Abstract
Tumor suppressor gene p53 functions as the guardian of the human genome and mutations in p53 contribute to cancer development. However, studies that investigated the potential of p53 as a prognostic marker in osteosarcoma patients have yielded inconclusive results. Based on recommendation of the Cochrane Collaboration, this meta-analysis was conducted using data from the 17 published studies to evaluate the association of p53 alterations with clinical outcome of osteosarcoma patients. Different databases, including MEDLINE, PsycINFO, Scopus, EMBASE, and the Cochrane Central Register of Controlled Trials (CENTRAL) were searched. Prognostic value of p53 alterations was determined by risk ratio (RR). The data showed that p53-positive immunostaining tended to associate with decreased 2-year survival rates (RR, 1.94; 95% CI, 1.43 to 2.64; p < 0.0001, I2 = 10%). However, the prediction value of RR was smaller with p53 expression than with p53 mutations. Moreover, patients who received neoadjuvant chemotherapy and surgery tended to have a stronger association between p53-positive staining and 2-year mortality compared to the patients treated with surgery only. However, p53-positive staining was not associated with 3-year (RR, 1.64; 95% CI, 0.84 to 3.20; P = 0.15; I2 = 56%) and 5-year survival (RR, 1.25; 95% CI, 0.78 to 2.01; P = 0.36; I2 = 70%). The data from the current study suggest that p53-positive osteosarcoma only predicted a decreased short-term survival rate, but not 3- or 5-year survival.
Keywords: p53, osteosarcoma, survival, meta-analysis
Introduction
Osteosarcoma (OS) is an aggressive malignant bone tumor occurring frequently in children and adolescents. To date, the incidence of osteosarcoma remains high, accounting for the eighth most common childhood cancer and the sixth leading cancer in children under age 15, although recent advances in treatment of osteosarcoma have led to significant improvements in patient outcome [1]. Tumor metastasis frequently occurs in approximately 40% of such patients, indicating tumor resistance to cytostatic chemotherapy [2]. Thus, it is urgently needed to develop and identify biomarkers to predict prognosis and treatment outcome for these patients.
Tumor suppressor gene p53 functions as the guardian of the human genome and mutations in p53 contribute to human carcinogenesis [3]. p53 is localized at chromosome 17 band p13.1 where loss of heterozygosity, deletion, and mutation frequently occur [3]. p53 functions to maintain the stability of the genome [4] and acts as “the guardian of DNA”, especially when cells are under stress (such as DNA damage, aberrant proliferative signals, heat shock, or hypoxia) [5]. The wild-type p53 protein regulates genes that are involved in DNA repair, cell cycle checkpoint, and apoptosis [6-8]. In early studies, p53 was found to be frequently mutated in osteosarcoma [9] and subsequent studies investigated the clinical significance of p53 mutations or overexpression of p53 protein in osteosarcoma [10-18]. For example, previous studies showed that p53 expression was associated with a poor response to chemotherapy and worsened survival of patients [11,16], whereas in other studies the data were inconclusive [15,18]. In 2004, Pakos et al. conducted a meta-analysis, which suggested that p53 alterations might be associated with a poor survival of osteosarcoma patients [19]. However, this controversy continued with the emergence of more recent studies [20-27]. We therefore conducted an updated meta-analysis of all available studies for association of p53 expression or p53 mutations with clinical outcome of osteosarcoma patients.
Methods
Identification of eligible and relevant studies
Based on the recommendations of the Cochrane Collaboration, we performed this meta-analysis. To do so, we considered all studies for association of p53 expression and/or p53 alterations with osteosarcoma outcomes. We searched different electronic databases, including MEDLINE (January 1980 to December 2013), PsycINFO (January 1980 to December 2013), Scopus (January 1980 to December 2013), EMBASE (January 1980 to December 2013), and the Cochrane Library (Issue 11 of 12, Dec 2013). The search was limited to human studies in all languages and types of publications. The search terms used were: osteosarcoma, p53, TP53, p53 protein, p53 mutation, and 17p13 gene and the full search strategy were illustrated in Figure 1 for numbers of studies reviewed and analyzed. Such strategy was developed for MEDLINE and was adapted for the other electronic databases. References of retrieved studies were screened and we then contacted the investigators to request additional data when key information relevant to the meta-analysis was missing. All studies on the relationship between TP53 status and clinical outcome (death) were eligible for this meta-analysis, regardless of the method of detection [immunohistochemistry (IHC) for measuring protein levels and reverse transcription-PCR (RT-PCR) techniques for identifying mutations or other gene changes].
Figure 1.
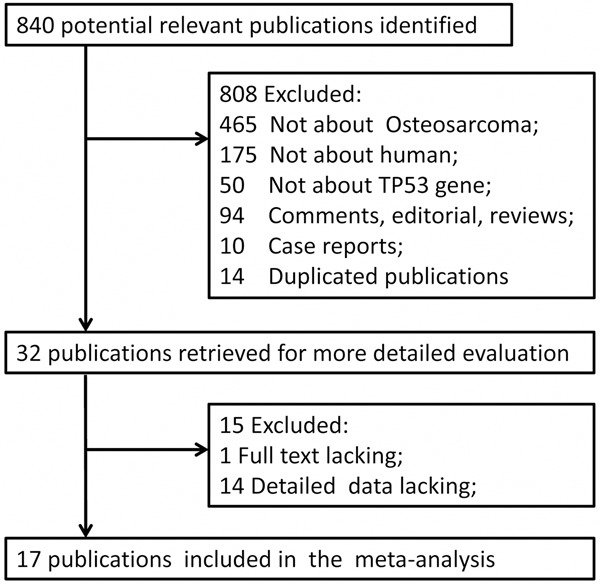
The flow chart of included studies.
