Abstract
The influence of environmental chemicals including arsenic, a type 1 carcinogen, on the composition and function of the human-associated microbiota is of significance in human health and disease. We have developed a suite of bioinformatics and visual analytics methods to evaluate the availability (presence or absence) and abundance of functional annotations in a microbial genome for seven Pfam protein families: As(III)-responsive transcriptional repressor (ArsR), anion-transporting ATPase (ArsA), arsenical pump membrane protein (ArsB), arsenate reductase (ArsC), arsenical resistance operon transacting repressor (ArsD), water/glycerol transport protein (aquaporins), and universal stress protein (USP). These genes encode function for sensing and/or regulating arsenic content in the bacterial cell. The evaluative profiling strategy was applied to 3,274 genomes from which 62 genomes from 18 genera were identified to contain genes for the seven protein families. Our list included 12 genomes in the Human Microbiome Project (HMP) from the following genera: Citrobacter, Escherichia, Lactobacillus, Providencia, Rhodococcus, and Staphylococcus. Gene neighborhood analysis of the arsenic resistance operon in the genome of Bacteroides thetaiotaomicron VPI-5482, a human gut symbiont, revealed the adjacent arrangement of genes for arsenite binding/transfer (ArsD) and cytochrome c biosynthesis (DsbD_2). Visual analytics facilitated evaluation of protein annotations in 367 genomes in the phylum Bacteroidetes identified multiple genomes in which genes for ArsD and DsbD_2 were adjacently arranged. Cytochrome c, produced by a posttranslational process, consists of heme-containing proteins important for cellular energy production and signaling. Further research is desired to elucidate arsenic resistance and arsenic-mediated cellular energy production in the Bacteroidetes.
Keywords: arsenic, arsenate, arsenite, Bacteroides, Bacteroidetes, bioinformatics, genomes, gut microbiota, heavy metal transport, Human Microbiome Project, human symbiont, mercuric transport, secondary data analysis, visual analytics
Introduction
High-throughput technologies for assaying biological macromolecules and metabolites are providing wealth of data on the structure, function, and condition-induced changes within host-associated microbial communities.1 The influence of environmental chemicals including arsenic, a type 1 carcinogen, on the composition and function of the human-associated microbiota is of significance in human health and disease.2 The data from the Human Microbiome Project (HMP) include genome sequences and functional annotations for over 1000 microbial isolates obtained from diverse body sites of healthy adults.3–5 There is an urgent need for data analytics (modeling and simulation, statistical analysis, and visual analytics) of the wealth of data on the human microbiome for new types of treatment as well as mechanisms of chronic diseases.6–8 The results from data analytics of microbiome data hold promise to advance knowledge of how the human microbiota at body sites respond to ubiquitous environmental chemicals such as arsenic.
The overall theme of our research is to identify and evaluate microbial gene clusters that are equipped for stress response.9 In this article, we report the integration of data on the availability and abundance of genes for arsenic stress response in microbial genomes. We reason that the integration of availability (presence or absence) and abundance of genes for functions can be informative on the microbe’s potential to perform the functions. In the case of influences of arsenic on the human microbiome, knowledge on the availability and abundance of arsenic stress response genes will guide further research on the pre-systemic metabolism of arsenic by the microbiota at the body site.
Arsenic is a naturally occurring environmental chemical, and drinking water and dietary intake are two main routes through which human beings are exposed to it.10,11 Pentavalent (arsenate) and trivalent (arsenite) inorganic arsenic species perturbs the normal cell function.12 The ubiquitous natural occurrence of arsenic means that cells from all domains of life must develop molecular and phenotypic mechanisms to respond to arsenic-induced stress.13–15 Ingested arsenic is a cause of cancers of the skin, lungs, bladder, and kidneys.16 Gut microbial metabolism of arsenate produces the more absorbable and toxic arsenite. The genome sequences of single isolates and microbial communities encode mechanisms by which gut microbiota transforms ingested arsenic to more toxic trivalent methylated and thiolated arsenicals prior to their metabolism in human cells.17,18 Therefore, to make progress on elucidating pre-systemic metabolism of arsenic, it is necessary to identify microbes of the human microbiota with genes for sensing and regulating arsenic.
Exposure of the human microbiota to arsenic presents an unfavorable environment to microbial cells. In microbial genomes, several genes function in the sensing and regulation of inorganic arsenic. We are interested in the genes encoding arsenic resistance operon, the aquaporins, and the universal stress proteins (USPs). These genes encode function for sensing and/or regulating (resistance) arsenic content in the bacterial cell.19 The best-characterized arsenic genes include As(III)-responsive transcriptional repressor (arsR), anion-transporting ATPase (arsA), arsenical pump membrane protein (arsB), arsenate reductase and related proteins, glutaredoxin family (arsC arsenical resistance operon trans-acting repressor (arsD), arylsulfatase family, member H (arsH), putative membrane permease (ArsP), and As(III)-S-adenosylmethionine methyltransferase (arsM).20 The genes for conversion of arsenate to arsenite and arsenite extrusion from the cell are typically organized as operons, such as arsRBC, arsRABC, and arsRDABC, but the genes can also exist alone.20 Proteins for water and/or glycerol transport across cellular membranes termed aquaporins can also function in arsenic transport.21 The USP family is a protein family known to enable bacteria, archaea, fungi, viridiplantae, and certain metazoans that respond to stresses.22–24 The USP family includes proteins that contain 140–160 amino acid (aa) USP domain [PF00582 (or Pfam00582) in the Pfam database].22–24 The domain architecture of USPs can be (i) one USP domain, (ii) two USP domains in tandem, or (iii) one or two USP domains together with other functional domains including transporters, kinases, permeases, transferases, and bacterial sensor.24,25 In Exiguobacterium sp. PS, a Gram-positive bacteria that lacks arsenic reductase activity, a USP was induced by arsenate stress.26 The uspA of Escherichia coli has been evaluated as a sensor to detect chemical toxicants.27
The availability of diverse data from HMP allows for secondary data analytics including constructing profiles of functional annotations for genes involved in arsenic sensing and regulation in the HMP genomes collection. Therefore, we report the development of a genome profiling scheme based on the availability of functional annotations for seven Pfam protein families, including known arsenic resistance operon proteins, aquaporins, and USPs. A list of 62 genomes from 18 genera was identified including 12 genomes in the HMP genomes collection. Several noteworthy findings could be a basis for further investigations. For example, in multiple Bacteroides genomes, a gene for arsenic binding and transfer (arsD) is adjacent to a gene for cytochrome c biogenesis protein. Cytochrome c, produced by a posttranslational process, consists of heme-containing proteins important for cellular energy production and signaling.28,29 Previous reported research on Bacteroides and arsenic appears to be limited to phenotypic characterization of susceptibilities to arsenate in which 25% of strains in the Bacteroides fragilis group, which included Bacteroides thetaiotaomicron, were resistant to 0.01 M arsenate.30 Further research is desired to elucidate arsenic resistance and arsenic-mediated cellular energy production in the Bacteroidetes.
