Abstract
Background
Francisella isolates from patients suffering from tularemia in Germany are generally strains of the species F. tularensis subsp. holarctica. To our knowledge, no other Francisella species are known for Germany. Recently, a new Francisella species could be isolated from a water reservoir of a cooling tower in Germany.
Results
We identified a Francisella sp. (isolate W12-1067) whose 16S rDNA is 99% identical to the respective nucleotide sequence of the recently published strain F. guangzhouensis. The overall sequence identity of the fopA, gyrA, rpoA, groEL, sdhA and dnaK genes is only 89%, indicating that strain W12-1067 is not identical to F. guangzhouensis. W12-1067 was isolated from a water reservoir of a cooling tower of a hospital in Germany. The growth optimum of the isolate is approximately 30°C, it can grow in the presence of 4–5% NaCl (halotolerant) and is able to grow without additional cysteine within the medium. The strain was able to replicate within a mouse-derived macrophage-like cell line. The whole genome of the strain was sequenced (~1.7 mbp, 32.2% G + C content) and the draft genome was annotated. Various virulence genes common to the genus Francisella are present, but the Francisella pathogenicity island (FPI) is missing. However, another putative type-VI secretion system is present within the genome of strain W12-1067.
Conclusions
Isolate W12-1067 is closely related to the recently described F. guangzhouensis species and it replicates within eukaryotic host cells. Since W12-1067 exhibits a putative new type-VI secretion system and F. tularensis subsp. holarctica was found not to be the sole species in Germany, the new isolate is an interesting species to be analyzed in more detail. Further research is needed to investigate the epidemiology, ecology and pathogenicity of Francisella species present in Germany.
Keywords: Francisella isolate, W12-1067, Cooling tower, Genome sequence, Pathogenicity island FPI, Environmental, Legionella, Germany
Background
Francisella tularensis is an facultative intracellular pathogen that causes tularemia in humans and a wide range of animals [1]. Strains of F. tularensis subsp. (Ft.) tularensis can be lethal to humans and are mostly associated with cases of tularemia in the U.S.A. Doses as low as 10–20 bacteria can be infective [1]. Transmission mostly occurs via aerosol, alimentary ingestion or skin inoculation. In addition, F. tularensis is suspected as a potential bacterial biological weapon [2]. The species Ft. novicida is almost avirulent for humans in contrast to mice, and is thought to be an opportunistic pathogen [3,4]. Ft. novicida is assumed to constitute an environmental lineage along with F. philomiragia. In rare cases the latter has also been associated with human disease in immunocompromised individuals [4,5].
Human infections caused by F. tularensis are rare in Germany, but seroprevalence studies in wild animals revealed a high seroprevalence of F. tularensis in wildlife in eastern Germany [6-8]. In Germany, Ft. holarctica is generally identified in affected animals or humans as well as in known vectors (like ticks and other arthropods) [1,9-11]. Other as yet known species of the genus Francisella are F. hispaniensis[12,13], F. halioticida[14], F. piscida[15], F. noatunensis[16], F. asiatica[16], F. noatunensis subsp. orientalis[17] and F. philomiragia subsp. noatunensis[18,19]. Very recently, a new Francisella species (F. guangzhouensis) was isolated from a cooling tower in China, which had not been reported before [20].
However, to our knowledge, no species other than Ft. holarctica has been identified in Germany until now. Therefore, our new isolate W12-1067 is the first aquatic isolate identified in Germany which does not belong to the species F. tularensis and is closest related to F. guangzhouensis.
Methods
Strains, media and growth conditions
Strains used in this study were Ft subsp. holarctica LVS (ATCC 29684), Ft. novicida U112 (ATCC 15482), F. philomiragia (ATCC 25015), Legionella pneumophila Paris (CIP 107629) and the new environmental Francisella isolate W12-1067. Francisella strains were cultivated in medium T [21] (1% brain heart infusion broth [Difco Laboratories, Inc., Sparks, MD, USA], 1% bacto tryptone [Difco], 1% technical casamino acids [Difco], 0.005 g of MgSO4, 0.01% FeSO4, 0.12% sodium citrate, 0.02% KCl, 0.04% K2HPO4, 0.06% L-cysteine and 1.5% glucose) or on enriched cystine-heart agar (CHA [Difco], 1% brain heart infusion broth, 1% proteose-peptone, 1% D-glucose, 0.5% NaCl, 0.1% L-cystine, 1.5% agar and 1% hemoglobin). W12-1067 and L. pneumophila Paris were cultivated in ACES-buffered yeast extract (AYE) broth [1% N-(2-acetamido)-2-aminoethanesulfonic acid (ACES), 1% yeast extract, 0.04% L-cysteine and 0.025% ferric pyrophosphate, adjusted to pH 6.8 with 3 M potassium hydroxide (KOH) and sterile filtrated], on ACES-buffered charcoal–yeast extract (BCYE) agar [22] or on GVPC agar plates (Heipha Dr. Müller GmbH, Eppelheim, Germany, BCYE agar supplemented with 80,000 IE polymyxin B, 1 mg/l of vancomycin and 80 mg/l of cycloheximide). Isolate W12-1067 was initially cultivated on GVPC agar plates. The U937 human macrophage-like cell line ATCC CRL-1593.2 and the mouse macrophage cell line J774A.1 were cultivated in RPMI 1640 + 10% FCS medium (PAA/GE Healthcare Europe GmbH, Freiburg, Germany) at 37°C and 5% CO2.
Phenotypic assays
Growth without additional cysteine was done on BCYE agar plates without additional cysteine. Physiological characteristics of analyzed strains were determined by using API ZYM (bioMérieux Deutschland GmbH, Nürtingen, Germany).
Chitinase activity tests were done on 1.5% agarose plates containing 0.1% deacetylated glycol chitin. Chitinase activity experiments were done as described earlier [23], with the modification that the deacetylated glycol chitin was suspended in 0.01 M sodium phosphate (pH 5.5) by heating. In short, bacteria were grown in medium T overnight. The supernatant was precipitated by isopropanol and the protein pellet was resuspended in PBS (concentrated 40-fold). 50 μl were inoculated into agar plates (as described earlier [24]) and incubated for two days at 37°C. Degrading activity was visualized by incubation with 0.01% Calcofluor Brightener 28 (Sigma-Aldrich Chemie GmbH, Munich, Germany) for 10 min, washing two times with water and then incubation at room temperature (RT) overnight.
The NaCl sensitivity assays were done in 96-well plates in a total volume of 200 μl of medium T and ~2 × 107 bacterial cells. Plates were incubated 2–3 days at 37°C and 5% CO2, and then the optical density (OD) at 600 nm was measured using an Infinite 200 reader (Tecan Deutschland GmbH, Crailsheim, Germany).
Intracellular multiplication of Francisella in host cells
To determine which amoeba strain may be suitable to be used for replication assays, isolate W12-1067 (~1010 cells) was suspended in 1 ml of dH2O and 100 μl were plated onto NN-agar plates (14 g/l of agar in dH2O). The amoeba strain (15 μl, A. castellanii ATCC 30010, A. castellanii ATCC 30234, A. castellanii 50739, A. lenticulata 45 ATCC 50703, A. lenticulata 118 ATCC 50706, Hartmannella vermiformis OS101, Hartmannella vermiformis ATCC 50256 and Naegleria gruberi ATCC 30244, respectively) was dropped onto the centre of the plates and incubated at RT or at 30°C for 7 days. The plates were inspected daily for movement and replication of the amoeba. All amoeba tested were motile and not killed by isolate W12-1067. Therefore, no further experiments were performed using amoebae as host cells.
For differentiation into macrophage-like cells, U937 cells were adjusted to 3 × 105 cells/ml and transferred into 100 ml of fresh RPMI medium containing 10% fetal calf serum (10% FCS), and PMA (phorbol-12-myristate-13-acetate, 1 mg/ml in dH2O [P-8139; Sigma-Aldrich Chemie]) was added at a concentration of 1:20,000. After incubation for 36 h at 37°C and 5% CO2, the supernatant was discarded and adherent cells were washed once with 10 ml of 0.2% EDTA in PBS. Cells were mechanically detached from the flask bottom with RPMI + 10% FCS, transferred into 50 ml tubes and centrifuged at 800 g for 10 min. All cells were counted after trypan blue staining in a Neubauer counting chamber and adjusted to 5 × 105 cells/ml with RPMI + 10% FCS. To each well of a 24-well plate 1 ml of the cell suspension was added and incubated for adhesion during 2 h at 37°C and 5% CO2. Macrophage-like mouse cell line J774A.1 was also grown in fresh RPMI medium containing 10% fetal calf serum and treated as described above, but without the differentiation step (no PMA treatment).
For both cell lines, stationary phase bacteria grown for 3 days on CHA or BCYE agar were diluted in plain RPMI medium and the infection was done with a multiplicity of infection (MOI) of 1, 10 or 100 (time point 0 h) for 2 h at 37°C and 5% CO2. Cells were washed three times with RPMI and incubated with 50 μg/ml of Gentamycin for 1 h to kill extracellular bacteria. Cells were washed again three times with RPMI and covered with 1 ml of RPMI + 10% FCS. For colony-forming unit (CFU) determination at various time points of infection, coincubations of cells and bacteria were lysed by addition of 10 μl of 10% Saponin (S4521, Sigma-Aldrich Chemie) for 5 min, and serial dilutions were plated on BCYE agar. In a control experiment we showed that Saponin treatment did not affect the number of remaining CFU of strain W12-1067 (data not shown).