Definitions and standardizations
For consistency, “p53” stands for the gene, while p53 is for protein, and “p53 status” is to cover both the gene and protein as a marker. Nuclear accumulation of mutant p53 protein, which are induced by p53 alterations, can be detected by immunohistochemistry (IHC) [10]. However, accumulation of p53 protein detected by IHC does not necessarily correspond to p53 mutations measured by RT-PCR [28]. Thus, an overall analysis was considered for all data, regardless of whether protein expression or mutation was being evaluated. For example, for studies using IHC only, we used prespecified rules to standardize the p53 status as much as possible to define a positive p53 status based on different cut-off thresholds. We defined positive p53 protein expression as nuclear staining in at least 10% of tumor cells, a standard used by most studies [27]. When different definitions were used, we accepted the cutoff point closest to the 10% level [19]. The clinical outcome used was mortality of the patient. Clinical outcomes were standardized to include 24, 36, or 60 months follow-up in all studies.
Inclusion criteria
Original studies were considered for inclusion in this meta-analysis if they met with the following criteria: i) The patients were diagnosed pathologically as osteosarcoma; ii) treatments of patients included radiotherapy, chemotherapy, surgery, or a combination of both; iii). The 2-year, 3-year or 5-year survival rates were reported; and iv). The comparison between patients with low or undetectable p53 and patients with upregulated p53 was performed in terms of the survival rate.
Data extraction
Two investigators (D. Y. and J. C.) independently screened the titles and abstracts of all potentially eligible studies. The full text articles were then assessed independently by two other investigators (Y-P. L. and G-H. C.) to determine whether the articles met the inclusion criteria. After that, three other investigators (S-X. W., R.L. and G-H. C.) independently extracted data (study characteristics and results) using data extraction forms and then the collected data were entered into Rev-Man 5.1 using the double-entry system. Point estimates for selected variables were extracted and checked by the other two reviewers. In case of disagreement between these two reviewers, a consensus was achieved through discussion among all of the reviewers. A record of reasons for excluding studies was kept.
Data collection and analysis
We collected the following data from each study: i). General study information, such as title, authors, publication source and publication year; ii). Characteristics of study population (e.g. sample size, patient age, and osteosarcoma classification); iii). Treatment data, such as neoadjuvant chemotherapy and surgery; iv). p53 status, such as expression or mutation; v). Detection of p53 status methods (e.g. IHC, antibody used, IHC cut-off point, and PCR amplification of the exons).
After that, the meta-analyses were performed using Rev-Man analyses software (Rev-Man 5.1) according to Cochrane Handbook for Systematic Reviews of Interventions [29]. Data on the predictive ability of p53 overexpression or p53 alterations for outcome were combined from all 17 studies using RR for 2-year, 3-year and 5-year mortality. Measurements of the graphs published in the articles were used if we could not get the raw data from the authors. When only the standard error was reported, it was converted into standard deviation [29]. I2 statistics were used to measure heterogeneity of the studies. If the I2 value was less than 50%, a fixed-effects meta-analysis was applied, whereas if the I2 value was 50% or more, the random-effects meta-analysis was performed [29]. Sensitivity analyses were performed and aisual assessment of the funnel plot calculated by RevMan Analyses software was used to investigate the potential publication bias.
Results
Study selection
In this study, we first searched MEDLINE, PsycINFO, Scopus, EMBASE, and the CENTRAL databases and reviewed a total of 840 published studies (Figure 1). Initially, we excluded 808 publications, 175 of which contained animal experiments, 465 of which were not in osteosarcoma, 50 of which were not for p53, and 118 of which were excluded because they were either comments, editorials, reviews, case reports, or duplicated publications. We obtained 32 publications that met our inclusion criteria, but additional 15 publications were eventually excluded because of lack of full text [30] or detailed data [31-44]. Finally, we obtained the remaining 17 studies for this meta-analysis [10-18,20-27].
Description of included studies
The detailed characteristics of the included studies were shown in Table 1. Overall, 595 patients were included in this analysis. The median or mean age of patients was 24.6 years old, ranging between 15 years [13,16,20] and 67 years old [12]. Seven studies [11-13,15,17,18,20] were conducted on osteosarcoma in high histological grades, while three studies [10,21,27] were conducted on osteosarcoma in low or intermediate histological grades and four studies [14,22,24,25] were on osteosarcoma in all histological grades. However, there was no grade data in three studies [16,23,26]. Patients in 5 of these 17 studies received surgery treatment only [12,18,20,24,26], whereas patients in 12 studies were treated with neoadjuvant chemotherapy and surgery. p53 status was shown as p53 gene/protein expression in 13 studies, while remaining 4 studies only showed p53 mutation [13,16,18,26].
Table 1.
