NAD for their deacetylation reaction. Name sirtuin originates from the discovery of the yeast gene, silent information regulator 2 (Sir2), which was originally described as regulators of transcriptional silencing of mating-type loci, ribosomal DNA and lifespan of yeast5. Subsequently, as more isoforms of this gene were identified, they were named together as sirtuins. Because of dependency of sirtuins to NAD and their ability to deacetylate histones, they are considered sensors of cellular energy status and effectors of gene transcription by controlling acetylation of histones5. With identification of more isoforms of sirtuins it did not take long to realize that sirtuins not only deacetylate histones, but also a wide variety of transcription factors, metabolic enzymes and signaling kinases, and thereby controlling their activity.
The mammalian genome encodes seven sirtuin isoforms (SIRT1 to SIRT7), which vary in their tissue specificity, subcellular localization, enzymatic activity and targets6. SIRT1 is the prototype member of this class which is studied the most. SIRT1 is localized in the nucleus and cytoplasm7, 8. Recent studies suggest that, to a lesser extent, SIRT1 is also localized in the plasma membrane, where it up regulates insulin signaling9. SIRT1 is implicated in the control of cell survival, apoptosis, autophagy and metabolism10. SIRT2 is a cytoplasmic deacetylase which deacetylates tubulin and regulates cytoskeletal reorganization, autophagy and metabolism11-13. The sirtuins SIRT3, SIRT4 and SIRT5 are localized in the mitochondria, though a lesser amount of SIRT3 is also present in the nucleus, where it participates in gene regulation14, 15. These three isoforms of sirtuins are implicated in regulating several mitochondrial-dependent metabolic pathways, including oxidative phosphorylation, ROS synthesis, urea-cycle, ATP synthesis and apoptosis14. SIRT6 is a chromatin associated enzyme involved in deacetylating H3K9 and H3K56, thereby regulating gene expression, cellular metabolism and the inflammatory response16-19. SIRT7 is localized in the nucleolus and up-regulates the RNA polymerase I dependent gene transcription20-23. Each of these seven sirtuin isoforms has been knocked out in mice. The results indicated that while most inbred SIRT1 knockout mice die postnatally, outbred mice survive to adulthood with retarded body size. SIRT6 knockout mice die nearly one month after birth with characteristics of multi-organ pre-mature aging19, 24. Similar to SIRT1, outbred SIRT6 knockout mice survive to adulthood and have retarded body size. Cardiac phenotypes of all the SIRT knockout and transgenic mice studied so far are summarized in table-1.
Table 1. Cardiac phenotype of Akt and Sirtuins knockout and transgenic mice.
| Mice genotype | Cardiac Phenotype | Refs |
|---|---|---|
| Akt1-KO | Akt1-KO mice show reduced cardiac muscle mass and display defects in physiological growth. Mice develop an exacerbated form of cardiac hypertrophy when subjected to pressure overload. | 25-27 |
| Akt2-KO | Akt2-deficiency does not affect cardiac mass, but induces insulin resistance, mitochondrial and cardiac contractile dysfunction. Akt2-KO mice are more sensitive to myocardial injury as evidenced by increased myocyte death and inflammatory cardiomyopathy. | 25, 28-32 |
| Akt3-KO | No obvious cardiac phenotype noted. | 33 |
| Akt1-transgenic | Cardiomyocyte specific Akt expression increases myocyte size and induces cardiac hypertrophy. But Akt1 transgenic mice are protected from injury caused by transient cardiac ischemia. | 34-39 |
| Akt3-transgenic | Cardiac-specific over expression of Akt3 induces cardiac hypertrophy, and contractile dysfunction together with increasing susceptibility of the heart to cardiac injury. | 40 |
| Nuclear-Akt1 transgenic | Transgenic hearts exhibit increased number of smaller cardiomyocytes that relates with cardiomyocyte survival and enhanced contractile function. | 41, 42 |
| SIRT1-KO | Whole-body SIRT1-deficient mice show reduced heart size and are resistant to develop cardiac hypertrophy. Contrary, cardiomyocyte specific SIRT1-KO mice do not show any cardiac abnormalities at basal conditions; however, they are more susceptible to ischemic injury. Partial deficiency of SIRT1 attenuated pressure overload induced cardiac hypertrophy and failure. | 43-45 |
| SIRT2-KO | No basal cardiac abnormalities are noted, but SIRT2-KO mice are protected from ischemic injury due to attenuated programmed apoptosis in the heart. | 46 |
| SIRT3-KO | SIRT3-KO mice show no basal cardiac abnormalities at birth, but develop hypertrophy, fibrosis and contractile dysfunction with age. SIRT3 deficient mice are highly susceptible to cardiac hypertrophic stimuli. | 47-49 |
| SIRT6-KO | Massive concentric cardiac hypertrophy and heart failure were observed in the whole-body and cardiac-specific SIRT6 deficient mice. | 50 |
| SIRT7-KO | SIRT7-deficiency induces spontaneous cardiac hypertrophy and inflammatory cardiomyopathy in mice. | 51 |
| SIRT1-transgenic | Low to moderate over expression of SIRT1 attenuates age-dependent decline in cardiac functions in mice. However, high level of SIRT1 expression induces cardiac hypertrophy and heart failure. | 43, 44, 52 |
| SIRT3-transgenic | Cardiac specific over expression of SIRT3 protects the heart from hypertrophic stimuli by preserving mitochondrial function. | 48 |
| SIRT6-transgenic | Cardiac specific over expression of SIRT6 protects the heart from hypertrophic stimuli by blocking activation of Akt signaling at the level of chromatin. | 50 |
In the last several years, sirtuins received significant attention because of their roles in regulating aging process, and their responsiveness to calorie restriction1. Calorie restriction and physical exercise robustly increase expression levels of SIRT1, SIRT3 and SIRT625-28. Among them, the expression levels of SIRT3 and SIRT6 have been linked with longevity of mammals, whereas the role of SIRT1 in this process is equivocal29-32. Same as for their roles in the aging process, SIRT3 and SIRT6 expression blocks the development of cardiac hypertrophy and heart failure, but not SIRT19, 33, 34. Although SIRT1 activation protects cardiomyocytes from apoptosis and ischemia-reperfusion injury, overexpression of SIRT1 in mice leads to development of cardiac hypertrophy and heart failure35, 36. Each one of these sirtuin isoform has been found to target Akt signaling to produce their specific cellular response9, 33, 34. Before we discuss how sirtuins control Akt activation, a brief description of Akt and its mechanism of activation is discussed below.
Akt isoforms and their functions
Akt, also called protein kinase B due to its similarity with protein kinase A and C, is a serine/threonine kinase involved in the regulation of a variety of cellular functions including metabolism, glucose uptake, proliferation and protein synthesis, all assigned towards a single goal of cell survival37, 38. Mammals have three isoforms of Akt, designated as Akt1, Akt2 and Akt3, all having greater than 80% homology at the amino acid level39. In vivo function of these isoforms is deduced by generating mouse mutants that lack each one of these isoforms or in combination. Akt1 null mouse is growth retarded with proportional decrease in organ size and shows shorter lifespan due to exacerbated apoptosis when subjected to oxidative stress40, 41. Akt2 deficient mice show reduced insulin sensitivity, whereas Akt3 null mice exhibit a 20-25% reduction in brain size and weight, partly due to a significant reduction in cell size and number42, 43. Combined deficiency of Akt1 and 2 in mice results in neonatal lethality, severe growth deficiency, muscle atrophy and defects in adipogenesis as well as in skin and bone development44. Mice deficient in both Akt1 and Akt3 are embryonically lethal, and show defects in the development of nervous system, cardiovascular system and vasculature45. Akt2 and Akt3 null mice have normal embryonic development but are growth retarded with a smaller brain and testis size. They also have impaired glucose metabolism46, 47. These observations underscore the unique function as well as functional redundancy among the three Akt isoforms. For additional information we have summarized the cardiac phenotype of Akt knockout and transgenic mice in table-1.