Electron microscopy (EM)
J774A.1 cells were infected with Francisella strain W12-1067 (MOI of 10) at 37°C as described above. Cells were fixed 96 h post infection with 2.5% glutaraldehyde in 0.05 M HEPES buffer. Bacteria cultivated in medium T were fixed with 4% paraformaldehyde 5% glutaraldehyde in 0.05 M HEPES buffer for 2 h at RT. All samples were post-fixed with osmium tetroxide (1% in distilled water) and uranyl acetate (2% in distilled water), dehydrated stepwise in a graded ethanol series and embedded in LR White resin (Science Services GmbH, Munich, Germany) which was polymerized at 60°C overnight. Thin sections were prepared with an ultramicrotome (UC-T; Leica, Vienna, Austria) and counterstained with uranyl acetate and lead citrate.
Samples were examined using a transmission electron microscope (EM 902; Carl Zeiss Microscopy GmbH, Jena, Germany) at 80 kV, and the images were recorded using a slow-scan charge-coupled-device camera (Pro Scan elektronische Systeme GmbH, Lagerlechfeld, Germany).
Genome sequencing, ORF finding and annotation
Genome sequencing of chromosomal DNA of isolate W12-1067 was performed by Eurofins MWG Operon (Eurofins Medigenomix GmbH, Ebersberg, Germany): (i) Short insert shotgun library (FLX + library): 1 μg of DNA was fragmented using a Covaris E210 instrument (Covaris Inc., Woburn, MA) according to manufacturer’s instructions. End-repair, dA-tailing and ligation of barcoded adapter were performed following New England Biolabs’ instructions (New England Biolabs GmbH, Frankfurt/Main, Germany). Emulsion-based clonal amplification (emPCR amplification) was performed following Roche’s instructions (Roche Diagnostics GmbH, Mannheim, Germany). FLX + Sequencing: Sequencing was performed on an FLX + platform according the manufacturer’s instructions using 1/8 plate. The sequencing process was controlled by the Roche 454 software gsRunProcessor v2.8 (shotgun signal processing pipeline). (ii) 8 kb mate-pair-like library (Long-Jumping-Distance library): Creation of the 8 kb mate-pair-like library was done at Eurofins MWG Operon (Ebersberg, Germany) using their proprietary protocol. (iii) Illumina Sequencing: For sequencing, the library was loaded on an Illumina MiSeq machine. Cluster generation and paired-end sequencing was performed using the manufacturer’s instructions. MiSeq Control Software 2.2.0 was used for sequencing. For processing of raw data RTA version 1.17.28 and CASAVA 1.8.2 were used to generate FASTQ-files. (iv) Data analysis: A two-step hybrid de novo assembly was conducted using the sequencing data of the two libraries. First, the FLX + data (long reds, single-end) has been assembled separately using the Roche 454 software Newbler (v2.6). The resulting contigs as well as the Illumina long-jumping-distance pairs (Illumina mate-pair-like) were then assembled together with a hardware-accelerated assembly pipeline based on the Convey hardware and software tools (http://www.conveycomputer.com) that mimic a standard de novo assembly using the Velvet assembler (Eurofins proprietary assembly pipeline) [25,26]. The draft genome (all contigs) was annotated by using the RAST server, freely available at http://www.patricbrc.org[27].
Phylogenetic analysis
16S rDNA gene and the multi-gene locus (in frame gene sequence) of genes fopA, gyrA, rpoA, groEL, sdhA and dnaK of strain W12-1067 and available homologous sequences from Francisella species and L. pneumophila Paris (obtained from GenBank) were used for nucleotide comparison. The multi-gene locus of L. pneumophila Paris exhibited no fopA gene because no homolog is present within the genome. Phylogenetic analysis (phylogenetic tree) was generated by using the ClustalW program (ClustalW 2.1; http://www.patricbrc.org).
Accession number
This whole genome shotgun project has been deposited at DDB/EMBL/GenBank under accession AWHF00000000. The version described in this paper is version [AWHF01000000].
Results and discussion
During 2012 health authorities of the city of Heilbronn (Germany) observed some coincident spots of Legionnaires’ disease (LD). Therefore, different putative sources were screened for the presence of Legionella species using GVPC (glycine-vancomycin-polymyxin-cycloheximide) agar plates. The investigation of a water reservoir of a cooling tower led to the isolation of strain W12-1067. The isolate W12-1067 was first thought to belong to the genus Legionella because of its habitus and growth on GVPC agar plates, but PCR analysis did not support this finding. Preliminary 16S rDNA PCR analysis performed by the German Consultant Laboratory for Legionella (Dresden, Germany) revealed that this strain may belong to the genus Francisella. The identified isolate was not involved in the LD outbreak. However, the strain was send for further analysis to the Centre for Biological Threats and Special Pathogens at the Robert Koch Institute (Berlin, Germany). Here, W12-1067 was identified to be the first German isolate of the genus Francisella which did not belong to the species F. tularensis.
Analysis of strain W12-1067
First we performed 16S rDNA PCR and sequenced the PCR product. The phylogenetic analysis of the 16S rDNA revealed that isolate W12-1067 is a close representative of the recently identified new environmental Francisella species F. guangzhouensis[20]. The phylogenetic tree of 16S rDNA of different Francisella strains is given in Figure 1A, corroborating the close relationship of isolate W12-1067 with F. guangzhouensis and other Chinese cooling tower isolates of this species (99% identity). The other Francisella strains analyzed revealed DNA identities of 16S rDNA sequences of 94–95% with 16S rDNA of isolate W12-1067 and 83% identity with the 16S rRNA gene of L. pneumophila Paris.
Figure 1.
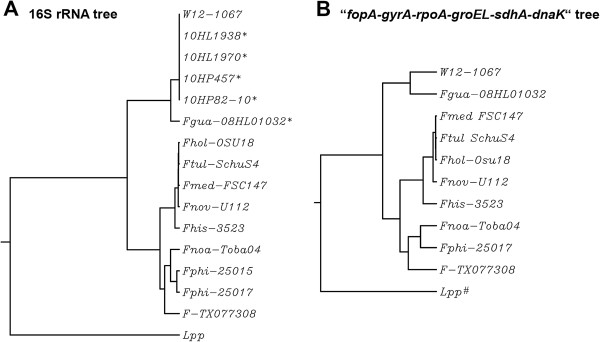
Phylogenetic tree analysis. (A) Phylogenetic tree analysis of different Francisella strains using 16S rDNA or (B) 6-loci concatenated DNA sequences. Name and function of genes used for the 6-loci concatenated sequences are given in Table 2. *, 16S rDNA sequences of different isolates of F. guangzhouensis published by [20]; #, Concatenated sequence of L. pneumophila Paris (Lpp) did not exhibit a fopA gene, because no homolog of this gene is present within the genome sequence. Fhol-OSU18, Ft. holartica strain OSU18; Ftul-SchuS4, Ft. tularensis strain SchuS4; Fmed-FSC147, F. mediasiatica strain FSC147, Fnov-U112, Ft. novicida strain U112; Fhis-3523, F. hispaniensis (Ft. novicida-like strain 3523); Fnoa-Toba04, F. noatunensis strain Toba04; Fphi-25015 and 25017, F. philomiragia strain ATCC 25015 and ATCC 25017; F-TX077308, Francisella isolate TX077308.
We then sequenced the whole genome of W12-1067, resulting in an annotated draft genome of this strain. We used the sequences of the genes fopA, gyrA, rpoA, groEL, sdhA and dnaK (see Table 1) to build a multi-gene locus (9918 bp for strain W12-1067) for each strain analyzed. With these 6-loci concatenated sequences we performed a further phylogenetic analysis (Figure 1B). The overall DNA identity was only 89%, indicating that W12-1067 is not identical to strain F. guangzhouensis, but that F. guangzhouensis is the closest relative identified to date. The overall DNA identity of the gene cluster of isolate W12-1067 to other Francisella strains analyzed was 80–81% and that to L. pneumophila Paris 67% (Figure 1B).
Table 1.
Genes used for phylogenetic tree analysis
| Gene name | Peg Nr. | Feature | % DNA similarity to F. gua -08HL01032 T |
|---|---|---|---|
| 16S rRNA |
- |
Small subunit ribosomal RNA |
99% |
| 23S rRNA |
- |
Large subunit ribosomal RNA |
98% |
|
fopA |
25 |
Francisella outer membrane protein A |
87% |
|
gyrB |
513 |
DNA gyrase subunit B |
88% |
|
rpoA |
853 |
DNA-directed RNA polymerase A |
90% |
|
groEL |
1260 |
Heat shock protein 60, chaperone |
91% |
|
sdhA |
1002 |
Succinate dehydrogenase subunit A |
88% |
| dnaK | 687 | Heat shock protein K, chaperone | 92% |
Type strain of F. guangzhouensis sp. nov (Qu et al. [20]).
Similar to strain W12-1067, F. guangzhouensis strains had been isolated from water of air conditioning systems of cooling towers in China, during a routine investigation to detect Legionella[20]. The growth optimum of this species ranged between 25 and 28°C and it showed growth on BCYE-alpha (minus cysteine) Legionella-agar plates. Furthermore, no virulence to mice was found for this strain [20,28]. No further information about virulence properties of this species was available yet.
