Abstract
Multi-omics research is a key ingredient of data-intensive life sciences research, permitting measurement of biological molecules at different functional levels in the same individual. For a complete picture at the biological systems level, appropriate statistical techniques must however be developed to integrate different ‘omics’ data sets (e.g., genomics and proteomics). We report here multivariate projection-based analyses approaches to genomics and proteomics data sets, using the case study of and applications to observations in kidney transplant patients who experienced an acute rejection event (n=20) versus non-rejecting controls (n=20). In this data sets, we show how these novel methodologies might serve as promising tools for dimension reduction and selection of relevant features for different analytical frameworks. Unsupervised analyses highlighted the importance of post transplant time-of-rejection, while supervised analyses identified gene and protein signatures that together predicted rejection status with little time effect. The selected genes are part of biological pathways that are representative of immune responses. Gene enrichment profiles revealed increases in innate immune responses and neutrophil activities and a depletion of T lymphocyte related processes in rejection samples as compared to controls. In all, this article offers candidate biomarkers for future detection and monitoring of acute kidney transplant rejection, as well as ways forward for methodological advances to better harness multi-omics data sets.
Introduction
Multi-omics research has been at the epicenter of the post-genomics research agenda, raising both promises and challenges, to extract the best value out of such data-intensive form of life sciences inquiry (Altman, 2013; Gomez-Cabrero et al., 2014), not to mention the pressing need for novel methods for analysis of multi-omics data sets (Gomez-Cabrero et al., 2014).
Indeed, recent advances in high throughput ‘omics’ technologies now enable quantitative measurements of expression or abundance of biological molecules of a whole biological system and at different cellular levels. The improvement of analytical techniques to quantify different levels of gene products (mRNA, proteins, metabolites) permits understanding of cell metabolism as one ‘integrated system’ rather than as a combination of different parts (Zhang et al., 2010). Whilst single omics analyses are commonly performed to detect between-groups difference from either static or dynamic experiments, the integration or combination of multi-layer information is required to more fully unravel the complexities of a biological system. Data integration relies on the currently accepted biological assumption that each functional level is related to each other. Therefore, considering all the biological entities (genes, proteins, metabolites) as part of a whole biological system is crucial to unravel the complexity of living organisms.
The analysis of high throughput ‘omics’ approaches generates large amounts of data that require significant statistical and computational breakthroughs to decipher complex biological systems. Several statistical approaches have been proposed in the literature for the integration of two or more high-throughput data sets. These include projection-based multivariate approaches for biological exploration and ensemble classifiers for biomarker development and medical decision making (Günther et al., 2012; Lê Cao et al., 2007).
Ensemble classifiers combine separately developed, platform-specific classifiers using different combination rules (Polikar, 2006). A popular rule is majority vote where the predicted class is simply the one that is called by the majority of classifiers in an ensemble. Integration of information from different biological entities in ensemble classifiers happens after platform-specific analyses are performed. This is in contrast to projection-based multivariate approaches that are discussed in this article, which integrate data from different platforms at the analysis level.
Projection-based multivariate approaches are computationally efficient to handle large data sets, where the number of biological features is much larger than the number of samples, by projecting the data into a smaller subspace while capturing the largest sources of variation in the biological studies. During this statistical integration process, these approaches produce a snapshot of the data and highlight the largest sources of variation. However, when there is a large number of biological entities to summarize each functional level, a projection of the data in a smaller subspace might not be sufficient to extract relevant information (i.e., ‘which genes, which proteins are relevant and are acting in concert?’).
In recent years, several variants of these statistical integrative approaches have been proposed to perform variable selection and highlight those contributing to the largest variation in the data (Chun and Keleş, 2010; Lê Cao et al., 2008, 2009; Parkhomenko, et al., 2009; Waaijenborg, et al., 2008). The approaches are based on the Partial Least Squares regression methodology (PLS), which enables the integration of two data sets in a statistical sense: each data set is projected into a smaller subspace so that the covariance or the correlation between both data sets is maximized. The improvement that these authors propose is to perform variable selection, so that biological entities from both data sets that are correlated with each other are directly extracted from the methods. However, very few approaches have been proposed so far to both integrate more than two data sets and to select variables. Witten and Tibshirani (2009) proposed to concatenate all data sets with an appropriate weight applied to each of them. Recently, a promising approach based on regularized generalised Canonical Correlation Analysis (rGCCA) was proposed by (Tenenhaus and Tenenhaus, 2011) as a generalization to the PLS approaches for more than two data sets by maximizing the sum of the correlation in a pairwise fashion between two data sets at a time, followed by a variant that enables variable selection (Tenenhaus, et al., 2014.
In this article, we illustrate the usefulness and biological relevance of selected multivariate approaches from Lê Cao et al., (2008; 2011) and Tenenhaus et al., (2014) on a clinically relevant biological example, which is an acute renal allograft rejection study from the Biomarkers in Transplantation study. Kidney transplantation is a means to restore kidney function in patients with kidney failure. Acute kidney rejection after transplantation is a complication due to the recipient's immune responses to the foreign organ. Acute kidney rejection is observed in ∼10% of renal transplant patients. It is an important clinical problem with consequences for long-term graft survival. While current clinical practice in post-transplant care and detection of acute rejection relies on additional sources of information such as tissue biopsy and metabolic markers such as creatinine, the potential for blood-based biomarkers has been demonstrated using data from a single genomics or proteomics platform (Freue et al., 2010; Günther et al., 2009). If one were able to diagnose acute rejection accurately based on a simple blood sample draw, one could reduce the need for renal biopsies, which are invasive, risky, and costly procedures. The main aim of the kidney transplant study is to diagnose organ transplant rejection using plasma protein and gene expression data in blood samples from organ transplant patients. However, there exists a knowledge gap on how to best combine multi-platform data sets to improve biological understanding and accuracy of diagnosis. We show how the application of projection-based multivariate statistical approaches can help to better understand the information contained in the data sets and to identify gene and protein signatures that can be used to predict rejection status.
Material and Methods
The renal allograft study
Presentation of the study
Acute kidney allograft rejection was studied in a cohort of 40 patients, 20 of whom experienced an acute rejection (AR-patients) within 30 days post-transplant and 20 controls who also received a transplant but did not experience an acute rejection for at least 6-months post-transplant (NR-patients). Blood samples were collected at multiple scheduled time points and at the time of suspected rejection for the AR-patients. Control samples from the NR-patients were time-matched with biopsy-confirmed AR-samples (i.e., for each AR-sample there was a NR-sample from the same time-point). See Supplementary Figure S1 for an overview of the blood sample collection). Demographic and clinical information was considered in Control-sample selection to minimize the impact of potential confounders across the two groups (Table 1).
Table 1.
Summary of Demographics for 20 AR and 20 NR Patients Used in Study
| AR (n=20) | NR (n=20) | |
|---|---|---|
| Age at Transplant (s.d.) | 45.74 (12.10) | 49.04 (9.30) |
| Gender | ||
| Female | 6 (30%) | 8 (40%) |
| Male | 14 (70%) | 12 (60%) |
| Race | ||
| White | 19 (95%) | 16 (80%) |
| Asian | 1 (5%) | 3 (15%) |
| Other | 0 (0%) | 1 (5%) |
| Donor Type | ||
| Living donor | 13 (65%) | 13 (65%) |
| Deceased donor | 7 (35%) | 7 (35%) |
Description of the genomics and proteomics experiments
Peripheral blood samples were drawn into PAXgene tubes (BD, Oakville, Canada) for genomics, and EDTA tubes for proteomics analysis, stored at −80°C, and further analyzed on genomics and proteomics platforms. All 40 patients had genomics and proteomics experiments run on blood samples obtained at the same time. This prospective study was conducted at the University of British Columbia. All subjects provided written consent. The consent form and the study was approved by the UBC-Providence Health Care Research Ethics Board, and the UBC-Clinical Research Ethics Board.