Mechanism of Akt activation
Akt activation is a multistep process. It involves binding of Akt to membrane lipids, recruitment of Akt to the plasma membrane and phosphorylation of Akt by the upstream kinase PDK1 which is also localized in the plasma membrane. Structurally, Akt consists of three domains, an N-terminal PH domain followed by a kinase domain and a hydrophobic C-terminal regulatory domain. For its basal activation, Akt needs to be phosphorylated at T308 by PDK148. When Akt is inactive, intra molecular interaction between the PH and kinase domains prevents accessibility of PDK1 to T30849. In order for PDK1 to access the kinase domain of Akt, the latter needs to undergo a significant conformational change. This happens only when the PH domain of Akt binds to PIP3 molecule, which is generated from PIP2 by activation of PI3-kinase (PI3K)50. PIP3 generation takes place in the inner leaflet of the plasma membrane. Once the PH domain of Akt binds to PIP3, it undergoes a conformational change, exposing its kinase domain to its upstream kinase PDK1, resulting in T308 phosphorylation and a 100 fold increase in Akt activity51, 52. For maximal activation, Akt needs to be phosphorylated at yet another site S473 by mTORC-253. mTORC2 is a multi-protein complex that consists of mTOR, Rictor, mSIN1 and Protor-154. Phosphorylation of Akt at S473 further increases its activity by 10 fold. Thus cumulatively, T308 and S473 phosphorylation augments Akt activity by 1000 fold from the basal level in response to growth factor stimulation, and customarily these phosphorylation sites are regarded as the surrogate markers of Akt activity55, 56.
Factors that prime Akt phosphorylation
Two critical steps occur prior to PDK1-dependent phosphorylation and activation of Akt. These are (1) Transportation of Akt to the plasma membrane and (2) binding of Akt to PIP3. The PH domain of Akt regulates both these steps. One of the seminal studies that linked defects in Akt PH domain to disease conditions is the finding that a mutation (E17K) in the PH domain increases the affinity of Akt for PIP3 and enhances Akt membrane localization. These changes render Akt hyperactive even in un-stimulated NIH3T3 cells, and hence promoting their proliferation and survival57. The E17K mutation of Akt induces leukemia in mice, and in humans this is associated with breast, colon and ovarian cancers57. A recent study elaborated these observations to the role of lysine ubiquitination in the activation of Akt58.
Ubiquitination recruits Akt to the plasma membrane
Ubiquitination is a reversible posttranslational modification that covalently attaches ubiquitin protein to lysine residues of the target protein. This reaction was originally associated with protein recycling as ubiquitin labeled proteins are directed to proteasome-mediated degradation. Recent studies impart degradation independent functions to ubiquitination including kinase activation57. TRAF6 an E3 ubiquitin ligases was shown to monoubiquitinate Akt in the PH domain in response to IGF stimulation of cells. This modification helps to recruit Akt to the plasma membrane58. Activation of Akt by TRAF6-mediated ubiquitination was however independent of its ability to bind to PIP3, suggesting that ubiquitination does not regulate Akt binding to PIP3. In this report, it was also shown that the enhanced membrane trafficking of E17K mutants of Akt is due to the TRAF6-mediated ubiquitination of this additional lysine residue, leading to overall hyper-ubiquitination of Akt, thus promoting its tumorigenic activity58. More recently another E3 ligase, the Skp2 was found to be critical for ErbB-receptor-mediated Akt ubiquitination and membrane recruitment in response to EGF stimulation of cells, thus suggesting that different growth factors target distinct E3 ligases for ubiquitination and activation of Akt59.
SIRT1-mediated deacetylation regulates Akt-binding to PIP3 and hence activation
Another post translational modification that regulate Akt activity is reversible acetylation. Under basal conditions, Akt is acetylated in various tissues, including heart, liver, brain, and skeletal muscle, and this modification suppresses Akt activity. The amino acids that underwent acetylation were identified as K14 and K20, both located in the PH domain of Akt (Figure 1). Deacetylation of these lysines by SIRT1 is necessary for the binding of Akt to PIP3 and for its membrane localization and activation9. In this study, it was also shown that the PH domain of another kinase, PDK1 is acetylated. This modification hampered the binding of PDK1 to PIP3, whereas deacetylated form of PDK1 displayed opposite results, thus suggesting that acetylation-dependent regulation could be a common mechanism controlling activity of the membrane-lipid binding proteins. On a similar note, another study found insulin receptor substrate 2 (IRS2) as a constitutively acetylated protein60. IRS2 is a receptor associated downstream effector of IGF-1R signaling. Lysine acetylation inhibits IRS2 activity and SIRT1-dependent deacetylation increases its activity, and thereby increasing the activity of Akt. SIRT1-mediated deacetylation is also necessary for the phosphorylation of IRS2 by the IR kinase in hepatocytes60. These findings suggest that SIRT1 upregulates insulin signaling and Akt activation at multiple levels. A model describing roles of the PH domain acetylation and ubiquitination for regulating Akt activation is presented in Figure 2.
Figure 1. Reversible lysine acetylation in the PH domain regulates Akt activation.
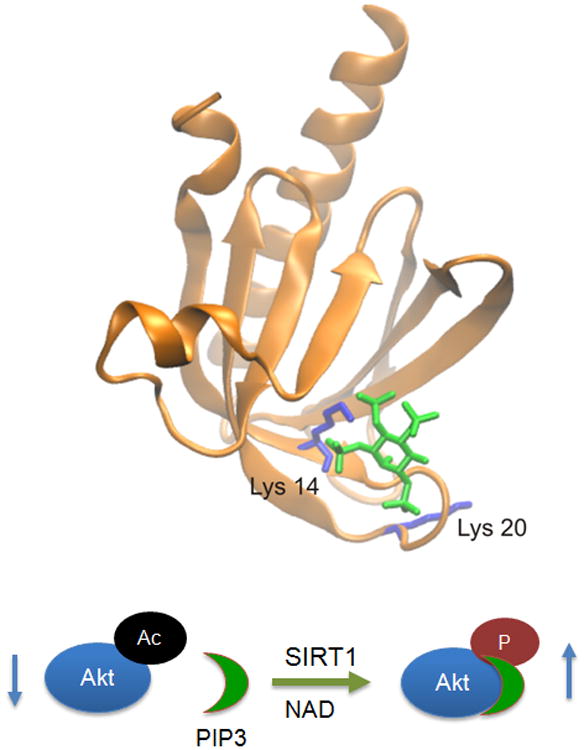
Upper panel: crystal structure of the PH domain of Akt. Acetylated lysine residues (Lys14 and Lys20) are shown in purple and PIP3 in green. Both lysines are in close proximity to the binding pocket for PIP3. Lower panel shows schematic activation of Akt by SIRT1. Lysine acetylation of the PH domain makes Akt incapable to bind to PIP3, leading to inactivation of the kinase. SIRT1-dependent deacetylation promotes Akt-PIP3 binding and hence phosphorylation and activation of Akt by the upstream kinases (for details see ref 9).
Figure 2. A model illustrating the role of SIRT1 and SIRT3 in regulating Akt activation.