Methods
Construction of functional annotation profiles for arsenic stress response of microbial genomes
We assembled a list of protein family functions (Pfam) that are known to participate in the metabolism of arsenic and in stress response in bacteria and archaea. The Pfam identifiers, names, and common abbreviation of the proteins are Pfam02374 [anion-transporting ATPase (ArsA)]; Pfam02040 [arsenical pump membrane protein (ArsB)]; Pfam03960 [arsenate reductase and related proteins, glutaredoxin family (ArsC)]; Pfam06953 [arsenical resistance operon trans-acting repressor (ArsD)]; Pfam01022 [As(III)-responsive transcriptional repressor (ArsR)]; Pfam00230 [major intrinsic protein family (MIP/AQP)]; and Pfam00582 [universal stress protein domain (Usp)]. Genomes with a profile of interest were further grouped into relevance annotation (eg, agriculture, biotechnology, human pathogen, and medical) provided by the Integrated Microbial Genomes (IMG) system. When relevance is not annotated, we tagged the genome as “Not_Reported.” A binary matrix that encodes the presence (1) or absence (0) of a relevance annotation for selected genomes was constructed. The binary matrix was visualized with matrix2png.31
Integration of availability and abundance of genes for arsenic metabolism and USPs
Genes annotated for presence of the above seven protein families in microbial genomes were initially retrieved from the IMG system (http://img.jgi.doe.gov/) in December 2011 (IMG version 3.5).32 The datasets were downloaded as Excel spreadsheets and integrated in a visual analytics software (Tableau Desktop Professional, Seattle, WA). The dataset includes several fields including the genome name (Genome) and the gene object identifier (Gene Object ID). The visual analytics tool displayed the availability (presence or absence) and abundance (number of genes annotated with the Pfam function) of each microbial genome. A list of reference genomes sequenced by the HMP was obtained from the HMP catalog (http://www.hmpdacc.org/catalog/).
Because B. thetaiotaomicron is a dominant symbiont of the gut of humans and other mammals,33 we decided to determine the abundance of genes for the arsenic resistance operon (arsRABCD) in the Bacteroides genomes in the dataset. A visualization was also generated to provide an overview of the abundance of the arsenic resistance genes in Bacteroides genomes.
Functional associations of ArsD encoded transcription units in B. thetaiotaomicron
The gene content of the transcription unit with arsenic-associated genes was determined for B. thetaiotaomicron using the BioCyc database collection of pathway/genome databases (PGDBs).34 In BioCyc, a transcription unit is defined as a set of one or more genes that are transcribed to produce a single messenger RNA. Our particular interest is in multigene transcription units. Based on the observed functions in the transcription units, the presence of two Pfam functions in a chromosomal cassette was determined with the Cassette search tool of the IMG system.32 In the IMG system, a chromosomal cassette is defined as a stretch of protein coding genes with intergenic distance lesser than or equal to 300 base pairs.
Known and predicted functional associations of proteins encoded in a transcription unit were retrieved from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database.35 With this approach, we expect that the STRING database will provide a score for gene neighborhood evidence for genes in a transcription unit. Other types of evidence that are used in the STRING database to calculate a combined score are gene fusion, co-occurrence, co-expression, experiment, databases, text mining, and homology.35
Results
Arsenic stress response profiles for genomes
A seven-digit binary code was assigned to 3,274 genomes (119 archaea, 3033 bacteria, and 122 eukaryota) obtained from the IMG system. The order of the seven Pfam families in the binary code is ArsA, ArsB, ArsC, ArsD, ArsR, Aqp, and Usp (Table 1). As functional annotations of genes could change, we selected only genomes that have the complete profile (111111111). A total of 62 microbial genomes from 18 genera had a binary code in which all the seven Pfam families were present. We further grouped the genomes according to their relevance (eg, agriculture, biotechnology, human pathogen, and medical) to help direct further research. A subset of 57 genomes with the complete profile was mapped to 22 assignments of relevance. Additionally, five genomes did not have an assignment of relevance and their assignment was designated “Not_Reported”. A visualization of the matrix of 23-digit binary signatures was constructed for 62 genomes (Fig. 1).
Table 1.