Genomics data
Samples were prepared and processed as previously described (Günther et al., 2009). Each sample was then run on Affymetrix Human Genome U133 Plus 2.0 arrays and scanned. Quality of arrays was assessed by visual inspection of boxplots, RLE, Nuse and M/A-plots that were produced with the AffyPLM-package (Bolstad et al., 2005). The 40 samples were combined with 253 additional Microarray samples from the larger Biomarkers in Transplantation study, and all 293 samples together were background adjusted, normalized, and probe information was summarized into probe-sets using the Robust Multi-Array Average (RMA)-procedure from the RefPlus package in Bioconductor (Harbron, et al., 2007). A prefilter step based on inter-quartile range was applied to remove variables with little variation across samples independent of sample class. Half of the 54,613 probe-sets with an IQR below the median IQR were removed. The data are available through GSE46474.
Proteomics data
Plasma samples were collected in EDTA tubes. Plasma was depleted of the 14 most abundant proteins, followed by trypsin digestion and iTRAQ labeling. Samples were then analyzed with iTRAQ MALDI-TOF/TOF Mass Spectrometry and pre-processed as previously described (Freue et al., 2010). ProteinPilot™ was used to assemble identified peptide data into a list of identified proteins for each iTRAQ run. These groups can contain more than one protein (e.g., homologous or redundant proteins), or proteins belonging to the same family that could not be distinguished based on the available peptide. The Protein Group Code Algorithm (PGCA) developed at the PROOF Centre linked protein groups across different iTRAQ experiments. A protein group code (PGC) dictionary was created with data from 444 kidney plasma samples, including the 40 proteomics samples in this study.
The proteomics data set represents ratios of protein abundance relative to pooled controls for all protein group codes that were detected in at least one of the 40 samples. Different from raw genomics data, proteomics data can have missing values and a pre-filter was applied that required at least 75% non-missing protein ratios across all 40 analysis samples for the corresponding protein group (PG) to be included in the analysis. Missing values were imputed with NIPALS and the resulting imputed data were log2 transformed (Wold and Lyttkens, 1969). The data are available in Supplementary Material T1.
Statistical analysis
Overview
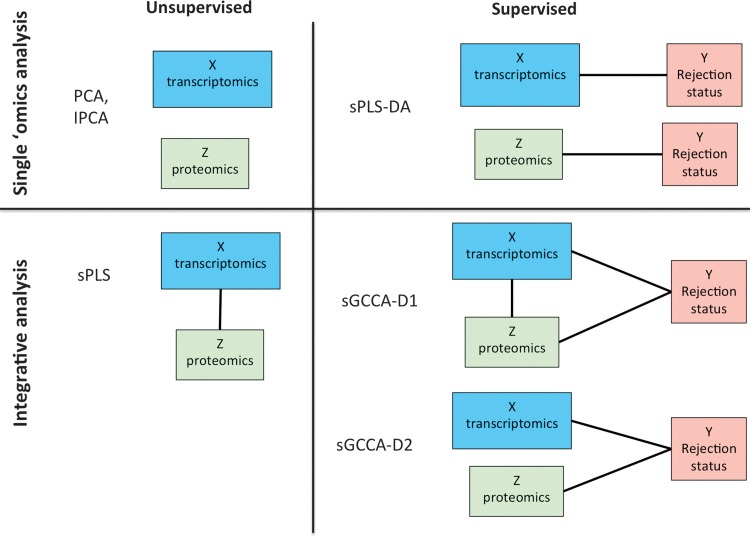
The exploratory statistical methods that were applied in this report are projection-based methods, where the aim is to summarize the main characteristics of each data set. From those approaches we can benefit from insightful built-in graphical outputs. These dimension reduction methods, project the data into a much smaller subspace than the original space (which for omics data is typically highly dimensional), while capturing the largest sources of variation in the data. The exploration and visualization of the structure of the data in these new subspaces enable us to (1) assess if the samples separate by phenotype (rejection status of the patients), (2) identify possible outliers, (3) reveal artificial separation of data due to confounding variables, laboratory or platform effects, and (4) identify relevant features in these high dimensional data sets. To answer a wide variety of biological questions, we have performed two types of analyses using several multivariate approaches. Figure 1 summarizes the different analyses that were performed. The statistical methods are described in the Supplementary Material M1.
FIG. 1.
Summary of the exploratory (Part 1) and integrative (Part 2) statistical approaches applied to the kidney transplant study. The different sources of data are represented by blocks, and black lines between blocks illustrate the relationship that has been modeled between the data sets.
Analysis of each data set separately
Principal Component Analysis (Jolliffe, 2002) and Independent Principal Component Analysis (Yao, et al., 2012) are both unsupervised techniques that do not take into account any prior biological knowledge regarding the sample groups. This preliminary step is particularly useful to understand the origin of the largest sources of the variation in the data, and the similarities and differences that can be observed between the independent samples given the expression or abundance data sets. A supervised analysis was also performed with sparse Partial Least Squares Discriminant Analysis (Lê Cao et al., 2011) by including prior knowledge regarding the groups of patients (rejection status) in the analysis. The aim is to extract and combine discriminative features (genes or proteins) that best separate the different outcomes. Supervised analysis is often performed for the identification of potential biomarkers and the development of clinical classification models (Günther et al., 2012).
Integrative analysis of two data sets
The second part of the results presents a higher-level and novel analysis to integrate several sources of data simultaneously (genomics, proteomics, but also rejection status of the patient). The aim of these analyses is to gain additional insights that could not be obtained by analyzing each data set alone, by identifying biological features (genes, proteins) whose expression or abundance is highly correlated across patients, in a supervised or unsupervised framework. Different integrative modeling scenarios were investigated, building on the results from the first part of the analyses. Sparse Partial Least Squares regression (Lê Cao et al., 2008) integrates genomics and proteomics data in an unsupervised way, whereas sparse Generalized Canonical Correlation Analysis (sGCCA, (Tenenhaus et al., 2014)) enables the integration of different sources of data in a supervised framework. Different designs were considered to model the relationship between the different data sets (subsequently called sGCCA-D1 and sGCCA-D2). These integrative approaches are also part of the projection-based methodologies and the resulting graphical outputs enable to visualize sample clustering, outlier samples, and bias in the data.
Insightful outputs from the different approaches
The statistical methods considered in this study are all based on similar principles to PCA and output components (also called ‘scores’) which are linear combinations of the original variables. The weights in the linear combinations are determined according to a given criterion, for example, maximizing the variance in the data in PCA, or the covariance in PLS and sGCCA methods. The components represent the data projected into a smaller subspace and lead to insighful graphical outputs to highlight similarities between samples.
Other graphical outputs can also be obtained based on the variable weights in the linear combinations to represent the relationships between features when integrating two data sets. In this output, a pair-wise similarity matrix is directly computed from sPLS or sGCCA components on the basis of the selected features only, instead of computationally intensive calculation of Pearson correlation coefficients between each pair of variables in a high dimensional setting. We have demonstrated in González, et al. (2012) that the pair-wise association values can be seen as a robust approximation of Pearson correlation. Classical visualization outputs are then obtained based on the pair-wise similarity matrix using relevance networks and Clustered Image Maps.
In addition, the sparse variants that we have applied enable the selection of relevant features with respect to the criterion that is maximized in each approach. In this article, we also provide biological insight about the genes and proteins that were selected with these multivariate approaches.
Results
Exploration and analysis of each data set
Unsupervised analysis of each separate data set
Principal Component Analysis
Three principal components explained 57.2% and 31.6% of the total variance on the genomics and proteomics data respectively (Supplementary Fig. S2). These rather low percentages give a first hint of the amount of information that can be summarized from each data source in a small number of components. The proteomics data seem to contain less relevant information than the genomics data with three principal components. No clear separation between the AR and the NR samples was observed in the PCA sample plots (Supplementary Fig. S3). Interestingly however, most of the late samples (>2 weeks post-transplantation) clustered separately from the corresponding early samples.