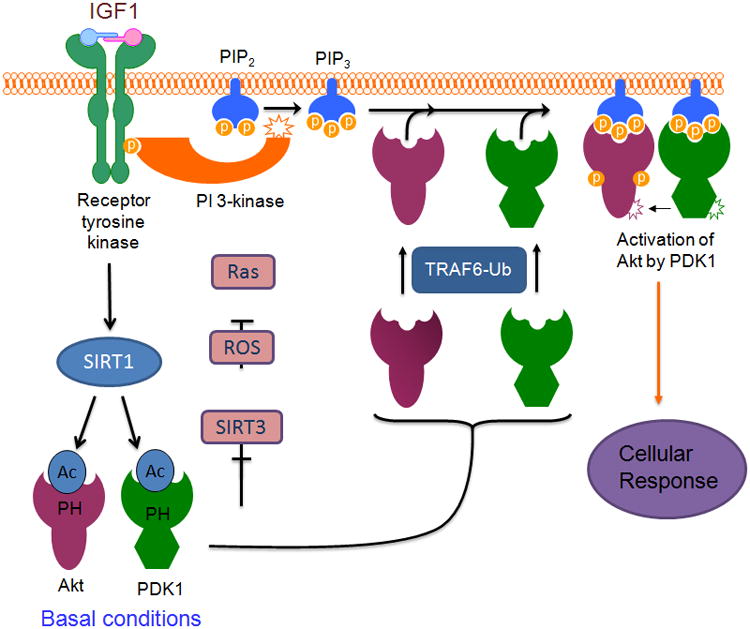
Under basal conditions the PH domains of Akt and PDK1 are acetylated, leading to inhibition of their binding to PIP3 and hence inactivation. During growth factor stimulation of cells, SIRT1 binds to and deacetylates Akt and PDK1 PH domians. This change enables them to bind to PIP3 which is generated from PIP2 by activation of PI3K during growth factor stimulation of cells. The PH domain deacetylation is likely to make lysine residues available for TRAF6-mediated ubiquitination, thereby facilitating ubiquitination-mediated transport of the protein to the plasma membrane (ubiquitination of PDK1 is not shown yet, but based on the PH domain acetylation studies, it seems also subject to ubiquitination, same as Akt). At the level of plasma membrane PDK1 phosphorylates and activates Akt leading to cellular response (for details see ref 9). Under oxidative stress Akt can be hyper-activated by ROS-mediated Ras activation, which activates PI3K. In this condition SIRT3 blocks Akt activation by suppressing ROS production from mitochondria (for details see ref 9 and 33).
SIRT3 blocks ROS-mediated hyper activation of Akt signaling
Another sirtuin analogue implicated in regulating Akt activity and the aging process is SIRT3. SIRT3 is a mitochondrial deacetylase regulating variety of mitochondrial functions and therefore considered to be a mitochondrial fidelity protein61. SIRT3 knockout mice do not show any noticeable phenotype at birth, but they are sensitive to stress stimuli. Because of this reason it is believed that SIRT3 does not play a role in the development, but rather it fine tunes the activity of mitochondrial substrates by lysine deacetylation to protect cells from stress. SIRT3 regulates activity of several mitochondrial enzymes including antioxidant MnSOD and enzymes of the electron transport chain, NDUFA9 in complex I and SDHA in complex II62-65. SIRT3KO mice manifests nearly 50% reduced cellular ATP and increased ROS levels in many tissues including liver and heart63. Since increased ROS levels are known to activate Ras oncogene, which indirectly activates Akt via activation of PI3K and increased synthesis of PIP3, in SIRT3KO hearts robust activation of Ras and Akt was found33. These hearts also exhibited robust cardiac hypertrophic response following infusion of hypertrophy agonist. On the other hand SIRT3 over expressing transgenic hearts were resistant to hypertrophic stimuli and showed no signs of Ras-Akt activation33. Thus SIRT3 indirectly controls hyperactivation of Akt by regulating mitochondrial ROS production and ROS-mediated Ras-PI3K-Akt activation (Figure 2).
SIRT6 negatively regulates Akt signaling at the level of chromatin
Recently, yet another sirtuin analogue SIRT6 received considerable importance for its role in maintaining cellular homeostasis and regulating aging and associated diseases. SIRT6KO mice have shortened lifespan with metabolic defects19. H3K9 and H3K56 are the two histone substrates of SIRT666,67, 68. By deacetylating H3K9, SIRT6 controls the expression of genes including telomere maintenance, DNA repair, inflammation and metabolism66, 69-71. SIRT6 binds to NF-kB and HIF1α transcription factors to negatively regulate their target gene transcription70, 71. Most recently, it was shown that SIRT6 directly controls IGF/Akt signaling at the level of chromatin through deacetylation of H3K934. SIRT6 knockout mice spontaneously developed cardiac hypertrophy by 2-3 months of age. Consistent with this observation, SIRT6 levels were reduced in different mouse models of cardiac failure as well as in human failing hearts. All these hearts showed robust activation of many transcription/translational factors and growth factors and their receptors (R), related to IGF/Akt signaling, including, IGF-1R, IR, IGF-2R, IGF-2, IRS1/2, Akt, Foxo1, mTOR, GSK3, myc, β-catenin, Elf4E, p70S6P and S6P (Figure 3). The IGF-1 levels were, however, downregulated in SIRT6 deficient hypertrophied hearts. Increased activation of IGF/Akt signaling in these hearts was due to increased binding of IGF-2, which can bind to IGF-1R, IGF-2R and insulin receptor (IR). In SIRT6-deficient hearts, SIRT1 was also elevated, which is needed for deacetylation and activation of Akt. Further studies provided evidence that SIRT6 physically interacts with c-Jun, recruiting it to the chromatin and suppressing transcriptional activity of c-Jun. Under stress and pathological conditions, cellular SIRT6 levels are reduced, leading to de-repression of c-Jun activity and thereby increasing expression of IGF-Akt signaling related genes harboring c-Jun binding sites in their promoters (Figure 3). In accordant with this finding, another study reported the incidence of chronic inflammation in SIRT6 knockout mice by 7-8 months of age as a result of increased activity of c-Jun72. Another recent report by Kanfi et al observed a 15% increase in median lifespan in male transgenic mice over expressing SIRT630. This enhanced longevity of male mice was again linked to alterations in IGF/Akt signaling related genes. All these studies provided strong evidence that SIRT6 is an endogenous negative regulator of IGF/Akt signaling at the level of chromatin. These studies together demonstrated that sirtuins act as master regulators of IGF/Akt signaling by establishing their control both at the transcriptional and posttranslational levels. Other factors which activate or terminate Akt signaling are summarized in a supplement table (see supplement table).
Figure 3. Schematic model showing SIRT6-depdendent transcriptional regulation of IGF/Akt signaling-related genes.
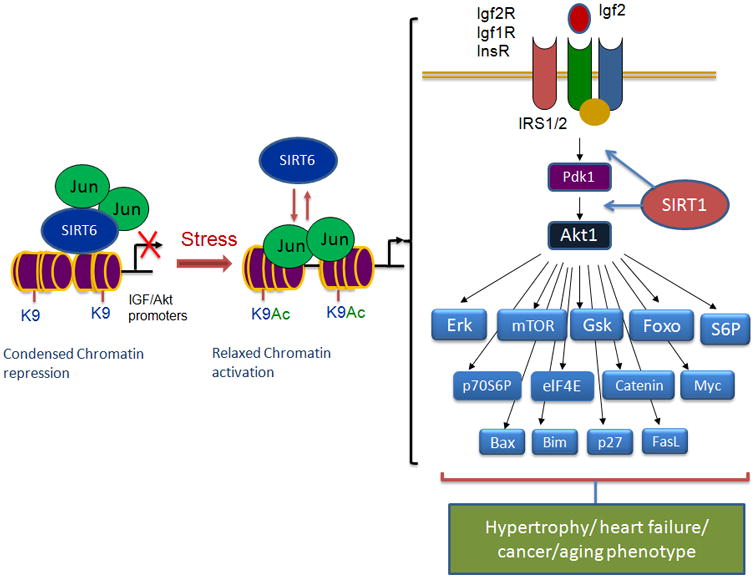
Under basal conditions SIRT6 binds to chromatin as a complex with c-jun and deacetylates histones (H3K9), leading to chromatin condensation and repression of the IGF/Akt signaling-related genes. Under pathological stress, SIRT6 levels are reduced leading to increased acetylation of H3K9 at the promoters of IGF/Akt signaling related genes and c-jun mediated transcriptional activation. IGF/Akt signaling related genes which are activated in SIRT6KO hearts are shown on the right side of the figure (For details see ref 34).
