Abstract
Somatic mutations reported in large-scale breast cancer (BC) sequencing studies primarily consist of protein coding mutations. mRNA splicing mutation analyses have been limited in scope, despite their prevalence in Mendelian genetic disorders. We predicted splicing mutations in 442 BC tumour and matched normal exomes from The Cancer Genome Atlas Consortium (TCGA). These splicing defects were validated by abnormal expression changes in these tumours. Of the 5,206 putative mutations identified, exon skipping, leaky or cryptic splicing was confirmed for 988 variants. Pathway enrichment analysis of the mutated genes revealed mutations in 9 NCAM1-related pathways, which were significantly increased in samples with evidence of lymph node metastasis, but not in lymph node-negative tumours. We suggest that comprehensive reporting of DNA sequencing data should include non-trivial splicing analyses to avoid missing clinically-significant deleterious splicing mutations, which may reveal novel mutated pathways present in genetic disorders.
Large-scale DNA sequencing studies have attempted to elucidate the genomic landscapes of breast cancer tumours to identify mutated genes and genomic variation that contribute to tumour development and progression1,2,3,4,5. Typically, somatic mutations within gene coding regions are identified and then filtered for rare or novel variants predicted to affect protein structure or function6,7,8,9. Frequently mutated genes are cataloged, with the goal of inferring defective genes that are more likely to contribute to tumour phenotypes. However, there does not appear to be a consistent set of somatic driver mutations in most breast cancer cases. For instance, in 100 cases, 73 different combinations of abnormal gene sequences were reported4.
Some established cancer genes are enriched for mutations (i.e. TP53, PIK3CA, PTEN, MAP3K1, AKT1, CDH1, GATA3, MLL3 and RB1), in addition to genes that were not previously associated with breast cancer (including CBFB, RUNX1, TBX3, NF1 and SF3B1)1,2,3,4,5. At least 49 genes (including known breast cancer genes) have been found to be significantly mutated, 16 of these reproducibly across multiple studies, and the majority were mutated in <10% of tumours.
Inconsistencies in mutation composition among different tumours present significant challenges to understanding the underlying etiology of tumour phenotypes. As a result of epistasis, mutations in genes with linked biochemical functions would be expected to reveal dysfunctional pathways in tumours10. Focusing analyses to one molecular subtype of breast cancer can also be useful in delineating dysregulated pathways that define the basis of tumour phenotypes3. Significant insight into tumour biology has come from selecting tumours with specific clinical identifiers, for example, by limiting mutation catalogs in metastatic tumours10,11.
Somatic mutation analyses of tumour exomes have focused on alteration of amino acid sequences, or highly conserved dinucleotides adjacent to exons, which usually impact mRNA splicing. Since these variants most likely comprise only a fraction of the total mutational load, the pathways inferred to be dysregulated in these tumours may be incomplete. For example, in familial breast cancer, variants of unknown significance have been explained by both experimental validation and in silico predictions of defects in BRCA 1/2 mRNA splicing12,13. Typically, genomic studies have used tools that predict splicing mutations based on the highly conserved dinucleotide sequences at mRNA 5′ donor and 3′ acceptor sites8,14. There are other well established methods that can identify splicing mutations beyond those directly at natural sites15,16,17, but these approaches have not been applied to genome-scale cancer studies, until recently18. Published studies have revealed only a small fraction of reported somatic mutations in cancer to be splicing mutations, accounting for only 2% of those reported1,2,3,4,5. The present study considers the possibility that many somatic splicing mutations may be overlooked or are undetected by the conservative approaches currently used in analyses of tumour genomes.
Splicing mutations frequently lead to changes in the sequence and structure of the encoded protein, which are usually distinguishable from those generated by normal alternative splice isoforms. Constitutive splicing mutations are frequently deleterious and are a major cause of inherited and acquired diseases19. In cancer, aberrant splicing (including alternative isoforms that are not a result of cis mutation) is known to cause or promote tumour propagation20, and has been described as an additional hallmark of the disease21. RNA analyses can detect the effect of many splicing mutations directly22,23. In this paper, we comprehensively analyze predicted splicing mutations in breast cancer tumours using DNA sequencing data from The Cancer Genome Atlas (TCGA)5. We then use tumour-matched RNA sequencing data to statistically validate aberrant splicing patterns of expressed genes in these tumours that result from these mutations24. We extended our splicing mutation analyses beyond molecular breast cancer subtypes and identified other clinical parameters associated with specific mutation pathways. We suggest that DNA sequencing analyses that incorporate in-depth splicing mutation studies reveal additional mutant genes and biochemical pathways, which may contribute to breast cancer etiology.