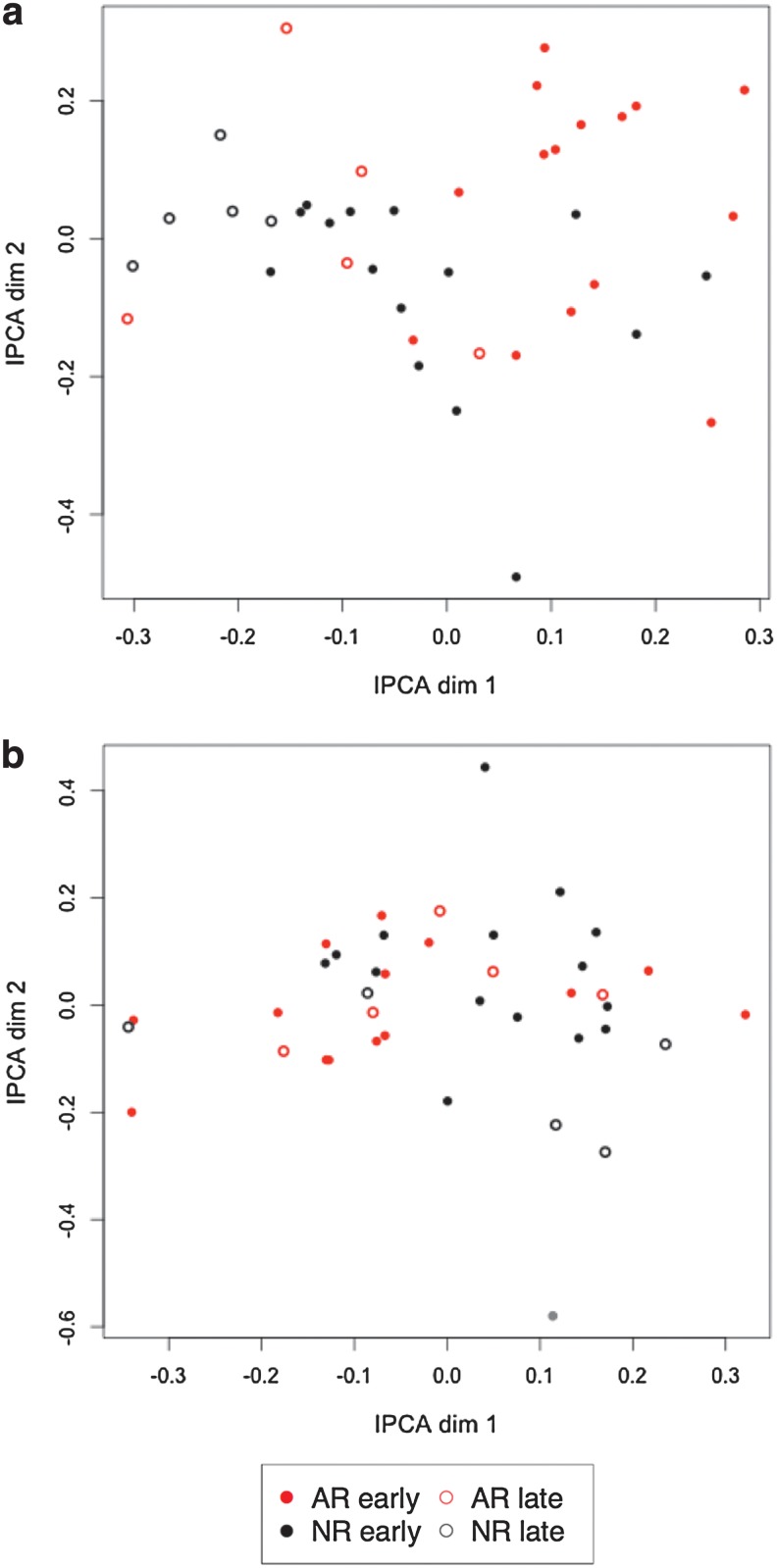
Independent Principal Component Analysis
IPCA was able to separate the two groups of samples in a better way than PCA and with less components (Fig. 2). The main separation between AR vs. NR can be observed on the first component in the genomics data but less so in the proteomics data. Most importantly, the IPCA results support the earlier observation in that the later samples cluster separately from the earlier ones within their respective group (Supplementary Fig. S3).
FIG. 2.
IPCA sample representation. Samples were projected on the first two independent principal components for the genomics data (a) and for the proteomics data (b).
Supervised analysis of each separate data set
Training and testing phases
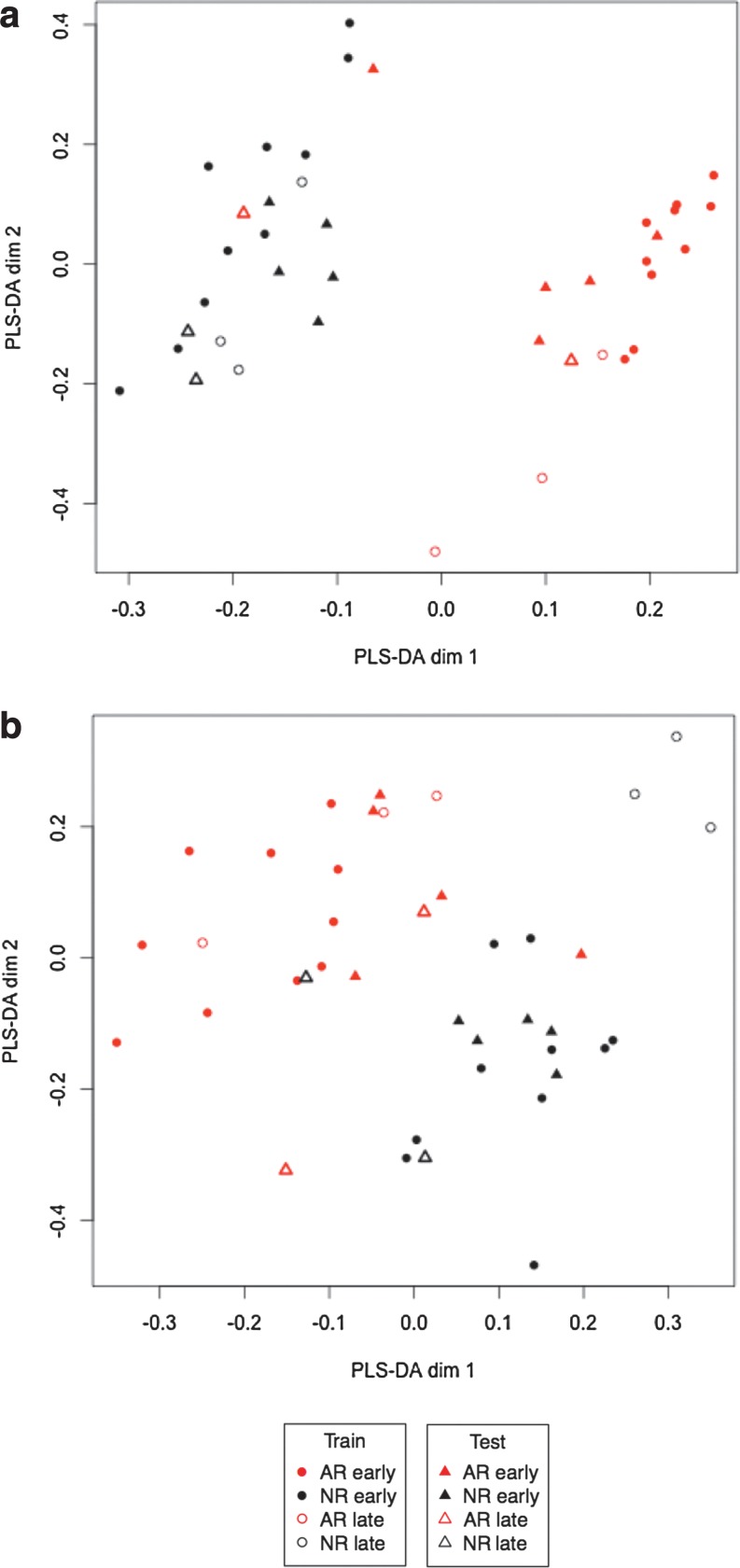
As a way of assessing the discriminative power of a selection of genes and proteins to predict the rejection status of external patients, we divided the original data set into a training and a testing set (see Supplementary Material M1). The parameter tuning indicated an optimal selection of 90 probe-sets and 21 protein groups (see Supplementary Fig. S4) Based on these selected variables, the trained model could predict the class of new test samples. Figure 3 shows that the separation between the two groups is more accentuated for the genomics data than for the proteomics data.
