Abstract
Background
In this work we predict enzyme function at the level of chemical mechanism, providing a finer granularity of annotation than traditional Enzyme Commission (EC) classes. Hence we can predict not only whether a putative enzyme in a newly sequenced organism has the potential to perform a certain reaction, but how the reaction is performed, using which cofactors and with susceptibility to which drugs or inhibitors, details with important consequences for drug and enzyme design. Work that predicts enzyme catalytic activity based on 3D protein structure features limits the prediction of mechanism to proteins already having either a solved structure or a close relative suitable for homology modelling.
Results
In this study, we evaluate whether sequence identity, InterPro or Catalytic Site Atlas sequence signatures provide enough information for bulk prediction of enzyme mechanism. By splitting MACiE (Mechanism, Annotation and Classification in Enzymes database) mechanism labels to a finer granularity, which includes the role of the protein chain in the overall enzyme complex, the method can predict at 96% accuracy (and 96% micro-averaged precision, 99.9% macro-averaged recall) the MACiE mechanism definitions of 248 proteins available in the MACiE, EzCatDb (Database of Enzyme Catalytic Mechanisms) and SFLD (Structure Function Linkage Database) databases using an off-the-shelf K-Nearest Neighbours multi-label algorithm.
Conclusion
We find that InterPro signatures are critical for accurate prediction of enzyme mechanism. We also find that incorporating Catalytic Site Atlas attributes does not seem to provide additional accuracy. The software code (ml2db), data and results are available online at http://sourceforge.net/projects/ml2db/ and as supplementary files.
Background
Previous research has already been very successful in predicting enzymatic function at the level of the chemical reaction performed, for example in the form of Enzyme Commission numbers (EC) or Gene Ontology terms. A much less researched problem is to predict by which mechanism an enzyme carries out a reaction. Differentiating enzymatic mechanism has important applications not only for biology and medicine, but also for pharmaceutical and industrial processes which include enzymatic catalysis. For example, biological and pharmaceutical research could leverage different mechanisms in host and pathogen for drug design, or to evaluate if antibiotic resistance is likely to appear in certain micro-organisms. And enzymes that perform the same reaction but require less costly cofactors can be more interesting candidates for industrial processes. Predicting the existence of a mechanism of interest in a newly sequenced extremophile, for example, could lead to applications in medicine or industry and to significant cost savings over non-biological industrial synthesis.
An enzyme is any protein able to catalyse a chemical reaction. In this work we do not focus on the questions associated with defining or assigning enzyme mechanisms, but rather take our definitions and assignments directly from the MACiE (Mechanism, Annotation and Classification in Enzymes) database [1-3]. Version 3.0 of the MACiE database contains detailed information about 335 different enzymatic mechanisms. Thanks to this information manually derived from literature, it is possible in MACiE to compare exemplars of enzymes that accept the same substrate and produce the same product, but do so using a different chemical mechanism, intermediate activation step or cofactor. Unfortunately, relatively few proteins are annotated with MACiE identifiers because confirming the exact mechanism of an enzyme requires significant effort by experimentalists and study of the literature by annotators.
Given the limited available examples, the aim of this work is to verify whether prediction of enzyme mechanism using machine learning is possible, and to evaluate which attributes best discriminate between mechanisms. The input is exclusively a protein sequence. The output, or predicted class labels, comprises zero or more MACiE mechanism identifiers, while the attributes used are sequence identity, InterPro [4] sequence signatures and Catalytic Site Atlas (CSA) site matches [5].
InterPro sequence signatures are computational representations of evolutionarily conserved sequence patterns. They vary from short, substitution-strict sets of amino acids representing binding sites to longer and substitution-relaxed models of entire functional domains or protein families. The Catalytic Site Atlas sites are akin to InterPro patterns, but they do not provide an evolutionary trace, more a record of an individual catalytic machinery, derived from a single Protein Data Bank [6] 3D structure which is transformed into a strict sequence pattern containing only the catalytic amino acids.
Only three proteins in our data have more than one mechanism label, because the current dataset privileges simple, one catalytic site enzymes. However, here we use a multi-label (and not only multi-class) machine learning scheme to be able to predict real life enzymes with multiple active sites or alternative mechanisms. Multi-label learning also provides flexibility by allowing seamless integration of additional labelling schemes. For example, Enzyme Commission numbers or Gene Ontology terms could be predicted together with mechanism. We evaluate the method by training a classifier on enzymes with known mechanisms. The classifier learns from the available attributes (for example sequence signatures) and then attempts to predict the mechanisms of a previously unseen test sequence. The quality of the predictions on the test set is evaluated using a number of metrics such as accuracy, precision, recall and specificity.
Previous work
To our knowledge, no previous research has attempted bulk prediction of enzymatic mechanism from sequence. However, past research has proved that the Enzyme Commission class of enzymes can be successfully predicted even for distantly related sequences using exclusively InterPro signatures [7-9]. Traube et al.[10] used QSAR and enzyme mechanism to predict and design covalent inhibitors for serine and cysteine proteases. Their method, like ours, does not require a solved protein structure, but its mechanism predictions are aimed at drug design and not easily portable to enzymes other than proteases. Choi et al.[11] use sequence to predict the existence and position of probable catalytic sites (grouped and aligned by Enzyme Commission number) with about 25% accuracy (approximately 8% better than random) but their prediction does not specify which mechanism the enzyme might be using in that active site. Other work tried to predict whether an amino acid is catalytic, and could in principle lead towards mechanism identification, but in practice has not been used to infer mechanism, only enzyme reaction. Using 3D structural information, Chea et al.[12] used graph theory to predict whether an amino acid is catalytic, followed by filtering using solvent accessibility and compatibility of residue identity since some amino acids are less likely to be involved in active catalysis. But their output is a binary label (catalytic or not) and not a prediction of mechanism. Using only sequence, Mistry et al.[13] have developed a strict set of rules to transfer experimentally determined active site residues to other Pfam family proteins, achieving a 3% FP rate, 82% specificity and 62% sensitivity. However, again, they do not link the active site residues to the mechanism performed.
Methods
Database sources and datasets
Data were taken from MACiE (Mechanism, Annotation and Classification in Enzymes database) [3] version 3.0, EzCatDb (Enzyme Catalytic-mechanism Database) [14], SFLD (Structure Function Linkage Database) [15], UniProtKB [16], InterPro [4] and Expasy Enzyme [17] in September 2013.
The complete data set includes 540 proteins that have been manually annotated with a MACiE mechanism in either MACiE, EzCatDb or SFLD, corresponding to 335 different MACiE mechanisms and 321 Enzyme Commission numbers. Three of these enzymes, the beta lactamases having UniProt entry name BLAB_SERMA from Serratia marcescens (beta-lactamase IMP-1, UniProt accession P5269), BLA1_STEMA from Stenotrophomonas maltophilia (metallo-beta-lactamase L1, P52700) and BLAB_BACFG from Bacteroides fragilis (beta-lactamase type II, P25910) have two MACiE mechanism labels in our dataset, due to the fact that EzCatDb does not distinguish between MACiE mechanisms M0015 and M0258. Both mechanisms are class B beta lactamase reactions, but performed with different catalytic machinery: M0015 uses an Asn residue, while M0258 uses Asp and Tyr. So the need for multi-label prediction is not strong for our dataset, however, multi-label classification is essential for mechanism prediction of real life multi-domain proteins. UniProt Swiss-Prot already contains 12,456 enzymes with more than one Enzyme Commission number. As just one example, the replicase polyprotein 1ab of the bat coronavirus (UniProt name R1AB_BC279 or accession number P0C6V) is cleaved into fifteen different chains, several of which are enzymes with one or more EC numbers, thus totalling nine Enzyme Commission numbers for a single transcript, varying from cysteine endopeptidase to RNA-directed RNA polymerase activities.