Results
Derivation of mutations
Somatic mutations in 472 breast cancer tumours from 445 breast cancer patients were called using matched tumour-normal DNA exome sequencing data from TCGA5 (Supplementary Table S1). There were 149,959 single-nucleotide variants (SNVs) and 10,000 insertion/deletions (indels) detected using the variant caller, Strelka6 (see Supplementary Material section I for results from an alternative variant caller and reasons for our selection of Strelka). Protein coding mutations were annotated by ANNOVAR8 and splicing mutations with the Shannon Human Splicing Pipeline18 (Table 1, see Supplementary Tables S2–4 for a list of all mutations). The Shannon Pipeline predicted significantly more splicing mutations than reported by TCGA, because the information-theoretic method employed enables analyses of variants beyond exon boundaries that alter mRNA splicing. 948 variants were found to affect both protein coding and splicing in 747 genes, among 319 tumours. DYNC2H1, TP53 and PASD were the most commonly mutated of this group, containing 21, 11, and 9 exonic variants, respectively. Alteration of mRNA splicing was predicted as a result of 213 substitutions at synonymous codons among 139 tumours. Reanalysis of coding changes confirmed high concordance with the validated TCGA SNVs, however indels were less reproducible (Supplementary Table S5). Overall, 82.1% (n = 21,041) of protein coding mutations, and 86.5% (n = 371) of splicing mutations reported by TCGA were confirmed. A small subset of protein coding TCGA substitutions that were missed occurred within genes commonly mutated in breast cancer (35 TP53, 13 MLL3, 22 GATA3, 25 MAP3K1, 11 CDH1 and 10 PIK3CA; see Supplementary Table S6), however all splicing-associated SNVs found by TCGA in cancer-related genes were detected.
Table 1. Single nucleotide variant summaries by mutation type.
| Type | Mutation Count |
|---|---|
| Protein coding | |
| Synonymous | 14,717 |
| Nonsynonymous | 40,649 |
| Stop gain or loss | 2,587 |
| Total protein coding variants | 57,953 |
| Splicing | |
| Cryptic | 1,130 |
| Inactivating | 1,355 |
| Leaky | 2,721 |
| Total splicing variants | 5,206 |
| Protein coding mutations also predicted to affect splicing | |
| Synonymous | 213 |
| Nonsynonymous | 664 |
| Stop gain or loss | 71 |
| Total | 948 |
| Synonymous also splicing | 1.4473% |
| Nonsynonymous also splicing | 1.6335% |
| Stop gain or loss also splicing | 2.7445% |
Significantly mutated genes
Significantly mutated genes were identified with the Mutational Significance in Cancer (MuSiC) software suite25. There were 225 genes with false discovery rates (FDR) of <0.05, based on the Fisher's combined P-value (FCPT), convolution (CT) and likelihood ratio (LRT) tests. These results were compared with the 49 genes previously identified as significantly mutated1,2,3,4,5 (Supplementary Table S7). Among the previous genes reported by TCGA, TP53, CDH1, MAP3K1, and MLL3 were significantly mutated in this study by all tests, and AFF2, SF3B1, and CBFB were significant for the CT and LRT tests only. We additionally identified ARID1A as significantly mutated, concordant with an independent, large-scale, breast cancer genomics study4.
Validating predicted splicing mutations
Changes in mRNA splicing from the predicted mutations were validated with Veridical24, which corroborates predicted, aberrant splice isoforms by assessing mutation-derived sequence reads in tumour RNA relative to their abundance in controls lacking the mutation. Controls comprised tumours lacking a particular mutation (usually, n = 414) plus additional normal samples (n = 106). Of all variants analyzed from the 415 tumours with RNA-Seq data (n = 4,952), 988 variants (~20%) in 819 genes caused one or more splicing aberrations at significantly higher levels than in controls (p ≤ 0.05; i.e. intron inclusion, exon skipping, or cryptic splicing). Predicted natural splice site mutations (822 of 3,863, or 21.3%), were validated by abnormal mRNA isoforms more often than cryptic splice site mutations (166 of 1,089 or 15.2% variants). A total of 309 mutations were found to cause exon skipping, of which 163 (53%) led to expected frameshift mutations.