FIG. 3.
sPLS-DA sample representations of the genomics (a) and proteomics (b) data. sPLS-DA was first trained on each data set [selection of 90 probe-sets in (a) and 21 proteins in (b)] on the 26 training samples (circles). For illustrative purposes, a second dimension was added but this second component is not relevant to discriminate the two classes. The testing samples (triangles) were then overlaid on the plot based on the prediction given by the sPLS-DA model. Late samples are shown with open symbols.
Final gene and protein selection
Final genomics and proteomics models were trained on the full 26-sample training set for the chosen parameters. The list of the selected genes and protein groups can be found in Supplementary Tables T2 and T3. Performance was assessed on the 14 test samples by means of classification error, sensitivity, specificity, and AUC and compared with PLS-DA which includes all variables in the model (Table 2). For both platforms, the sparse version of PLS performed better than the non sparse version. In the proteomics data, the PLS-DA method produced a high CE of 43% and an AUC of close to 0.5, which is no better than a random classifier compared to a CE of 29% (AUC=0.76) with 21 selected proteins. The genomics classifiers performed better than the proteomics classifiers (AUC=0.90 with the selected 90 genes).
Table 2.
Summary of Classification Performance for sPLS-DA and PLS-DA Classifiers Trained on Full 26-Sample Training Set as Determined on a 14-Sample Testing Set (7AR and 7NR)
| Classifier | Number of selected variables | Classification Error rate (CE) | Sensitivity | Specificity | Area under curve | |
|---|---|---|---|---|---|---|
| Genomics | sPLS-DA | 90 | 0.14 | 0.71 | 1 | 0.90 |
| PLS-DA | 27,306 (all) | 0.21 | 0.57 | 1 | 0.82 | |
| Proteomics | sPLS-DA | 21 | 0.29 | 0.57 | 0.86 | 0.76 |
| PLS-DA | 133 (all) | 0.43 | 0.43 | 0.71 | 0.55 |
Integration of the two data sets
Even though genomics and proteomics data were extracted from blood samples, the two data sources are derived from different compartments in the peripheral blood (leukocyte cellular RNA in whole blood and proteins in plasma). Previous analyses on the kidney biomarker in transplantation data found that genomics and proteomics biological rejection signals differ from each other, while being consistent with the current understanding and pathogenesis of acute rejection injury (Freue et al., 2010; Günther et al., 2009). In an effort to unravel useful information about acute kidney rejection that could not be obtained by analyzing each data set alone, we investigated two integrative approaches, sPLS (Lê Cao et al., 2008) which is an unsupervised approach to extract common signal between two omics data sets (i.e., correlated genes and protein expression across patients) and its supervised variant sGCCA (Tenenhaus et al., 2014).
Unsupervised analysis of the integrated data sets with sPLS
Variable selection
We ran a stability analysis as described in Supplementary Material M1. The stability plot from Supplementary Figure S5 (b) shows that the selection of proteins was more stable than the genomics data (probably due to the higher number of variables in the genomics data set), with a small number of protein groups consistently being selected. Our final sPLS selection included 33 stable genes and 38 stable proteins with a cutoff of 0.7 (see Supplementary Tables T4 and T5).
Graphical representations
Samples were projected on the first two sPLS components for visualization purposes [Supplementary Fig. S6 (a)]. Correlation circle plots are graphical representations of the correlations between the selected variables and the sPLS components. These plots (whose interpretation is detailed in González et al., (2012) help to visualize the correlation between the two types of selected variables [clusters of features close to the circle of radius 1, Supplementary Fig. S6 (b)]. Our observations were twofold: first, we did not observe a very strong correlation between genes and proteins; second, the sample representation does not highlight a clear separation between the two groups of patients, rather, the first sPLS dimension seemed to separate the samples according to the time of rejection (early vs. late).
Supervised analysis of the integrated data sets with sGCCA
Two types of design were investigated with sGCCA. The number of components and features to choose for each data set were tuned using the stability criterion described in Supplementary Material M1.
sGCCA-Design 1: All blocks are connected
The first design is represented in Figure 1. The sample representations of each data set highlighted a separation between the different groups of patients, with a somewhat stronger separation in the genomics space. Late samples are shifted along component 1 [Supplementary Fig. S9 (a) and (b)]. The stability analysis of the variables selected produced higher frequencies compared to sPLS (see Supplementary Fig. S7), which is understandable given the additional constraints posed in the sGCCA model (the connection between blocks). Based on the stability analysis results, sGCCA selected 46 genes and 64 proteins that were stable. The genes and protein from this analysis are further discussed in the biological interpretation section.
sGCCA-Design 2: Genomics and the proteomics blocks are connected separately to outcome status
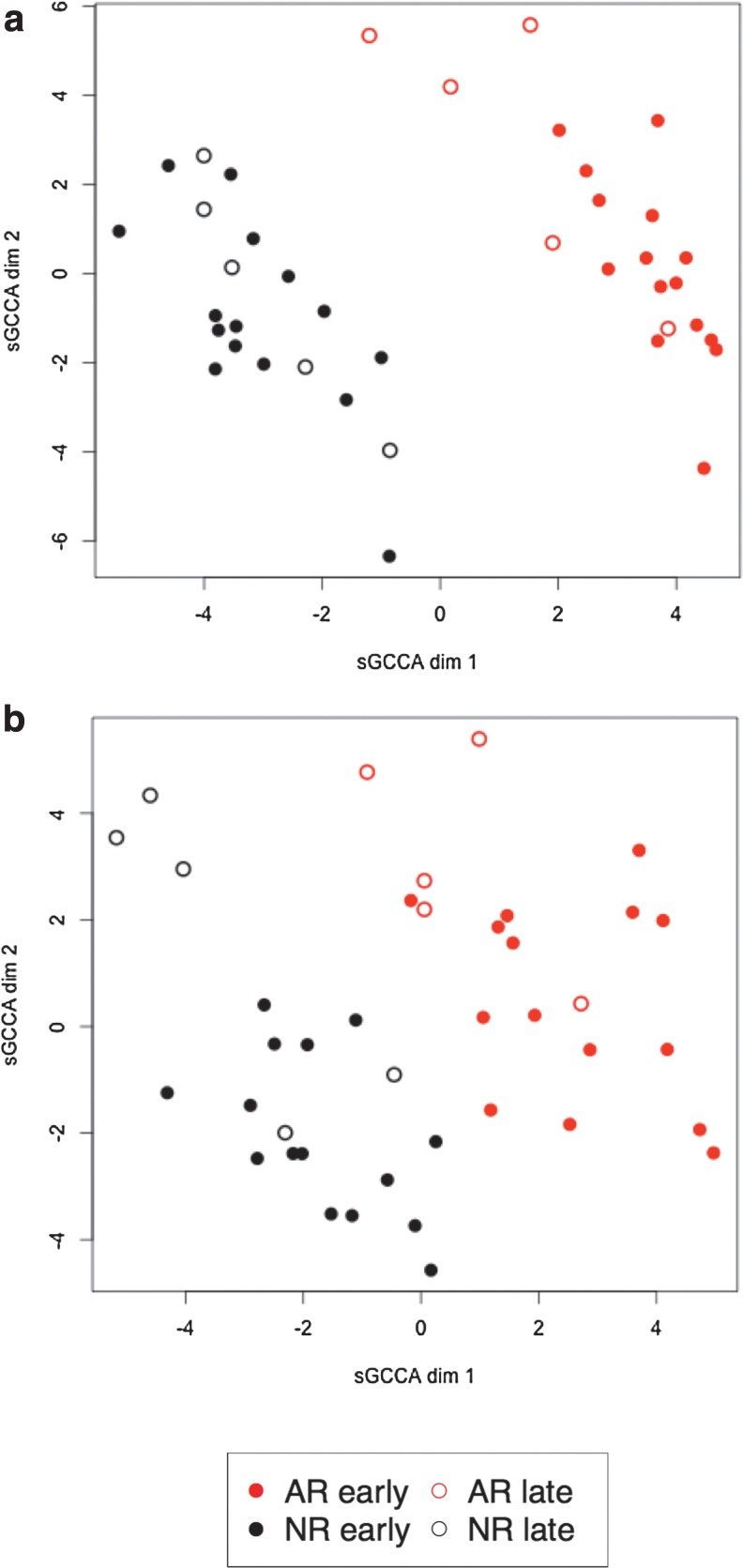
Compared to design 1, a much clearer separation between the two groups of samples is obtained on the genomics side and is also better on the proteomics side [Fig. 4 (a) and (b)], for a selection of 41 genes and 60 proteins that were stable (Supplementary Fig. S8). The genes and proteins from this analysis can be found in Supplementary Tables T6 and T7.