We investigated the growth of strain W12-1067 on different agar plates and within different liquid media. Francisella sp. strain W12-1067 grew well on BCYE, GVPC and CHA plates. The isolate grew faster in medium T than in AYE medium, and it did not grow in the cell culture medium RPMI (data not shown). Whereas L-cysteine within BCYE agar plates is necessary for the growth of L. pneumophila Paris, it stimulates the growth of F. philomiragia, and the growth of strain W12-1067 was nearly similar with or without additional cysteine (see Additional file 1: Figure S1A). Growth in medium T revealed that the growth optimum of strain W12-1067 is about 30°C (Figure 2A), but growth of strains F. philomiragia and Ft. novicida in general was faster and reached a higher cell density compared to that of the growth of strain W12-1067 (Figure 2B–D). Growth of strain W12-1067 in media with NaCl was reduced from a concentration of 4–5% NaCl, which was comparable to growth of strains F. philomiragia and Ft. novicida (up to 6–7% NaCl), but more resistant than the non-halotolerant strain Ft. holarctica LVS (see Additional file 1: Figure S1B). Using the API ZYM assay kit (bioMérieux), W12-1067 showed a profile similar to F. guangzhouensis (data not shown). In contrast to F. guangzhouensis, isolate W12-1067 was negative for alkaline phosphatase activity and showed only very low activity of the acid phosphatase, which is in good agreement with only one putative phosphatase encoding gene (peg_768) present within the genome sequence (see below).
Figure 2.
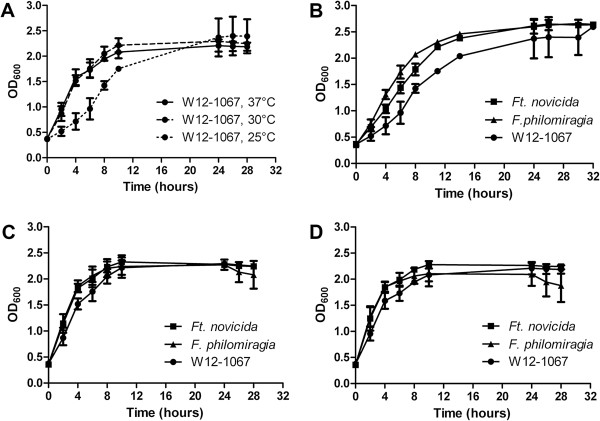
Growth of different Francisella strains at 25, 30 and 37°C in medium T. (A) Comparison of growth of Francisella isolate W12-1067 at different growth temperatures. (B–D) Comparison of growth of Francisella isolate W12-1067 with the growth of strains Ft. novicida U112 (Ft. novicida) and F. philomiragia 25015 (F. philomiragia ATCC 25015) at 25°C (B), 30°C (C) and 37°C (D). Results are mean standard deviations of three independent experiments of duplicate samples.
These experiments were followed by co-culture studies using macrophage-like cell lines of human (U937) or mice (J774A.1) origin as host cells. We found that strain W12-1067 was able to persist within U937 cells (data not shown), and it replicated intracellularly in J774A.1 cells (Figure 3). Replication within J774A.1 was slower than that of L. pneumophila Paris. However, W12-1067 multiplied about 16-fold within 4 days of co-incubation, with a 24 h lag-phase at the beginning of the infection (Figure 3), indicating that the new isolate is able to infect and multiply within eukaryotic cells. Cells of strain W12-1067 grown in medium T at 37°C possessed a rod-shaped, slightly pleomorphic morphology (Figure 4A). EM analysis of the infection assay revealed that strain W12-1067 was localized intracellularly within a vacuole 96 h after infection (Figure 4B–D). The data indicate that W12-1067 is able to replicate intracellularly, but from EM analysis it was not clear yet whether the bacteria were able to escape from the vacuole. Experiments are under way to investigate this question further.
Figure 3.
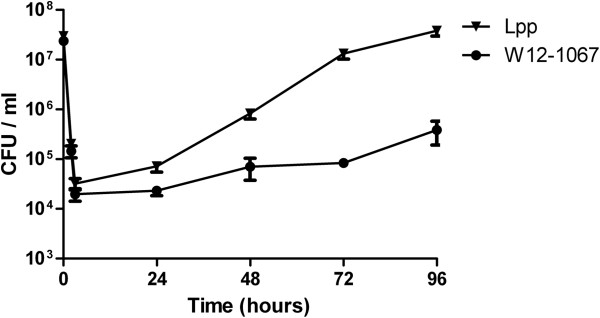
Infection assays of Francisella strain W12-1067 and L. pneumophila Paris (Lpp) using the J774A.1 mouse cell line. Growth curve over a period of four days. Cells were infected with bacteria at an MOI of 10. Cells were washed three times with RPMI and incubated with 50 μg/ml of Gentamycin for 1 h to kill extracellular bacteria. Cells were washed again three times with RPMI and covered with 1 ml of RPMI + 10% FCS. The number of CFU per well was determined by plating on CHA agar plates. Results are mean standard deviations of three independent experiments of duplicate samples.
Figure 4.
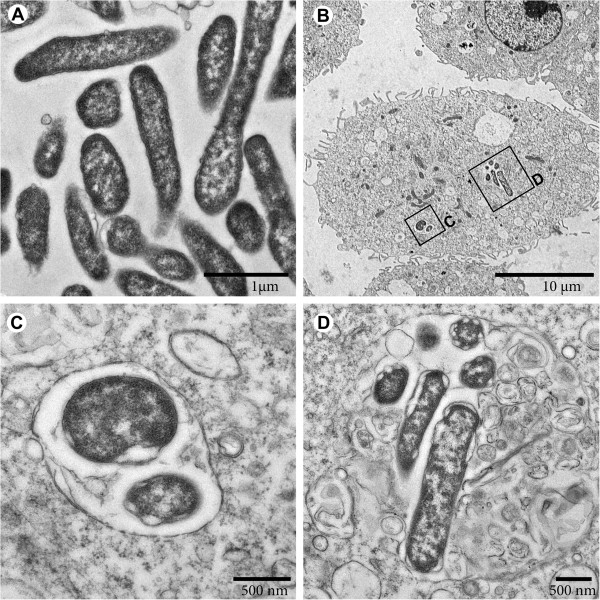
Thin-section EM of Francisella strain W12-1067. (A) Bacteria cultivated in medium T at 37°C possess a rod-shaped, slightly pleomorphic morphology. (B-D) Incubation of J774A.1 cells with bacteria (MOI 10). (B) Overview of host cells with two compartments containing bacteria (96 h post infection). Rectangles mark regions shown at higher magnification in C and D, respectively. (C) Two bacteria in a membrane-bound compartment. (D) Several bacteria in a compartment which shows a clearly discernible membrane at least in some regions.
Whole genome sequencing and sequence analysis of strain W12-1067
General features
The genomic DNA was sequenced using 454 shotgun reads and paired-end Illumina reads (for details, see Methods), followed by de novo assembly of the sequence, which was done by Eurofins (Ebersberg, Germany), resulting in 73 contigs and a chromosomal size of approximately 1,704,745 bp. The draft genome was annotated by using the RAST server (freely available at http://rast.nmpdr.otg). All genes encoded by the draft genome sequence of isolate W12-1067 (GenBank AWHF01000000) are given in the Additional file 2: Table S1. The draft genome exhibits 1,541 protein-coding genes (peg), three copies of the 16S-23S-5S rRNA gene locus (plus one additional 5S rRNA gene) and 37 tRNA genes (Table 2). The G + C content is 32.2% and therefore not significantly different from various other Francisella strains already sequenced [12,29-37]. However, the number of protein-coding genes and of tRNA genes seemed to be lower compared with other Francisella strains (Table 2), but this is not surprising since the W12-1067 genome is of draft quality and the others are finished. The genes encoding the tRNAs are present in four (tRNA-Leu), three (tRNA-Ala, -Ile, -Met, -Arg, -Gly, -Ser) and two (tRNA-Val, -Thr) copies. Like in other Francisella genomes, W12-1067 genome exhibits only one copy of the other tRNA genes (Asn, Asp, Cys, Gln, Glu, His, Lys, Phe, Pro, Trp and Tyr). The tRNA genes for Ala and Ile are only present within the ribosomal RNA locus, which is also true for the other Francisella strains and for most other bacteria [12,31,38,39]. Genome sequence comparison using the MAUVE program (http://www.DNASTAR.com) demonstrated that the genome of strain W12-1067 is poorly similar to the genomes of Ft. holarctica, Ft. novicida, F. philomiragia or F. hispaniensis AS02-814 (Ft. novicida-like 3523), indicating the evolutionary distance between these strains (data not shown). The MAUVE alignment of the draft genome sequence of strain W12-1067 with the genome sequence of Ft. novicida U112 is shown in the Additional file 3: Figure S2. The genome sequence of strain F. guangzhouensis 08HL01032T was not yet available for a comparative analysis, but a G + C content of about 32.5% of its draft genome was reported [20].
Table 2.
Genome features of various Francisella genome sequences
| Genome feature | W12-1067* | Fnov -U112 | Fphi -25017 | F-TX077308 |
|---|---|---|---|---|
| Chromosome size (bp) |
1,704,745 |
1,910,031 |
2,045,775 |
2,035,931 |
| Nr. of protein-coding genes |
1,541 |
1,733 |
1,915 |
1,976 |
| GC content (%) |
32.2 |
32.48 |
32.57 |
32.9 |
| 5S rRNA genes |
3 + 1 |
3 + 1 |
3 + 1 |
3 + 1 |
| 16S RNA genes |
3 |
3 |
3 |
3 |
| 23S RNA genes |
3 |
3 |
3 |
3 |
| tRNA genes |
37 |
39 |
39 |
39 |
| GenBank accession number | AWHF01000000 | CP000439 | CP000937 | CP002872 |
*Draft genome (73 contigs); Fnov-U112, F. tularensis subsp. novicida U112; Fphi-25017, F. philomiragia strain ATCC25017; F-TX077308, Francisella spp. strain TX077308.