Class labels
An instance in our datasets is composed of a protein identifier (a UniProt accession number), a set of attributes (for example, the absence or presence of a sequence feature or the sequence identity with other sequences), and zero or more class labels representing the MACiE mechanisms of the enzyme, where available. Several MACiE mechanism entries can exist for one Enzyme Commission number. A MACiE mechanism identifier corresponds to a detailed mechanism entry modelled on one PDB [18] 3D structure and its associated literature. The entry describes not only the enzyme reaction, but also the catalytic machinery (reactive amino acids, organic and metal cofactors) used to perform the catalysis, down to the role of the individual amino acids, cofactor and molecular intermediates in each reaction step (such as proton or electron donor or acceptor and others) and the chemical mechanism steps (such as bond breaking, bond formation, electron transfer, proton transfer, tautomerisation and others) in temporal order.
A detailed analysis of the false positives generated by an initial prediction test highlighted the presence of distinct and diverse enzyme moieties labelled with the same MACiE mechanism code. For example, MACiE code M0013 (amine dehydrogenase) is used in MACiE only to annotate the methylamine dehydrogenase light chain of Paracoccus denitrificans (DHML_PARDE, P22619). However, in the database EzCatDb, the Paracoccus denitrificans heavy chain (DHMH_PARDE, P29894) is also annotated with MACiE code M0013, possibly because the holoenzyme is a tetramer of two light and two heavy chains (with the light chain hosting the active site). There is little or no similarity between each light and heavy chain (sequence identity < 12%), while the light chains are highly conserved within related organisms (sequence identity > 90%).
We thus proceeded to examine our training set to decide when the original MACiE mechanism code could be enriched with two or more sub-labels providing a better description of the underlying organisation of the enzyme chains. For all MACiE labels we did the following: 1. if the label annotates two or more proteins, we examined the "subunit structure" section of each UniProt protein, 2. if the section contained words such as heterodimer, heterotetramer or complex, we proceeded to split the MACiE label into two or more labels according to the enzyme complex subunits, and 3. we then re-annotated each protein with one of the new and more appropriate MACiE + subunit labels. We would like to stress that during this process the original MACiE mechanism annotations remain unchanged. The additional subunit information improves the learning, but, if the user so wishes, can easily be ignored simply by discarding any text beyond the 5th character (thus transforming, for example, M0314_component_I into M0314).
To give an example of the procedure to generate the new labels, MACiE label M0314 (anthranilate synthase) annotates two proteins in MACiE: TRPE_SULSO from the bacterium Sulfolobus solfataricus (anthranilate synthase component I, Q06128) and TRPG_SULSO (anthranilate synthase component II, Q06129) also from Sulfolobus solfataricus. In addition, the database EzCatDb uses the same MACiE label to annotate the corresponding component I and II of another bacterium, Serratia marcescens (EzCatDb identifier D00526, UniProt accessions TRPE_SERMA, P00897 and TRPG_SERMA, P00900). The "subunit structure" section of these four proteins in UniProt specifies: "Subunit structure: tetramer of two components I and two components II". We thus proceed to re-annotate the four proteins as M0314_component_I (Sulfolobus Q06128 and Serratia P00897, both described as anthranilate synthase component I) and M0314_component_II (Sulfolobus Q06129 and Serratia P00900, both described as anthranilate synthase component II).
The set of the old MACiE labels which did not require splitting and the new split labels (such as M0314_component_I, M0314_component_II, M0013_light_chain, M0013_heavy_chain etc.) is referred to as MACiE + subunit labels or simply mechanism labels.
As previously noted, in our current data most mechanisms only have one annotated protein exemplar and hence cannot be used for cross-validation or leave-one-out validation: the protein would always be either exclusively in the training set or exclusively in the testing set. This leaves us with only 82 MACiE + subunit mechanisms (corresponding to 73 classic MACiE mechanisms) having at least two protein examples, thus providing 248 enzyme sequences usable for cross-validation. This dataset is from now on referred to as the mechanism dataset.
However, the proteins belonging to mechanisms having only one exemplar can still be pooled together and used as negative examples for the other mechanisms (negative dataset), and the resulting false positive predictions can be analysed to assess why the method makes certain mistakes.
Also, in nearest neighbours algorithms, an instance must necessarily have a closest neighbour. An instance having no attributes in common with any other instance will "gravitate" towards the shortest available instance in the set (the instance with the fewest attributes). In order to avoid these artefacts, two empty instances (instances with no attributes and no class labels) have been added to the mechanism dataset for the training-testing experiments.
The set of UniProt Swiss-Prot proteins lacking Enzyme Commission annotation has also been used (swissprot-non-EC) as a "negative" test set. This set contains 226,213 proteins (as of September 2013) which are most probably non-enzymes (or have a yet unknown catalytic activity or an enzymatic activity which was mistakenly overlooked by curators). Of these, only 68,677 share at least one InterPro signature with a protein in the mechanism or negative datasets and could hence be mispredicted as enzymes (all the other proteins in the swissprot-non-EC set are, by definition, automatically and correctly predicted as not having a mechanism when using the InterPro attributes).
Attributes
Once defined the mechanism class labels to be predicted, we analysed which sequence-based attributes or features could be used for learning. More specifically, we have compared the accuracy of enzyme mechanism predictions when various different sets of attributes are used. The InterPro set of attributes includes the presence (1) or absence (0) of each InterPro signature for each sequence in the given protein dataset. InterPro is an extensive database of conserved sequence signatures and domains [4] that can be computed from sequence data alone and for any sequence using the publicly available InterProScan algorithm [4,19]. The 248 proteins in the mechanism dataset, for example, have 444 distinct InterPro attribute values, with an average of 4.4 InterPro signatures per protein.
InterPro signatures are composed of one or several sub-signatures provided by its repositories: GENE3D [20], HAMAP [21], PANTHER [22], Pfam [23], PIRSF [24], PRINTS [25], ProDom [26], PROSITE patterns and profiles [27], SMART [28], SUPERFAMILY [29] and TIGRFAM [30]. One or more of these sub-signatures usually correspond to one InterPro signature. However, some of these sub-signatures have not been integrated into InterPro because they provide too many false positives, do not have enough coverage or do not pass other criteria fixed by InterPro. We have tried using all these sub-signatures (integrated or not) as attributes for learning, to understand if they could provide a more powerful and finely grained alternative to the classic InterPro signatures.
Another set of attributes represents the presence (or absence) of a sequence match versus one of the Catalytic Site Atlas active sites (CSA 2D or simply CSA attributes). Each CSA 2D site is a tuple of active amino acids that must match the given sequence both for position and amino acid type.
In order to compare learning by sequence with learning based on structure, we matched our dataset also against the Catalytic Site Atlas three dimensional templates [31] (CSA 3D). CSA templates store the geometrical position of exclusively the atoms of the residues involved in a catalytic site. A residue is considered catalytic if it is chemically involved in the catalysis, if it alters the pKa of another residue or water molecule, if it stabilises a transition state or if it activates a substrate, but not if it is involved solely in ligand binding. Each CSA template is matched against the protein structure using the JESS algorithm [32].
To generate CSA 3D templates matches we first selected an exemplar (best) PDB X-ray structure for each UniProt protein in the mechanism dataset. To select the exemplar structure we collected all PDB structures for each UniProt record and chose the structures that covered the longest stretch of the protein sequence. If several structures of identical coverage and resolution existed, we chose the structure(s) with the best (highest) resolution. If several structures still existed, we chose the last when ordered alphabetically by PDB structure identifier. We then used the ProFunc service [33] to scan each exemplar PDB against CSA 3D templates (CSA 3D data set). For evaluation we also compare "best" matches against the MACiE dataset (having an E value below 0.1) versus all matches provided by ProFunc (E value below 10.0).