FIG. 4.
sGCCA analysis with design 2. Sample representation on the genomics space (a) and the proteomics space (b) for the first two dimensions.
Variable representations
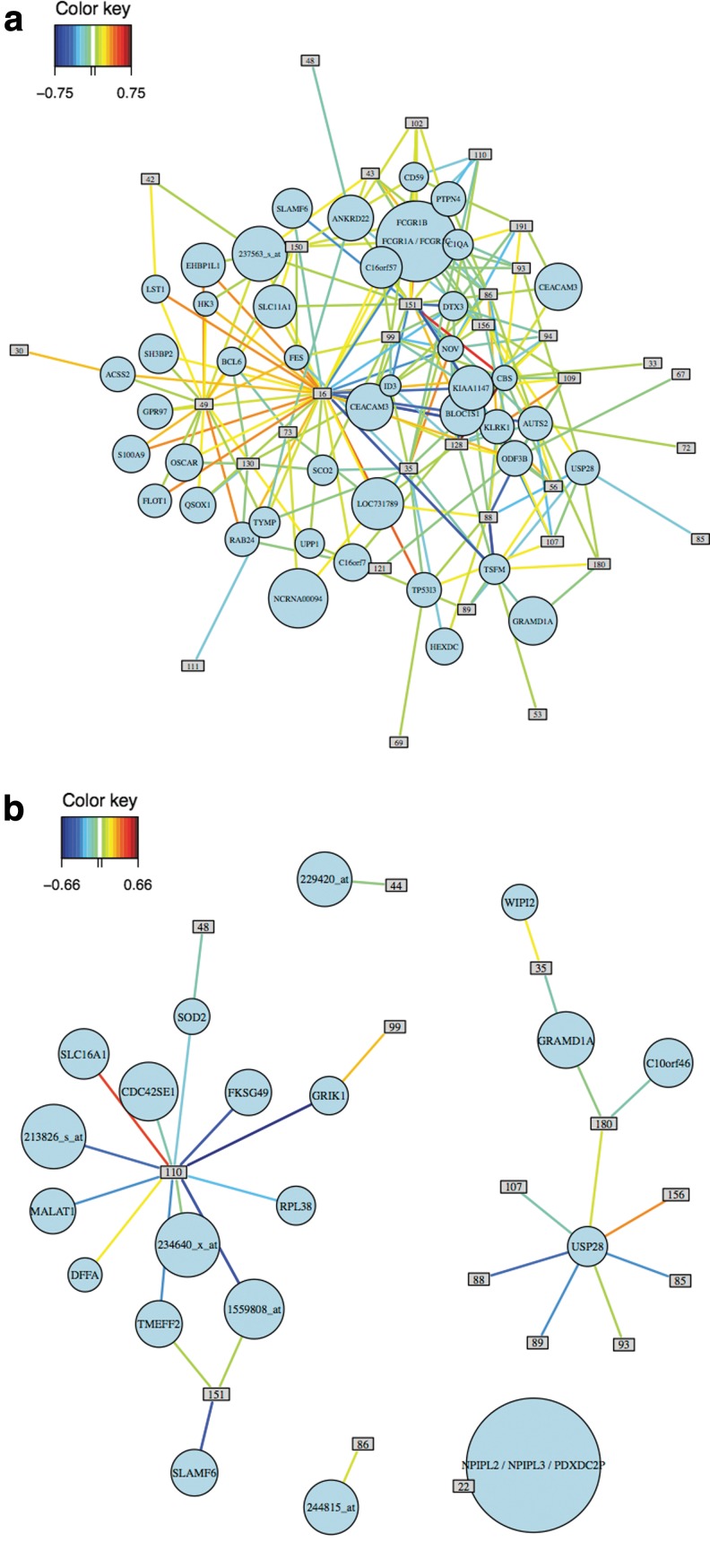
The relevance network for design 1 shows many more correlations between genes and protein groups than the corresponding network for design 2 (Fig. 5). This is not surprising since design 1 implicitly required correlations between the two data sets while design 2 did not include this constraint. The same conclusions can be drawn from the Clustered Image Maps (Supplementary Fig. S10).
FIG. 5.
Relevance networks of genes and protein groups selected with sGCCA-design 1 (a) and with sGCCA-design 2 (b). Only associations with an absolute correlation greater than 0.5 are represented.
Comparison between the different lists
Tables 5 and 6 show the overlap between genes and between proteins for all three integrative analyses. Also included for comparison are the 90 probe-sets and 21 protein group lists returned by sPLS-DA (see Supplementary Tables T2 and T3) when applied to the genomics and proteomics data set separately. For the genomics data, we observed little overlap between the lists for sPLS and the sGCCA models, and some overlap with sPLS-DA, which is to be expected as these methodologies focus on different characteristics of the features. The concordance tables showed that there was more overlap between the protein lists than the gene lists, which is not surprising since the protein data set is much smaller than the gene data set, and proteins tend to be consistently found by the different approaches. Regarding sPLS-DA, the same pattern as for the genes can be observed (i.e., sPLS-DA had most overlap with sGCCA-D2 and least with sPLS).
Table 5.
Overlap of Probe-sets in Gene Lists Selected by sPLS, sGCCA-D1, sGCCA-D2, and sPLS-DA
| Genomics | sPLS | sGCCA D1 | sGCCA D2 | sPLS-DA |
|---|---|---|---|---|
| sPLS | 33 | 5 | 0 | 0 |
| sGCCA D1 | 5 | 46 | 3 | 5 |
| sGCCA D2 | 0 | 3 | 41 | 24 |
| sPLS-DA | 0 | 5 | 24 | 90 |
The diagonal indicates the total number of variables selected by each approach.
Table 6.
Overlap of Protein Groups in Protein Lists Selected by sPLS, sGCCA-D1, sGCCA-D2 and sPLS-DA
| Proteomics | sPLS | sGCCA D1 | sGCCA D2 | sPLS-DA |
|---|---|---|---|---|
| sPLS | 38 | 35 | 23 | 8 |
| sGCCA D1 | 35 | 64 | 43 | 11 |
| sGCCA D2 | 23 | 43 | 60 | 17 |
| sPLS-DA | 8 | 11 | 17 | 21 |
The diagonal indicates the total number of variables selected by each approach.
Biological interpretation
In this section we particularly focused on the genes and proteins returned by sGCCA-D1. To ensure a robust gene analysis, we only considered the differential probe sets with a median log2 expression higher than 6 (Welsh's t-test, AR vs NR, FDR <0.05). This resulted in 51 genes upregulated in AR and 17 genes down-regulated in AR (Table 3).
Table 3.