Sufficient expression levels for each gene, based on RNA-Seq coverage, were required for validation of mutations. An expression heat map, clustered by BC subtype, is shown in Supplementary Fig. S1. Variants occurring within significantly expressed genes (defined as an average of ≥20 reads per base) were statistically validated for 862 (27%) of 3,156 variants (p ≤ 0.05). Of 263 variants reported by TCGA in genes with at least this level of expression, 156 (59%) were validated by exon skipping (26 variants), by intron inclusion (80 variants), and by the combination of both types of evidence (50 variants, p ≤ 0.05).
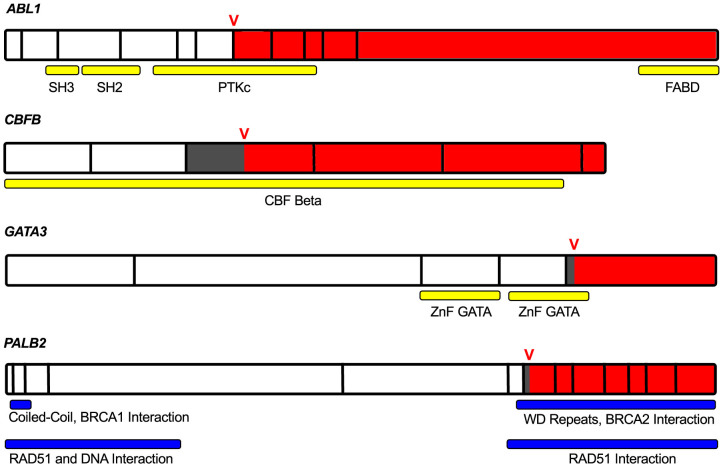
Predicted cryptic splicing mutations were confirmed based on the presence of unique junction-spanning reads corresponding the ectopically spliced isoforms in GATA3, PALB2, CBFB, ABL1, C2CD2L, ENSA, NASP, NOP9, and TFE3 (Supplementary Fig. S2). Four of these genes have been linked to tumourigenesis: ABL1, an oncogene, GATA3 and PALB2, which are associated with familial breast cancer26,27, and CBFB has been recently implicated in breast cancer by TCGA5 and others1,2. These cryptic splicing mutations lead to short exonic deletions that alter the reading frame, and likely affect the activity of the gene products (Fig. 1). The GATA3 cryptic isoform is the only detectable transcript in the majority of controls, although it is substantially more abundant in the tumour sample (Supplementary Fig. S3).
Figure 1. Cancer genes with validated cryptic splicing.
mRNA of ABL1, CBFB, GATA3 and PALB2, which each have validated cryptic splicing mutations confirmed using tumour-matched RNA-Seq data. Full gene lengths are displayed with vertical black bars outlining exon boundaries. The location of the cryptic variant is denoted by the red V, and the variant consequence is highlighted by white (wild type), dark grey (exonic deletion), and red (frameshift mutation). Conserved domains and protein interactions are labeled by the yellow and blue horizontal bars. In ABL1, the catalytic and C-terminal F-actin binding domains are disrupted. In PALB2, the region that interacts with BRCA2 is truncated. In the GATA3 aberrant transcript, the second zinc finger domain and a conserved motif crucial for DNA binding and protein function are affected by the altered reading frame.
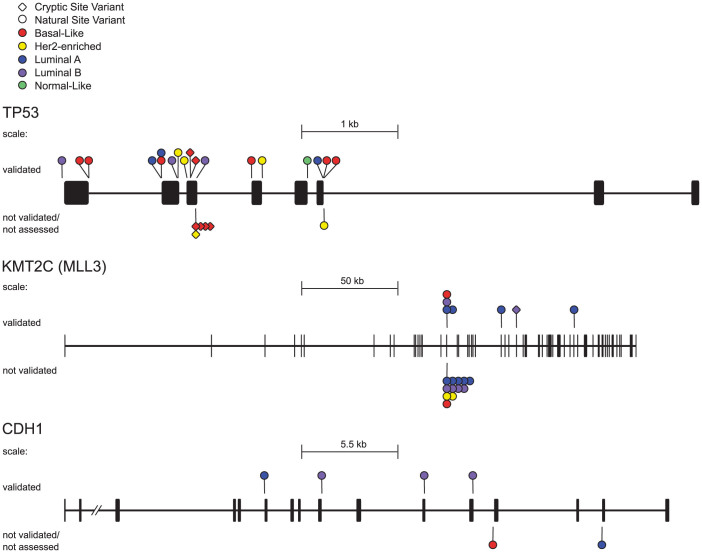
The most commonly mutated genes with splicing mutations were also found by MuSiC to be significantly mutated in these tumours (n = 13, FDR < 0.05), and at least one third of the mutations were validated with RNA-Seq data (Table 2). In TP53, which exhibited the highest density of splicing mutations (Fig. 2), 18 of 23 (78%) predicted variants were validated to cause aberrant splicing (p ≤ 0.05). All of the validated mutations exhibited statistically significant intron inclusion, and, in three instances, also resulted in exon skipping.