Patient characteristics in each study
| Author (yrs.) | N | Age (mean yrs.) | HG (Grade, N) | Treatment | TP53 | Method | IHC antibody | IHC cutoff | PCR Exons | Death in 2 yrs. n (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Papai (1997) [10] | 21 | 20 | 17 (Grade IIb) | NC + surgery | expression | IHC | DO-7 | 5% | 8 (38) | |
| 4 (Grade IIa) | ||||||||||
| Goto (1998) [11] | 32 | 16 | 23 (Grade III) | NC + surgery | expression | IHC/PCR | DO-7/Rsp53 | > 0% | MS | 14 (44) |
| 9 (Grade IV) | ||||||||||
| Jensen (1998) [12] | 25 | 67 | 9 (Grade III) | surgery | expression | IHC | DO-7 | 10% | 11 (44) | |
| 16 (Grade IV) | ||||||||||
| Yokoyama (1998) [13] | 17 | 15 | 8 (Grade III) | NC + surgery | mutation | PCR* | 4-8 | 1 (6) | ||
| 7 (Grade IV) | ||||||||||
| Gorlick (1999) [14] | 53 | 17 | 11 (Grade I) | NC + surgery | expression | IHC | 1801/DO-7 | > 0% | 16 (30) | |
| 24 (Grade II) | ||||||||||
| 10 (Grade III) | ||||||||||
| 8 (Grade IV) | ||||||||||
| Tsuchiya (2000) [16] | 27 | 15 | NA | NC + surgery | mutation | PCR* | 5-9 | 11 (41) | ||
| Uozaki (2000) [17] | 70 | 16 | 43 (Grade III) | NC + surgery | expression | IHC | DO-7 | 10% | 17 (24) | |
| 27 (Grade IV) | ||||||||||
| Oda (2000) [15] | 25 | 17 | 6 (Grade III) | NC + surgery | expression | IHC | NR | 10% | 4 (16) | |
| 19 (Grade IV) | ||||||||||
| Kawaguchi (2002) [18] | 23 | 55 | 8 (Grade III) | surgery | mutation | IHC/PCR | 1801 | 10% | 5-9 | 12 (52) |
| 15 (Grade IV) | ||||||||||
| Tsai (2004) [21] | 22 | 16 | 3 (Grade II) | NC + surgery | expression | IHC | DO-7 | 5% | 9 (41) | |
| 11 (Grade IIa) | ||||||||||
| 8 (Grade III) | ||||||||||
| Ferrari (2004) [20] | 19 | 15 | NA | surgery | expression | IHC | DO-7 | 25% | 3 (16) | |
| Kaseta (2008) [22] | 35 | 30 | 4 (Low grade) | NC + surgery | expression | IHC | DO-7 | 25% | NA | |
| 31 (High grade) | ||||||||||
| Ozger (2009) [23] | 45 | 20 | NA | NC + surgery | expression | IHC | DO-7 | 10% | 6 (13) | |
| Boulytcheva (2010) [24] | 40 | NA | 4 (Grade I) | surgery | expression | IHC | DO-7 | 10% | NA | |
| 22 (Grade II) | ||||||||||
| 2 (Grade III) | ||||||||||
| 12 (Grade IV) | ||||||||||
| Hu (2010) [25] | 44 | 25 | 5 (Grade I) | NC + surgery | expression | IHC | NR | 10% | NA | |
| 16 (Grade II) | ||||||||||
| 16 (Grade III) | ||||||||||
| 7 (Grade IV) | ||||||||||
| Seidinger (2011) [26] | 41 | NA | NA | surgery | mutation | IHC/PCR | DO-7 | NR | 10 | 12 (29) |
| Wu (2012) [27] | 56 | Range (13-37) | 56 (Grade IIb) | NC + surgery | expression | IHC | DO-7 | 10% | NA |
Note: HG, histological grades; N, number; NC, neoadjuvant chemotherapy;
PCR/single-strand conformational polymorphism.
p53 gene/protein expression was analyzed by using immunohistochemistry, while p53 mutation was assessed using PCR. Eight studies used 10% as the cutoff value for p53 protein positivity, whereas different thresholds (0-25%) were used in the remaining reports (Table 1). In most studies, clone DO-7 antibody was used immunohistochemically to detect expression of p53 protein. Two-year survival rates differed significantly (P < 0.001) across the thirteen eligible studies (ranged between 6% and 52%), which may be due to differences in patient populations (e.g., tumor grade and stage) and/or treatment options.
The meta-analyses
Based on Cohen categories for evaluating the magnitude of effect sizes, p53-positive status tended to be associated with a poor 2-year survival rate and a higher risk of death within 2 years (Figure 2A, RR, 1.94; 95% CI, 1.43 to 2.64; P < 0.0001). The test for heterogeneity showed that these studies were not heterogeneous (I2 = 10%, Figure 2A). RR was smaller in studies of p53 protein expression than in studies of p53 alterations (Table 2). These studies showed that patients received neoadjuvant chemotherapy and surgery tended to have a stronger association of p53-positive status with 2-year mortality when compared to patients treated with surgery only.
Figure 2.