The various sets of attributes above have been evaluated, alone or in combination, for their ability to predict enzyme mechanism in the datasets presented. Combining attribute sets such as InterPro and CSA (as in the InterPro+CSA attribute set) means that the dataset matrix will have, for each protein row, all CSA columns and all InterPro columns filled with either 1 (signature match) or 0 (no match). This provides a sparse data matrix particularly suitable for large datasets of millions of protein sequences.
Considering though that our current dataset is not large, we have also created two more computationally intensive attribute sets. The first set (minimum Euclidean distance) involves calculating the Euclidean distance in the InterPro space between the protein of interest and all other proteins (sets of InterPro attributes). An attribute vector is then built with as many values as there are mechanisms. As each attribute value (that is, for each mechanism) we keep only the minimum Euclidean distance between the protein of interest and the proteins having that mechanism, giving:
where a is the vector of attribute values composed of one value am for each of the M mechanisms in the data, p is the protein of interest and pm is a protein having a mechanism m. The function Euclidean distance(p,pm) returns the Euclidean distance between the InterPro set of signatures of protein p and the InterPro set of signatures of another protein pm having mechanism m. We can also note that the k-Nearest Neighbour algorithm must calculate Euclidean distances, but, with the simpler aim of finding the closest instances, it does not usually need to store and manipulate the distances for every protein and mechanism combination.
The second set of attributes (maximum sequence identity) is even more computationally intensive because it substitutes distance with sequence identity. It thus requires an alignment between each pair of proteins in the dataset. The sequence identity of each protein versus every other protein in the mechanism and negative datasets was calculated by downloading the FASTA sequences from UniProt in September 2013 and aligning each pair using the Emboss [34] implementation of the Needleman-Wunsch algorithm [35]. The algorithm was run with the default substitution matrix EBLOSUM62 with gap opening penalty of 10 and gap extension penalty of 0.5. The resulting maximum sequence identity vector of attributes is given by:
where b is the vector of attribute values in the data (composed of one value bm for each of the M mechanisms), p is the protein of interest and pm is a protein having a mechanism m. The function sequence identity(p,pm) returns the sequence identity between the protein sequence p and another protein sequence pm having mechanism m (the emitted value can span from zero, if no amino acids could be aligned, to one, if the two sequences are identical).
Algorithm
Several algorithms [36-51] were evaluated by comparing their precision, recall, accuracy and run time on a leave-one-out prediction of the mechanism dataset (see Additional file 1 for full results). The top two algorithms for accuracy (about 96%) and speed (about 24 seconds for 248 instances) are instance-based learning algorithms (Mulan’s [46] BRkNN [45] and Weka’s [50] IBk [36] with a label powerset multi-label wrapper). The Mulan Hierarchical Multi Label Classifier (HMC) [47] also performs well (96% accuracy, 28 seconds). Support vector machine [39,42,43] and Homer (Hierarchy Of Multi-labEl leaRners) [47] are only slightly less accurate (about 95%), but significantly slower (from 13 to 90 minutes), and they are followed by random forest [37] with about 94% accuracy and between 1 and 44 minutes run time.
We have thus used throughout this work the BRkNN [45] nearest neighbours implementation (as in our previous work on predicting Enzyme Commission classes [9]), using the implementation available in the Mulan software library version 1.4 [46]. The nearest neighbours algorithm also provides an immediate visual representation of the clustering of the protein labels and their attributes. BRkNN is a multi-label adaptation of the classic k-Nearest Neighbour algorithm. The best parametrisation for the data is k = 1, that is, only the closest ring of neighbour instances are used to predict the label of an instance. This suggests a pattern of local similarity among the instances causing efficient but local learning. Our ml2db Java code uses queries to generate a Mulan datafile from MySQL database. A Mulan datafile consists of an XML file for the class labels and a Weka ARFF (Attribute Relation File Format) file for the protein instances and their attributes. Where possible, a sparse ARFF format, parsimonious of disk space and computational power, was used. This was possible for the InterPro, CSA and InterPro+CSA attribute sets, given that most attribute values are zero for these attributes (most signatures have no match in a given sequence).
We present results produced using the Euclidean distance in the chosen attribute space. Instances with exactly the same attribute set will have distance 0 (for example, two proteins having exactly the same InterPro features, if the attribute set of choice is InterPro signatures). If the instances differ in one attribute they will have a distance of one; if the two instance differ in x attributes, they will have a distance of
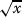 . The Jaccard distance
[52] was also used but produces slightly worse accuracy (data not shown).
. The Jaccard distance
[52] was also used but produces slightly worse accuracy (data not shown).
Evaluation
Due to the limited number of examples available, we performed leave-one-out validation on the mechanism dataset (n-fold cross-validation with n equal to the number of instances). In short, we trained on all proteins but one, predicted the mechanism for the omitted protein, and then compared the predicted label(s) with the protein’s true label(s). Considering the known shortcomings of leave one out validation (causing high variance when few instances are available for each class label [53]), in a second experiment the entire mechanism dataset has also been used for training followed by testing on the negative set to examine the false positive cases in more detail. Also, the mechanism dataset together with all the non-enzymes in Swiss-Prot (swissprot-non-EC set) have been used in two-fold cross validation.
To compare the predictive strength of the various attribute sets, we present the average value of the classification accuracy (also called subset accuracy), a strict measure of prediction success, as it requires the predicted set of class labels to be an exact match of the true set of labels [49]:
| (1) |
where I(true) = 1, I(false) = 0 and D is a dataset with |D| multi-label examples (proteins), each with a set Yi of labels (enzyme mechanisms) taken from the set of all labels (MACiE mechanisms) L: (xi,Yi), i = 1…|D|, Yi ⊆ L. If we define as Zi the set of mechanisms predicted by the model h (for example the BRkNN classifier or direct assignment rule) for the ith protein (xi): Zi = h(xi), then the classification accuracy represents the percentage of proteins for which the model predicted the true, whole set of mechanisms.
We also report micro and macro metrics (precision and recall) for completeness since the mechanism classes are long tail distributed. Consider a binary evaluation measure M(TP,FP,TN,FN) that is calculated based on the number of true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN), such as or . Let TPλ, FPλ, TNλ and FNλ be the number of TP, FP, TN and FN after binary evaluation for a label λ. The macro-averaged and micro-averaged versions of measure M become:
| (2) |
| (3) |
In this context micro averaging (averaging over the entire confusion matrix) favours more frequent mechanisms, while macro averaging gives equal relevance to both rare and frequent mechanism classes. Hence a protein will affect the macro-averaged metrics more if it belongs to a rare mechanism. Micro and macro specificity are not presented because these metrics never fall below 99.7%. For binary classification, , hence, because of the hundreds of possible mechanism labels, most prediction methods provide a very high proportion of true negatives in comparison with false positives, making specificity very close to 100% for any reasonable method and thus not particularly informative. All metrics are further defined and discussed in [46,49,54]. The best achievable value of all these measures is 100% when all instances are correctly classified.
Software code and graph layout
All experiments were run under a Linux operating system (Ubuntu 12.04 Precise Pangolin) using Oracle Java version 1.7, Python 2.7 and MySQL 5.5. All the Java code (ml2db) and data files used in this paper are available online at http://sourceforge.net/projects/ml2db/ and as Additional file 2 (code) and Additional file 3 (ARFF and XML data files). The full MySQL database dump of all the data and results is available on request. The graphs in Additional file 4 and Additional file 5 have been generated with PyGraphviz, a Python programming language interface to the Graphviz graph layout and visualization package, coded by Aric Hagberg, Dan Schult and Manos Renieri.