Table 2. Genes Most Commonly Mutated with Splicing Mutations.
| Gene Symbol* | # Splicing Mutations | # Validated | % Validated |
|---|---|---|---|
| TP53 | 24 | 18 | 75 |
| HMCN1 | 19 | 9 | 47 |
| KMT2C (MLL3) | 19 | 7 | 37 |
| FHAD1 | 12 | 4 | 33 |
| RAB3GAP1 | 11 | 4 | 36 |
| BCLAF1 | 11 | 3 | 27 |
| ANKEF1 | 10 | 6 | 60 |
| RRM1 | 8 | 4 | 50 |
| RPRD1A | 7 | 2 | 29 |
| SCAMP5 | 7 | 2 | 29 |
| CDH1 | 6 | 4 | 67 |
| ACTR3 | 6 | 2 | 33 |
*FDR < 0.05 for all genes from MuSiC (Fisher's combined P-value, convolution and likelihood ratio tests).
Figure 2. Significantly mutated breast cancer genes with validated splicing mutations.
TP53, KMT2C and CDH1 gene lengths are displayed with both exons (thick lines/boxes) and introns (thin horizontal lines), along with the location of all splicing mutations. Diamond markers denote cryptic mutations, natural splice site mutations are indicated by a circle and the colour of the marker corresponds with breast cancer tumour subtype. Mutations validated by Veridical are found above the gene, and those mutations not assessed or not validated are below.
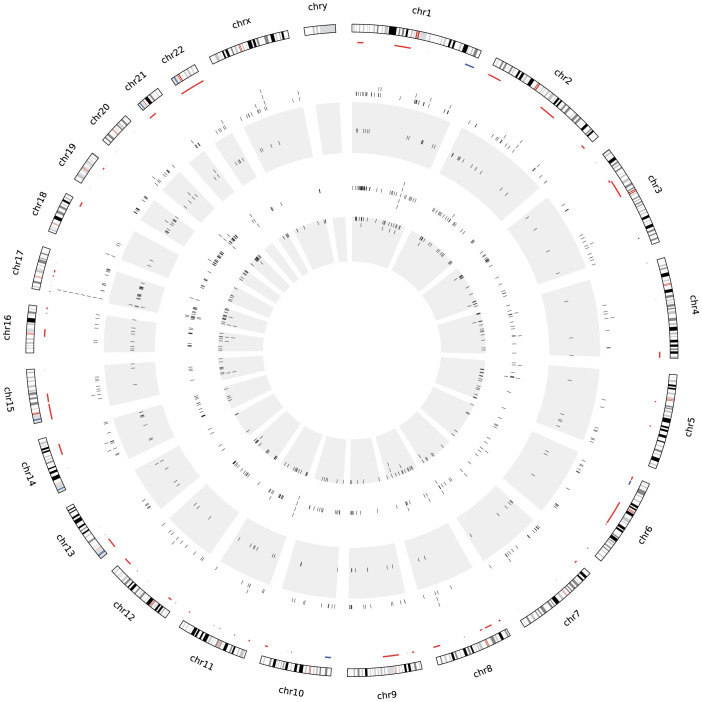
Copy number analysis of mutated genes
The validated mutations are organized and segregated by tumour subtype on a Circos plot28 (Fig. 3). Copy number changes portray the genomic locations of deletions or amplifications that coincide with these variants. Validated splicing mutations exhibit a relatively uniform genomic distribution, except for significantly mutated genes, such as TP53 on chromosome 17 and HMCN1 on chromosome 1. We investigated variants in regions showing copy number losses, which may constitute the “second hit” in oncogenesis. Of the 49 genes found to be significantly mutated in breast cancer1,2,3,4,5, five contained splicing mutations (BRCA1 (2 tumours), PTEN (2 tumours), MAP2K4 (4 tumours), MAP3K1 (4 tumours) and KMT2C (7 tumours; also known as MLL3)) and also recurred within commonly deleted intervals. Of all genes with validated mutations in deleted regions, 9 harbored more than 2 variants: 1 had three, 4 had four, and only KMT2C possessed more than 4 variants.