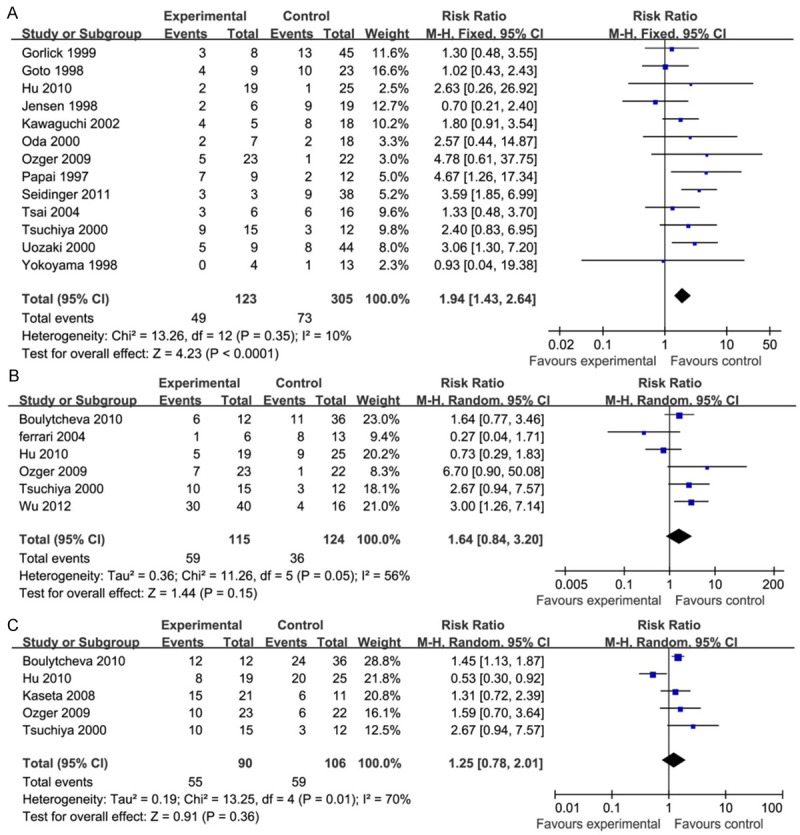
The meta-analysis of p53 status association with patients’ risk of death. A: Within two years. B: Within three years. C: Within five years.
Table 2.
Association of TP53 expression with patient 24 months mortality
| Studies | Cases/total cases | I2 (%) | Risk ratio (95% CI) |
|---|---|---|---|
| All | 13/428) | 13.26 | 1.94 [1.43, 2.64] |
| IHC only | 8/288 | 7 | 2.05 [1.33, 3.15] |
| Studies on expression | 9/320 | 14 | 1.81 [1.24, 2.66] |
| Studies on mutation | 4/108 | 0 | 2.28 [1.38, 3.77] |
| Treatment: NC + surgery | 10/339 | 0 | 2.06 [1.41, 3.02] |
| Treatment: surgery only | 3/89 | 72 | 1.84 [0.75, 4.52] |
| Sensitivity analyses | |||
| Specific 10% cutoff | 7/256 | 12 | 2.22 [1.46, 3.40] |
Note: If the I2 value was less than 50%, a fixed-effects meta-analysis was applied. If the I2 value was 50% or more, the random-effects meta-analysis was used. Abbreviations: CI, confidence interval; NC, neoadjuvant chemotherapy.
However, our further analysis showed that p53-positive status was not associated with 3-year survival (Figure 2B, RR, 1.64; 95% CI, 0.84 to 3.20; P = 0.15) and 5-year survival (Figure 2C, RR, 1.25; 95% CI, 0.78 to 2.01; P = 0.36). The test for heterogeneity showed that these studies were more heterogeneous (I2 = 56%, Figure 2B; I2 = 70%, Figure 2C). Nevertheless, there was no further layer analysis conducted when considering the small size effect and limited number of included studies. In this case, only the random-effects model was performed when the I2 value was 50% or more.
Risk of bias in these included studies
Sensitivity analyses were performed to assess the effect of limitations on the evaluation of studies using the 10% IHC cut-off point for p53 expression. The survival difference was somewhat stronger and formally statistically significant in studies using the 10% IHC cut-off value (Table 2). Moreover, the funnel plots of analysis of p53-positive status for association with 2-year mortality confirmed a symmetric distribution and suggested that there was non-publication bias (Figure 3).
Figure 3.
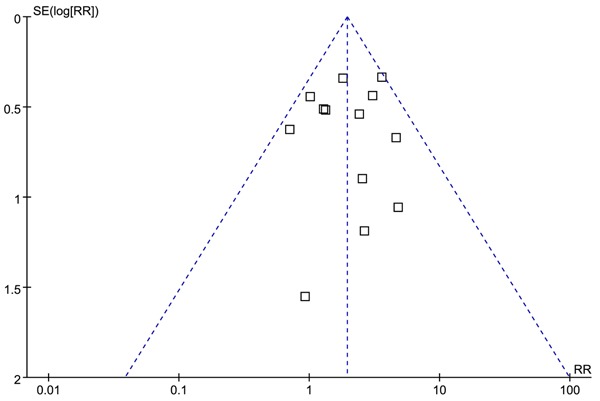
Analysis of the publication bias.