Mobile elements
In F. tularensis strains six different insertion sequences (IS) elements (ISFtu1–ISFtu6) have been described. IS elements are important elements of F. tularensis genomes and are thought to be generally stable among different isolates [36,40]. Within the genome of Francisella isolate W12-1067, we identified several mobile elements and transposases (Table 3). For the element ISFw3 (1,177 bp in length) we identified 18 copies within the genome sequence. The element encodes a putative integrase of 359 amino acids with 70% amino acid identity to the protein encoded by the IS481 element of Wolbachia endosymbiont of Drosophila simulans. All other elements are only present as a single copy. There is only one element (ISFw7) with significant similarity to one of the ISFtu elements of F. tularensis. ISFw7 encodes a putative transposase exhibiting 78% amino acid identity to ISFtu1 of Ft. holarctica strains. Of the 14 mobile elements identified, six (ISFw4, 5, 7, 9, 10 and peg_1255) exhibit homologs within Francisella strains (Table 3).
Table 3.
Mobile elements and transposases of Francisella strain W12-1067
| Name | Peg Nr.* | Copies | Feature | Closest homolog |
|---|---|---|---|---|
| ISFw1 |
1, 2 |
1 |
DDE_4 SF |
ISRin1, can. Regiella insecticola |
| ISFw2 |
129 |
|
DDE_Tnp_1, IS4, |
IS4, Nitrosomas sp. |
| ISFw3 |
100, 130, 195, 232, 260, 274, 556, 647, 738, 756, 758, 896, 930, 965, 1146, 1212, 1298, 1415 |
18 |
Pfam_rve, integrase |
IS481, wHa_02240 Wolbachia endosymbiont of Drosophila simulans |
| ISFw4 |
277 |
1 |
Tra8, IS30, integrase |
Fphi_0709 |
| ISFw5 |
331 |
1 |
transposase, partial |
Fphi_0985 |
| ISFw6 |
568 |
1 |
DDE_4_2 |
Aasi_1822, can. Amoebophilus asiaticus 5o2 |
| ISFw7 |
822 |
1 |
DDE_3, transposase ISFtu1 |
FTH_0348, Francisella holarctica OSU18 |
| ISFw8 |
895 |
1 |
DDE_4_2 |
Aasi_1822, can. Amoebophilus asiaticus 5o2 |
| ISFw9 |
1118 |
1 |
transposase, partial |
Fphi_1490 |
| ISFw10 |
1214 |
1 |
DDE_Tnp_1, IS4/5 |
Fphi_0257 |
| ISFw11 |
1295 |
1 |
DDE_4_2, partial |
OTT_1632, Orientia tsutsugamushi str. Ikeda |
| Tp |
701 |
1 |
DDE_Tnp_1_2, truncated |
WRi_008070 Wolbachia sp. wRi |
| Tp |
898 |
1 |
HTH_Tnp_IS630, |
ISRin2, ORFA, can. Regiella insecticola |
| Tp | 1255 | 1 | IS4 | NE061598_00570 F. tularensis NEO |
*see Additional file 2: Table S1.
Virulence factors and secretion systems
A number of genes involved in Francisella pathogenesis have been identified in various different studies, including in vivo negative selection screens of transposon mutant libraries of Francisella strains [41-44], reviewed in [45] and [46]. We therefore looked for homologs of these virulence genes within the genome sequence of Francisella strain W12-1067. Some of them are shown in Table 4, and others, like LPS, capsule, type IV pili or FPI-associated genes, are described below. Various known virulence factors of Francisella exhibit homologs in strain W12-1067. For the most important virulence factors of F. tularensis, encoded by the genes of the Francisella Pathogenicity Island (FPI), no close homologs could be identified within the genome. However, we identified an FPI-like island at the genome sequence which will be discussed later.
Table 4.
(Putative) virulence factors of strain W12-1067
| Name | Peg Nr. | Feature | Closest homolog |
|---|---|---|---|
| MglA |
693 |
Macrophage growth locus protein |
FTN_1290 (82%) |
| CapBCA |
74-76 |
Capsular biosynthesis |
Fphi_1486-88 (nd) |
| FeoB |
1092 |
Ferrous iron transport protein |
FTN_0066 (82%) |
| PilT |
1248 |
Twitching motility protein |
FTN_1622 (94%) |
| DeoB |
802 |
Phosphopentomutase |
FN3523_1666 (80%) |
| BipA |
325 |
GTP-binding protein |
Fphi_0048 (91%) |
| SurA |
543 |
Peptidyl-prolyl cis-trans isomerase |
FTN_0559 (69%) |
| MviN |
843 |
Flippase |
FTN_0276 (61%) |
| HlyB |
1149 |
Toxin secretion ABC transporter |
FTN_1693 (74%) |
| Phospholipase |
743 |
Lecithinase/hemolysin |
FTN_0436 (75%) |
| Lysophospholipase |
167 |
Lysophospholipase |
Fphi_1625 (67%) |
| HlyC/CorC |
182 |
CBS domain, putative hemolysin |
FTN_1006 (80%) |
| ClpB |
1114 |
ClpB chaperone domain |
FTN_1743 (89%) |
| Chitinase 1 (372 aa) |
490 |
GH18_chitinase-like superfamily |
Fphi_0209 (83%) |
| Chitinase 2 (590 aa) |
816 |
Chitinase_glyco-hydro_19 domain, PP location, CBM |
Fphi_0512 (69%) |
| Chitinase 3 (437 aa) |
818 |
Chitinase_glyco-hydro_19 domain, SP |
Fphi_0512 (70%) |
| Chitinase 4 (979 aa) |
1009 |
GH18_chitinase-like superfamily, SP, two internal repeats |
FTW_0142 (55%) |
| Chitinase 5 (731 aa) |
1044 |
GH18_chitinase-like superfamily, SP, EC location, 2 × CBMn |
FN3523_1814 (49%) |
| Chitinase 6 (843 aa) |
1477 |
GH18_chitinase-like superfamily, SP, EC location, 2 × CBM |
Fphi_0208 (55%) |
| Hypothetical protein |
523 |
Ankyrin repeat, Ank_2 domain |
--- |
| Hypothetical protein |
567 |
Ankyrin repeat, Ank_2 domain |
--- |
| Hypothetical protein |
1109 |
Ankyrin repeat, Ank_4 domain |
--- |
| Hypothetical protein |
173 |
TPR domain |
FTW_0991 (63%) |
| Hypothetical protein |
1024 |
TPR domain |
Fphi_0624 (69%) |
| Hypothetical protein | 1030 | TPR domain, TPR_16 | FTM_1557 (72%) |
SP, signal peptide; EC, extracellular; PP, periplasmic; CBM, carbohydrate binding motif; TPR, tetratricopeptide repeat.
Interestingly, the genome of isolate W12-1067 exhibit six different chitinases of different sizes and putative location (cytoplasmic, periplasmic or extracellular) (Table 4), which was in good agreement with the phenotypically identified chitinase activity within the supernatant of Francisella W12-1067 cells (see Additional file 1: Figure S1C). Four chitinases (peg_818, 1009, 1044 and 1477) exhibit a signal peptide and therefore could be secreted by the general secretion system (Sec). Two of the chitinases (peg_0816 and peg_0818) are separated by a gene encoding a putative DNA-invertase (peg_0817). We could not identify a homolog of this protein in the available genome sequences of Francisella strains. We were unable to determine whether the invertase is involved in DNA inversion in Francisella and whether this may influence the expression of the nearby chitinase genes. Chitinase peg_0816 is 76% identical to chitinase peg_0818. Both chitinases exhibit a homolog in F. philomiragia 25017 (Fphi_0512) and 25015, but not in the other Francisella strains, whereas homologs of the chitinases 1, 4 and 5 could be found in the genomes of Ft. tularensis, F. holarctica and F. philomiragia.
Chitinases (FTN_0627 and FTN_1744) have been detected as essential virulence factors of Ft. novicida and for biofilm formation [42,44,47]. In L. pneumophila a chitinase was also found to be involved in the infectivity of Legionella for mice [48]. It would be interesting to further analyze the role of the different chitinases in strain Francisella W12-1067.
Furthermore, we identified three hypothetical proteins (peg_523, 567 and 1109) containing ankyrin-repeat domains. These proteins did not exhibit significant homology to any known protein (Table 4). For L. pneumophila it was shown that ankyrin-repeat-containing proteins are involved in the pathogen–host interplay during intracellular replication [49,50].
Furthermore, we looked for genes encoding putative antibiotic resistance proteins. We identified ten putative multidrug resistance proteins (peg_126, 152, 681, 682, 683, 764, 1134, 1430, 1444 and 1445). In addition, we identified a Chloramphenicol acetyltransferase (peg_183), a Chloramphenicol phosphotransferase (peg_1413) and a Streptomycin-6 kinase (peg_1416) without a homolog in any of the available Francisella genomes. We performed growth inhibition experiments with the isolate W12-1067 and found that levels of resistance to erythromycin, chloramphenicol and streptomycin were comparable to those of F. philomiragia (data not shown). We also identified a putative Acriflavin resistance protein (peg_810) exhibiting 85% amino acid identity to the AcrB protein (Fphi_1007) of F. philomiragia 25017.
Since we identified putative signal peptides at the N-terminus of some of the chitinases and chitinase activity within the supernatant, we searched the genome sequence for genes of the general Sec system. The identified proteins are given in the Additional file 4: Table S2. We identified all proteins necessary for a putative functional Sec system plus two signal sequence recognition proteins (SRP) and three different signal peptidases. Therefore, strain W12-1067 seems to encode a functional Sec system for the transport of proteins across the inner membrane. We could not detect genes encoding homologs of a type-II secretion system (T2SS), but proteins (HlyB, HlyD, TolC2) of a putative T1SS (Table S2). We were also able to identify a putative functional Tol-Pal system generally involved in vitamin B12 or colicin translocation, but also in capsule synthesis and outer membrane vesicle formation [51,52]. The presence of a putative T6-like SS will be discussed in the next section.