Implications of Akt/SIRT interaction in cardiac hypertrophy
Akt represents one of the most potential therapeutic targets to meet clinical needs of medicine today. We have discussed how sirtuins act as master regulators of IGF/Akt signaling by regulating its activity at the transcriptional and post-translational levels. Here, we discuss more about how sirtuin/Akt interaction influences cardiac hypertrophic phenotype. Additionally, we discuss how sirtuin/Akt interplay modulates angiogenesis, apoptosis, autophagy and aging, four conditions which influence the disease aggressiveness in cardiac hypertrophy.
The role of SIRT1 in cardiac hypertrophy is complex. SIRT1 levels are upregulated in response to pressure overload and oxidative stress. High levels (12.5 fold) of SIRT1 expression induced cardiac hypertrophy and heart failure, whereas, low level of SIRT1 (7.5 fold) attenuated age-dependent increase in cardiac hypertrophy73. In the pressure overload model of cardiac hypertrophy, haploinsufficiency of SIRT1 was found to be protective and over expression of SIRT1 exacerbated the cardiac dysfunction74. We also observed increased cardiac protection in SIRT1 knockout mice in response to agonist induced cardiac hypertrophy75. This effect is associated with reduced Akt signaling in the heart.
SIRT3 and SIRT6 are two other sirtuins, whose role in cardiac hypertrophy is elucidated. SIRT3 knockout mice spontaneously developed cardiac hypertrophic phenotype at adult hood33, 76. Over expression of SIRT3 or maintenance of endogenous SIRT3 levels by treating mice with NAD blocked the agonist induced cardiac hypertrophic response in mice33, 77. As mentioned above lack of SIRT3 or its reduced activation was associated with increased ROS levels and activation of Akt signaling33, 77. Similar to SIRT3, SIRT6 also acts as an antihypertrophic molecule. Cardiac specific over expression of SIRT6 protected mice from pressure overload and agonist-induced hypertrophy. This was achieved by down regulation the IGF/Akt signaling by the interaction of SIRT6 with c-Jun, resulting in deacetylation of histone 3 at Lys9 (H3K9)34. These findings reinforce the possible interplay between sirtuins and Akt in modulating cardiac hypertrophic response.
Role of SIRT/Akt in angiogenesis
Growth and development of an organ is dependent on the coordinated reinforcement of new vasculature to the newly formed cells necessary for providing essential nutrients, macromolecules and oxygen78. When cells proliferate or grow, oxygen demand also increases79. If the supply of oxygen is less, hypoxic tissues secrete growth factors and chemokines that stimulate endothelial cells to proliferate, differentiate and migrate, a process termed as sprouting and branching80, 81. The SIRT1 and Akt pathways play a cardinal role in this process82. In the heart, during development of physiologic hypertrophy even though cardiomyocytes grow in size, they are adequately nourished by the development of new capillaries. Contrary to this, during pathologic cardiac hypertrophy, cardiomyocyte growth outweighs capillary density, resulting in the supply of less nutrients and oxygen to the growing cardiomyocyte83. SIRT1 plays a critical role in regulating sprouting angiogenesis and vascular growth. SIRT1 deficient mice displayed impaired ability to develop new blood vessels in response to angiogenic signals84. Similarly, SIRT1 deficient zebra fish also showed dys-regulated endothelial sprouting, vessel navigation and vascular patterning84. Although the role of SIRT1 in cardiac angiogenesis has not been studied, acute activation Akt in the heart induces angiogenesis whereas chronic activation inhibits the same83.
One of the key factors participating in vasculature development and growth is nitric oxide. Nitric oxide synthesized from endothelial cells by endothelial nitric oxide synthase (eNOS), promotes vasodilatation and protects vessels from atherosclerotic stimuli. eNOS is a target of both Akt and SIRT1. Akt activates eNOS by phosphorylation and SIRT1 does the same by deacetylation84, 85, thereby functionally linking SIRT1 with Akt for maintaining the endothelial cellular function and agiogenesis86.
Although the role of other sirtuins in angiogenesis is not yet explored, studies utilizing MEFs and cancer cell lines demonstrate that SIRT3 destabilizes HIF1α during hypoxia to reduce transcription of its pro-angiogenic gene VEGF-A87. Also, a recent study implicated the role of SIRT6 in the regulation of endothelial cell function. Depletion of SIRT6 reduced the proliferation and increased the senescence of endothelial cells. This effect of SIRT6 is again associated with lower levels of eNOS mRNA and protein, thus suggesting that same as for IGF/AKT related genes, SIRT6 may also regulate the expression of eNOS at the level of chromatin88.
Role of SIRT/Akt in apoptosis
Proper development of an organism is dependent on the balance between cell death and cell growth. Apoptosis or programmed cell death is a well-orchestrated gene regulated suicide program by which unwanted or harmful cells are removed from the system89. Corollary, defects in apoptotic pathways are associated with a variety of human diseases like cancer, neurodegeneration and cardiac hypertrophy89-91. Apoptosis plays an imperative role in the development of heart failure. Studies carried out using rabbit as a model system has demonstrated that ischemia reperfusion injury is associated with extensive apoptosis (14%) of cardiomyocytes92. In human failing hearts, apoptosis rate ranging from 0.12 to 0.70% is reported93. This small level of apoptosis is considered sufficient to cause heart failure, based on the observation that in the hearts with conditionally active caspase 3, even very low level of apoptosis (23 myocytes/105) was sufficient to induce dilated cardiomyopathy and heart failure94.
About the role of sirtuins in cardiomyocyte apoptosis, SIRT1 plays an anti-apoptotic role and contributes to hearts tolerance to oxidative stress. This effect of SIRT1 seems to be governed by its ability to shuttle between nucleus and cytoplasm under stress conditions. It is the nuclear SIRT1, rather than the cytoplasmic, that has the antiapoptotic activity8. Increased nuclear SIRT1 levels were observed in the cardiomyocytes of TO-2 hamster failing hearts, rat model of myocardial infarction, and in dilated cardiomyopathy patients as a compensatory mechanism to protect cells from death stimuli. In another study, reduced levels of nuclear SIRT1 were reported in aging hearts, and this was associated with impaired SIRT1 activation and reduced protection of the heart from I/R injury95. In agreement with this, nuclear Akt also appeared to be antiapoptotic. In cardiomyocytes nuclear expression of Akt blocked apoptosis induced by staurosporine, deoxyglucose and hypoxia. Besides, mice over expressing nuclear Akt were also protected against ischemia-reperfusion injury96.
Studies conducted to explore the mechanism behind cytoprotective effects of nuclear SIRT1 have shown that it upregulates activity of antioxidants and downregulates proapoptotic molecules35. SIRT1 upregulates the expression of cardioprotective molecules including MnSOD, TrX1 and Bcl-xL35. In addition, SIRT1-mediated deacetylation can negatively regulate the activity of proapoptotic molecules including Bax and p5335, 97. Both SIRT1 and SIRT3 can deacetylate Ku70 to sequester Bax away from mitochondria thus inhibiting apoptosis98, 99. In this process, Akt may help to maintain cellular Ku70 levels by preventing its Hdm2-mediated degradation100.
Another step where SIRT1 and Akt can cooperate to regulate cellular survival is modification of the activity p53. P53 is an acetylated protein and this post-translational modification is indispensable for its function101. Deacetylation of p53 by SIRT1 renders it inactive101. Deacetylated p53 binds to Mdm2, an E3 ubiquitin ligase which promotes the proteasome-mediated degradation of p53. Akt acts synergistically in this process by phosphorylating Mdm2 at S166 and S186 and promoting its association with p53102. Another sirtuin which has been studied for its in role in regulating cardiac myocyte survival is SIRT2. In contrast to the antiapoptotic role of SIRT1, ablation of SIRT2 was found to be beneficial in ischemia/reperfusion models. The hearts of SIRT2KO mice or wild-type mice treated with AKG2, a specific pharmacologic inhibitor of SIRT2, were protected from ischemic injury103. These studies suggest the contrasting roles of sirtuins in the regulation of cardiomyocyte apoptosis.