Discussion
A previously published meta-analysis showed a significant association of p53 alterations (p53 gene mutation or loss of heterozygosity) with 2-year survival [19], while a meta-analysis published in 2013 showed that high p53 expression associated with a poorer prognosis for patients with osteosarcoma [45]. Our current study is remarkably different from these previous studies and we assessed that i). In contrast to the previous studies, we have paid more attention to evaluating the association between TP53 status and long-term survival of osteosarcoma patients and ii). We included more studies; in this meta-analysis, we found that p53 alterations (either p53 protein overexpression detected by IHC or p53 mutation detected by RT-PCR) associated with poor 2-year survival of osteosarcoma patients, particularly in studies that evaluated p53 mutations. However, our current data showed p53 alterations didn’t have any associations with 3-year survival or 5-year survival of the patients. Thus, our current data demonstrated that p53 alterations could only predict short clinical outcome, but not the longer-term survival of osteosarcoma patients.
It is true that the outcome of osteosarcoma patients has significantly improved throughout the last two decades and 5-year overall survival rate has reached between 50% and 70% [46-48]. In this regards, research in the field would pay more attention to predict 3 or 5-year survival of the patients [20,22-25,27]. In terms of biomarker study using p53 status, the prognostic value of p53 status for long-term survival of osteosarcoma patients seemed to be more controversial, whereas positive results were shown for 2-year survival rate [20,22-25,27]. In our current study, the prognostic value of p53 for long-term survival of osteosarcoma patients was limited. Further studies using a larger sample size are needed to confirm it.
p53 alterations contribute to tumorigenesis as an early event and the detection of p53 alterations could help determine the features of osteosarcoma (such as tumor grade, type, aggressiveness, and metastatic potential) [19,49]. However, our current results showed that patients who received neoadjuvant chemotherapy and surgery seemed to have a stronger association between TP53-positive status and 2-year mortality when compared to patients who were treated with surgery only. This piece of data was inconsistent with others [19,49]. The reason for this controversial data may be due to the limitation of patients who were treated with surgery only.
Furthermore, in the current meta-analysis, we found that association of poor 2-year survival of patients with presence of p53alterations was observed stronger than that with the IHC data. It is true that IHC can only detect protein in the case of p53 alterations for p53 point mutation, but IHC can’t detect protein in the case of p53 deletion, frame-shift mutation, or early stop codon mutations [50]. Thus, there is no straightforward correlation between IHC and RT-PCR [51]. Other investigators have suggested that the combination of IHC and RT-PCR data may provide complementary prognostic information [52]. However, this has not been accomplished over time; most association studies continued using IHC or PCR individually rather than in combination in the recent 10 years. Thus, studies on p53 mutation could be more reliable.
However, our current meta-analysis does have its limitations. First, there was significant heterogeneity in the results for association between long-term survival (3-year/5-year survival) and p53 status. Considering the small size effect and limited number of included studies, we did not apply a further layer of analysis and only performed the random-effect model analysis. Secondly, although we had made our best effort to get the full text of all published studies, there were still some studies that failed to be included in our meta-analysis due to the lack of detailed data. Thirdly, some statistical methods used in our current study may be limited, such as using I2 to assess the amount of heterogeneity in random-effects meta-analysis [53] and visual assessment of the funnel plot for excluding a publication bias. Fourthly, we didn’t assess the association between p53 alterations and some osteosarcoma features (such as tumor type, aggressiveness, and metastatic potential), which may be related to osteosarcoma outcomes.
But, our current meta-analysis did obtain the following data: i). p53 alterations positive status associated with poor short-term survival of patients with osteosarcoma, particularly in osteosarcoma with p53 mutations; ii). p53 alterations didn’t associate with the long-term survival of the patients; and iii). Patients received neoadjuvant chemotherapy and surgery had a stronger association of p53 alterations with a 2-year mortality when compared to those treated with surgery only. However, further studies with a larger sample size will confirm the prognostic value of p53 for long-term survival of patients with osteosarcoma and detection of p53 mutations could be the better choice for future study of p53 alterations.
Acknowledgements
This work was supported by grants from Program for Shaanxi Province Key Research Team of Science and Technology Innovation (2012KCT-14).
Disclosure of conflict of interest
None.