A new putative homolog of the Francisella pathogenicity island (FPI) and regulatory proteins
T6SS are widely distributed amongst diverse Gram-negative species. It is a complex molecular machine which injects effector proteins to target cells or bacteria [53]. T6SS are involved in virulence and in eukaryotic cell targeting. They are also reported to have antibacterial activity [54,55]. Most systems are able to function “anti-eukaryotically” or “anti-bacterially”, and one system has been reported to be able to do both [53,56,57]. It was also proposed that T6SS may target and defend predatory eukaryotes in the environment [58]. For an overview see [53].
Francisella strains exhibit an FPI encoding a functional T6SS, needed for preventing phagolysosomal fusion and escape into the cytosol, which is therefore essential for intracellular replication and pathogenicity of Francisella[59-63]. The island is ~33 kb in length, encodes for 15–19 open reading frames (ORFs) and is present in Ft. tularensis strains, Ft. novicida and F. philomiragia (Figure 5A). In strains of F. tularensis this island is duplicated [36,64-66]. However, it is still unclear yet whether both copies are needed for full virulence of these strains. Genes of the locus were named igl (intracellular growth locus, [66]) and pdp (pathogenicity determinant proteins, [65]). The complete locus was identified in 2004, and it seems to be acquired via horizontal gene transfer because of a lower G + C content compared with the core genome [60,64,65].
Figure 5.
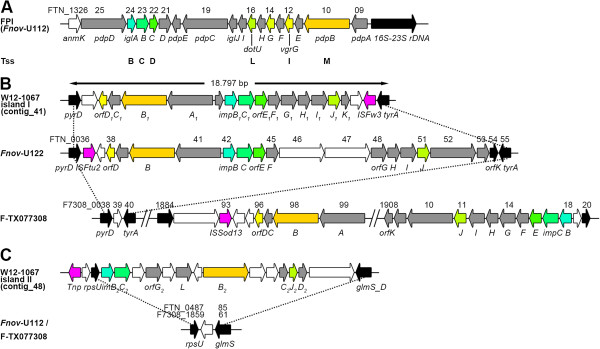
Genetic organization of T6 secretion systems in Francisella. Organization of the FPI island of Ft. novicida U112 (A) and of genomic islands I (B) and II (C) encoding putative T6-like secretion systems of Francisella isolate W12-1067. Genomic island I is integrated between genes pyrD and tyrA in W12-1067 and Ft. novicida U112, whereas it is integrated between F7308_1884 und F7308_1920 in Francisella strain TX077308 (F-TX077308). Genomic island II is integrated between rpsU and glmS in W12-1067 and is not present in Ft. novicida U112 and Francisella strain TX077308. Genes (ORFs) are indicated by arrows. Gene names are given below the genes, and the protein-encoding gene (peg) numbers are given in Table 5. Genes encoding homologous proteins are boxed in the same color, with the exception of pink (mobile elements) and black (conserved core genome genes).
We were not able to identify a close homolog of the FPI within the genome of strain W12-1067. However, we identified two FPI-like clusters which seem to encode a further putative T6SS (Figure 5). Clusters I (Figure 5B) and II (Figure 5C) each code for 16 ORFs. Homologs of iglABC, dotU and pdbB are present, and the whole island I is similar to an FPI-like element (FTN_0038-0054) recently identified within the genome of Ft. novicida, but not yet discussed further [60,64]. Within the genome sequence, island I is also localized between the pyrD and tyrA genes and island II between rpsU and glmS (Figure 5B and C). All genes, features of the ORFs and closest homologs of both systems identified within the genome of W12-1067 are given in Table 5. In a recently published model of an FPI-encoded T6SS-like apparatus IglAB (TssBC), IglC (TssD), DotU (TssL), PdpB (TssM) and VgrG (TssI) were identified as core proteins of the T6SS [67]. We identified similar proteins within the identified loci of strain W12-1067. In strain F-TX077308, the FPI-like gene cluster is localized at a different site within its genome sequence (Figure 5B). Altogether these findings led us to hypothesize that these loci may encode a further T6-like SS in Francisella strain W12-1067. As mentioned above, infection assays and EM analysis indicateed that strain W12-1067 is able to replicate intracellularly (Figures 3 and 4), which also suggest that there should be a functional T6SS in this strain. However, since the EM analysis did not yet clearly demonstrate whether the strain is released into the cytosol, further experimental data are needed to verify this hypothesis.
Table 5.
Genes of the putative T6SSs I and II of Francisella isolate W12-1067
| Name (T6SS I) | Peg Nr. (aa) | kDa | Feature | Closest homolog (% aa identity) |
|---|---|---|---|---|
| OrfD1 |
950 (92) |
9.9 |
DUF2345, VgrG-like, Rhs family |
FTN_0038 (57%) |
| OrfC1 |
951 (138) |
15.7 |
SP, lipid attachment site (IglE-like) |
FTN_0039 (66%) |
| OrfB1 |
952 (1097) |
127.1 |
SP, TM domain (PdpB-like/TssM) |
FTN_0040 (58%) |
| OrfA1 |
953 (735) |
84.6 |
Cl* |
FTN_0041 (63%) |
| Orf48 |
954 (48) |
5.8 |
SP |
--- |
| ImpB1 |
955 (179) |
20.8 |
homologous to IglA/TssB |
FTN_0042 (96%) |
| ImpC1 |
956 (508) |
57.7 |
homologous to IglB/TssC |
FTN_0043 (91%) |
| OrfE1 |
957 (214) |
23.1 |
helical bundle domain (putative TssD) |
FTN_0044 (77%) |
| OrfF1 |
958 (356) |
41.3 |
Cl |
FTN_0045 (51%) |
| OrfG1 |
059 (406) |
47.3 |
Cl, T6SS-associated like protein |
FTN_0048 (72%) |
| OrfH1 |
960 (274) |
31.9 |
C-terminal TM domain |
FTN_0049 (63%) |
| OrfI1 |
961 (501) |
58.6 |
Cl |
FTN_0050 (72%) |
| OrfJ1 |
962 (203) |
23.8 |
DUF2077 (putative DotU/TssL) |
FTN_0051 (63%) |
| OrfK1 |
963 (126) |
13.1 |
DUF4280 |
FTN_0054 (82% |
| Orf374 |
964 (374) |
43.3 |
Cl |
Hyp. protein (33%) Flavobacterium |
| IglD3 |
1538 (398) |
46.1 |
DUF876 (TssK) |
IglD (49%) F-TX077308 |
|
Name (T6SS II) |
Peg Nr. |
kDa |
Feature |
Closest homolog (% aa identity) |
| ImpB2 |
1376 (177) |
19.9 |
Cl, homologous to IglA/TssB, DUF770 |
FTN_0042 (42%) |
| ImpC2 |
1375 (493) |
55.5 |
Cl, homologous to IglB/TssC, COG3517 |
FTN_0043 (47%) |
| Orf204 |
1374 (204) |
22.4 |
Cl, |
--- |
| OrfG2 |
1373 (405) |
47.1 |
Cl, IglD-like |
F7308_1914 (28%) |
| Orf368 |
1372 (368) |
43.4 |
Cl |
--- |
| OrfL |
1371 (629) |
74.6 |
Hypothetical protein |
F7308_1916 (33%) |
| Orf96 |
1370 (96) |
11.1 |
2 TM domains |
Hyp. protein (49%) Prevotella histicola |
| OrfB2 |
1369 (928) [1020] |
106.3 |
Cytoplasmic membrane, SP |
FTN_0040 (20%) |
| Orf457 |
1368 (457) |
53.4 |
Hypothetical protein |
--- |
| Orf254 |
1367 (254) |
30.3 |
Cl |
--- |
| OrfC2 |
1366 (146) |
17 |
SP |
FTN_0039 (24%) |
| OrfJ2 |
1365 (195) |
23.1 |
DUF2077 (putative DotU/TssL) |
DotU, (29%) Desulfonatronospira |
| OrfD2 |
1364 (111) |
12.4 |
Cl (DUF2345, vgrG) |
--- |
| ORF705 | 1363 (705) | 77.7 | Cl | --- |
*determined by using “psortb” (http://www.psort.org/psortb); Cl, cytoplasmic localization; TM, transmembrane domain; SP, signal peptide; (aa), number of amino acids.
Various regulatory proteins are known for Francisella, and the regulators MglAB, SspA, PmrA, FevR, MigR and Hfq were identified to be involved in the regulation of genes of the FPI [59,61,68-76]. In F. tularensis the genes of the FPI are upregulated during intracellular growth within macrophages [61,68,77-80]. In the genome of strain W12-1067 we identified only homologs of MglAB and Hfq, but also two further response regulator proteins (OmpR1 and R2) as well as two sigma factors (Sig-70 and Sig-32) and homologs of IscR, ArsR, Crp and Fur (see Additional file 5: Table S3). In addition, proteins involved in the stringent response could be identified (SspB, SpoT and RelA).
Surface structures: The wbt locus, capsule and type IV pili
(i) LPS. The lipid A core portion of the LPS anchors the lipopolysaccharide structure to the outer membrane, whereas the O-polysaccharide chain is the predominant epitope recognized by the immune system and specifies antigenicity. Ft. tularensis subspecies-specific antisera have been generated and applied to antigen detection in F. tularensis[81]. Furthermore, LPS is used as an antigen in seroprevalence studies and for diagnostic of human tularemia [6,82,83]. However, the LPS produced by F. tularensis is less endotoxic compared to other Gram-negative bacteria, such as E. coli, a phenotype also known for L. pneumophila LPS [84-86]. Genes probably involved in the biosynthesis of the O-polysaccharide chain are given in supplementary Table 4 (see Additional file 6: Table S4). Obviously, there is one cluster of genes (peg_0636-0646) for which homologs were found in Ft. novicida and F. philomiragia, whereas for genes peg_0609-0615 and peg_0628-0631 the homologs were only found in F. philomiragia or Ft. novicida U112, respectively. In addition, there is another cluster of genes (peg_0616-0627) which seems to have no homologs in F. tularensis or F. philomiragia, but homologs were identified in different bacterial species, as in Vibrio, Pseudomonas, Sulfurovum or Acinetobacter (see Additional file 6: Table S4). The LPS structure of strain W12-1067 has not yet been analyzed further.