Position of Pfam protein family annotation in genome binary profile.
| Pfam ID | Pfam NAME AND ABBREVIATION | POSITION IN BINARY DIGIT |
|---|---|---|
| Pfam02374 | Anion-transporting ATPase [ArsA] | 1 |
| Pfam02040 | Arsenical pump membrane protein [ArsB] | 2 |
| Pfam03960 | Arsenate reductase and related proteins, glutaredoxin family [ArsC] | 3 |
| Pfam06953 | Arsenical resistance operon trans-acting repressor, [ArsD] | 4 |
| Pfam01022 | As(III)-responsive transcriptional repressor [ArsR] | 5 |
| Pfam00230 | Major intrinsic protein family [MIP/AQP] | 6 |
| Pfam00582 | Universal stress protein domain [Usp] | 7 |
Figure 1.
Visualization of binary-encoded matrix for relevance of genomes with genes for arsenic operon, aquaporin, and universal stress protein. Data were obtained from the Integrated Microbial Genomes. Black square, relevance annotated for genome; white square, relevance not annotated for genome. When relevance data were not available, we entered a “Not_Reported” for data processing.
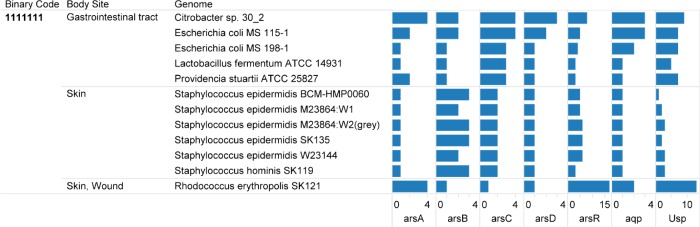
The 12 HMP reference genomes with the complete profile were grouped in the body sites: gastrointestinal tract, skin, and skin wound. The genomes with the complete binary profile from the human gastrointestinal tract were Citrobacter sp. 30_2, E. coli MS 115-1, E. coli MS 198-1, Lactobacillus fermentum ATCC 14931, and Providencia stuartii ATCC 25827. Additionally, the selected genomes based on the complete seven-digit profile and isolated from the skin were five strains of Staphylococcus epidermidis, Staphylococcus hominis SK119, and Rhodococcus erythropolis SK121. The locus tags for genes encoding the protein families are presented in Table 2. The locus tags are presented in the sequence of the arsRDABC operon (Table 2). The arrangement of the genes with reference to their function is presented in Table 3. In Citrobacter sp. 30_2, two arsenic resistance gene clusters were identified with the same gene order of arsRDABC. Overall, a pattern in Table 3 is that arsA and arsD when present are adjacent for all the genomes from the four Gram-negative bacteria. However, in L. fermentum ATCC 14931, the gene cluster order was arsRABDC.
Table 2.
Locus tags for genes encoding selected arsenic-associated protein families in five Human Microbiome Project reference genomes.
| Pfam FAMILY | CITROBACTER SP. 30_2 | ESCHERICHIA COLI MS 115-1 | ESCHERICHIA COLI MS 198-1 | LACTOBACILLUS FERMENTUM ATCC 14931 | PROVIDENCIA STUARTII ATCC 25827 |
|---|---|---|---|---|---|
| arsR (Pfam01022) | CSAG_00049 | HMPREF9540_00434 | HMPREF9552_00168 | HMPREF0511_0214 | PstuA_020100015920 |
| CSAG_00058 | HMPREF9540_00675 | HMPREF9552_02803 | HMPREF0511_1131 | PstuA_020100016320 | |
| CSAG_00761 | HMPREF9540_01104 | HMPREF9552_02903 | HMPREF0511_1475 | PstuA_020100017025 | |
| CSAG_02502 | HMPREF9540_04804 | HMPREF9552_02908 | |||
| CSAG_04185 | HMPREF9540_04813 | ||||
| CSAG_04189 | |||||
| CSAG_04238 | |||||
| CSAG_04297 | |||||
| arsD (Pfam06953) | CSAG_00050 | HMPREF9540_04807 | HMPREF9552_02907 | HMPREF0511_1134 | PstuA_020100016315 |
| CSAG_00055 | HMPREF9540_04812 | ||||
| CSAG_04239 | |||||
| arsA (Pfam02374) | CSAG_00051 | HMPREF9540_04808 | HMPREF9552_02906 | HMPREF0511_1132 | PstuA_020100007500 |
| CSAG_00054 | HMPREF9540_04811 | PstuA_020100016310 | |||
| CSAG_04240 | |||||
| CSAG_04243 | |||||
| arsB (Pfam02040) | CSAG_00052 | HMPREF9540_00433 | HMPREF9552_02905 | HMPREF0511_1133 | PstuA_020100016305 |
| CSAG_04241 | HMPREF9540_04810 | ||||
| arsC (Pfam03960) | CSAG_00053 | HMPREF9540_00432 | HMPREF9552_01835 | HMPREF0511_0280 | PstuA_020100013100 |
| CSAG_02267 | HMPREF9540_04809 | HMPREF9552_01863 | HMPREF0511_0923 | PstuA_020100013225 | |
| CSAG_02283 | HMPREF9540_05016 | HMPREF9552_02904 | HMPREF0511_0962 | PstuA_020100016300 | |
| CSAG_04242 | HMPREF9540_05044 | ||||
| Aqp (Pfam00230) | CSAG_01847 | HMPREF9540_02867 | HMPREF9552_00087 | HMPREF0511_1378 | PstuA_020100002768 |
| CSAG_01948 | HMPREF9540_03890 | HMPREF9552_03971 | |||
| CSAG_04569 | HMPREF9540_04727 | ||||
| Usp (Pfam00582) | CSAG_00404 | HMPREF9540_00348 | HMPREF9552_01620 | HMPREF0511_0613 | PstuA_020100008480 |
| CSAG_00475 | HMPREF9540_00443 | HMPREF9552_02920 | HMPREF0511_1339 | PstuA_020100010015 | |
| CSAG_01459 | HMPREF9540_00492 | HMPREF9552_03271 | HMPREF0511_1387 | PstuA_020100010755 | |
| CSAG_01471 | HMPREF9540_01169 | HMPREF9552_03967 | HMPREF0511_1569 | PstuA_020100010765 | |
| CSAG_01741 | HMPREF9540_02863 | HMPREF9552_04168 | HMPREF0511_1702 | PstuA_020100011480 | |
| CSAG_03714 | HMPREF9540_04247 | HMPREF9552_05110 | PstuA_020100015380 | ||
| CSAG_03977 | HMPREF9540_04414 | HMPREF9552_05202 | PstuA_020100019714 | ||
| CSAG_04126 | |||||
| CSAG_00328 | |||||
Table 3.