Role of SIRT/Akt in Autophagy
Autophagy is a catabolic response, where cells degrade their own components through lysosomes. This process removes dysfunctional proteins and organelles104. Under stress situation, autophagy serves as a mechanism to maintain cellular metabolism by degrading damaged proteins, organelles as well as undamaged components that are not essential for cell survival under a given circumstance to generate amino acids and fatty acids for ATP production. Autophagy involves several sequential steps including autophagosome nucleation, elongation, lipidation and degradation which are controlled by autophagy related genes (Atgs)104. SIRT1 can directly interact with and deacetylate several Atg proteins, including Atg5, Atg7 and Atg8, leading to activation of these proteins105. In cardiomyocytes, glucose deprivation upregulates the activity of SIRT1 and its downstream target FOXO1, and both these factors are needed for increased autophagic flux106. Cardiac-specific overexpression of a FOXO mutant which cannot interact with SIRT1, or cardiac-specific deletion of FOXO1 significantly reduced autophagic flux, thus suggesting a role of SIRT1 in regulating autophagy in the heart106.
The role of autophagy in heart is complex; however, evidence suggests that autophagy may be an adaptive mechanism under most conditions107. Autophagy is found to be up-regulated in human failing hearts caused by dilated cardiomyopathy resulting from valvular diseases or ischemic heart disease108. The results obtained from use of animal models of cardiac diseases have shown contrasting results in terms of the role of autophagy in cardiac protection. Autophagosome nucleation requires beclin1 (Atg6)109. In the heart, beclin1 heterozygous knockout mice showed reduced autophagy and displayed blunted pathologic cardiac remodeling in response to aortic banding as well as to ischemia reperfusion injury110, 111. Beclin1 is shown to be down regulated in the SIRT1 knockout mice, thus again indicating the possible role of SIRT1 in regulating the autophagy process112. Contrary to this, cardiac-specific deletion of ATG5, another target of SIRT1, lead to development of cardiac hypertrophy and failure, and dominant negative ATG5 mutant abolished the cardioprotective effects of autophagy inducing drug, chloramphenicol113, 114. In the rat myocardial infarction model, blocking autophagy by use of bafilomycin led to exacerbated cardiac dysfunction115. In another study, glucose deprivation or ischemia induced autophagy helped to promote cell survival110. Also intermittent fasting, an intervation known to induce SIRT1, helped to reduce infarct size by 2 fold in the rat myocardial infraction model116. Based on these reports it appears that increased autophagy is a physiological or pathological response to promote myocardial cell survival largely depends on the nature and extend of the cellular stress.
A direct role of sirtuins other than SIRT1 in the regulation of autophagy is not studied so far. But evidence suggests that autophagy may be associated with increased activation of SIRT6, because the transcriptional factors, NFkB and AP1, whose activity is negatively regulated by SIRT6, are shown to be positive regulators of autophagy117, 118. Regarding the possible connection of sirtuins with Akt, recent reports show that chronic Akt activation worsens aging-induced cardiac hypertrophy and myocardial contractile function through loss of autophagic regulation119. Further studies using cardiomyocytes are needed to elucidate the conditions where sirtuins and Akt crossover to regulate autophagy.
Sirtuins, Akt and Aging
Calorie restriction is the only proven approach to lessen the aging process1. Both, SIRT1 and IGF/Akt signaling pathways are regulated by nutrition supply and both pathways are suggested to be involved in regulation of lifespan in many organisms. Many reports suggest that the health benefits of calorie restriction are mediated through activation of sirtuins; however a role of SIRT1 in this process is disputed. SIRT1 knockout mice failed to increase physical activity during calorie restriction120. Also, calorie restriction exacerbated the decreased survivability of SIRT1 null mice, suggesting a positive role of SIRT1 in mediating effects of calorie restriction121. In contrast, over expression of SIRT1 did not extend replicative lifespan of human fibroblasts or prostrate epithelial cells, rather caused replicative senescence in response to cellular stress7, 122. Also calorie restriction and/or mutations in the yeast Akt homologue; Sch9 causes dramatic chronological lifespan extension in yeast lacking Sir2123.
One of the family of transcription factors whose activity is regulated by SIRT1 and which play a role in the aging process is Foxo124, 125. Consistent with the ambiguous role of SIRT1 in lifespan extension, SIRT1 can positively and negatively regulate activity of the Foxo family of factors. SIRT1 activates Foxo1 and Foxo3 by deacetylation, which promotes nuclear localization of these factors126, 127. Contrary to this, SIRT1 can also hamper Foxo3a activity by making it a target for skp2-mediated ubiquitination and degradation128. In this process Akt can synergies with SIRT1 by phosphorylating Foxo isoforms which prevents their translocation to the nucleus, thereby abolishing their transcriptional function129.
In our studies we found that SIRT1-mediated deacetylation positively regulates the activity of Akt upon growth factor stimulation of cells9. We therefore propose that in the presence of growth (insulin) signaling, SIRT1 activates Akt, resulting in the phosphorylation of Foxo. This event will expel Foxo from the nucleus thereby inhibiting its activity. In the absence of insulin signaling lack of Akt-mediated phosphorylation and SIRT1-mediated deacetylation will facilitate localization of Foxo into the nucleus, where it promotes transcription of genes involved in promoting endurance, stress resistance and longevity, thus suggesting that SIRT1 may promote longevity under calorie-restricted or growth factor depleted conditions. But in conditions where nutrients are ample, SIRT1 promotes Akt signaling and cellular senescence. It should be also noted that apart from direct activation of Akt, SIRT1 can activate IGF signaling by release of insulin from pancreas or by decreasing the expression of IGFBP, an inhibitory modulator of IGF signaling130, 131.
As for the role of other sirtuins is concerned, health benefits of calorie restriction were also found to be mediated through activation of SIRT3 and SIRT6. Mice lacking SIRT3 failed to show benefits of calorie restriction with regard to aging associated hearing loss132. Similarly, protective effects of calorie restriction on oxidative stress were diminished in SIRT3 knockout mice due to reduced activity of MnSOD26. SIRT3 activation has been also linked with life-span extension in humans as polymorphism in the SIRT3 gene prompter which causes gene activation was found associated with longevity of the man29, 133.
So far, SIRT6 is the only sirtuin whose increased expression conclusively extends the lifespan of mammals. Whole body SIRT6 knockout mice develop aging phenotype, and SIRT6 over expressing mice have an extended lifespan, compared to their wild-type littermates30, 70. Interestingly, SIRT6 increases longevity by inhibiting IGF signaling. Transgenic mice over expressing SIRT6 shows lower serum levels of IGF-1, which caused reduced activation of the IGF-1 signaling pathway, including reduced activity of Akt and reduced phosphorylation of Foxo1 and Foxo330. In the heart SIRT6 can suppress the expression of IGF/Akt signaling-related genes by interacting with c-Jun and deacetylating histone H3K934. Through this mechanism, SIRT6 blocked cardiac hypertrophic response in various mouse models of cardiac hypertrophy (Figure 3). Similarly, by inhibiting c-Jun, SIRT6 was reported to block expression of pro-inflammatory genes72. For the reasons that cardiac hypertrophy and inflammation are associated with aging, it is conceivable to believe that SIRT6 is an anti-aging sirtuin whose up-regulation may help to impede the development of many diseases.