(ii) Capsule. Electron microscopy of strain W12-1067 grown on agar plates or in medium revealed the absence of a capsule (Figure 4A and data not shown). However, we identified three ORFs encoding homologs of capBCA genes (Table 4). The capBCA locus of Francisella is similar to determinants encoding the poly-gamma-glutamic capsule in Bacillus anthracis[87]. These genes have been shown to be essential for the virulence of F. tularensis in a murine model of tularemia [44,87]. Further experiments will be needed to analyze the role and structure of the capsule of Francisella strain W12-1067 and to identify conditions necessary for the putative induction of capsule gene expression.
(iii) Type IV pili. Electron microscopy of strain W12-1067 did not show any pili on the surface of the bacteria grown in medium at 37°C (Figure 4A) or on agar plates (data not shown). However, we identified homologs of the type IV pilus (Tfp) encoding loci of F. philomiragia ATCC 25017 in the genome sequence of W12-1067 (see Additional file 7: Table S5) [88]. We could not identify a PilA homolog, a homolog of the second PilW protein Fphi_0522 and of the two additional PilA/PilE pilus assembly proteins (Fphi_0424/0449). Tfp systems are known to be involved in bacterial pathogenesis, bacterial adhesion and twitching motility [89]. In L. pneumophila Tfp pili are required for twitching motility, natural competence, biofilm formation and are involved in the attachment to host cells [90-93]. Tfp have been observed on the surface of Ft. novicida and Ft. holarctica[94,95], and Tfp are involved in the pathogenicity of Francisella[96-98].
Toxin–antitoxin systems
We identified three different type II toxin–antitoxin systems (peg_0599-0600, peg_0704-0705 and peg1296-1297). In type II systems, the antitoxin (small unstable protein) sequesters the toxin (stable protein) through protein complex formation (reviewed in [99]). Peg_0599 encodes a protein (84 amino acids, putative toxin), exhibiting a Pfam_Plasmid_Txe domain and 67% amino acid identity to the YoeB toxin of Pleurocapsa sp. PCC7319. peg_0600 encodes the respective putative antitoxin (83 amino acids), exhibiting a Pfam_PhdYeFM (type II toxin–antitoxin) and shows 60% amino acid identity to the prevent-host-death protein of Methylocystis rosea. The second system is composed of protein Peg_0704 (96 amino acids), exhibiting a HTH-XRE motif and a HigA-antidote (VapI) domain and shows 65% amino acid identity to protein LLO_065 of Legionella longbeachae NSW150. The respective putative toxin (HigB, 97 amino acids) is encoded by peg_0705, exhibiting a Pfam_plasmid killer domain, and shows 62% amino acid identity to the plasmid maintenance system killer protein of Deferribacter desulfurricans SSM1. Both described systems seemed to have no homolog in the sequences of Francisella available so far and are localized on contig_34 of the draft genome of strain W12-1067.
The third system is localized on contig_46. peg_1296 encodes for a protein (85 amino acids, putative antitoxin), exhibiting a Pfam_PhdYeFM domain (type II toxin–antitoxin system), and shows 66% amino acid identity to the plasmid-encoded (pF243) protein F243_0001 of F. philomiragia ATCC 25017 and 65% identity to the Phd protein (pFNL10_p3) of Ft. novicida. The respective putative toxin (84 amino acids) is encoded by peg_1297, exhibits a Pfam_Plasmid_Txe (YoeB) domain and shows 79% amino acid identity to F243_0002 of F. philomiragia. Plasmid pF243 is 5,072 bp long and was predicted to encode six putative ORFs [100]. ORFs F243_0001 and F243_0002 are organized in an operon that is similar to the phd-doc post-segregation killing system operon of pFNL10 [100,101]. This post-segregation killing mechanism relies on the difference in stability of the antitoxin and toxin. In the daughter cells the plasmid-free bacteria will be killed by the activity of the toxin [102,103]. Chromosomally encoded toxin–antitoxin systems may stabilize chromosomal regions during evolution and seem to be involved in host regulatory networks or fitness advantages [102]. Less is known so far about toxin–antitoxin systems in Francisella, but they have been used to construct plasmids which are stable without a selective marker gene [101,104].
Conclusions
The isolation of strain W12-1067 in Germany indicates for the first time the presence of a close homolog in Europe of the new species F. guangzhouensis recently identified in China. In addition, to our knowledge this is the first report of a Francisella species other than F. tularensis isolated in Germany. Further research is needed to analyze the spectrum of Francisella species present in natural habitats in Germany.
The growth optimum of the isolate is approximately 30°C, it is able to grow without additional cysteine within the medium and the strain is halotolerant. The analysis of the genome sequence of the new isolate revealed a lot of known Francisella virulence factors, but also the absence of FPI, the major virulence factor of Francisella strains. Instead, the isolate seems to exhibit a putative new T6SS, and W12-1067 is able to replicate within eukaryotic host cells. Therefore, the isolate seems to be an interesting species to be analyzed further.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KH performed the annotation, the comparative and phylogenetic analysis and drafted most of the manuscript. EB generated the end version of the draft-genome which was submitted to NCBI/GenBank. KR achieved the phenotypic analysis, growth experiments and infection assays. TS performed the MAUVE alignments and generated figures. CL and JF were involved in the isolation and preliminary typing of strain W12-1067. GH performed the EM analysis of strain W12-1067. RG participated in writing the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Phenotypic analysis of Francisella isolate W12-1067, Ft. holarctica strain LVS (Ft. LVS), Ft. novicida, F. philomiragia and L. pneumophila Paris (Lpp). Growth on BCYE agar plates with (+) and without (-) additional cysteine (Cys) (A). The results are representative of three independent experiments. Growth in medium T in the presence of different amounts of NaCl (B). Results are mean standard deviations of three independent experiments of duplicate samples. Chitin degradation by the supernatant of different strains grown in medium T and then incubated on agarose plates containing 0.1% deacetylated glycol chitin (C). Halos around the inoculation site revealed the presence of degrading activity after 2 days of incubation at 37°C. The results represented are representative of three independent experiments.
Annotated genes (peg), rRNAs and tRNAs.
MAUVE alignment of strain W12-1067 and Ft. novicida U112. The colored boxes represent homologous segments free of genomic rearrangements. Homologous regions are connected by lines between genomes. Non-boxed regions lack homology between genomes. White areas indicate that the sequences are specific to a genome. (The synteny between both genomes was not estimated, since the genome of W12-1067 is a draft genome).
Genes of the Sec, type I and Tol secretion systems.
Regulatory proteins.
The wbt gene cluster of strain Francisella W12-1067.
Type IV pili encoding genes.
Contributor Information
Kerstin Rydzewski, Email: rydzewskik@rki.de.
Tino Schulz, Email: schulzt@rki.de.
Elzbieta Brzuszkiewicz, Email: ebrzusz@gwdg.de.
Gudrun Holland, Email: hollandg@rki.de.
Christian Lück, Email: christian.lueck@tu-dresden.de.
Jens Fleischer, Email: jens.fleischer@rps.bwl.de.
Roland Grunow, Email: grunowr@rki.de.
Klaus Heuner, Email: heunerk@rki.de.
Acknowledgements
We thank U. Erikli for her careful review of the manuscript. This work was supported by the Robert Koch Institute (ZBS 2) and grant 1369-364 from the Robert Koch Institute to CL.