Comparison of gene function order in arsenic resistance operon in selected Human Microbial Project reference genomes.
| GENOME | GENE CLUSTER IDENTIFIER* | GENE ORDER | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Citrobacter sp. 30_2 | CSAG_00049-CSAG_00053 | arsR | arsD | arsA | arsB | arsC |
| CSAG_00054-CSAG_00055 | arsA | arsD | ||||
| CSAG_04238-CSAG_04242 | arsR | arsD | arsA | arsB | arsC | |
| CSAG_04243-CSAG_04243 | arsA | |||||
| Escherichia coli MS 115-1 | HMPREF9540_00432-HMPREF9540_00434 | arsC | arsB | arsR | ||
| HMPREF9540_04807-HMPREF9540_04810 | arsD | arsA | arsC | arsB | ||
| HMPREF9540_04811-HMPREF9540_04813 | arsA | arsD | arsR | |||
| Escherichia coli MS 198-1 | HMPREF9552_02903-HMPREF9552_02907 | arsR | arsC | arsB | arsA | arsD |
| HMPREF9552_02908-HMPREF9552_02908 | arsR | |||||
| Lactobacillus fermentum ATCC 14931 | HMPREF0511_1131-HMPREF0511_1134 | arsR | arsA | arsB | arsD | |
| Providencia stuartii ATCC 25827 | PstuA_020100016300-PstuA_020100016320 | arsC | arsB | arsA | arsD | arsR |
Note:
Start and end genes are used to identify gene clusters.
Integration of availability and abundance of genes for arsenic sensing and regulation
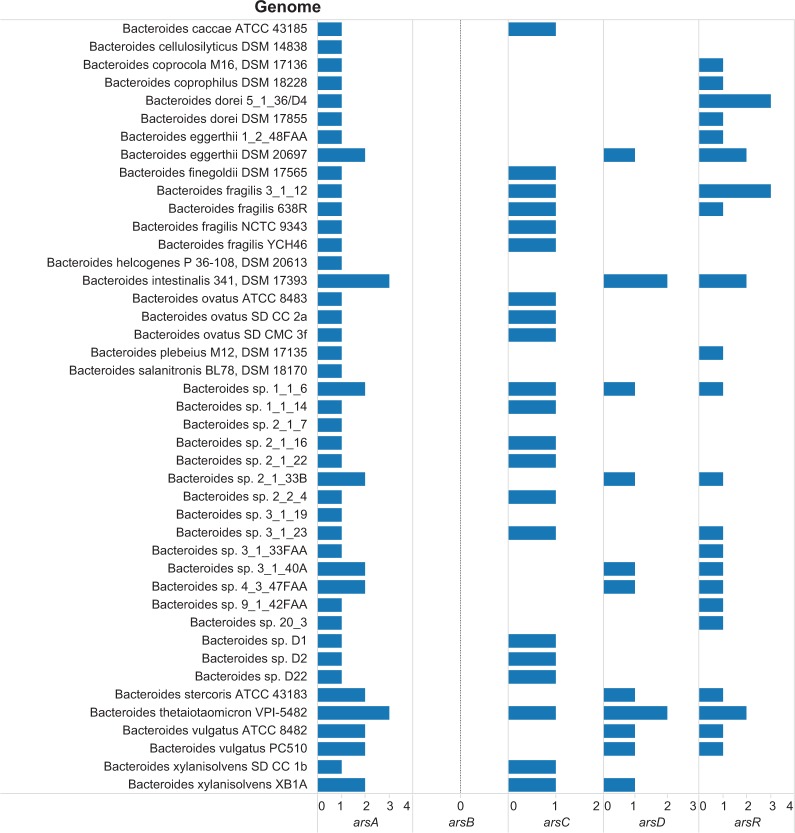
The integration of data fields from several data sources was accomplished through visual analytics tasks. Figure 2 is a visualization that integrates the data on binary code; body site; genome; and the availability of Pfam annotations and abundance (number of genes) for 12 reference genomes sequenced by the HMP. Several patterns can be identified from Figure 2. As gastrointestinal pre-systemic metabolism is an essential step of arsenic metabolism in humans, we further investigated the abundance of arsenic resistance genes in 43 Bacteroides genomes (Fig. 3). There were several noteworthy findings from the visualization including (i) multiple copies of arsA, arsD, and arsR were observed in the genomes of Bacteroides intestinalis 341, DSM 17393, and B. thetaiotaomicron VPI-5482; (ii) all the Bacteroides genomes did not include the annotation for arsB (Pfam2040, arsB); and (iii) in 21 Bacteroides genomes, only one arsC gene per genome was annotated. Clearly, the B. intestinalis and B. thetaiotaomicron strains have multiple arsA, arsD, and arsR, which is indicative of the presence of at least two arsenic resistance operons. The genomic context of the genes in the arsenic operons of B. thetaiotaomicron is presented in Figure 4. Further analysis of the Pfam domain composition of the three genes annotated with the Pfam for the arsA gene revealed that two genes (BT_0116 and BT_0802) had only the Pfam02374 annotation, whereas BT_3895 had a protein domain annotation of Pfam02374 (arsA) and Pfam10609 (ParA/MinD ATPase like).