Gene Selected with sGCCA-Design 1
| Probe Set ID | Gene title | Gene symbol | Regulation in AR |
|---|---|---|---|
| 234312_s_at | acyl-CoA synthetase short-chain family member 2 | ACSS2 | Up |
| 228758_at | B-cell CLL/lymphoma 6 | BCL6 | Up |
| 202592_at | biogenesis of lysosomal organelles complex-1, subunit 1 | BLOC1S1 | Up |
| 204495_s_at | chromosome 15 open reading frame 39 | C15orf39 | Up |
| 1554016_a_at | chromosome 16 open reading frame 57 | C16orf57 | Up |
| 235568_at | chromosome 19 open reading frame 59 | C19orf59 | Up |
| 212463_at | CD59 molecule, complement regulatory protein | CD59 | Up |
| 208052_x_at | carcinoembryonic antigen-related cell adhesion molecule 3 | CEACAM3 | Up |
| 210789_x_at | carcinoembryonic antigen-related cell adhesion molecule 3 | CEACAM3 | Up |
| 219183_s_at | cytohesin 4 | CYTH4 | Up |
| 214017_s_at | DEAH (Asp-Glu-Ala-His) box polypeptide 34 | DHX34 | Up |
| 201536_at | dual specificity phosphatase 3 | DUSP3 | Up |
| 222483_at | EF-hand domain family, member D2 | EFHD2 | Up |
| 221755_at | EH domain binding protein 1-like 1 | EHBP1L1 | Up |
| 216950_s_at | Fc fragment of IgG, high affinity Ia, receptor (CD64) /// Fc fragment of IgG, high affinity Ic, receptor (CD64) | FCGR1A /// FCGR1C | Up |
| 214511_x_at | Fc fragment of IgG, high affinity Ib, receptor (CD64) | FCGR1B | Up |
| 205418_at | feline sarcoma oncogene | FES | Up |
| 208749_x_at | flotillin 1 | FLOT1 | Up |
| 210142_x_at | flotillin 1 | FLOT1 | Up |
| 220404_at | G protein-coupled receptor 97 | GPR97 | Up |
| 224807_at | GRAM domain containing 1A | GRAMD1A | Up |
| 205936_s_at | hexokinase 3 (white cell) | HK3 | Up |
| 39402_at | interleukin 1, beta | IL1B | Up |
| 210184_at | integrin, alpha X (complement component 3 receptor 4 subunit) | ITGAX | Up |
| 210629_x_at | leukocyte specific transcript 1 | LST1 | Up |
| 211581_x_at | leukocyte specific transcript 1 | LST1 | Up |
| 211582_x_at | leukocyte specific transcript 1 | LST1 | Up |
| 215633_x_at | leukocyte specific transcript 1 | LST1 | Up |
| 218376_s_at | microtubule associated monoxygenase, calponin and LIM domain containing 1 | MICAL1 | Up |
| 205323_s_at | metal-regulatory transcription factor 1 | MTF1 | Up |
| 219862_s_at | nuclear prelamin A recognition factor | NARF | Up |
| 233072_at | netrin G2 | NTNG2 | Up |
| 238327_at | outer dense fiber of sperm tails 3B | ODF3B | Up |
| 1554503_a_at | osteoclast associated, immunoglobulin-like receptor | OSCAR | Up |
| 219394_at | phosphatidylglycerophosphate synthase 1 | PGS1 | Up |
| 219066_at | phosphopantothenoylcysteine decarboxylase | PPCDC | Up |
| 201482_at | quiescin Q6 sulfhydryl oxidase 1 | QSOX1 | Up |
| 225251_at | RAB24, member RAS oncogene family | RAB24 | Up |
| 217762_s_at | RAB31, member RAS oncogene family | RAB31 | Up |
| 240862_at | RAS guanyl releasing protein 4 | RASGRP4 | Up |
| 217728_at | S100 calcium binding protein A6 | S100A6 | Up |
| 203535_at | S100 calcium binding protein A9 | S100A9 | Up |
| 205241_at | SCO cytochrome oxidase deficient homolog 2 (yeast) | SCO2 | Up |
| 209370_s_at | SH3-domain binding protein 2 | SH3BP2 | Up |
| 211250_s_at | SH3-domain binding protein 2 | SH3BP2 | Up |
| 210569_s_at | sialic acid binding Ig-like lectin 9 | SIGLEC9 | Up |
| 220371_s_at | solute carrier family 12 (potassium/chloride transporters), member 9 | SLC12A9 | Up |
| 204099_at | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 | SMARCD3 | Up |
| 204858_s_at | thymidine phosphorylase | TYMP | Up |
| 203234_at | uridine phosphorylase 1 | UPP1 | Up |
| 229743_at | zinc finger protein 438 | ZNF438 | Up |
| 1552497_a_at | SLAM family member 6 | SLAMF6 | Down |
| 1553681_a_at | perforin 1 (pore forming protein) | PRF1 | Down |
| 202206_at | ADP-ribosylation factor-like 4C | ARL4C | Down |
| 205171_at | protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) | PTPN4 | Down |
| 205291_at | interleukin 2 receptor, beta | IL2RB | Down |
| 205758_at | CD8a molecule | CD8A | Down |
| 210606_x_at | killer cell lectin-like receptor subfamily D, member 1 | KLRD1 | Down |
| 210972_x_at | T cell receptor alpha locus /// T cell receptor alpha constant /// T cell receptor alpha joining 17 /// T cell receptor alpha variable 20 | TRA@ /// TRAC /// TRAJ17 /// TRAV20 | Down |
| 211597_s_at | HOP homeobox | HOPX | Down |
| 212656_at | Ts translation elongation factor, mitochondrial | TSFM | Down |
| 216920_s_at | TCR gamma alternate reading frame protein /// T cell receptor gamma constant 2 | TARP /// TRGC2 | Down |
| 219034_at | poly (ADP-ribose) polymerase family, member 16 | PARP16 | Down |
| 221011_s_at | limb bud and heart development homolog (mouse) | LBH | Down |
| 222482_at | single stranded DNA binding protein 3 | SSBP3 | Down |
Only the most differentially expressed between AR and NR for a p value<0.05 (FDR correction).
Using InnateDB (Lynn et al., 2008), we examined significantly up- and downregulated genes in order to identify over-represented biological pathways or processes (Supplementary Table T8). We identified that significantly upregulated genes in AR compared to NR samples are representative of biological processes involved in metabolic pathways and the hematopoietic cell lineage. Conversely, significantly downregulated genes represent components of the T cell receptor (TCR) pathway and downstream signaling pathways in naïve CD8+T cells.
Furthermore, performing an over-represented GO term analysis of the upregulated genes using GOstat (Beißbarth and Speed, 2004) revealed that negative regulation of lymphocyte activation, negative regulation of T-helper 2 cell differentiation, and negative regulation of B cell apoptotic process (Supplementary Table T9) are the most significant biological processes represented by these genes. These biological processes correlate with the downregulated lymphocyte-mediated pathways revealed by the over-represented pathway analysis described above. In addition, upregulated biological processes include the positive regulation of interleukin-6 production, leukocyte chemotaxis, actin cytoskeleton reorganization, regulation of integrin biosynthetic processes, regulation of cytokine production, and response to wounding, fever generation, and phagocytosis. Together these biological pathways are representative of immune responses, including systemic inflammation and innate immune activities.