Results
Data statistics
Table 1 summarises the composition of the data sets used in terms of number of instances, attributes and class labels. As already described in the Methods section, each sequence in the mechanism + negative dataset (all the available MACiE mechanism annotations) was aligned with every other sequence and the percentage of sequence identity calculated. The resulting 126,499 couples are presented in Figure 1, which provides an overview of the sequence identity and Euclidean distance (in the InterPro attribute space) for each protein couple. As expected, most protein couples have low sequence identity (between 0% and 30%) and Euclidean distance between two and four, that is, have between four and sixteen differences in their InterPro signatures. This area seems to represent a very frequent sequence distance for protein couples with different function (triangle markers), but also contains a few couples of enzymes having the same mechanism (circle markers).
Table 1.
Datasets statistics
| Dataset | Instances | Attributes | Class labels |
|---|---|---|---|
|
Mechanism set with CSA |
248 |
134 |
82 |
|
Mechanism set with Maximum sequence identity |
248 |
82 |
82 |
|
Mechanism set with Minimum Euclidean distance (InterPro) |
248 |
82 |
82 |
|
Mechanism set with InterPro + CSA |
248 |
456 |
82 |
|
Mechanism set with Max seq. Id. + min Eucl. Dist. (InterPro) |
248 |
162 |
82 |
|
Mechanism set with all InterPro sub-signature matches |
248 |
743 |
82 |
|
Mechanism set with InterPro signatures |
248 |
322 |
82 |
|
Negative set with InterPro attributes |
290 |
917 |
290 |
|
Mechanism set + Swiss-Prot non-EC with InterPro attributes |
35,171 |
4418 |
82 |
| Swiss-Prot non-EC set with InterPro attributes | 68,667 (226,213) | 4,825 | 0 |
The table presents the number of instances (proteins), attributes (signatures or sequence identity values) and class values (mechanisms) for the datasets used in this work; for the swissprot-non-EC set we present the instances that need prediction (the ones sharing a signature with the mechanism set), while the total number of instances is shown between parentheses.
Figure 1.
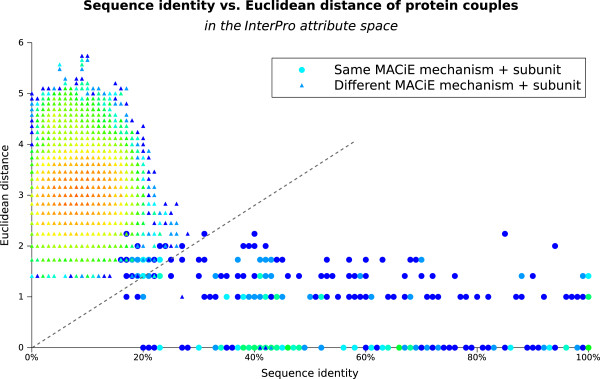
The sequence identity and Euclidean distance of enzymes with the same and different mechanism. The diagram presents, for every pair of proteins in the mechanism + negative datasets, the percentage of identity between the two proteins’ sequences and also the Euclidean distance between their signature sets (in the InterPro attribute space). Protein couples having the same MACiE mechanism are represented as circles, while those with different MACiE mechanisms as triangles. The colour scale is logarithmic increasing from blue (for one instance) to light blue (2-3 instances), green (4-9), yellow (70-100), orange (250) and red (up to 433 instances) and represents the number of protein couples having that sequence identity and Euclidean distance. The dashed grey line shown, with equation Euclidean distance = 7 × sequence identity, separates most same-mechanism couples (on its right) from an area dense with different-mechanism couples on its left.
The figure shows how enzymes having different mechanisms (triangle markers) concentrate in the upper left area of the plot, mostly having both low sequence identity (<30%) and high Euclidean distance between their signature sets (1.4 to 6, between 2 and 36 different signatures). In contrast, enzymes having the same mechanism form a long band across the figure, showing an extensive range of sequence identity, from about 18% to 100% but a lower and less varied Euclidean distance (0 to 2.2, that is, from having the same signatures to having 5 different signatures).
Mechanism prediction from sequence identity and Euclidean distance
Using the data in Figure 1 we evaluated whether a simple line separator could tell when a protein has the same label as another protein. To evaluate this simple form of learning (binary predictions in the form "same mechanism" or "different mechanism") we used a line passing through the origin and we varied the angle of the line between zero and ninety degrees, recording the number of correct and incorrect predictions for each line. As it is often the case, there is no absolute best line, some maximise precision, others recall. However, to give an example, the line passing through the origin with equation: Euclidean distance = 7 × sequence identity provides a recall of 93.5%, while still conserving an accuracy of 99.8% and a precision of 93.2%. For this binary case accuracy is calculated with the usual formula , precision is , and recall (or sensitivity) is .
Another way to read the equation Euclidean distance = 7 × sequence identity is that for two proteins differing in two signatures, at least about 20% sequence identity is necessary for the proteins to have the same mechanism (about 25% sequence identity for three differences, 29% for four differences and so on). In addition, while the equation suggests that proteins having exactly the same signatures can have any level of sequence identity, in practice the sequence identity for couples having the same mechanism never falls below 18% in the data, possibly because two random sequences (of approximately the same length as our sequences) will have a minimum number of identical amino acids by chance alone. The couples having the same mechanism are almost homogeneously scattered above this 18% threshold, but with several couples having about 40% sequence identity and few having very high sequence identity (80% to 100%). The same result structure holds when sequence similarity is used instead of sequence identity (data not shown).
Mechanism prediction with InterPro and Catalytic Site Atlas sequence attributes
In this section we use machine learning (k-Nearest Neighbour) to compare the ability of InterPro signatures and Catalytic Site Atlas (CSA) matches to predict enzyme mechanism on the basic mechanism dataset. Figure 2 presents an overview of the performance of different set of attributes in predicting the mechanism dataset. As an indicative baseline for prediction we used the labels predicted when mechanism is assigned simply by the presence of a certain set of InterPro domains (InterProdirect transfer). For example, protein ODPB_GEOSE of Geobacillus stearothermophilus (pyruvate dehydrogenase E1 component subunit beta, P21874) is part of the dataset and has MACiE mechanism M0106 (pyruvate dehydrogenase) and InterPro IPR005475, IPR005476, IPR009014 and IPR015941. Hence, if we use direct transfer of mechanism labels, another protein such as ODBB_HUMAN (2-oxoisovalerate dehydrogenase subunit beta mitochondrial, P21953) which has exactly the same InterPro signatures will receive a M0106 label, thereby introducing an error, since ODBB_HUMAN’s mechanism is in fact M0280 (or 3-methyl-2-oxobutanoate dehydrogenase). If several proteins in the training set have exactly the same InterPro attributes, the given test protein will be assigned all of their mechanism labels. The direct transfer method achieves 99.9% accuracy and 95.7% precision on the mechanism set, but only 76.6% recall. That is, when it assigns a label, it tends to be correct, but about a quarter of the proteins do not find another protein with exactly the same InterPro signatures in the training set, and so do not receive a prediction. The low recall is thus mainly caused by false negatives.
Figure 2.
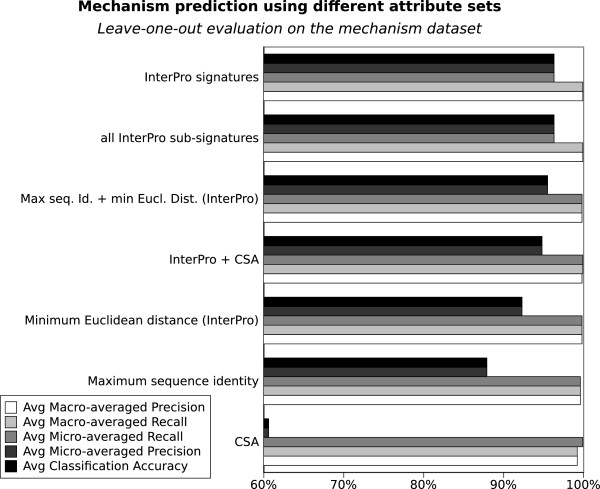
Predicting mechanism using InterPro and Catalytic Site Atlas attributes. A comparison of the predictive performance of various sets of attributes in a leave one out evaluation of the mechanism dataset. The x axis starts at 60% to better highlight the small differences between the top methods.