Figure 2.
Genomes in the Human Microbiome Project (HMP) genomes collection with genes for arsenic operon, aquaporin, and universal stress protein.
Notes: Binary code is based on the presence of seven protein families: Pfam02374 [anion-transporting ATPase (ArsA)]; Pfam02040 [arsenical pump membrane protein (ArsB)]; Pfam03960 [arsenate reductase and related proteins, glutaredoxin family (ArsC)]; Pfam06953 [arsenical resistance operon trans-acting repressor (ArsD)]; Pfam01022 [As(III)-responsive transcriptional repressor (ArsR)]; Pfam00230 [major intrinsic protein family (MIP/AQP)]; and Pfam00582 [universal stress protein domain (Usp)].
Figure 3.
Abundance of genes for arsenic-associated genes in genomes of Bacteroides species.
Notes: The horizontal axis has the count for arsenic-associated genes in the genome. The scale for each Pfam family varies according to the maximum abundance observed in the genomes evaluated. The genes and their Pfam encoding are arsA, Pfam02374 (anion-transporting ATPase); arsB, Pfam02040 (arsenical pump membrane protein); arsC, Pfam03960 (arsenate reductase and related proteins, glutaredoxin family); arsD, Pfam06953 (arsenical resistance operon trans-acting repressor); and arsR, Pfam01022 (As(III)-responsive transcriptional repressor).
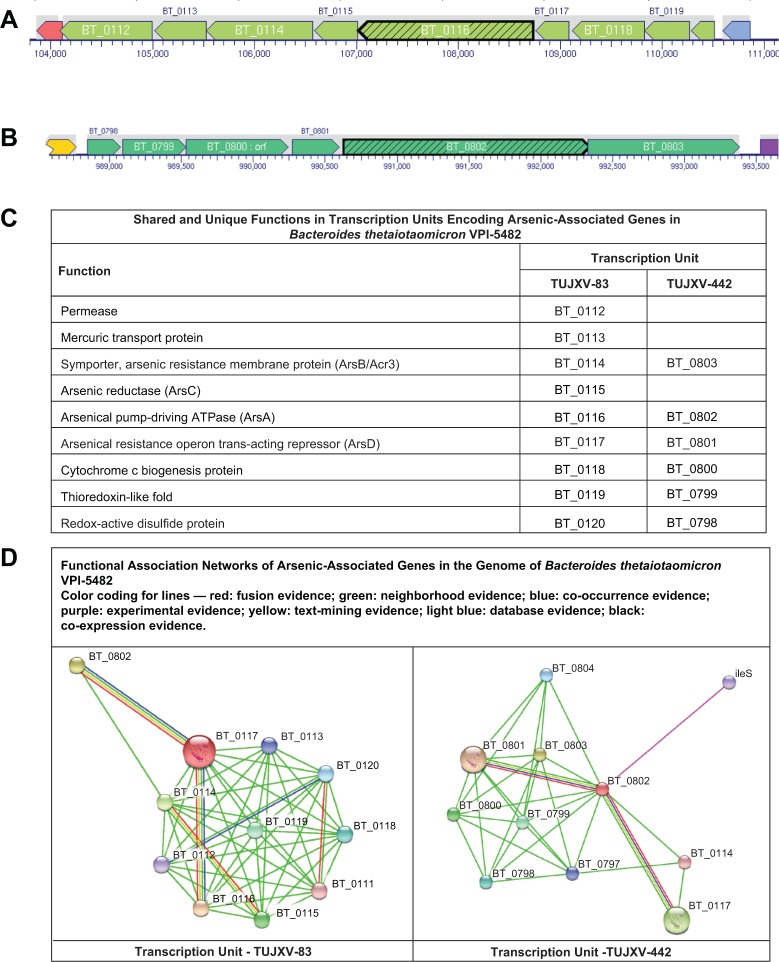
Figure 4.
Transcription units and functional associations of arsenic resistance operon in Bacteroides thetaiotaomicron VPI-5482. The web pages for the transcription units are http://biocyc.org/BTHE226186/NEW-IMAGE?type=OPERON&object=TUJXV-83 and http://biocyc.org/BTHE226186/NEW-IMAGE?type=OPERON&object=TUJXV-442.
Functional associations of ArsD-encoded transcription units in B. thetaiotaomicron
Figure 4 provides an overview of the two transcription units, the predicted function of proteins, and the predicted protein–protein networks involving arsenic-associated genes for B. thetaiotaomicron VPI-5482. The two arsD genes (BT_0117 and BT_0801) in B. thetaiotaomicron VPI-5482 are located on two transcription units, which are, respectively, labeled as TUJXV-83 and TUJXV-442 in BioCyc, respectively. These transcription units have nine and six genes, respectively (Fig. 4A and B). As shown in Figure 4C, both transcription units have the genes for homologs of arsA (BT_0116; BT_0802), acr3 [homologous to arsB] (BT_0114; BT_0803), and arsD (BT_0117; BT_0801). TUJXV-83 is unique for a permease (BT_0113), mercuric transport protein (BT_0114), and arsenic reductase (BT_0115). Three additional proteins encoded in both transcription units are cytochrome c biogenesis protein (BT_0118; BT_0800), protein with thioredoxin-like fold (BT_0119; BT_0799), and redox-active disulfide protein (BT_0120; BT_0798).