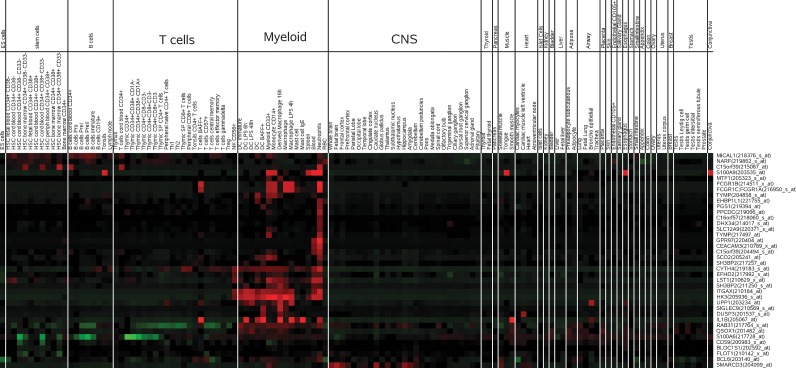
To examine the modulated genes across various cell and tissue types, we took advantage of the gene enrichment profiles created by Benita et al., (2010). Interestingly, both the significantly up- and downregulated genes showed blood cell specificity, as shown in Figures 6 and 7. Upregulated genes are significantly enriched in myeloid cells, especially neutrophils, compared to other blood cell types and various other tissues. Conversely downregulated genes appear to be significantly enriched in T-lymphocytes. Overall, these results reveal that increases in innate immune responses, neutrophil activities, and accumulation in the system, and depletion of T lymphocyte-related processes are present in ARs patient samples, as compared to NRs samples.
FIG. 6.
Gene enrichment profile of upregulated genes in sGCCA-D1. Red cells indicate genes that are enriched (known to be highly expressed) in the corresponding tissue cell-types.
FIG. 7.
Gene enrichment profile of downregulated genes in sGCCA-D1. Red cells indicate genes that are enriched (known to be highly expressed) in the corresponding tissue cell-types.
We also examined the proteins in this network that correlated with the genes addressed above (Table 4). Most notably, among the 13 most significantly downregulated proteins there are three members of the serpin (serine protease inhibitor) family of proteins, namely SERPINA5, SERPINC1, and SERPING1. These blood plasma proteins are involved in regulation of blood coagulation and complement cascades and inflammation.
Table 4.
Top Down- and Upregulated Proteins in sGCCA-Design 1
| geneSymbol | refseq | Regulation in AR |
|---|---|---|
| SERPING1 | NM_000062 | down |
| F13A1 | NM_000129 | |
| APOA4 | NM_000482 | |
| SERPINC1 | NM_000488 | |
| F2 | NM_000506 | |
| GC | NM_000583 | |
| SERPINA5 | NM_000624 | |
| AFM | NM_001133 | |
| F13B | NM_001994 | |
| ADIPOQ | NM_004797 | |
| SERPINA4 | NM_006215 | |
| PROC | NM_000312 | |
| SHBG | NM_001040 | |
| CST3 | NM_000099 | up |
| PON1 | NM_000446 | |
| CANX | NM_001024649 | |
| LBP | NM_004139 | |
| ARNTL2 | NM_020183 |
Discussion
In an effort to explore relationships between genes and proteins in acute kidney rejection, we have applied recently developed variants of multivariate projection-based approaches to genomics and proteomics data sets from kidney transplant patients that experienced an acute rejection event versus nonrejecting controls.
The unsupervised analyses PCA and IPCA pinpointed an interesting phenomenon regarding the separation between the early and late samples, which has also been observed in a related study that focused on longitudinal genomics data only (Shin et al., 2014). In our study, the time of rejection was defined as “early” (within 2 weeks post-tx) and “late” (more than 2 weeks post-tx) based on input from clinicians. Most acute rejection episodes in renal transplant patients in our Vancouver, Canada cohort happened within the first 2 weeks post-transplant. This is not necessarily true for other countries where different medication regimens may be used pre- and post-transplant. Australia and India show a similar behavior, while numbers reported for the USA indicate that acute renal rejection episodes occur later. It should be stressed that both early and late rejection samples represent biopsy-confirmed acute rejection and, as such, are phenotypically different from the non-rejection samples. The observed clustering of late AR with early NR in analysis sGCCA-D1 highlights the importance of the time of rejection relative to transplantation date, rather than a rejection signal. It is possible that transplantation surgery and accompanying immunosuppressive therapy can cause responses in the immune system that are likely to be picked up in gene expression experiments. These changes then overlay any acute rejection signal at times close to transplantation.
A better separation between the two groups of samples was observed in IPCA than in PCA. IPCA is a combined variant of both ICA and PCA approaches. In previous studies, IPCA was found to summarize the information in a smaller number of components than PCA [see (Yao et al., 2012) for more details]. The separation was not clear enough for definite conclusions and led us to investigate supervised approaches instead.
Genomics- and proteomics sPLS-DA classifiers were able to separate AR and NR groups. The genomics classifier based on 90 probe-sets performed better than the proteomics classifier based on 21 protein groups (Supplementary Tables T2 and T3). The predictive ability of the genes was higher than the proteins, a fact that might not come as a surprise considering that iTRAQ technology is a semi-quantitative approach and subsequent estimate of protein abundance is noisy and error-prone. Other technologies such as MRM will likely work better but are typically targeted approaches that are useful when a small list of candidate protein markers is available.
The overlap between the different lists of genes and proteins selected by the various multivariate approaches are not surprising from a modeling perspective. As expected, we observed large overlaps between supervised approaches (sPLS-DA and sGCCA-D2) and approaches that focus on the maximisation of the covariance between the genomics and proteomics data sets (sPLS and sGCCA-D1). sPLS solely focuses on maximizing the covariance between the omics data sets, whereas sGCCA-D1 focuses on maximizing the covariance between the omics as well as discriminating the groups of samples, and sGCCA–D2 focuses on the discriminative properties of the omics data to separate the groups of samples. The approaches sPLS and sGCCDA-D1 did not display a clear separation between AR and NR samples which points towards the hypothesis of a lack of relationship between the genomics and the proteomics data.
Even though sGCCA-D2 was expected to find a better separation between the AR and NR groups than sGCCA-D1 due to fewer constraints, this does not necessarily mean that sGCCA-D2 has a better performance than sGCCA-D1 when tested in an independent cohort. Design 2 allows selection of features in a less constrained way but might include noisy features that happen to have good individual discrimination ability. We should note that for biomarker studies that go beyond exploration, validation in an independent testing set is required. Studies in which genes and proteins are integrated for better understanding of underlying biology as presented in this article would also benefit from validation studies in the future.
We found that among the most downregulated proteins in the plasma in AR were SERPINA5, SERPINC1, and SERPING1. These proteins are known to have anti-inflammatory functions (Eror et al., 1999) through the inhibition of blood coagulation factors as well as complement components. It follows that the downregulation of these serpin family proteins in AR patient samples might be an indication of increased systemic inflammation and vascular permeability. This proposal correlates with the known activities of the genes identified in this network discussed above.
It has been shown that ischemic reperfusion tissue injury might prevent immunosuppression-medicated acceptance of allograft tissue and neutrophil accumulation and activity is linked with the inhibition of the graft acceptance (de Perrot, et al., 2003; Kreisel, et al., 2011a, b). The overall upregulation of inflammation and innate immune responses, including neutrophil activation and accumulation, that we observe in AR patient samples as compared to NR sample might therefore be due to ischemic reperfusion tissue injuries from the kidney transplantation.
It has also been shown that the complement activation is associated with ischemic tissue injuries that result in the production of a number of inflammatory mediators. For instance, C1 inhibitor, SERPINC1, is found to have protective activities on ischemic injuries (Buerke, et al., 1995) that treatment of intestinal ischemic injuries with C1 inhibitor limits tissue damages and improves survival by inhibition of complement activation as well as inhibition of neutrophil infiltration, which leads to a reduction in the local inflammatory response (Lu et al., 2008). Also, complement depletion is shown to prevent neutrophil recruitment in a model of myocardial ischemia (Buerke et al., 1995). The downregulation of SERPIN proteins in AR patients and the potential decrease in complement activation as a consequence might therefore indicate reduced protective effects of these proteins on ischemic tissue injuries.
Alternatively, downregulation of lymphocyte activities in AR patient samples might be due to uremic conditions brought on by kidney failure, as is implied by the increase in patient creatinine levels. The association between uremia and depletion of lymphocyte activities and proliferation is well documented (Hauser et al., 2008; Kato et al., 2008; Nakai, et al., 1992).