References
- 1.Qu JT, Wang M, He HL, Tang Y, Ye XJ. The prognostic value of elevated vascular endothelial growth factor in patients with osteosarcoma: a meta-analysis and systemic review. J Cancer Res Clin Oncol. 2012;138:819–25. doi: 10.1007/s00432-012-1149-7. [DOI] [PubMed] [Google Scholar]
- 2.Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, Barany F, Ladanyi M, Gorlick R, Levine AJ. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci U S A. 2003;100:11547–52. doi: 10.1073/pnas.1934852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride OW, Merry D, Givol D. The gene for human p53 cellular tumor antigen is located on chromosome 17 short arm (17p13) Proc Natl Acad Sci U S A. 1986;83:130–4. doi: 10.1073/pnas.83.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine AJ, Perry ME, Chang A, Silver A, Dittmer D, Wu M, Welsh D. The 1993 Walter Hubert Lecture: the role of the p53 tumour-suppressor gene in tumorigenesis. Br J Cancer. 1994;69:409–16. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vousden KH. Activation of the p53 tumor suppressor protein. Biochim Biophys Acta. 2002;1602:47–59. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- 6.Soussi T, Beroud C. Significance of TP53 mutations in human cancer: a critical analysis of mutations at CpG dinucleotides. Hum Mutat. 2003;21:192–200. doi: 10.1002/humu.10189. [DOI] [PubMed] [Google Scholar]
- 7.Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Wild-type p53 can inhibit oncogene-mediated focus formation. Proc Natl Acad Sci U S A. 1989;86:8763–7. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–93. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 9.Masuda H, Miller C, Koeffl er HP, Battifora H, Cline MJ. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci U S A. 1987;84:7716–9. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papai Z, Féja CN, Hanna EN, Sztán M, Oláh E, Szendrôi M. P53 Overexpression as an Indicator of Overall Survival and Response to Treatment in Osteosarcomas. Pathol Oncol Res. 1997;3:15–19. doi: 10.1007/BF02893346. [DOI] [PubMed] [Google Scholar]
- 11.Goto A, Kanda H, Ishikawa Y, Matsumoto S, Kawaguchi N, Machinami R, Kato Y, Kitagawa T. Association of loss of heterozygosity at the p53 locus with chemoresistance in osteosarcomas. Jpn J Cancer Res. 1998;89:539–47. doi: 10.1111/j.1349-7006.1998.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lidang Jensen M, Schumacher B, Myhre Jensen O, Steen Nielsen O, Keller J. Extraskeletal osteosarcomas: a clinicopathologic study of 25 cases. Am J Surg Pathol. 1998;22:588–94. doi: 10.1097/00000478-199805000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama R, Schneider-Stock R, Radig K, Wex T, Roessner A. Clinicopathologic implications of MDM2, p53 and K-ras gene alterations in osteosarcomas: MDM2 amplification and p53 mutations found in progressive tumors. Pathol Res Pract. 1998;194:615–21. doi: 10.1016/s0344-0338(98)80096-4. [DOI] [PubMed] [Google Scholar]
- 14.Gorlick R, Huvos AG, Heller G, Aledo A, Beardsley GP, Healey JH, Meyers PA. Expression of HER2/erbB-2 correlates with survival in osteosarcoma. J. Clin. Oncol. 1999;17:2781–8. doi: 10.1200/JCO.1999.17.9.2781. [DOI] [PubMed] [Google Scholar]
- 15.Oda Y, Naka T, Takeshita M, Iwamoto Y, Tsuneyoshi M. Comparison of histological changes and changes in nm23 and c-MET expression between primary and metastatic sites in osteosarcoma: a clinicopathologic and immunohistochemical study. Hum Pathol. 2000;31:709–16. doi: 10.1053/hupa.2000.8230. [DOI] [PubMed] [Google Scholar]
- 16.Tsuchiya T, Sekine K, Hinohara S, Namiki T, Nobori T, Kaneko Y. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet Cytogenet. 2000;120:91–8. doi: 10.1016/s0165-4608(99)00255-1. [DOI] [PubMed] [Google Scholar]
- 17.Uozaki H, Ishida T, Kakiuchi C, Horiuchi H, Gotoh T, Iijima T, Imamura T, Machinami R. Expression of heat shock proteins in osteosarcoma and its relationship to prognosis. Pathol Res Pract. 2000;196:665–73. doi: 10.1016/S0344-0338(00)80118-1. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi K, Oda Y, Sakamoto A, Saito T, Tamiya S, Iwamoto Y, Tsuneyoshi M. Molecular analysis of p53, MDM2, and H-ras genes in osteosarcoma and malignant fibrous histiocytoma of bone in patients older than 40 years. Mod Pathol. 2002;15:878–88. doi: 10.1097/01.MP.0000024264.48690.EA. [DOI] [PubMed] [Google Scholar]
- 19.Pakos EE, Kyzas PA, Ioannidis JP. Prognostic significance of TP53 tumor suppressor gene expression and mutations in human osteosarcoma: a meta-analysis. Clin Cancer Res. 2004;10:6208–14. doi: 10.1158/1078-0432.CCR-04-0246. [DOI] [PubMed] [Google Scholar]