In both the transcription units, the arsD gene was adjacent to the gene for a cytochrome c biogenesis protein DsbD_2 (Pfam13386) and an anion-transporting ATPase arsA (Pfam06953). Using the IMG system, a search of chromosomal cassettes with Pfam06953 and Pfam13386 in 2,841 finished bacterial genomes identified cassettes in the following nine genomes: B. thetaiotaomicron VPI-5482; Bacteroides vulgatus ATCC 8482; Bacteroides xylanisolvens XB1A; Porphyromonas asaccharolytica VPI 4198, DSM 20707; Prevotella melaninogenica ATCC 25845; Shewanella putrefaciens 200; S. putrefaciens CN-32; Shewanella sp. ANA-3; and Shewanella sp. W3-18-1. It was only in the Bacteroidetes genomes (Bacteroides, Poryphyromonas, and Prevotella) that genes for ArsD and DsbD_2 were adjacent.
The gene for ArsD proteins (BT_0117 and BT_0801) of B. thetaiotaomicron VPI-5482 was selected as input proteins for generation of protein–protein interaction network. As expected, the generated networks (Fig. 4D) include the genes in the transcription units that had the neighborhood evidence (green line). The interaction between BT_0117 (arsD) and BT_0116 (arsA) has multiple types of evidence and is expected. A predicted interaction of BT_0120 (redox-active disulfide protein) had co-occurrence and fusion evidence types with BT_0112 (a permease) and BT_0110 (hypothetical protein), respectively. There was experimental evidence for the interaction between homologs of BT_0802 (arsA) and ileS (BT_0806; isoleucyl-tRNA synthetase).
Discussion
We have provided an integrated view of relevance of 62 genomes from 18 genera with genes for arsenic operon, aquaporin, and USPs (Fig. 2). A set of 12 bacteria genomes in the HMP collection5 was identified to have genes encoding seven protein families defined in this research as relevant to arsenic sensing and regulation (Table 1). The following bacteria in the HMP reference genomes are associated with the gastrointestinal tract: Citrobacter sp. 30_2, E. coli MS 115-1, E. coli MS 198-1, L. fermentum ATCC 14931, and P. stuartii ATCC 25827. Citrobacter sp. 30_2 is a Gram-negative isolate from an intestinal biopsy specimen of patient with Crohn’s disease.36
The Citrobacter genus are rod shaped, motile, non-spore forming members of the family Enterobacteriaceae that use citrate as their sole source of carbon.37,38 In terms of arsenic metabolism, Citrobacter sp. NC-1 isolated from soil contaminated with arsenic at levels as high as 5,000 mg. As kg−1 was able to reduce 20 mM arsenate within 24 h.39 A comparison of Citrobacter sp. 30_2 and Citrobacter UC1CIT strains from a premature infant revealed the presence of an arsenic operon unique to strain 30_2.36 The two E. coli strains identified are part of the project on human gut microbiota and Crohn’s disease (http://genome.wustl.edu/projects/). An indication that the genomic composition of E. coli MS 115-1 is equipped for environmental fitness is the presence of the clpK gene for thermal resistance in Klebsiella pnuemoniae.36 E. coli MS 115-1 is one of the two E. coli strains with the clpK gene. In Table 3, we observed that arsA and arsD were adjacent for all the four Gram-negative bacteria. However, in L. fermentum ATCC 14931, the arsB and arsD were adjacent. A systematic evaluation of more than 19,000 bacterial genomes could provide additional examples of this gene adjacency. Functional analysis with molecular techniques could also elucidate impact of the adjacency on arsenic extrusion.
We have included the USPs as markers for arsenic exposure because of (i) prior research that support induction of USPs by arsenic and (ii) proximity of arsenic-associated genes and genes for USPs. Arsenite early exposure (15 minutes) induced the transcription of two usp genes in Herminiimonas arsenicoxydans, a bacterium isolated from arsenic-contaminated sludge.40 A usp gene and an arsenic resistance operon are located on an antibiotic-resistant island in the genome of Acinetobacter baumannii, an opportunistic pathogen that causes nosocomial infections.41 We have observed that the genome of Bacillus cereus Q1 contains a usp gene that is adjacent and in the same transcription direction with a gene with predicted function for HTH ArsR-type DNA-binding domain (Inter-Pro Database Identifier: IPR001845).9 Further research is needed to better define the relationship between expression of USP genes and the level of arsenic exposure. Additionally, investigations are desired in the context of arsenic sensing to compare the speed of expression of USP genes and arsenic resistance operon (ars) genes.
In the Gram-negative colon inhabiting B. thetaiotaomicron VPI-5482, two transcription units include arsenic resistance genes (ars) (Fig. 4). Investigations to confirm these genes would help define mechanisms for arsenic sensing and regulation by B. thetaiotaomicron VPI-5482, which is able to acquire and utilize indigestible dietary polysaccharides.42 Multiple copies of ars genes in B. thetaiotaomicron VPI-5482 are consistent with expansion of paralogous genes and the species environmental sensing abilities needed to adapt to changing ecosystems.33 Only genomes categorized under the phylum Bacteroidetes contain genes encoding for the cytochrome c biogenesis protein adjacent to the arsD gene protein is adjacent to the arsD gene only genomes of the Bacteroidetes phylum (Fig. 3). Cytochrome c, produced by a posttranslational process, consists of heme-containing proteins important for cellular energy production and signaling.28,29 The arsD controls the maximal expression of the arsenic-resistant operon (arsRDABC).43 The ArsD metallochaperone protein delivers arsenite to ArsA efflux pump.44,45 The significance of the adjacency of cytochrome c biogenesis protein and the metallochaperone needs further investigation. In Shewanella putrefaciens strain CN-2, a subunit of c-type cytochrome (CymA) that is present in anaerobic conditions functions in conjunction with a known respiratory arsenate reductase.46
Through additional functional annotation data curation, we noted the presence of a gene for mercuric transport protein (Locus Tag: BT_0113; UniProt Accession: Q8ABJ7) in one of the BioCyc transcription units (TUJXV-83) of the B. thetaiotaomicron VPI-5482 (Fig. 4). Predictions available at OrthoDB indicate that the gene encodes a mercuric transport protein (http://cegg.unige.ch/orthodb/results?searchtext=Q8ABJ7).47 Genes BT_0112 and BT_0114 encode transport functions. The proteins encoded by the genes BT_0112, BT_0113, and BT_0114 could be investigated for mechanisms of heavy metal transport in B. thetaiotaomicron.