Future Perspective
Akt and sirtuins regulate the very basis of cellular functioning and alterations in their functions have potentially lethal effects on the organism. Although both Akt and SIRT1 complement each other in function, the consequence of their interaction and its implications is not yet fully understood. Other than T308, components of mTORC complex could also be regulated by SIRT1 at the posttranslational level. How acetylation modulates these phosphorylation events will provide a deeper insight into the acetylation-mediated regulation of Akt activity. Furthermore, acetylation is known to regulate the activity of phosphatases, SIRT1 may have yet another regulatory function at the level of phosphatases which need to be studied. More than 250 mammalian proteins possess the PH domain, and the findings showing that acetylation regulates the activity of two PH domain proteins, Akt and PDK1, could be a prelude to the existence of a similar mechanism in other PH domain proteins. The presence of SIRT1 in the plasma membrane also suggests that many other molecules in the membrane could be a target of SIRT1. Acetylation and ubiquitination counter balance each other as both modifications occur in lysine residues, and acetylated lysine residues are immune to ubiquitination. This suggests the existence of an intricate interplay between acetylation and ubiquitination in the activation of Akt. Activation of SIRT1 also seems to be a complex process since we found that SIRT1 activates Akt only in the presence of growth factors. It is well established that SIRT1 gets activated in conditions where growth factors are depleted. How the same deacetylase performs contradictory functions under different cellular conditions, is intriguing and worth further studying? SIRT1 and SIRT6 seem to contradict each other in cell signaling pathways associated with cellular growth. It will be fascinating to study their relationship in conditions like calorie restriction and diseases associated with abnormal cellular growth. Implications of a direct role of SIRT6 in cellular processes like apoptosis, angiogenesis and autophagy needs to be studied with the aim that inhibitors or activators of these molecules will emerge as promising drugs for the treatment of cancer and cardiac hypertrophy. The synergistic effects of SIRT1 and Akt inhibitors and SIRT6 activator could have a profound effect in the management of malignant diseases. From a cardiac hypertrophy perspective, where cell growth needs to be regulated without cell death, it seems that magnitude and cellular distribution of SIRT1 could influence cardiac phenotype. We believe that activation of SIRT1 in the presence of growth factor signaling can exacerbate hypertrophic response, hence warrants caution while taking food supplements to enhance its activity.
Supplementary Material
Acknowledgments
Sources of funding: These studies were supported from NIH grants RO1 HL117041, HL111455, HL 83423 and HL77788 to MP Gupta.
Non-Standard abbreviations and Acronyms
- IGF
insulin like growth factor
- IGF-1R
insulin like growth factor receptor
- PI3K
Phosphatidylinositide 3-kinase
- HAT
histone acetyltransferases
- SIRT
silent information regulator
- ROS
Reactive oxygen species
- PDK1
3-phosphoinositide dependent protein kinase-1
- PIP3
Phosphatidylinositol (3,4,5)-triphosphate
- mTOR
mammalian target of rapamycin
- TRAF6
TNF receptor associated factor
- Skp2
S-phase kinase-associated protein 2
- PH domain
Pleckstrin homology domain
- IRS2
Insulin receptor substrate 2
- MnSOD
Manganese Superoxide dismutase 2
- NDUFA9
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 9
- SDHA
Succinate dehydrogenase complex, subunit A
- Ras
Rat sarcoma
- NF-kB
nuclear factor kappa-light-chain-enhancer of activated B cells
- HIF1α
Hypoxia-inducible factor 1-alpha
- H3K9
histone 3 at Lys9
- eNOS
endothelial nitric oxide synthase
- VEGF-A
Vascular endothelial growth factor A
- TrX1
Redox status of thioredoxin-1
- Bcl-xL
B-cell lymphoma-extra large
- Bax
Bcl2 associated X protein
- KO
Knock out
- Atgs
autophagy related genes
- FOXO1
Forkhead box protein O1
- AP1
activator protein 1
- IGFBP
Insulin-like growth factor-binding protein
Footnotes
Disclosures: None
References
- 1.Heilbronn LK, Ravussin E. Calorie restriction and aging: Review of the literature and implications for studies in humans. The American journal of clinical nutrition. 2003;78:361–369. doi: 10.1093/ajcn/78.3.361. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Anzo M, Cohen P. Control of aging and longevity by igf-i signaling. Experimental gerontology. 2005;40:867–872. doi: 10.1016/j.exger.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 3.van Heemst D. Insulin, igf-1 and longevity. Aging and disease. 2010;1:147–157. [PMC free article] [PubMed] [Google Scholar]
- 4.Longo VD. Linking sirtuins, igf-i signaling, and starvation. Experimental gerontology. 2009;44:70–74. doi: 10.1016/j.exger.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nature reviews Molecular cell biology. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic sir2-like proteins. Biochemical and biophysical research communications. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 7.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human sirt proteins. Molecular biology of the cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the nad+-dependent histone deacetylase sirt1. The Journal of biological chemistry. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 9.Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP. The deacetylase sirt1 promotes membrane localization and activation of akt and pdk1 during tumorigenesis and cardiac hypertrophy. Science signaling. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 10.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. Sirt2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 12.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting pepck1 degradation via recruiting the ubr5 ubiquitin ligase. Molecular cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gal J, Bang Y, Choi HJ. Sirt2 interferes with autophagy-mediated degradation of protein aggregates in neuronal cells under proteasome inhibition. Neurochemistry international. 2012;61:992–1000. doi: 10.1016/j.neuint.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 14.He W, Newman JC, Wang MZ, Ho L, Verdin E. Mitochondrial sirtuins: Regulators of protein acylation and metabolism. Trends in endocrinology and metabolism: TEM. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Iwahara T, Bonasio R, Narendra V, Reinberg D. Sirt3 functions in the nucleus in the control of stress-related gene expression. Molecular and cellular biology. 2012;32:5022–5034. doi: 10.1128/MCB.00822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. Sirt6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu SY, Michishita E, Park JY, Nunez N, Hursting SD, Barrett C, Horikawa I. Analysis of mammalian sirt6, a homolog of the yeast sir2 protein, during adipogenesis and calorie-restriction. Cancer Epidem Biomar. 2005;14:2731s–2731s. [Google Scholar]
- 18.Liszt G, Ford E, Kurtev M, Guarente L. Mouse sir2 homolog sirt6 is a nuclear adp-ribosyltransferase. Journal of Biological Chemistry. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 19.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian sirt6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 20.Barber MF, Michishita-Kioi E, Xi YX, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen KF, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. Sirt7 links h3k18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–+. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of sirt7 with chromatin remodeling complexes and expands its role in regulation of rna polymerase i transcription. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of sirt7 in resumption of rdna transcription at the exit from mitosis. J Cell Sci. 2009;122:489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford E, Voit R, Liszt G, Magin C, GrumMt I, Guarente L. Mammalian sir2 homolog sirt7 is an activator of rna polymerase i transcription. Genes & development. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian sir2alpha protein has a role in embryogenesis and gametogenesis. Molecular and cellular biology. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, Cohen HY. Regulation of sirt6 protein levels by nutrient availability. FEBS letters. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by sirt3-mediated sod2 activation. Cell metabolism. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and sirt3 trigger global reprogramming of the mitochondrial protein acetylome. Molecular cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canto C, Auwerx J. Caloric restriction, sirt1 and longevity. Trends in endocrinology and metabolism: TEM. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel vntr enhancer within the sirt3 gene, a human homologue of sir2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin sirt6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 31.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baur JA, Chen D, Chini EN, Chua K, Cohen HY, de Cabo R, Deng C, Dimmeler S, Gius D, Guarente LP, Helfand SL, Imai S, Itoh H, Kadowaki T, Koya D, Leeuwenburgh C, McBurney M, Nabeshima Y, Neri C, Oberdoerffer P, Pestell RG, Rogina B, Sadoshima J, Sartorelli V, Serrano M, Sinclair DA, Steegborn C, Tatar M, Tissenbaum HA, Tong Q, Tsubota K, Vaquero A, Verdin E. Dietary restriction: Standing up for sirtuins. Science. 