- 20.Ferrari S, Bertoni F, Zanella L, Setola E, Bacchini P, Alberghini M, Versari M, Bacci G. Evaluation of P-glycoprotein, HER-2/ErbB-2, p53, and Bcl-2 in primary tumor and metachronous lung metastases in patients with high-grade osteosarcoma. Cancer. 2004;100:1936–42. doi: 10.1002/cncr.20151. [DOI] [PubMed] [Google Scholar]
- 21.Tsai JY, Aviv H, Benevenia J, Chang VT, Patterson F, Aisner S, Hameed M. HER-2/neu and p53 in osteosarcoma: an immunohistochemical and fl uorescence in situ hybridization analysis. Cancer Invest. 2004;22:16–24. doi: 10.1081/cnv-120027577. [DOI] [PubMed] [Google Scholar]
- 22.Kaseta MK, Khaldi L, Gomatos IP, Tzagarakis GP, Alevizos L, Leandros E, Papagelopoulos PJ, Soucacos PN. Prognostic value of bax, bcl-2, and p53 staining in primary osteosarcoma. J Surg Oncol. 2008;97:259–66. doi: 10.1002/jso.20913. [DOI] [PubMed] [Google Scholar]
- 23.Ozger H, Eralp L, Atalar AC, Toker B, Esberk Ateş L, Sungur M, Bilgiç B, Ayan I. [The effect of resistance-related proteins on the prognosis and survival of patients with osteosarcoma: an immunohistochemical analysis] . Acta Orthop Traumatol Turc. 2009;43:28–34. doi: 10.3944/AOTT.2009.028. [DOI] [PubMed] [Google Scholar]
- 24.Boulytcheva IV, Soloviev YN, Kushlinskii NE, Mahson AN. Expression of molecular markers in the tumor and survival prognosis in osteosarcoma. Bull Exp Biol Med. 2010;150:237–42. doi: 10.1007/s10517-010-1114-x. [DOI] [PubMed] [Google Scholar]
- 25.Hu X, Yu AX, Qi BW, Fu T, Wu G, Zhou M, Luo J, Xu JH. The expression and significance of IDH1 and p53 in osteosarcoma. J Exp Clin Cancer Res. 2010;29:43. doi: 10.1186/1756-9966-29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidinger AL, Mastellaro MJ, Paschoal Fortes F, Godoy Assumpção J, Aparecida Cardinalli I, Aparecida Ganazza M, Correa Ribeiro R, Brandalise SR, Dos Santos Aguiar S, Yunes JA. Association of the highly prevalent TP53 R337H mutation with pediatric choroid plexus carcinoma and osteosarcoma in southeast Brazil. Cancer. 2011;117:2228–35. doi: 10.1002/cncr.25826. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Cai ZD, Lou LM, Zhu YB. Expressions of p53, c-MYC, BCL-2 and apoptotic index in human osteosarcoma and their correlations with prognosis of patients. Cancer Epidemiol. 2012;36:212–6. doi: 10.1016/j.canep.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–40. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 29.Higgins JPT, Green S, editors. Cochrane Hand-book for Systematic Reviews of Interventions. Version 5.0.2. The Cochrane Collaboration; 2009. [updated September 2009]. www.cochrane-handbook.org. [Google Scholar]
- 30.Serra M, Maurici D, Scotlandi K, Barbanti-Brodano G, Manara MC, Benini S, Picci P, Bertoni F, Bacci G, Sottili S, Baldini N. Relationship between P-glycoprotein expression and p53 status in high-grade osteosarcoma. Int J Oncol. 1999;14:301–7. doi: 10.3892/ijo.14.2.301. [DOI] [PubMed] [Google Scholar]
- 31.Ueda Y, Dockhorn-Dworniczak B, Blasius S, Mellin W, Wuisman P, Böcker W, Roessner A. Analysis of mutant P53 protein in osteosarcomas and other malignant and benign lesions of bone. J Cancer Res Clin Oncol. 1993;119:172–8. doi: 10.1007/BF01229533. [DOI] [PubMed] [Google Scholar]
- 32.Radig K, Schneider-Stock R, Haeckel C, Neumann W, Roessner A. p53 gene mutations in osteosarcomas of low-grade malignancy. Hum Pathol. 1998;29:1310–6. doi: 10.1016/s0046-8177(98)90263-5. [DOI] [PubMed] [Google Scholar]
- 33.Kakar S, Mihalov M, Chachlani NA, Ghosh L, Johnstone H. Correlation of c-fos, p53, and PCNA expression with treatment outcome in osteosarcoma. J Surg Oncol. 2000;73:125–6. doi: 10.1002/(sici)1096-9098(200002)73:2<125::aid-jso14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 34.Entz-Werle N, Schneider A, Kalifa C, Voegeli AC, Tabone MD, Marec-Berard P, Marcellin L, Pacquement H, Terrier P, Boutard P, Meyer N, Gaub MP, Lutz P, Babin A, Oudet P. Genetic alterations in primary osteosarcoma from 54 children and adolescents by targeted allelotyping. Br J Cancer. 2003;88:1925–31. doi: 10.1038/sj.bjc.6600968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junior AT, de Abreu Alves F, Pinto CA, Carvalho AL, Kowalski LP, Lopes MA. Clinicopathological and immunohistochemical analysis of twenty-five head and neck osteosarcomas. Oral Oncol. 2003;39:521–30. doi: 10.1016/s1368-8375(03)00017-4. [DOI] [PubMed] [Google Scholar]