The focus of the evaluative profiling scheme was limited to seven Pfam annotations. Thus, certain functions that are arsenic associated would not be evaluated. For example, in the Bacteroides genomes, annotation for ArsB (Pfam02040; arsB) was not observed (Fig. 3). The annotation available in the genomes was the ACR3 form. Furthermore, in L. fermentum ATCC 14931, arsC (HMPREF0511_1135) in the arsenic resistance operon (HMPREF0511_1131 to HMPREF0511_1135) was not annotated with the Pfam family Pfam0396, but the annotation observed was Pfam01451. Our evaluative scheme assessed the arsenic reductase genes annotated with Pfam03960. Further development of the evaluative profiling for arsenic sensing and regulation would be more comprehensive using the arsenic-related gene families: cytoplasmic AsV reduction (ars), periplasmic AsV reduction (arr), arsenite oxidation (aio), and arsenite methylation (arsM).48 Finally, the evaluative profiles will account for instances where multiple Pfam families map to a gene as with arsC and arsB.
Conclusion
In conclusion, we have developed a suite of bioinformatics and visual analytics methods to evaluate the availability (presence or absence) and abundance of functional annotations in a microbial genome for seven Pfam protein families: As(III)-responsive transcriptional repressor (ArsR); anion-transporting ATPase (ArsA); arsenical pump membrane protein (ArsB); arsenate reductase (ArsC); arsenical resistance operon trans-acting repressor (ArsD); water/glycerol transport protein (aquaporins); and USP. We identified 62 genomes from 18 genera that have genes for all the seven protein families. Our list included 12 genomes in the HMP reference genomes from the following genera Citrobacter, Escherichia, Lactobacillus, Providencia, Rhodococcus, and Staphylococcus. The use of visual analytics methods makes it possible to include additional arsenic-associated protein families in the profiling scheme. Finally, investigations are desired on the arsenic sensing and regulatory systems in members of the Bacteroidetes phylum.
Footnotes
Author Contributions
Conceived and designed the experiments: RDI, UKU, SSS, ALH, AEC, and ON. Analyzed the data: RDI, UKU, SSS, ALH, AEC, SAO, OAH, ZAK, OAE, DEF, and ON. Wrote the first draft of the manuscript: UKU, RDI, and SSS. Contributed to the writing of the manuscript: RDI, UKU, SSS, ALH, AEC, SAO, OAH, ZAK, OAE, DEF, and ON. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: Raul Rivas, Editor in Chief
FUNDING: This work was funded by Office of the Provost, Bethune-Cookman University, National Institutes of Health (NIH/NIMHD 1P20MD006899, NIH/NIGMS 5T36GM095335, NIH/NIMHD G12MD007581), National Science Foundation (EPS-0903787, DBI-1062057),and U.S. Department of Homeland Security Science and Technology Directorate: 2011-ST-062-000048. The H3Africa Bioinformatics Network (H3ABioNet) is funded by NIH Common Fund Award/NHGRI Grant Number U41HG006941. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the funding agencies.
COMPETING INTERESTS: Authors disclose no potential competing interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties.
REFERENCES
- 1.Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74(3):453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu K, Mahbub R, Cable PH, et al. Gut microbiome phenotypes driven by host genetics affect arsenic metabolism. Chem Res Toxicol. 2014;27(2):172–174. doi: 10.1021/tx400454z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449(7164):804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science. 2010;328(5981):994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace BD, Redinbo MR. The human microbiome is a source of therapeutic drug targets. Curr Opin Chem Biol. 2013;17(3):379–384. doi: 10.1016/j.cbpa.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 8.Kong LC, Tap J, Aron-Wisnewsky J, et al. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am J Clin Nutr. 2013;98(1):16–24. doi: 10.3945/ajcn.113.058743. [DOI] [PubMed] [Google Scholar]
- 9.Williams BS, Isokpehi RD, Mbah AN, et al. Functional annotation analytics of Bacillus genomes reveals stress responsive acetate utilization and sulfate uptake in the biotechnologically relevant Bacillus megaterium. Bioinform Biol Insights. 2012;6:275–286. doi: 10.4137/BBI.S7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas A, Deb D, Ghose A, et al. Dietary arsenic consumption and urine arsenic in an endemic population: response to improvement of drinking water quality in a 2-year consecutive study. Environ Sci Pollut Res. 2014;21(1):609–619. doi: 10.1007/s11356-013-1947-8. [DOI] [PubMed] [Google Scholar]
- 11.Johnson MO, Cohly HH, Isokpehi RD, Awofolu OR. The case for visual analytics of arsenic concentrations in foods. Int J Environ Res Public Health. 2010;7(5):1970–1983. doi: 10.3390/ijerph7051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beauchamp EM, Serrano R, Platanias LC. Regulatory effects of arsenic on cellular signaling pathways: biological effects and therapeutic implications. In: Kumar R, editor. Nuclear Signaling Pathways and Targeting Transcription in Cancer. New York: Springer; 2014. pp. 107–119. [Google Scholar]
- 13.Sharples JM, Meharg AA, Chambers SM, Cairney JW. Mechanism of arsenate resistance in the ericoid mycorrhizal fungus Hymenoscyphus ericae. Plant Physiol. 2000;124(3):1327–1334. doi: 10.1104/pp.124.3.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol. 1995;177(8):2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu SF, Chen PY, Nguyen QT, et al. Transcriptome profiling of genes and pathways associated with arsenic toxicity and tolerance in Arabidopsis. BMC Plant Biol. 2014;14(1):94. doi: 10.1186/1471-2229-14-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberoi S, Barchowsky A, Wu F. The global burden of disease for skin, lung and bladder cancer caused by arsenic in food. Cancer Epidemiol Biomarkers Prev. 2014;23(7):1–8. doi: 10.1158/1055-9965.EPI-13-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alava P, Tack F, Laing GD, Van de Wiele T. Arsenic undergoes significant speciation changes upon incubation of contaminated rice with human colon micro biota. J Hazard Mater. 2013;262:1237–1244. doi: 10.1016/j.jhazmat.2012.05.042. [DOI] [PubMed] [Google Scholar]