References
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K. Working Group on Civilian Biodefense. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Clarridge JE 3rd, Raich TJ, Sjosted A, Sandstrom G, Darouiche RO, Shawar RM, Georghiou PR, Osting C, Vo L. Characterization of two unusual clinically significant Francisella strains. J Clin Microbiol. 1996;34:1995–2000. doi: 10.1128/jcm.34.8.1995-2000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CW, Brenner DJ. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J Clin Microbiol. 1989;27:1601–1608. doi: 10.1128/jcm.27.7.1601-1608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger JD, Hollis DG, Weaver RE, Baker CN, Brown GR, Brenner DJ, Broome CV. Infection caused by Francisella philomiragia (formerly Yersinia philomiragia). A newly recognized human pathogen. Ann Intern Med. 1989;110:888–892. doi: 10.7326/0003-4819-110-11-888. [DOI] [PubMed] [Google Scholar]
- Kuehn A, Schulze C, Kutzer P, Probst C, Hlinak A, Ochs A, Grunow R. Tularaemia seroprevalence of captured and wild animals in Germany: the fox (Vulpes vulpes) as a biological indicator. Epidemiol Infect. 2013;141:833–840. doi: 10.1017/S0950268812001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenzora A, Jansen A, Ranisch H, Lierz M, Wichmann O, Grunow R. Seroprevalence study of Francisella tularensis among hunters in Germany. FEMS Immunol Med Microbiol. 2008;53:183–189. doi: 10.1111/j.1574-695X.2008.00408.x. [DOI] [PubMed] [Google Scholar]
- Otto P, Chaignat V, Klimpel D, Diller R, Melzer F, Muller W, Tomaso H. Serological investigation of wild boars (Sus scrofa) and red foxes (Vulpes vulpes) as indicator animals for circulation of francisella tularensis in germany. Vector Borne Zoonotic Dis. 2014;14:46–51. doi: 10.1089/vbz.2013.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A, Franke J, Schmoock G, Pauliks K, Kramer A, Straube E. Diversity and coexistence of tick-borne pathogens in central Germany. J Med Entomol. 2011;48:651–655. doi: 10.1603/me10254. [DOI] [PubMed] [Google Scholar]
- Muller W, Hotzel H, Otto P, Karger A, Bettin B, Bocklisch H, Braune S, Eskens U, Hormansdorfer S, Konrad R, Nesseler A, Peters M, Runge M, Schmoock G, Schwarz BA, Sting R, Myrtennäs K, Karlsson E, Forsman M, Tomaso H. German Francisella tularensis isolates from European brown hares (Lepus europaeus) reveal genetic and phenotypic diversity. BMC Microbiol. 2013;13:61. doi: 10.1186/1471-2180-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnvik A, Priebe HS, Grunow R. Tularaemia in Europe: an epidemiological overview. Scand J Infect Dis. 2004;36:350–355. doi: 10.1080/00365540410020442. [DOI] [PubMed] [Google Scholar]
- Siddaramappa S, Challacombe JF, Petersen JM, Pillai S, Hogg G, Kuske CR. Common ancestry and novel genetic traits of Francisella novicida-like isolates from North America and Australia as revealed by comparative genomic analyses. Appl Environ Microbiol. 2011;77:5110–5122. doi: 10.1128/AEM.00337-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B, Escudero R, Busse HJ, Seibold E, Scholz HC, Anda P, Kampfer P, Splettstoesser WD. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int J Syst Evol Microbiol. 2010;60:1887–1896. doi: 10.1099/ijs.0.015941-0. [DOI] [PubMed] [Google Scholar]
- Brevik OJ, Ottem KF, Kamaishi T, Watanabe K, Nylund A. Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantea) in Japan. J Appl Microbiol. 2011;111:1044–1056. doi: 10.1111/j.1365-2672.2011.05133.x. [DOI] [PubMed] [Google Scholar]
- Ottem KF, Nylund A, Isaksen TE, Karlsbakk E, Bergh O. Occurrence of Francisella piscicida in farmed and wild Atlantic cod, Gadus morhua L., in Norway. J Fish Dis. 2008;31:525–534. doi: 10.1111/j.1365-2761.2008.00930.x. [DOI] [PubMed] [Google Scholar]
- Mikalsen J, Colquhoun DJ. Francisella asiatica sp. nov. isolated from farmed tilapia (Oreochromis sp.) and elevation of Francisella philomiragia subsp. noatunensis to species rank as Francisella noatunensis comb. nov., sp. nov. Int J Syst Evol Microbiol. 2009. Epub 2009 Sep 25, doi:10.1099/ijs.0.002139-0. [DOI] [PubMed]
- Ottem KF, Nylund A, Karlsbakk E, Friis-Moller A, Kamaishi T. Elevation of Francisella philomiragia subsp. noatunensis Mikalsen et al. (2007) to Francisella noatunensis comb. nov. [syn. Francisella piscicida Ottem et al. (2008) syn. nov.] and characterization of Francisella noatunensis subsp. orientalis subsp. nov., two important fish pathogens. J Appl Microbiol. 2009;106:1231–1243. doi: 10.1111/j.1365-2672.2008.04092.x. [DOI] [PubMed] [Google Scholar]
- Mikalsen J, Olsen AB, Rudra H, Moldal T, Lund H, Djonne B, Bergh O, Colquhoun DJ. Virulence and pathogenicity of Francisella philomiragia subsp. noatunensis for Atlantic cod, Gadus morhua L., and laboratory mice. J Fish Dis. 2009;32:377–381. doi: 10.1111/j.1365-2761.2008.00987.x. [DOI] [PubMed] [Google Scholar]
- Mikalsen J, Olsen AB, Tengs T, Colquhoun DJ. Francisella philomiragia subsp. noatunensis subsp. nov., isolated from farmed Atlantic cod (Gadus morhua L.) Int J Syst Evol Microbiol. 2007;57:1960–1965. doi: 10.1099/ijs.0.64765-0. [DOI] [PubMed] [Google Scholar]
- Qu PH, Chen SY, Scholz HC, Busse HJ, Gu Q, Kampfer P, Foster JT, Glaeser SP, Chen C, Yang ZC. Francisella guangzhouensis sp. nov., isolated from air-conditioning systems. Int J Syst Evol Microbiol. 2013;63:3628–3635. doi: 10.1099/ijs.0.049916-0. [DOI] [PubMed] [Google Scholar]
- Pavlovich NV, Mishan'kin BN. [Transparent nutrient medium for culturing Francisella tularensis] Antibiot Med Biotekhnol. 1987;32:133–137. [PubMed] [Google Scholar]
- Edelstein PH. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981;14:298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez L, Hammerschmidt R. Development of a method for the detection and quantification of total chitinase activity by digital analysis. J Microbiol Methods. 2004;59:7–14. doi: 10.1016/j.mimet.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Herrmann V, Eidner A, Rydzewski K, Bladel I, Jules M, Buchrieser C, Eisenreich W, Heuner K. GamA is a eukaryotic-like glucoamylase responsible for glycogen- and starch-degrading activity of Legionella pneumophila. Int J Med Microbiol. 2011;301:133–139. doi: 10.1016/j.ijmm.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010;Chapter 11(Unit 11):15. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P, Deng X, Zhang J, Chen J, Zhang J, Zhang Q, Xiao Y, Chen S. Identification and characterization of the Francisella sp. strain 08HL01032 isolated in air condition systems. Wei Sheng Wu Xue Bao. 2009;49:1003–1010. [PubMed] [Google Scholar]
- Barabote RD, Xie G, Brettin TS, Hinrichs SH, Fey PD, Jay JJ, Engle JL, Godbole SD, Noronha JM, Scheuermann RH, Zhou LW, Lion C, Dempsey MP. Complete genome sequence of Francisella tularensis subspecies holarctica FTNF002-00. PLoS One. 2009;4:e7041. doi: 10.1371/journal.pone.0007041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstrom-Sternberg SM, Auerbach RK, Godbole S, Pearson JV, Beckstrom-Sternberg JS, Deng Z, Munk C, Kubota K, Zhou Y, Bruce D, Noronha J, Scheuermann RH, Wang A, Wei X, Wang J, Hao J, Wagner DM, Brettin TS, Brown N, Gilna P, Keim PS. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PLoS One. 2007;2:e947. doi: 10.1371/journal.pone.0000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, Borowsky M, Young S, Koehrsen M, Engels R, Pearson M, Howarth C, Larson L, White J, Alvarado L, Forsman M, Bearden SW, Sjöstedt A, Titball R, Michell SL, Birren B, Galagan J. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Ren CP, Desmond L, Vincent GA, Silman NJ, Brehm JK, Elmore MJ, Hudson MJ, Forsman M, Isherwood KE, Gurycová D, Minton NP, Titball RW, Pallen MJ, Vipond R. Genome sequencing shows that European isolates of Francisella tularensis subspecies tularensis are almost identical to US laboratory strain Schu S4. PLoS One. 2007;2:e352. doi: 10.1371/journal.pone.0000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, Karp PD, Larsson E, Liu Y, Michell S, Prior J, Prior R, Malfatti S, Sjöstedt A, Svensson K, Thompson N, Vergez L, Wagg JK, Wren BW, Lindler LE, Andersson SG, Forsman M, Titball RW. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Modise T, Ryder C, Mane SP, Bandara AB, Jensen RV, Inzana TJ. Genomic comparison between a virulent type A1 strain of Francisella tularensis and its attenuated O-antigen mutant. J Bacteriol. 2012;194:2775–2776. doi: 10.1128/JB.00152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino JF, Xiang Q, Karpathy SE, Jiang H, Yerrapragada S, Liu Y, Gioia J, Hemphill L, Gonzalez A, Raghavan TM, Uzman A, Fox GE, Highlander S, Reichard M, Morton RJ, Clinkenbeard KD, Weinstock GM. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J Bacteriol. 2006;188:6977–6985. doi: 10.1128/JB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, Gallagher LA, Manoil C, Ernst RK, Drees B, Buckley D, Haugen E, Bovee D, Zhou Y, Chang J, Levy R, Lim R, Gillett W, Guenthener D, Kang A, Shaffer SA, Taylor G, Chen J, Gallis B, D'Argenio DA, Forsman M. et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytun A, Malfatti SA, Vergez LM, Shin M, Garcia E, Chain PS. Complete genome sequence of Francisella philomiragia ATCC 25017. J Bacteriol. 2012;194:3266. doi: 10.1128/JB.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K, Inui M, Yukawa H. Structure, expression and products of the ribosomal RNA operons of Rhodopseudomonas palustris No. 7. Mol Genet Genomics. 2001;265:778–790. doi: 10.1007/s004380100466. [DOI] [PubMed] [Google Scholar]
- Boyer SL, Flechtner VR, Johansen JR. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol. 2001;18:1057–1069. doi: 10.1093/oxfordjournals.molbev.a003877. [DOI] [PubMed] [Google Scholar]