- 36.Nakashima H, Nishida Y, Sugiura H, Katagiri H, Yonekawa M, Yamada Y, Iwata H, Nagasaka T, Ishiguro N. Telomerase, p53 and PCNA activity in osteosarcoma. Eur J Surg Oncol. 2003;29:564–7. doi: 10.1016/s0748-7983(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 37.Patino-Garcia A, Piñeiro ES, Díez MZ, Iturriagagoitia LG, Klüssmann FA, Ariznabarreta LS. Genetic and epigenetic alterations of the cell cycle regulators and tumor suppressor genes in pediatric osteosarcomas. J Pediatr Hematol Oncol. 2003;25:362–7. doi: 10.1097/00043426-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Park HR, Jung WW, Bertoni F, Bacchini P, Park JH, Kim YW, Park YK. Molecular analysis of p53, MDM2 and H-ras genes in low-grade central osteosarcoma. Pathol Res Pract. 2004;200:439–45. doi: 10.1016/j.prp.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Wunder JS, Gokgoz N, Parkes R, Bull SB, Eskandarian S, Davis AM, Beauchamp CP, Conrad EU, Grimer RJ, Healey JH, Malkin D, Mangham DC, Rock MJ, Bell RS, Andrulis IL. TP53 mutations and outcome in osteosarcoma: a prospective, multicenter study. J. Clin. Oncol. 2005;23:1483–90. doi: 10.1200/JCO.2005.04.074. [DOI] [PubMed] [Google Scholar]
- 40.Ghule P, Kadam PA, Jambhekar N, Bamne M, Pai S, Nair C, Banavali S, Puri A, Agarwal M. p53 gene gets altered by various mechanisms: studies in childhood sarcomas and retinoblastoma. Med Sci Monit. 2006;12:BR385–396. [PubMed] [Google Scholar]
- 41.Sorensen FB, Jensen K, Vaeth M, Hager H, Funder AM, Safwat A, Keller J, Christensen M. Immunohistochemical Estimates of Angiogenesis, Proliferative Activity, p53 Expression, and Multiple Drug Resistance Have No Prognostic Impact in Osteosarcoma: A Comparative Clinicopathological Investigation. Sarcoma. 2008;2008:874075. doi: 10.1155/2008/874075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakhshi S, Gupta A, Sharma MC, Khan SA, Rastogi S. Her-2/neu, p-53, and their coexpression in osteosarcoma. J Pediatr Hematol Oncol. 2009;31:245–51. doi: 10.1097/MPH.0b013e318197947e. [DOI] [PubMed] [Google Scholar]
- 43.Toffoli G, Biason P, Russo A, De Mattia E, Cecchin E, Hattinger CM, Pasello M, Alberghini M, Ferrari C, Scotlandi K, Picci P, Serra M. Effect of TP53 Arg72Pro and MDM2 SNP309 polymorphisms on the risk of high-grade osteosarcoma development and survival. Clin Cancer Res. 2009;15:3550–6. doi: 10.1158/1078-0432.CCR-08-2249. [DOI] [PubMed] [Google Scholar]
- 44.Jawad SN, Abdullah BH. Proliferative, apoptotic and angiogenic potentials in jaws and long bones osteosarcomas: a comparative immunohistochemical study. J Oral Pathol Med. 2010;39:681–6. doi: 10.1111/j.1600-0714.2010.00923.x. [DOI] [PubMed] [Google Scholar]
- 45.Jiang L, Tao C, He A. Prognostic significance of p53 expression in malignant bone tumors: a meta-analysis. Tumour Biol. 2013;34:1037–43. doi: 10.1007/s13277-012-0643-5. [DOI] [PubMed] [Google Scholar]
- 46.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, Zoubek A, Jürgens H, Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20:776–90. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 47.Petrilli AS, de Camargo B, Filho VO, Bruniera P, Brunetto AL, Jesus-Garcia R, Camargo OP, Pena W, Péricles P, Davi A, Prospero JD, Alves MT, Oliveira CR, Macedo CR, Mendes WL, Almeida MT, Borsato ML, dos Santos TM, Ortega J, Consentino E Brazilian Osteosarcoma Treatment Group Studies III and IV. Results of the Brazilian Osteosarcoma Treatment Group Studies III and IV: prognostic factors and impact on survival. J. Clin. Oncol. 2006;24:1161–8. doi: 10.1200/JCO.2005.03.5352. [DOI] [PubMed] [Google Scholar]
- 48.Ozger H, Eralp L, Atalar AC, Toker B, Ayan I, Kebudi R, Bağbek S, Başaran M, Ağaoğlu F, Dizdar Y, Bilgiç B. [Survival analysis and the effects of prognostic factors in patients treated for osteosarcoma] . Acta Orthop Traumatol Turc. 2007;41:211–9. [PubMed] [Google Scholar]
- 49.Shiraishi S, Tada K, Nakamura H, Makino K, Kochi M, Saya H, Kuratsu J, Ushio Y. Infl uence of p53 mutations on prognosis of patients with glioblastoma. Cancer. 2002;95:249–57. doi: 10.1002/cncr.10677. [DOI] [PubMed] [Google Scholar]
- 50.Borresen-Dale AL. TP53 and breast cancer. Hum Mutat. 2003;21:292–300. doi: 10.1002/humu.10174. [DOI] [PubMed] [Google Scholar]
- 51.Norberg T, Lennerstrand J, Inganäs M, Bergh J. Comparison between p53 protein measurements using the luminometric immunoassay and immunohistochemistry with detection of p53 gene mutations using cDNA sequencing in human breast tumors. Int J Cancer. 1998;79:376–83. doi: 10.1002/(sici)1097-0215(19980821)79:4<376::aid-ijc12>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Shahin MS, Hughes JH, Sood AK, Buller RE. The prognostic significance of p53 tumor suppressor gene alterations in ovarian carcinoma. Cancer. 2000;89:2006–17. doi: 10.1002/1097-0142(20001101)89:9<2006::aid-cncr18>3.3.co;2-z. [DOI] [PubMed] [Google Scholar]
- 53.Knapp G, Biggerstaff BJ, Hartung J. Assessing the amount of heterogeneity in random-effects meta-analysis. Biom J. 2006;48:271–85. doi: 10.1002/bimj.200510175. [DOI] [PubMed] [Google Scholar]