Conclusions
The availability of multiple –omics data sets that measure different biological properties of the same sample is becoming more common. As a consequence, there is a need for statistical analysis tools that are able to integrate and utilize information from all data sets beyond combination of results from analyses applied by each –omics data set separately. We have applied unsupervised and supervised methods to explore and integrate genomics and proteomics data to determine which genes and proteins play important roles in acute renal allograft rejection. Components along which groups of interest are separated were further investigated to understand which genes or proteins contribute most to that separation.
The exploratory unsupervised methods highlighted an interesting phenomenon regarding the time of rejection relative to transplantation date. The supervised sPLS-DA method was able to select a panel of diagnostic biomarkers that showed good performance in classifying our test samples. We have applied recently developed integrative methods that achieved a clear separation of the two sample groups while selecting promising genes and proteins candidates for the detection and monitoring of acute kidney transplant rejection.
Supplementary Material
Acknowledgments
This work was supported by a Burroughs Wellcome Fund Collaborative Research Travel Grant for Oliver Günther who visited the Queensland Facility for Advanced Bioinformatics (QFAB) in Brisbane, Australia for 6 weeks in 2011. The work of KALC is supported, in part, by the Wound Management Innovation CRC (established and supported under the Australian Government's Cooperative Research Centres Program) and the Australian Cancer Research Foundation (ACRF) for the Comprehensive Cancer Genomics Facility at The University of Queensland Diamantina Institute.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Altman RB. (2013). Personal genomic measurements: The opportunity for information integration. Clin Pharmacol Therapeut 93, 21–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beißbarth T, and Speed TP. (2004). GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20, 1464–1465 [DOI] [PubMed] [Google Scholar]
- Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, and Xavier RJ. (2010). Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115, 5376–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B, Collin F, Brettschneider J, et al. (2005). Quality assessment of Affymetrix GeneChip data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor, 33–47 [Google Scholar]
- Buerke M, Murohara T, and Lefer AM. (1995). Cardioprotective effects of a C1 esterase inhibitor in myocardial ischemia and reperfusion. Circulation 91, 393–402 [DOI] [PubMed] [Google Scholar]
- Chun H, and Keleş S. (2010). Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J Royal Stat Soc Series B, Statistical Methodology, 72, 3–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Perrot M, Liu M, Waddell TK, and Keshavjee S. (2003). Ischemia–reperfusion–induced lung injury. Am J Resp Crit Care Med 167, 490–511 [DOI] [PubMed] [Google Scholar]
- Eror AT, Stojadinovic A, Starnes BW, Makrides SC, Tsokos GC, and Shea-Donohue T. (1999). Antiinflammatory effects of soluble complement receptor type 1 promote rapid recovery of ischemia/reperfusion injury in rat small intestine. Clin Immunol 90, 266–275 [DOI] [PubMed] [Google Scholar]
- Freue GVC, Sasaki M, Meredith A, et al. (2010). Proteomic signatures in plasma during early acute renal allograft rejection. Mol Cell Proteomics 9, 1954–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cabrero D, Abugessaisa I, Maier D, et al. (2014). Data integration in the era of omics: Current and future challenges. BMC Systems Biol 8, I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González I, Lê Cao KA, Davis MJ, and Déjean S. (2012). Visualising associations between paired omics' data sets. BioData Mining 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther OP, Balshaw RF, Scherer A, et al. (2009). Functional genomic analysis of peripheral blood during early acute renal allograft rejection. Transplantation 88, 942–951 [DOI] [PubMed] [Google Scholar]
- Günther OP, Chen V, Freue GC, et al. (2012). A computational pipeline for the development of multi-marker bio-signature panels and ensemble classifiers. BMC Bioinformatics 13, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbron C, Chang KM, and South MC. (2007). RefPlus: An R package extending the RMA algorithm. Bioinformatics 23, 2493–2494 [DOI] [PubMed] [Google Scholar]
- Hauser AB, Stinghen AEM, Kato S, et al. (2008). Characteristics and causes of immune dysfunction related to uremia and dialysis. Periton Dialysis Intl 28, S183–S187 [PubMed] [Google Scholar]
- Jolliffe I. 2002. Principal Component Analysis. Springer Series in Statistics, Springer, New York [Google Scholar]
- Kato S, Chmielewski M, Honda H, et al. (2008). Aspects of immune dysfunction in end-stage renal disease. Clin J Am Soc Nephrol 3, 1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisel D, Sugimoto S, Tietjens J, et al. (2011a). Bcl3 prevents acute inflammatory lung injury in mice by restraining emergency granulopoiesis. J Clin Invest. 121, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisel D, Sugimoto S, Zhu J, et al. (2011b). Emergency granulopoiesis promotes neutrophil-dendritic cell encounters that prevent mouse lung allograft acceptance. Blood 118, 6172–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao KA, Besse P, and Goncalvez O. (2007). Selection of biologically relevant genes with a wrapper stochastic algorithm. Stat App Genetics Mol Biol 6, 29. [DOI] [PubMed] [Google Scholar]
- Lê Cao KA, Boitard S, and Besse P. (2011). Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao KA, González I, and Déjean S. (2009). IntegrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics 25, 2855–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao KA, Rossouw D, Robert-Granié C, and Besse P. (2008). A sparse PLS for variable selection when integrating omics data. Stat App Genetics Mol Biol 7, 35. [DOI] [PubMed] [Google Scholar]
- Lu F, Chauhan AK, Fernandes SM, Walsh MT, Wagner DD, and Davis III AE. (2008). The effect of C1 inhibitor on intestinal ischemia and reperfusion injury. Am J Physiol Gastrointest Liver Physiol 295, G1042–G1049 [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Winsor GL, Chan C, et al. (2008). InnateDB: Facilitating systems-level analyses of the mammalian innate immune response. Mol Systems Biol 4, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai I, Kaufman DB, Field MJ, Morel P, and Sutherland DE. (1992). Differential effects of preexisting uremia and a synchronous kidney graft on pancreas allograft functional survival in rats. Transplantation 54, 17–25 [DOI] [PubMed] [Google Scholar]
- Parkhomenko E, Tritchler D, and Beyene J. (2009). Sparse canonical correlation analysis with application to genomic data integration. Stat App Genetics Mol Biol 8, 1. [DOI] [PubMed] [Google Scholar]
- Polikar R. (2006). Ensemble based systems in decision making. Circuits Systems Mag IEEE, 6, 21–45 [Google Scholar]
- Shin H, Gunther O, Hollander Z, et al. (2014). Longitudinal analysis of whole blood transcriptomes to explore molecular signatures associated with acute renal allograft rejection. Bioinform Biol Insights 8, 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus A, Philippe C, Lê Cao KA, Grill J, and Frouin V. (2014). Variable selection for generalized canonical correlation analysis. Biostatistics 15, 569–583 [DOI] [PubMed] [Google Scholar]
- Tenenhaus A, and Tenenhaus M. (2011). Regularized generalized canonical correlation analysis. Psychometrika 76, 257–284 [DOI] [PubMed] [Google Scholar]
- Waaijenborg S, Verselewel PC, Hamer DW, and Zwinderman AH. (2008). Quantifying the association between gene expressions and DNA-markers by penalized canonical correlation analysis. Stat App Genetics Mol Biol 7, 3. [DOI] [PubMed] [Google Scholar]
- Witten DM, and Tibshirani RJ. (2009). Extensions of sparse canonical correlation analysis with applications to genomic data. Stat App Genetics Mol Biol 8, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold H, and Lyttkens E. (1969). Nonlinear iterative partial least squares (NIPALS) estimation procedures. Bull Intl Stat Instit 43, 29–51 [Google Scholar]
- Yao F, Coquery J, and Lê Cao KA. (2012). Independent Principal Component Analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinformatics 13, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li F,and Nie L. (2010). Integrating multiple “omics” analysis for microbial biology: Application and methodologies. Microbiology (Reading, England), 156, 287–301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.