Figure 3. Circos plot of validated splicing mutations.
From the outermost ring in, chromosomes are labeled clockwise with copy number data inside them that displays deletions in red and amplifications in blue, mutations validated by Veridical (indicated by black ticks) are then plotted by subtype with basal-like in the outer white ring, HER2-enriched in the outer grey ring, then luminal A (inner white) and luminal B (inner grey).
Analysis of pathways enriched in mutant genes
Mutated genes were clustered by pathway overrepresentation analysis29 for protein coding (Supplementary Table S8, n = 202) and splicing mutations (Supplementary Table S9, n = 452). There were 100 pathways were common to both mutation sets (Supplementary Table S10). Pathways associated with all types of mRNA splicing mutations include those that affect collagen structural genes and enzymes that modify or metabolize collagen (n = 14, Supplementary Table S11 #1–14), and several that involve the extracellular matrix (ECM, n = 4, Table S11 #15–18). Many of these pathways (n = 17, Table S11 #1–13,15–18) are also overrepresented by pathway analysis of protein coding mutations.
Relationship of mutation spectra to clinical findings
Segregating splicing mutations by patient lymph node status revealed significant differences in mutated pathways between the two groups. Biochemical pathways with overrepresented mutant genes in lymph node-negative (LN−) vs. lymph node-positive (LN+) tumours are indicated in Supplementary Tables S12 and S13, and compared in Supplementary Table S14. There are 94 pathways overrepresented in both LN+ and LN− (Table S14 #421–514), including 17 collagen (Table S14 #421–437), and 3 ECM (Table S14 #438–440) pathways. Ontologically-related pathways29,30 were grouped (Supplementary Table S15) and visualized as Word Clouds (Fig. 4). Pathway groups overrepresented (p < 0.05) in both tumour subsets included 17 pathways involving collagen-ECM protein phosphorylation pathways, metabolism, cell cycle, DNA repair, and cellular response to stress. However, 13 pathways involving collagen (Table S14 #1–13), and 9 pathways involving NCAM1 (Table S14 #17–25) were overrepresented uniquely in LN+ tumors, but not in LN− tumours.
Figure 4. Word Clouds demonstrating differences between overrepresented mutated pathways in lymph node-positive (a) and lymph node-negative (b) tumours.
The abstracted pathways (see methods) were plotted if present two or more times. The size of the words as well as the corresponding colours of the pathway names indicates the frequency of that abstracted pathway, and can be compared within and between the word clouds of each tumour subset.
NCAM1, or the neural cell adhesion molecule, is a member of the immunoglobulin super family with a role in cell-cell and cell-matrix interactions during development and cellular differentiation. Mutations in NCAM1 signaling genes for neurite outgrowth (Supplementary Table S16 #1) were still overrepresented in tumours with lymph node invasiveness, even after genes common to both tumour subsets were masked from the analysis, i.e. primarily collagen and ECM genes (Supplementary Tables S16–17). These include defects in NCAM1 interactions with FYN and GRB2, a ternary complex that participates in the conversion of RAS:GDP to RAS:GTP, which subsequently initiates the RAF/MAP kinase cascade.
We then reanalyzed these data after conservatively limiting the set of mutant genes to those containing the most deleterious mutations (Supplementary Table S18; stop-gain, stop-loss, frameshift/indel mutations, and validated splicing mutations). Four of the 8 sub-pathways of NCAM1 signaling for neurite outgrowth were overrepresented solely in LN+ tumours. Autophosphorylation/dephosphorylation of NCAM1- bound Fyn, as well as NCAM1-interactions with collagens were overrepresented. The most commonly mutated genes within these pathways are SPTA1, CACNA1D, COL6A5, NCAM1, and COL6A6 (Supplementary Table S19). CACNA1D is a voltage-dependent Ca2+ channel (VDCC) that associates with NCAM1 in growth cones at the sites of NCAM1 clustering29,30. In addition, 6 other channel genes that are expressed in breast tissue31 were found to be frequently mutated (CACNA1C, CACNA1D, CACNA1G, CACNA1H, CACNB1, CACNB3). Mutations interrupting these VDCC interactions may alter the NCAM-dependent Ca2+ influx. Collagen VI is expressed as supramolecular aggregates of composite structures of different chains and is among the most abundant components of the ECM32. Knockdown of NCAM significantly reduces expression of ECM components33, including collagen, weakening the ECM. Mutations in these ECM components may also diminish matrix integrity, possibly resulting in more porous structures34.