2010;329:1012–1013. doi: 10.1126/science.329.5995.1012. author reply 1013-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting foxo3a-dependent antioxidant defense mechanisms in mice. The Journal of clinical investigation. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, Cunningham JM, Deng CX, Lombard DB, Mostoslavsky R, Gupta MP. The sirtuin sirt6 blocks igf-akt signaling and development of cardiac hypertrophy by targeting c-jun. Nature medicine. 2012;18:1643–1650. doi: 10.1038/nm.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, Yamamoto T, Tian B, Sadoshima J. Pparalpha-sirt1 complex mediates cardiac hypertrophy and failure through suppression of the err transcriptional pathway. Cell metabolism. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sussman MA, Volkers M, Fischer K, Bailey B, Cottage CT, Din S, Gude N, Avitabile D, Alvarez R, Sundararaman B, Quijada P, Mason M, Konstandin MH, Malhowski A, Cheng Z, Khan M, McGregor M. Myocardial akt: The omnipresent nexus. Physiological reviews. 2011;91:1023–1070. doi: 10.1152/physrev.00024.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song G, Ouyang G, Bao S. The activation of akt/pkb signaling pathway and cell survival. Journal of cellular and molecular medicine. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the pten tumour suppressor. Nature reviews Molecular cell biology. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 40.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/pkbalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. The Journal of biological chemistry. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 41.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, Kadowaki T, Hay N. Growth retardation and increased apoptosis in mice with homozygous disruption of the akt1 gene. Genes & development. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu QW, Crenshaw EB, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase akt2 (pkb beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 43.Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, Lee VMY, Szabolcs M, de Jong R, Oltersdorf T, Ludwig T, Efstratiadis A, Birnbaum MJ. Role for akt3/protein kinase b gamma in attainment of normal brain size. Molecular and cellular biology. 2005;25:1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking akt1 and akt2. Genes & development. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang ZZ, Tschopp O, Di-Poi N, Bruder E, Baudry A, Dummler B, Wahli W, Hemmings BA. Dosage-dependent effects of akt1/protein kinase balpha (pkbalpha) and akt3/pkbgamma on thymus, skin, and cardiovascular and nervous system development in mice. Molecular and cellular biology. 2005;25:10407–10418. doi: 10.1128/MCB.25.23.10407-10418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dummler B, Hemmings BA. Physiological roles of pkb/akt isoforms in development and disease. Biochemical Society transactions. 2007;35:231–235. doi: 10.1042/BST0350231. [DOI] [PubMed] [Google Scholar]
- 47.Dummler B, Tschopp O, Hynx D, Yang ZZ, Dirnhofer S, Hemmings BA. Life with a single isoform of akt: Mice lacking akt2 and akt3 are viable but display impaired glucose homeostasis and growth deficiencies. Molecular and cellular biology. 2006;26:8042–8051. doi: 10.1128/MCB.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robey RB, Hay N. Is akt the “warburg kinase”?-akt-energy metabolism interactions and oncogenesis. Seminars in cancer biology. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingley E, Hemmings BA. Pleckstrin homology (ph) domains in signal transduction. Journal of cellular biochemistry. 1994;56:436–443. doi: 10.1002/jcb.240560403. [DOI] [PubMed] [Google Scholar]
- 50.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific pi3k signalling. Nature reviews Molecular cell biology. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 51.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase balpha. Current biology : CB. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 52.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter GF, Holmes AB, Gaffney PRJ, Reese CB, McCormick F, Tempst P, Coadwell J, Hawkins PT. Protein kinase b kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase b. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 53.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of akt/pkb by the rictor-mtor complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 54.Alessi DR, Pearce LR, Garcia-Martinez JM. New insights into mtor signaling: Mtorc2 and beyond. Science signaling. 2009;2:pe27. doi: 10.1126/scisignal.267pe27. [DOI] [PubMed] [Google Scholar]
- 55.Scheid MP, Marignani PA, Woodgett JR. Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase b. Molecular and cellular biology. 2002;22:6247–6260. doi: 10.1128/MCB.22.17.6247-6260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase b by insulin and igf-1. The EMBO journal. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 57.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of akt1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 58.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK. The e3 ligase traf6 regulates akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK. The skp2-scf e3 ligase regulates akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J. The direct involvement of sirt1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. The Journal of biological chemistry. 2007;282:34356–34364. doi: 10.1074/jbc.M706644200. [DOI] [PubMed] [Google Scholar]
- 61.Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, Aykin-Burns N, Spitz DR, Gius D, Kim HS. Sirt3, mitochondrial ros, ageing, and carcinogenesis. Int J Mol Sci. 2011;12:6226–6239. doi: 10.3390/ijms12096226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bao JJ, Scott I, Lu ZP, Pang LY, Dimond CC, Gius D, Sack MN. Sirt3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radical Bio Med. 2010;49:1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by sirt3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates mnsod activity in response to stress. Molecular cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. Sirt6 is a histone h3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone h3 lysine k56 by human sirt6. Cell cycle (Georgetown, Tex) 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin sirt6 deacetylates h3 k56ac in vivo to promote genomic stability. Cell cycle (Georgetown, Tex) 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. Sirt6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging. 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. Sirt6 links histone h3 lysine 9 deacetylation to nf-kappab-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase sirt6 regulates glucose homeostasis via hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, Reynolds D, Chen Q, Xu X, Young HA, Chen WJ, Gao B, Deng CX. Progression of chronic liver inflammation and fibrosis driven by activation of c-jun signaling in sirt6 mutant mice. The Journal of biological chemistry. 2012;287:41903–41913. doi: 10.1074/jbc.M112.415182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circulation research. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 74.Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, Yamamoto T, Tian B, Sadoshima J. Pparalpha-sirt1 complex mediates cardiac hypertrophy and failure through suppression of the err transcriptional pathway. Cell metabolism. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP. The deacetylase sirt1 promotes membrane localization and activation of akt and pdk1 during tumorigenesis and cardiac hypertrophy. Science signaling. 2011;4 doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 76.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CMK, Rosenzweig A, Sinclair DA. Regulation of the mptp by sirt3-mediated deacetylation of cypd at lysine 166 suppresses age-related cardiac hypertrophy. Aging-Us. 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, Gupta MP. Exogenous nad blocks cardiac hypertrophic response via activation of the sirt3-lkb1-amp-activated kinase pathway. Journal of Biological Chemistry. 2010;285:3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carmeliet P. Angiogenesis in health and disease. Nature medicine. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 79.Wachsberger P, Burd R, Dicker AP. Tumor response to ionizing radiation combined with antiangiogenesis or vascular targeting agents: Exploring mechanisms of interaction. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:1957–1971. [PubMed] [Google Scholar]
- 80.Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA. Vascular endothelial growth factor and angiogenesis. Pharmacological reviews. 2004;56:549–580. doi: 10.1124/pr.56.4.3. [DOI] [PubMed] [Google Scholar]
- 81.Jain RK. Molecular regulation of vessel maturation. Nature medicine. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 82.Karar J, Maity A. Pi3k/akt/mtor pathway in angiogenesis. Frontiers in molecular neuroscience. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shiojima I, Sato K, Izumiya Y, Schiekofer S, Ito M, Liao R, Colucci WS, Walsh K. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J Clin Invest. 2005;115:2108–2118. doi: 10.1172/JCI24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, Zeiher AM, Dimmeler S. Sirt1 controls endothelial angiogenic functions during vascular growth. Genes & development. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCabe TJ, Fulton D, Roman LJ, Sessa WC. Enhanced electron flux and reduced calmodulin dissociation may explain “calcium-independent” enos activation by phosphorylation. The Journal of biological chemistry. 2000;275:6123–6128. doi: 10.1074/jbc.275.9.6123. [DOI] [PubMed] [Google Scholar]