- 18.Van de Wiele T, Gallawa CM, Kubachka KM, et al. Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environ Health Perspect. 2010;118:1004–1009. doi: 10.1289/ehp.0901794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Jeon B, Sahin O, Zhang Q. Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Appl Environ Microbiol. 2009;75(15):5064–5073. doi: 10.1128/AEM.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castillo R, Saier MH. Functional promiscuity of homologues of the bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H-C, Cheng J, Finan TM, Rosen BP, Bhattacharjee H. Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti. J Bacteriol. 2005;187(20):6991–6997. doi: 10.1128/JB.187.20.6991-6997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mbah AN, Mahmud O, Awofolu OR, Isokpehi RD. Inferences on the biochemical and environmental regulation of universal stress proteins from Schistosomiasis parasites. Adv Appl Bioinform Chem. 2013;6:15. doi: 10.2147/AABC.S37191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isokpehi RD, Simmons SS, Cohly HH, Ekunwe SI, Begonia GB, Ayensu WK. Identification of drought-responsive universal stress proteins in viridiplantae. Bioinform Biol Insights. 2011;5:41–58. doi: 10.4137/BBI.S6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachin L, Nannmark U, Nyström T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol. 2005;187(18):6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvint K, Nachin L, Diez A, Nyström T. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol. 2003;6(2):140–145. doi: 10.1016/s1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 26.Sacheti P, Bhonsle H, Patil R, Kulkarni MJ, Srikanth R, Gade W. Arsenomics of Exiguobacterium sp. PS (NCIM 5463) RSC Adv. 2013;3(25):9705–9713. [Google Scholar]
- 27.Van Dyk TK, Smulski DR, Reed TR, Belkin S, Vollmer AC, LaRossa RA. Responses to toxicants of an Escherichia coli strain carrying a uspA’: lux genetic fusion and an E. coli strain carrying a grpE’: lux fusion are similar. Appl Environ Microbiol. 1995;61(11):4124–4127. doi: 10.1128/aem.61.11.4124-4127.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Travaglini-Allocatelli C. Protein machineries involved in the attachment of heme to cytochrome c: protein structures and molecular mechanisms. Scientifica. 2013;2013:505714. doi: 10.1155/2013/505714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavridou DA, Ferguson SJ, Stevens JM. Cytochrome c assembly. IUBMB Life. 2013;65(3):209–216. doi: 10.1002/iub.1123. [DOI] [PubMed] [Google Scholar]
- 30.Riley T, Mee B. Susceptibility of Bacteroides spp. to heavy metals. Antimicrob Agents Chemother. 1982;22(5):889–892. doi: 10.1128/aac.22.5.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19(2):295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 32.Markowitz VM, Chen IM, Palaniappan K, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(D1):D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comstock LE, Coyne MJ. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. Bioessays. 2003;25(10):926–929. doi: 10.1002/bies.10350. [DOI] [PubMed] [Google Scholar]
- 34.Caspi R, Altman T, Billington R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2014;42(D1):D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(D1):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morowitz MJ, Denef VJ, Costello EK, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci U S A. 2011;108(3):1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borenshtein D, Schauer DB. The genus Citrobacter. In: Dworkin M, et al., editors. The Prokaryotes. New York: Springer; 2006. pp. 90–98. [Google Scholar]
- 38.O’Hara CM, Roman SB, Miller JM. Ability of commercial identification systems to identify newly recognized species of Citrobacter. J Clin Microbiol. 1995;33(1):242–245. doi: 10.1128/jcm.33.1.242-245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang YC, Nawata A, Jung K, Kikuchi S. Isolation and characterization of an arsenate-reducing bacterium and its application for arsenic extraction from contaminated soil. J Ind Microbiol Biotechnol. 2012;39(1):37–44. doi: 10.1007/s10295-011-0996-6. [DOI] [PubMed] [Google Scholar]
- 40.Cleiss-Arnold J, Koechler S, Proux C, et al. Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics. 2010;11(1):709. doi: 10.1186/1471-2164-11-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299(5615):2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Rosen BP. Metalloregulatory properties of the ArsD repressor. J Biol Chem. 1997;272(22):14257–14262. doi: 10.1074/jbc.272.22.14257. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y-F, Yang J, Rosen BP. ArsD residues Cys12, Cys13, and Cys18 form an As (III)-binding site required for arsenic metallochaperone activity. J Biol Chem. 2007;282(23):16783–16791. doi: 10.1074/jbc.M700886200. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Salam A, Ajees A, Rosen BP. Genetic mapping of the interface between the ArsD metallochaperone and the ArsA ATPase. Mol Microbiol. 2011;79(4):872–881. doi: 10.1111/j.1365-2958.2010.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy JN, Saltikov CW. The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J Bacteriol. 2007;189(6):2283–2290. doi: 10.1128/JB.01698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41(D1):D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Zhang L, Wang G. Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PLoS One. 2014;9(3):e92236. doi: 10.1371/journal.pone.0092236. [DOI] [PMC free article] [PubMed] [Google Scholar]