If we use the BRkNN algorithm instead, as described in the Methods section, Figure 2 shows that InterPro attributes alone are very good predictors of mechanism and achieve 96.3% classification accuracy and micro-averaged precision, and with a 99.9% macro-averaged recall. Using all InterPro signatures (including the so called "non-integrated" signatures) does not significantly improve nor degrade the overall InterPro result. CSA attributes are significantly worse than InterPro attributes at predicting mechanism on this dataset (60.6% classification accuracy and micro-averaged precision, 99.2% macro-averaged recall). Combining CSA attributes with InterPro attributes (InterPro+CSA attribute set) causes a slight degradation compared with using InterPro alone, achieving only 94.8% accuracy.
Mechanism prediction from three-dimensional structure
Figure 3 presents an evaluation of predicting mechanism using Catalytic Site Atlas 3D template matches (CSA 3D), either alone or in combination with sequence based attributes. We note that CSA 3D attributes appear more accurate than CSA sequence attributes (CSA 2D) and that the integration of CSA sequence and 3D attributes generally improves prediction compared with using CSA 2D or CSA 3D alone. However, adding CSA 3D attributes to InterPro attributes does not provide an advantage and indeed degrades prediction.
Figure 3.
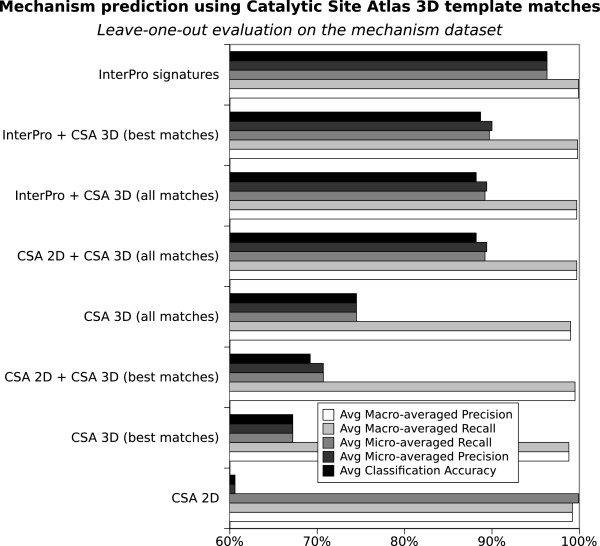
Predicting mechanism using Catalytic Site Atlas 3D attributes. A comparison of the predictive performance of various sets of sequence based (2D) and structure based (3D) Catalytic Site Atlas attributes in a leave one out evaluation of the mechanism dataset. The x axis starts at 60%.
The predictions based on CSA 3D templates mainly suffer from lack of coverage. The method generally predicts well, with few false positives, but it produces a high number of false negatives. This limitation is partly overcome by using all possible matches instead of only best matches (see Figure 3), but at the current state the method still appears to be less accurate than InterPro based methods. However, the current extension of CSA to CSA 2.0 [31], and any future extension in the number of 3D templates may improve its performance.
Statistical significance of the results
In order to define whether a set of attributes is a significantly better predictor than another set, we can imagine a random machine with characteristics similar to one of our predictors. Let us consider a method that emits either correct predictions with probability P or incorrect predictions with probability 1 - P. This method’s percentage of correct predictions will have mean 100 × P and standard deviation . If the machine predicts N = 250 protein-class label couples with P = 96.3% (1 - P = 3.7%) then the standard deviation equals . We can thus consider results with accuracy between 93.9% to 98.7% as being within two standard deviations and hence not significantly different.
Sequence identity and minimum Euclidean distance
Using only the maximum sequence identities as attributes (the maximum identity of the protein to be predicted when compared with the set of proteins having each mechanism) achieves 87.9% classification accuracy and micro-averaged precision and 99.6% macro-averaged recall. The results moderately improve when the minimum Euclidean distance is used (the minimum distance between the set of InterPro signatures of the protein to be predicted and the signatures of the proteins having each mechanism). The classification accuracy and micro-averaged precision grow from 87.9% to 92.3% and the macro-averaged recall from 99.6% to 99.8%. But it is the combination of the maximum sequence identity and minimum Euclidean distance that provides the best results within this style of data schema, with classification accuracy and micro-averaged precision reaching 95.5% while the macro-averaged recall remains at 99.8%. These results are not significantly worse than the results achieved by simply using InterPro signatures, but the method is much more computationally intensive.
Testing on negative sets
Here we assess the predictive performance of the best method (InterPro attributes + k-Nearest Neighbour) on a separate test set and we examine the type of false positive mistakes that the method produces. We use here the negative set, which contains 290 enzymes with known MACiE labels, but impossible to use for cross validation as they have only one protein per label. We thus train on the mechanism set plus the non-enzymes in Swiss-Prot (swissprot-non-EC), to provide training examples for both proteins having and not having the mechanisms of interest and we test on the separate negative set. If the method behaved in an ideal way, all the enzymes in the negative set would be predicted to be without labels, because none of the labels available in the training set is appropriate for the negative enzymes.
We also randomly partition the mechanism dataset into two folds (mech-fold1 and mech-fold2). Because many mechanisms in the mechanism set only have two proteins, we could not generate more than two folds without causing a further loss of mechanism labels and proteins. When training on fold 1 (mech-fold1 + half of swissprot-non-EC) and testing on fold 2 (mech-fold2 + the other half of swissprot-non-EC) there are only twelve false positive and twenty-three false negative predictions. Reversing the folds causes only six false positive and twenty-one false negative predictions. Thus even in such a vast test set, the mechanism training set only generates eighteen false positive predictions over more than 220,000 proteins. In addition, many of these false predictions are indeed very close to the mark. For example, Canis familiaris’ Inactive Pancreatic Lipase-related Protein 1 (LIPR1_CANFA, P06857) is predicted as having MACiE mechanism M0218_pancreatic_lipase. In fact, as recorded in Swiss-Prot’s annotation, this protein was originally thought to be a pancreatic lipase [55,56], but has been shown to lack lipase activity [57]. The same is true for the inactive pancreatic lipase-related proteins of Homo sapiens, Mus musculus and Rattus norvegicus which are also all predicted as M0218_pancreatic_lipase (UniProt accessions LIPR1_HUMAN/P54315, LIPR1_MOUSE/Q5BKQ4 and LIPR1_RAT, P54316 respectively). The method also predicts Legionella pneumophila’s Protein DlpA (DLPA_LEGPH, Q48806) as citrate synthase (MACiE M0078), and the protein is in fact highly related to the citrate synthase family, but lacks the conserved active His at position 264 which is replaced by an Asn residue.
Discussion
Sequence identity and Euclidean distance
The good accuracy, precision and recall obtained by the method are very encouraging but also highlight how similar in sequence many of the proteins belonging to one MACiE code are (as shown in Figure 1). This might be caused by strong conservation of many of these essential enzymes or, more prosaically, by a conservative manual annotation, which favours the transfer of labels among closely related orthologs. The consequence is a trusted but unchallenging data set for the methods presented.
In addition, even the performance of a simple line partition is reasonably high, provided that the Euclidean distance in the InterPro attributes space is used to further separate proteins, confirming the importance of using sequence signatures in addition to measures of sequence identity or similarity. Concluding, the InterPro based data schema seems to be essential to the good performance of: 1. machine learning over a sparse matrix (as presented using the k-Nearest Neighbour algorithm), 2. machine learning over a full matrix of sequence identity and Euclidean distance and even 3. simple regression (for example using the lines Euclidean distance = n × sequence identity).