- 86.Ota H, Eto M, Kano MR, Kahyo T, Setou M, Ogawa S, Iijima K, Akishita M, Ouchi Y. Induction of endothelial nitric oxide synthase, sirt1, and catalase by statins inhibits endothelial senescence through the akt pathway. Arteriosclerosis, thrombosis, and vascular biology. 2010;30:2205–2211. doi: 10.1161/ATVBAHA.110.210500. [DOI] [PubMed] [Google Scholar]
- 87.Bell EL, Emerling BM, Ricoult SJ, Guarente L. Sirt3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ros production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cardus A, Uryga AK, Walters G, Erusalimsky JD. Sirt6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovascular research. 2013;97:571–579. doi: 10.1093/cvr/cvs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Plati J, Bucur O, Khosravi-Far R. Apoptotic cell signaling in cancer progression and therapy. Integrative biology : quantitative biosciences from nano to macro. 2011;3:279–296. doi: 10.1039/c0ib00144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mattson MP. Apoptosis in neurodegenerative disorders. Nature reviews Molecular cell biology. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- 91.van Empel VP, De Windt LJ. Myocyte hypertrophy and apoptosis: A balancing act. Cardiovascular research. 2004;63:487–499. doi: 10.1016/j.cardiores.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 92.Yue TL, Ma XL, Wang X, Romanic AM, Liu GL, Louden C, Gu JL, Kumar S, Poste G, Ruffolo RR, Jr, Feuerstein GZ. Possible involvement of stress-activated protein kinase signaling pathway and fas receptor expression in prevention of ischemia/reperfusion-induced cardiomyocyte apoptosis by carvedilol. Circulation research. 1998;82:166–174. doi: 10.1161/01.res.82.2.166. [DOI] [PubMed] [Google Scholar]
- 93.van Empel VPM, Bertrand ATA, Hofstra L, Crijns HJ, Doevendans PA, De Windt LJ. Myocyte apoptosis in heart failure. Cardiovascular research. 2005;67:21–29. doi: 10.1016/j.cardiores.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 94.Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, Shirani J, Armstrong RC, Kitsis RN. A mechanistic role for cardiac myocyte apoptosis in heart failure. J Clin Invest. 2003;111:1497–1504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tong C, Morrison A, Mattison S, Qian S, Bryniarski M, Rankin B, Wang J, Thomas DP, Li J. Impaired sirt1 nucleocytoplasmic shuttling in the senescent heart during ischemic stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012 doi: 10.1096/fj.12-216473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shiraishi I, Melendez J, Ahn Y, Skavdahl M, Murphy E, Welch S, Schaefer E, Walsh K, Rosenzweig A, Torella D, Nurzynska D, Kajstura J, Leri A, Anversa P, Sussman MA. Nuclear targeting of akt enhances kinase activity and survival of cardiomyocytes. Circulation research. 2004;94:884–891. doi: 10.1161/01.RES.0000124394.01180.BE. [DOI] [PubMed] [Google Scholar]
- 97.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class iii histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circulation research. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 98.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the sirt1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 99.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. Sirt3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of ku70. Molecular and cellular biology. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gama V, Gomez JA, Mayo LD, Jackson MW, Danielpour D, Song K, Haas AL, Laughlin MJ, Matsuyama S. Hdm2 is a ubiquitin ligase of ku70-akt promotes cell survival by inhibiting hdm2-dependent ku70 destabilization. Cell death and differentiation. 2009;16:758–769. doi: 10.1038/cdd.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gottlieb TM, Leal JF, Seger R, Taya Y, Oren M. Cross-talk between akt, p53 and mdm2: Possible implications for the regulation of apoptosis. Oncogene. 2002;21:1299–1303. doi: 10.1038/sj.onc.1205181. [DOI] [PubMed] [Google Scholar]
- 103.Narayan N, Lee IH, Borenstein R, Sun JH, Wong R, Tong G, Fergusson MM, Liu J, Rovira II, Cheng HL, Wang GH, Gucek M, Lombard D, Alt FW, Sack MN, Murphy E, Cao L, Finkel T. The nad-dependent deacetylase sirt2 is required for programmed necrosis. Nature. 2012;492:199–+. doi: 10.1038/nature11700. [DOI] [PubMed] [Google Scholar]
- 104.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the nad-dependent deacetylase sirt1 in the regulation of autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hariharan N, Maejima Y, Nakae J, Paik J, DePinho RA, Sadoshima J. Deacetylation of foxo by sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circulation research. 2010;107:1470–+. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gottlieb RA, Mentzer RM., Jr Autophagy: An affair of the heart. Heart failure reviews. 2012 doi: 10.1007/s10741-012-9367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell death and differentiation. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 109.Funderburk SF, Wang QJ, Yue Z. The beclin 1-vps34 complex--at the crossroads of autophagy and beyond. Trends in cell biology. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsui Y, Takagi H, Qu XP, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion - roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circulation research. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 111.Zhu HX, Tannous P, Johnstone JL, Kong YL, Shelton JM, Richardson JA, Lei V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Powell MJ, Casimiro MC, Cordon-Cardo C, He X, Yeow WS, Wang C, McCue PA, McBurney MW, Pestell RG. Disruption of a sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer research. 2011;71:964–975. doi: 10.1158/0008-5472.CAN-10-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Giricz Z, Mentzer RM, Gottlieb RA. Cardioprotective effects of chloramphenicol are mediated by autophagy. J Am Coll Cardiol. 2011;57:E1015–E1015. [Google Scholar]
- 114.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nature medicine. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 115.Kanamori H, Takemura G, Goto K, Maruyama R, Ono K, Nagao K, Tsujimoto A, Ogino A, Takeyama T, Kawaguchi T, Watanabe T, Kawasaki M, Fujiwara T, Fujiwara H, Seishima M, Minatoguchi S. Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. Am J Physiol-Heart C. 2011;300:H2261–H2271. doi: 10.1152/ajpheart.01056.2010. [DOI] [PubMed] [Google Scholar]
- 116.Ahmet I, Wan R, Mattson MP, Lakatta EG, Talan M. Cardioprotection by intermittent fasting in rats. Circulation. 2005;112:3115–3121. doi: 10.1161/CIRCULATIONAHA.105.563817. [DOI] [PubMed] [Google Scholar]
- 117.Xiao G. Autophagy and nf-kappab: Fight for fate. Cytokine & growth factor reviews. 2007;18:233–243. doi: 10.1016/j.cytogfr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W. Ap1 is essential for generation of autophagosomes from the trans-golgi network. Journal of cell science. 2012;125:1706–1715. doi: 10.1242/jcs.093203. [DOI] [PubMed] [Google Scholar]
- 119.Hua Y, Zhang Y, Ceylan-Isik AF, Wold LE, Nunn JM, Ren J. Chronic akt activation accentuates aging-induced cardiac hypertrophy and myocardial contractile dysfunction: Role of autophagy. Basic research in cardiology. 2011;106:1173–1191. doi: 10.1007/s00395-011-0222-8. [DOI] [PubMed] [Google Scholar]
- 120.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 121.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. Sirt1 regulates energy metabolism and response to caloric restriction in mice. PloS one. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N, Murphy MM, Appella E, Alt FW. Mammalian sirt1 limits replicative life span in response to chronic genotoxic stress. Cell metabolism. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 123.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 124.Greer EL, Brunet A. Foxo transcription factors in ageing and cancer. Acta physiologica (Oxford, England) 2008;192:19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 125.Lin K, Dorman JB, Rodan A, Kenyon C. Daf-16: An hnf-3/forkhead family member that can function to double the life-span of caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 126.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of foxo transcription factors by the sirt1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 127.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of foxo3 by sirt1 or sirt2 leads to skp2-mediated foxo3 ubiquitination and degradation. Oncogene. 2012;31:1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 129.Salih DA, Brunet A. Foxo transcription factors in the maintenance of cellular homeostasis during aging. Current opinion in cell biology. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing ucp2 in pancreatic beta cells. PLoS biology. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lemieux ME, Yang X, Jardine K, He X, Jacobsen KX, Staines WA, Harper ME, McBurney MW. The sirt1 deacetylase modulates the insulin-like growth factor signaling pathway in mammals. Mechanisms of ageing and development. 2005;126:1097–1105. doi: 10.1016/j.mad.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 132.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the sirt3 gene, human silent information regulator sir2 homologue, and survivorship in the elderly. Experimental gerontology. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