- Thomas R, Johansson A, Neeson B, Isherwood K, Sjostedt A, Ellis J, Titball RW. Discrimination of human pathogenic subspecies of Francisella tularensis by using restriction fragment length polymorphism. J Clin Microbiol. 2003;41:50–57. doi: 10.1128/JCM.41.1.50-57.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlund MK, Ryden P, Sjostedt A, Stoven S. Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster. Infect Immun. 2010;78:3118–3128. doi: 10.1128/IAI.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R, Akimana C, Jones S, Abu Kwaik Y. Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ Microbiol. 2010;12:2587–2612. doi: 10.1111/j.1462-2920.2010.02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule MG, Monack DM, Schneider DS. Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 2010;6:e1001065. doi: 10.1371/journal.ppat.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimana C, Kwaik YA. Francisella-arthropod vector interaction and its role in patho-adaptation to infect mammals. Front Microbiol. 2011;2:34. doi: 10.3389/fmicb.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous RD, McCarthy TR, Zahrt TC. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol Mol Biol Rev. 2009;73:684–711. doi: 10.1128/MMBR.00028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JJ, El-Etr S, Joubert LM, Moore E, Robison R, Rasley A, Spormann AM, Monack DM. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl Environ Microbiol. 2010;76:596–608. doi: 10.1128/AEM.02037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A. 2006;103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. Comparative and functional genomics of legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front Microbiol. 2011;2:208. doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol. 2002;184:754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett. 1999;177:191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- Coulthurst SJ. The Type VI secretion system - a widespread and versatile cell targeting system. Res Microbiol. 2013;164:640–654. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata ST, Kitaoka M, Wieteska L, Frech C, Chen N, Pukatzki S. The Vibrio Cholerae Type VI Secretion System: Evaluating its Role in the Human Disease Cholera. Front Microbiol. 2010;1:117. doi: 10.3389/fmicb.2010.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- Broms JE, Sjostedt A, Lavander M. The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol. 2010;1:136. doi: 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Ludu JS, Nano FE. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 2007;7:1. doi: 10.1186/1471-2180-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Meyer L, Sun K, Lavander M, Sjostedt A. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One. 2012;7:e50473. doi: 10.1371/journal.pone.0050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Meyer L, Lavander M, Larsson P, Sjostedt A. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS One. 2012;7:e34639. doi: 10.1371/journal.pone.0034639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Elfsmark D, Svensson K, Wikstrom P, Forsman M, Brettin T, Keim P, Johansson A. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray CG, Cowley SC, Cheung KK, Nano FE. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol Lett. 2002;215:53–56. doi: 10.1111/j.1574-6968.2002.tb11369.x. [DOI] [PubMed] [Google Scholar]
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology. 2011;157:3483–3491. doi: 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998;29:247–259. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotcke A, Monack DM. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect Immun. 2008;76:3473–3480. doi: 10.1128/IAI.00430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guina T, Radulovic D, Bahrami AJ, Bolton DL, Rohmer L, Jones-Isaac KA, Chen J, Gallagher LA, Gallis B, Ryu S, Taylor GK, Brittnacher MJ, Manoil C, Goodlett DR. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. J Bacteriol. 2007;189:6580–6586. doi: 10.1128/JB.00809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerk CL, Duplantis BN, Wang D, Burke RD, Chou AY, Elkins KL, Ludu JS, Nano FE. Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology. 2009;155:1489–1497. doi: 10.1099/mic.0.025379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan BW, McCaffrey RL, Lindemann SR, Allen LA, Jones BD. Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect Immun. 2009;77:2517–2529. doi: 10.1128/IAI.00229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charity JC, Costante-Hamm MM, Balon EL, Boyd DH, Rubin EJ, Dove SL. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 2007;3:e84. doi: 10.1371/journal.ppat.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, Carlson RW, Gunn JS. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun. 2007;75:3305–3314. doi: 10.1128/IAI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, Forsberg A, Charbit A. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun. 2009;77:1866–1880. doi: 10.1128/IAI.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Ericsson M, Sandstrom G, Tarnvik A, Sjostedt A. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect Immun. 1997;65:2183–2189. doi: 10.1128/iai.65.6.2183-2189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine SM, Shen H, Kelly JF, Chen W, Sjostedt A, Conlan JW. Virulence comparison in mice of distinct isolates of type A Francisella tularensis. Microb Pathog. 2006;40:133–138. doi: 10.1016/j.micpath.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunow R, Splettstoesser W, McDonald S, Otterbein C, O'Brien T, Morgan C, Aldrich J, Hofer E, Finke EJ, Meyer H. Detection of Francisella tularensis in biological specimens using a capture enzyme-linked immunosorbent assay, an immunochromatographic handheld assay, and a PCR. Clin Diagn Lab Immunol. 2000;7:86–90. doi: 10.1128/cdli.7.1.86-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P, Splettstosser W, Porsch-Ozcurumez M, Finke EJ, Grunow R. A novel screening ELISA and a confirmatory Western blot useful for diagnosis and epidemiological studies of tularemia. Epidemiol Infect. 2005;133:759–766. doi: 10.1017/s0950268805003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsch-Ozcurumez M, Kischel N, Priebe H, Splettstosser W, Finke EJ, Grunow R. Comparison of enzyme-linked immunosorbent assay, Western blotting, microagglutination, indirect immunofluorescence assay, and flow cytometry for serological diagnosis of tularemia. Clin Diagn Lab Immunol. 2004;11:1008–1015. doi: 10.1128/CDLI.11.6.1008-1015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancuta P, Pedron T, Girard R, Sandstrom G, Chaby R. Inability of the Francisella tularensis lipopolysaccharide to mimic or to antagonize the induction of cell activation by endotoxins. Infect Immun. 1996;64:2041–2046. doi: 10.1128/iai.64.6.2041-2046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumeister B, Faigle M, Sommer M, Zahringer U, Stelter F, Menzel R, Schutt C, Northoff H. Low endotoxic potential of Legionella pneumophila lipopolysaccharide due to failure of interaction with the monocyte lipopolysaccharide receptor CD14. Infect Immun. 1998;66:4151–4157. doi: 10.1128/iai.66.9.4151-4157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahringer U, Knirel YA, Lindner B, Helbig JH, Sonesson A, Marre R, Rietschel ET. The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog Clin Biol Res. 1995;392:113–139. [PubMed] [Google Scholar]
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddaramappa S, Challacombe JF, Petersen JM, Pillai S, Kuske CR. Genetic diversity within the genus Francisella as revealed by comparative analyses of the genomes of two North American isolates from environmental sources. BMC Genomics. 2012;13:422. doi: 10.1186/1471-2164-13-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Sun H. Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect Immun. 2002;70:1–4. doi: 10.1128/IAI.70.1.1-4.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BJ, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BJ, Kwaik YA. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coil DA, Anne J. Twitching motility in Legionella pneumophila. FEMS Microbiol Lett. 2009;293:271–277. doi: 10.1111/j.1574-6968.2009.01532.x. [DOI] [PubMed] [Google Scholar]
- Stewart CR, Rossier O, Cianciotto NP. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J Bacteriol. 2009;191:1537–1546. doi: 10.1128/JB.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Chakraborty S, Liu J, Thanassi DG, Klose KE. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology. 2008;154:2139–2150. doi: 10.1099/mic.0.2008/018077-0. [DOI] [PubMed] [Google Scholar]
- Gil H, Benach JL, Thanassi DG. Presence of pili on the surface of Francisella tularensis. Infect Immun. 2004;72:3042–3047. doi: 10.1128/IAI.72.5.3042-3047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Monfett M, Maier TM, Benach JL, Frank DW, Thanassi DG. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect Immun. 2008;76:2852–2861. doi: 10.1128/IAI.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, Guina T. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol. 2006;62:227–237. doi: 10.1111/j.1365-2958.2006.05365.x. [DOI] [PubMed] [Google Scholar]
- Salomonsson EN, Forslund AL, Forsberg A. Type IV Pili in Francisella - A Virulence Trait in an Intracellular Pathogen. Front Microbiol. 2011;2:29. doi: 10.3389/fmicb.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Labadie RF, Mitchell J, Balachandran R, Fitzpatrick JM. Customized, rapid-production microstereotactic table for surgical targeting: description of concept and in vitro validation. Int J Comput Assist Radiol Surg. 2009;4:273–280. doi: 10.1007/s11548-009-0292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantsev AP, Golovliov IR, Ohara Y, Mokrievich AN, Obuchi M, Norqvist A, Kuoppa K, Pavlov VM. Genetic organization of the Francisella plasmid pFNL10. Plasmid. 2001;46:210–222. doi: 10.1006/plas.2001.1548. [DOI] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S, Nishimura Y, Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992;174:4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pihive E, Blaha D, Chenavas S, Thibault F, Vidal D, Valade E. Description of two new plasmids isolated from Francisella philomiragia strains and construction of shuttle vectors for the study of Francisella tularensis. Plasmid. 2009;62:147–157. doi: 10.1016/j.plasmid.2009.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic analysis of Francisella isolate W12-1067, Ft. holarctica strain LVS (Ft. LVS), Ft. novicida, F. philomiragia and L. pneumophila Paris (Lpp). Growth on BCYE agar plates with (+) and without (-) additional cysteine (Cys) (A). The results are representative of three independent experiments. Growth in medium T in the presence of different amounts of NaCl (B). Results are mean standard deviations of three independent experiments of duplicate samples. Chitin degradation by the supernatant of different strains grown in medium T and then incubated on agarose plates containing 0.1% deacetylated glycol chitin (C). Halos around the inoculation site revealed the presence of degrading activity after 2 days of incubation at 37°C. The results represented are representative of three independent experiments.
Annotated genes (peg), rRNAs and tRNAs.
MAUVE alignment of strain W12-1067 and Ft. novicida U112. The colored boxes represent homologous segments free of genomic rearrangements. Homologous regions are connected by lines between genomes. Non-boxed regions lack homology between genomes. White areas indicate that the sequences are specific to a genome. (The synteny between both genomes was not estimated, since the genome of W12-1067 is a draft genome).
Genes of the Sec, type I and Tol secretion systems.
Regulatory proteins.
The wbt gene cluster of strain Francisella W12-1067.
Type IV pili encoding genes.