At the current state of annotation, the small size of the training set makes the minimum Euclidean distance method look like a possible option for prediction. It is important to note though that a significant growth of the test or training sets will make a system based on alignments used to calculate the sequence identity (plus Euclidean distance calculation) much more computationally intensive than a machine learning algorithm (such as nearest neighbours) which relies on Euclidean distance alone.
Prediction quality
Additional file 4 is a graph of all enzymes in the mechanism dataset with their InterPro attributes and MACiE mechanism. The graph clearly shows that most clusters (proteins sharing a number of signatures) only have one MACiE mechanism, making predictions by k-Nearest Neighbour reasonably straightforward, as confirmed by the high accuracy, precision and recall of the leave one out evaluation on the mechanism dataset.
In fact, no false positive predictions appear when training on the negative dataset and testing on the mechanism dataset, but a small number of false positives (sixteen) appear when training on the mechanism set and testing on the negative, as shown in Table 2, which summarises the prediction errors for the training and testing evaluation experiments presented (a full list of the individual predictions can be found in Additional file 6).
Table 2.
Prediction statistics
| Training set | Test set | False positive | False negative |
|---|---|---|---|
|
mechanism +swissprot-non-EC fold 1 |
mechanism +swissprot-non-EC fold 2 |
12 |
23 |
|
mechanism +swissprot-non-EC fold 2 |
mechanism +swissprot-non-EC fold 1 |
6 |
21 |
|
mechanism |
negative |
16 |
n/a |
| negative | mechanism | 0 | n/a |
This table presents the number of false positive and false negative predictions for the training + testing evaluations (using InterPro attributes), a detailed list of the predictions is available in Additional file 6.
Additional file 5 contains a graph showing these sixteen false positive predictions in more detail. The clusters graphically show which protein neighbours caused the misprediction, and the signatures that these proteins share with the falsely predicted protein. For example, protein PABB_ECOLI has mechanism M0283: aminodeoxychorismate synthase (shown as a green oval), but it is predicted as M0314_component1_I: anthranilate synthase instead (shown as a red oval). The causes of the misprediction are signatures Anth_synth_I, ADC_synthase, Chorismate-bd_C and Anth_synth_I_N, which protein PABB_ECOLI shares with two anthranilate synthases (TRPE_SERMA and TRPE_SULSO). The two protein families are in fact very similar and the reactions’ Enzyme Commission numbers only differ in the last (substrate) digit: anthranilate synthase has EC 4.1.3.27 and aminodeoxychorismate lyase has EC 4.1.3.38.
One way to tease out the influence of these signatures that overlap across families is to introduce a larger sample of "negative" sequences. And this is what has been done in the two-fold cross evaluation experiment (mechanism plus swissprot-non-EC data sets). And, indeed, adding the non-enzymes keeps the number of false positive predictions extremely low (only 18 over 226,213 non-enzymes), but the split also somehow dilutes the informative signal of the mechanism dataset, causing a slightly larger number of false negative predictions (44 over 248 proteins).
Hence, in general, the methods seem to perform well. For the use of enzyme researchers we thus provide a list of all mechanism predictions for all Swiss-Prot enzymes (proteins having an Enzyme Commission number) as Additional file 7.
To provide correct neighbours for the instances currently receiving false negative or false positive predictions we would need to have either additional, more specific signatures in the set, or more proteins with the same signatures as the available instances. A detail of note is that the two best methods (InterPro and maximum sequence identity) label different proteins as false positives and false negatives. Hence by combining the predictions of the two methods (that is, accepting a label even if only one of the methods predicts it) we could reduce the number of false negatives to zero, but the number of false positive predictions would remain the same.
Conclusions
The machine learning method proposed can be applied to any sequenced protein and can assign a mechanism that cannot be immediately inferred from the InterPro signatures present in the sequence.
As future work it would be of interest to compare this approach with other representations of proteins, for example as discussed in [58] where protein sequences are described by fixed-length patterns with allowance for mutations, and the resulting mismatch string kernel is used with support vector machines to detect remote homology. These or other sequence features could be learned directly using a nearest neighbours algorithm or used as a kernel matrix for a support vector machine classifier, using a publicly available library such as libSVM which also allows for multi-label predictions.
The method presented is currently limited only by the lack of available data. Only 335 mechanisms have been described in detail in MACiE, the richest publicly available mechanism database, out of the more than 4,000 existing fourth level Enzyme Commission numbers, each of which could have one or more different mechanisms existing in nature. And only 540 proteins have been annotated with a specific MACiE mechanism. Additionally, most mechanisms only have one protein exemplar annotated within the MACiE, SFLD or EzCatDb databases, and cannot therefore be used for cross-validation.
Further validation will be needed when the dataset has grown, to clarify whether the best and fastest method remains the one we identified (InterPro attributes with k-Nearest Neighbour). However, the general indication is that mechanism prediction through sequence is possible, quick, accurate and produces a very limited number of false positives (just 0.00007% of 226,213 proteins) setting the foundations for further improvements to the methodology.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LDF and JBOM designed the work and analysed the results. LDF collected the data, ran the experiments and wrote the initial draft of the manuscript. Both authors read and approved the final manuscript.
Supplementary Material
Comparison of machine learning algorithms. Additional file comparison_of_machine_learning_algorithms.csv contains several evaluation metrics and run time for various machine learning algorithms when executing a leave-one-out experiment on the mechanism dataset.
Java code of ml2db. Additional file ml2db_code.tar.gz contains the Java source code to run the multi-label machine learning experiments and save the results to database. The code’s Javadoc is included.
ARFF and XML data files. Additional file arff_xml_files.tar.gz contains the ARFF and XML data files used in the machine learning experiments presented.
Neighbours clusters in the the mechanism dataset. Additional file mechanism_set_neighbours.pdf is a graph of all enzymes in the mechanism dataset with InterPro attributes. The proteins are shown as blue squares (containing their UniProt entry name) connected to their signatures (black ovals containing the InterPro signature short name) and their mechanisms (green ovals containing the MACiE entry number and name). The graph was generated with PyGraphviz, a Python interface to Graphviz.
Neighbours clusters of the false positive predictions when training on the mechanism set and testing on the negative set. Additional file graph_training_on_mechanism_testing_on_negative.pdf is a graph showing as red squares the proteins’ false positive labels when training on the mechanism set and testing on the negative set (using InterPro attributes, shown here as green rectangles). The green ovals represent the protein’s true mechanism, while the red ovals are the mistaken predictions. The yellow squares are neighbour proteins (proteins sharing some of the attributes) which caused the misprediction. The graph was generated with PyGraphviz, a Python interface to Graphviz.
List of false positive and false negative predictions. Additional file false_positive_and_negative_predictions.csv contains a comma separated set of tables with one row for each false positive or false negative prediction in the main training-testing experiments. The tables include each protein’s UniProt accession, entry name and species and the true and predicted mechanism identifiers and description (where relevant).
Mechanism predictions for all UniProt Swiss-Prot enzymes. Additional file swissprot_enzymes_mechanism_predictions.csv.tar.gz contains a comma separated file of the mechanism predictions for all Swiss-Prot enzymes (proteins having an Enzyme Commission number). The training set used for prediction includes all mechanism labelled proteins (the mechanism and negative sets) and all Swiss-Prot non-enzymes (the swissprot-non-EC set) with their InterPro attributes and MACiE mechanism class labels. The predicted mechanism is presented alongside the protein’s name, organism and Enzyme Commission number(s) taken from UniProt.
Contributor Information
Luna De Ferrari, Email: ldeferr@staffmail.ed.ac.uk.
John BO Mitchell, Email: jbom@st-andrews.ac.uk.
Acknowledgements
We thank the BBSRC for funding this research through grant BB/I00596X/1. JBOM thanks the Scottish Universities Life Sciences Alliance (SULSA) for financial support. Many thanks also to the Mulan mailing list members and to Lazaros Mavridis, Neetika Nath and to all the reviewers for their useful comments and suggestions. Special thanks to Roman Laskowski (EMBL-EBI) for his support in scanning structures using ProFunc and Catalytic Site Atlas 3D templates.
References
- Holliday GL, Bartlett GJ, Almonacid DE, O’Boyle NM, Murray-Rust P, Thornton JM, Mitchell JBO. MACiE: a database of enzyme reaction mechanisms. Bioinformatics. 2005;21(23):4315–4316. doi: 10.1093/bioinformatics/bti693. [ http://dx.doi.org/10.1093/bioinformatics/bti693] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday GL, Almonacid DE, Bartlett GJ, O’Boyle NM, Torrance JW, Murray-Rust P, Mitchell JBO, Thornton JM. MACiE (mechanism, annotation and classification in enzymes): novel tools for searching catalytic mechanisms. Nucleic Acids Res. 2007;35(Database issue):D515–D520. doi: 10.1093/nar/gkl774. [ http://dx.doi.org/10.1093/nar/gkl774] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday GL, Andreini C, Fischer JD, Rahman SA, Almonacid DE, Williams ST, Pearson WR. MACiE: exploring the diversity of biochemical reactions. Nucleic Acids Res. 2012;40(Database issue):D783–D789. doi: 10.1093/nar/gkr799. [ http://dx.doi.org/10.1093/nar/gkr799] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40(Database issue):D306–D312. doi: 10.1093/nar/gkr948. [ http://dx.doi.org/10.1093/nar/gkr948] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32(Database issue):D129–D133. doi: 10.1093/nar/gkh028. [ http://dx.doi.org/10.1093/nar/gkh028] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, Green RK, Goodsell DS, Prlic A, Quesada M, Quinn GB, Ramos AG, Westbrook JD, Young J, Zardecki C, Berman HM, Bourne PE. The RCSB protein data bank: new resources for research and education. Nucleic Acids Res. 2013;41(Database issue):D475–D482. doi: 10.1093/nar/gks1200. [ http://dx.doi.org/10.1093/nar/gks1200] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ. SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31(13):3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CZ, Han LY, Ji ZL, Chen YZ. Enzyme family classification by support vector machines. Proteins. 2004;55:66–76. doi: 10.1002/prot.20045. [ http://dx.doi.org/10.1002/prot.20045] [DOI] [PubMed] [Google Scholar]
- De Ferrari L, Aitken S, van Hemert J, Goryanin I. EnzML: Multi-label prediction of enzyme classes using InterPro signatures. BMC Bioinformatics. 2012;13:61. doi: 10.1186/1471-2105-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traube T, Vijayakumar S, Hirsch M, Uritsky N, Shokhen M, Albeck A. EMBM - a new enzyme mechanism-based method for rational design of chemical sites of covalent inhibitors. J Chem Inf Model. 2010;50(12):2256–2265. doi: 10.1021/ci100330y. [ http://dx.doi.org/10.1021/ci100330y] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim S. Sequence-based enzyme catalytic domain prediction using clustering and aggregated mutual information content. J Bioinform Comput Biol. 2011;9(5):597–611. doi: 10.1142/S0219720011005677. [DOI] [PubMed] [Google Scholar]
- Chea E. Livesay DR: How accurate and statistically robust are catalytic site predictions based on closeness centrality? BMC Bioinformatics. 2007;8:153. doi: 10.1186/1471-2105-8-153. [ http://dx.doi.org/10.1186/1471-2105-8-153] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007;8:298. doi: 10.1186/1471-2105-8-298. [ http://dx.doi.org/10.1186/1471-2105-8-298] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano N. EzCatDB: the enzyme catalytic-mechanism database. Nucleic Acids Res. 2005;33(Database issue):D407–D412. doi: 10.1093/nar/gki080. [ http://dx.doi.org/10.1093/nar/gki080] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Babbitt P. Using the structure-function linkage database to characterize functional domains in enzymes. Curr Protoc Bioinformatics. 2006;Chapter 2:Unit 2.10. doi: 10.1002/0471250953.bi0210s13. [ http://dx.doi.org/10.1002/0471250953.bi0210s13] [DOI] [PubMed] [Google Scholar]
- Consortium U. Update on activities at the universal protein resource (UniProt) in 2013. Nucleic Acids Res. 2013;41(Database issue):D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(Web Server issue):W597–W603. doi: 10.1093/nar/gks400. [ http://dx.doi.org/10.1093/nar/gks400] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The protein data bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112(3):535–542. doi: 10.1016/S0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- Lees JG, Lee D, Studer RA, Dawson NL, Sillitoe I, Das S, Yeats C, Dessailly BH, Rentzsch R, Orengo CA. Gene3D: Multi-domain annotations for protein sequence and comparative genome analysis. Nucleic Acids Res. 2014;42:D240–D245. doi: 10.1093/nar/gkt1205. [ http://dx.doi.org/10.1093/nar/gkt1205] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, Baratin D, Cuche BA, Bougueleret L, Poux S, Redaschi N, Xenarios I, Bridge A, Consortium U. HAMAP in 2013, new developments in the protein family classification and annotation system. Nucleic Acids Res. 2013;41(Database issue):D584–D589. doi: 10.1093/nar/gks1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41(Database issue):D377–D386. doi: 10.1093/nar/gks1118. [ http://dx.doi.org/10.1093/nar/gks1118] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [ http://dx.doi.org/10.1093/nar/gkt1223] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya AN, Arighi CN, Huang H, Barker WC, Wu CH. PIRSF family classification system for protein functional and evolutionary analysis. Evol Bioinform Online. 2006;2:197–209. [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Coletta A, Muirhead G, Pavlopoulou A, Philippou PB, Popov I, Romá-Mateo C, Theodosiou A, Mitchell AL. The PRINTS database: a fine-grained protein sequence annotation and analysis resource–its status in 2012. Database (Oxford) 2012;2012:bas019. doi: 10.1093/database/bas019. [ http://dx.doi.org/10.1093/database/bas019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru C, Courcelle E, CarrÃĺre S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33(Database issue):D212–D215. doi: 10.1093/nar/gki034. [ http://dx.doi.org/10.1093/nar/gki034] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41(D1):D344–D347. doi: 10.1093/nar/gks1067. [ http://dx.doi.org/10.1093/nar/gks1067] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(Database issue):D302–D305. doi: 10.1093/nar/gkr931. [ http://dx.doi.org/10.1093/nar/gkr931] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313(4):903–919. doi: 10.1006/jmbi.2001.5080. [ http://dx.doi.org/10.1006/jmbi.2001.5080] [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41(Database issue):D387–D395. doi: 10.1093/nar/gks1234. [ http://dx.doi.org/10.1093/nar/gks1234] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnham N, Holliday GL, de Beer TAP, Jacobsen JOB, Pearson WR, Thornton JM. The Catalytic Site Atlas 2.0: cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 2014;42(Database issue):D485–D489. doi: 10.1093/nar/gkt1243. [ http://dx.doi.org/10.1093/nar/gkt1243] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JA, Thornton JM. An algorithm for constraint-based structural template matching: application to 3D templates with statistical analysis. Bioinformatics. 2003;19(13):1644–1649. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33(Web Server issue):W89–W93. doi: 10.1093/nar/gki414. [ http://dx.doi.org/10.1093/nar/gki414] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [ http://www.sciencedirect.com/science/article/pii/0022283670900574] [DOI] [PubMed] [Google Scholar]
- Aha D, Kibler D. Instance-based learning algorithms. Mach Learn. 1991;6:37–66. [Google Scholar]
- Breiman L. Random Forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Fuernkranz J, Huellermeier E, Loza Mencia E, Brinker K. Multilabel classification via calibrated label ranking. Mach Learn. 2008;73(2):133–153. doi: 10.1007/s10994-008-5064-8. [DOI] [Google Scholar]
- Hastie T, Tibshirani R. Advances in Neural Information Processing Systems, Volume 10. Edited by Jordan MI, Kearns MJ, Solla SA. Cambridge, Massachusetts: MIT Press; 1998. Classification by pairwise coupling. [Google Scholar]
- Holte RC. Very simple classification rules perform well on most commonly used datasets. Mach Learn. 1993;11:63–90. doi: 10.1023/A:1022631118932. [DOI] [Google Scholar]
- John GH, Langley P. Eleventh Conference on Uncertainty in Artificial Intelligence. San Mateo: Morgan Kaufmann; 1995. Estimating Continuous Distributions in Bayesian Classifiers; pp. 338–345. [Google Scholar]
- Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. Improvements to Platt’s SMO Algorithm for SVM classifier design. Neural Comput. 2001;13(3):637–649. doi: 10.1162/089976601300014493. [ http://dx.doi.org/10.1162/089976601300014493] [DOI] [Google Scholar]
- Platt J. Advances in Kernel Methods - Support Vector Learning. Edited by Schoelkopf B, Burges C, Smola A. Cambridge, Massachusetts: MIT Press; 1998. Fast training of support vector machines using sequential minimal optimization. [ http://research.microsoft.com/en-us/um/people/jplatt/smo-book.pdf] [Google Scholar]
- Quinlan R. C4.5: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann Publishers; 1993. [Google Scholar]
- Spyromitros E, Tsoumakas G, Vlahavas I. Artificial Intelligence: Theories, Models and Applications. Berlin Heidelberg: Springer; 2008. An empirical study of lazy multilabel classification algorithms; pp. 401–406. [ http://dx.doi.org/10.1007/978-3-540-87881-0_40] [Google Scholar]
- Tsoumakas G, Katakis I, Vlahavas I. Mining Multi-label Data. US: Springer; 2010. [ http://mlkd.csd.auth.gr/publication_details.asp?publicationID=290] [Google Scholar]
- Tsoumakas G, Katakis I, Vlahavas I. Effective and efficient multilabel classification in domains with large number of labels. Proc. ECML/PKDD 2008 Workshop on Mining Multidimensional Data (MMD 2008) 2008. pp. 30–44.
- Tsoumakas G, Spyromitros-Xioufis E, Vilcek J, Vlahavas I. MULAN: A Java Library for Multi-Label Learning. J Mach Learn Res. 2011;12(Jul):2411–2414. [Google Scholar]
- Tsoumakas G, Vlahavas I. Random k -Labelsets: an ensemble method for multilabel classification. 2007. [ http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.97.5044&rep=rep1&type=pdf]
- Witten IH, Frank E. Data Mining - Practical machine learning tools and techniques with Java implementations. Morgan Kaufmann: San Francisco; 2005. [Google Scholar]
- Zhang ML, Zhou ZH. Multilabel neural networks with applications to functional genomics and text categorization. Knowl Data Eng IEEE Trans on. 2006;18(10):1338–1351. [Google Scholar]
- Jaccard P. The distribution of the flora in the alpine zone 1. New Phytologist. 1912;11(2):37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x. [ http://dx.doi.org/10.1111/j.1469-8137.1912.tb05611.x] [DOI] [Google Scholar]
- Dekel O, Shamir O. Proceedings of the 13 th International Conference on Artificial Intelligence and Statistics (AISTATS) 2010. Volume 9 of JMLR: W&CP. Chia Laguna Resort, Sardinia, Italy; 2010. Multiclass-multilabel classification with more classes than examples; pp. 137–144. [ http://machinelearning.wustl.edu/mlpapers/paper_files/AISTATS2010_DekelS10.pdf] [Google Scholar]
- Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Inf Process Manag. 2009;45(4):427–437. doi: 10.1016/j.ipm.2009.03.002. [ http://www.sciencedirect.com/science/article/pii/S0306457309000259] [DOI] [Google Scholar]
- Kerfelec B, LaForge KS, Puigserver A, Scheele G. Primary structures of canine pancreatic lipase and phospholipase A2 messenger RNAs. Pancreas. 1986;1(5):430–437. doi: 10.1097/00006676-198609000-00007. [DOI] [PubMed] [Google Scholar]
- Mickel FS, Weidenbach F, Swarovsky B, LaForge KS, Scheele GA. Structure of the canine pancreatic lipase gene. J Biol Chem. 1989;264(22):12895–12901. [PubMed] [Google Scholar]
- Roussel A, de Caro J, Bezzine S, Gastinel L, de Caro A, Carrière F, Leydier S, Verger R, Cambillau C. Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations. Proteins. 1998;32(4):523–531. doi: 10.1002/(SICI)1097-0134(19980901)32:4<523::AID-PROT10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Leslie CS, Eskin E, Cohen A, Weston J, Noble WS. Mismatch string kernels for discriminative protein classification. Bioinformatics. 2004;20(4):467–476. doi: 10.1093/bioinformatics/btg431. [ http://bioinformatics.oxfordjournals.org/content/20/4/467.abstract] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of machine learning algorithms. Additional file comparison_of_machine_learning_algorithms.csv contains several evaluation metrics and run time for various machine learning algorithms when executing a leave-one-out experiment on the mechanism dataset.
Java code of ml2db. Additional file ml2db_code.tar.gz contains the Java source code to run the multi-label machine learning experiments and save the results to database. The code’s Javadoc is included.
ARFF and XML data files. Additional file arff_xml_files.tar.gz contains the ARFF and XML data files used in the machine learning experiments presented.
Neighbours clusters in the the mechanism dataset. Additional file mechanism_set_neighbours.pdf is a graph of all enzymes in the mechanism dataset with InterPro attributes. The proteins are shown as blue squares (containing their UniProt entry name) connected to their signatures (black ovals containing the InterPro signature short name) and their mechanisms (green ovals containing the MACiE entry number and name). The graph was generated with PyGraphviz, a Python interface to Graphviz.
Neighbours clusters of the false positive predictions when training on the mechanism set and testing on the negative set. Additional file graph_training_on_mechanism_testing_on_negative.pdf is a graph showing as red squares the proteins’ false positive labels when training on the mechanism set and testing on the negative set (using InterPro attributes, shown here as green rectangles). The green ovals represent the protein’s true mechanism, while the red ovals are the mistaken predictions. The yellow squares are neighbour proteins (proteins sharing some of the attributes) which caused the misprediction. The graph was generated with PyGraphviz, a Python interface to Graphviz.
List of false positive and false negative predictions. Additional file false_positive_and_negative_predictions.csv contains a comma separated set of tables with one row for each false positive or false negative prediction in the main training-testing experiments. The tables include each protein’s UniProt accession, entry name and species and the true and predicted mechanism identifiers and description (where relevant).
Mechanism predictions for all UniProt Swiss-Prot enzymes. Additional file swissprot_enzymes_mechanism_predictions.csv.tar.gz contains a comma separated file of the mechanism predictions for all Swiss-Prot enzymes (proteins having an Enzyme Commission number). The training set used for prediction includes all mechanism labelled proteins (the mechanism and negative sets) and all Swiss-Prot non-enzymes (the swissprot-non-EC set) with their InterPro attributes and MACiE mechanism class labels. The predicted mechanism is presented alongside the protein’s name, organism and Enzyme Commission number(s) taken from UniProt.