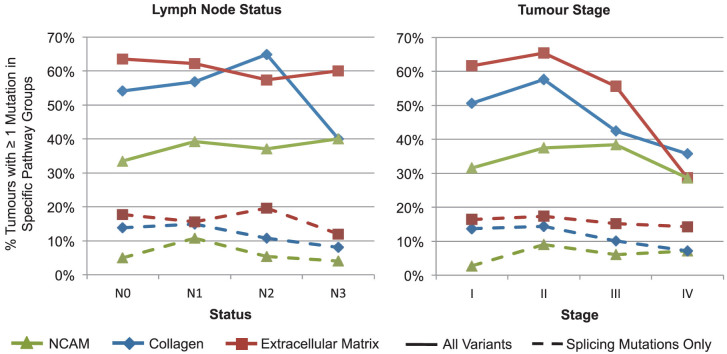
Elevation of NCAM1-related gene pathway mutations in lymph node-positive tumours
NCAM1, collagen, and ECM pathway mutations were assessed in tumours, stratifying by lymph node status and tumour stage (Fig. 5). The percentage of tumours with NCAM1-related pathway splicing mutations was increased in N0 (110 localized tumours) and N1 (84 tumours with lymph node involvement), as well as Stage I (37) and II tumours (140). Advanced lymph node involvement and tumour stage were not associated with increased numbers of collagen and ECM pathway splicing mutations, but rather a decrease in the percent of tumours with these pathways mutations in advanced stages was observed. A multiple factor analysis (MFA; Table 3) was performed to assess contributions of the number of NCAM1-related pathway mutations per tumour (both protein coding and splicing), clinical parameters including stage (AJCC tumour stage, lymph node status and metastasis stage), receptor status (HER2, PR, and ER positivity), and patient outcome (relapsed, living/deceased). NCAM1-related pathway mutations were either absent (n = 213), harbored a single mutation (n = 117), or two or more mutations (n = 112) per tumour. The MFA components containing NCAM1-related pathway mutations were moderately correlated with both tumour stage and receptor status, and accounted for 11% of the variance.
Figure 5. Percent of tumours with mutations by pathway group and clinical factors.
The percent of tumours with NCAM1 (red square), collagen (blue diamond), and ECM (green triangle) pathway mutations were plotted by lymph node status and tumour stage for all mutations (solid lines), and splicing mutations alone (dashed line).
Table 3. Multiple factor analysis of NCAM1 related pathway mutations and clinical parameters per tumour.
| Dim.1 | Dim.2 | Dim.3 | Dim.4 | Dim.5 | |
|---|---|---|---|---|---|
| A. No. Mutations in NCAM Pathways* | 0.103 | 0.892 | 0.910 | 0.367 | 0.321 |
| Stages | 0.804 | 0.459 | 0.381 | 0.833 | 0.725 |
| Receptor status | 0.379 | 0.356 | 0.406 | 0.471 | 0.641 |
| Patient status | 0.868 | 0.159 | 0.050 | 0.106 | 0.159 |
| % Variance explained | 7.618 | 5.699 | 5.635 | 4.944 | 4.694 |
| B. No. Mutations Unique to NCAM Pathways∧ | 0.264 | 0.899 | 0.894 | 0.304 | 0.300 |
| Stage | 0.791 | 0.413 | 0.380 | 0.877 | 0.752 |
| Receptor status | 0.389 | 0.427 | 0.411 | 0.429 | 0.610 |
| Patient status | 0.851 | 0.083 | 0.158 | 0.168 | 0.221 |
| % Variance explained | 7.716 | 5.816 | 5.534 | 4.941 | 4.743 |
*mutation count for all genes in NCAM pathways.
∧mutation count for genes unique to NCAM pathways, and not in collagen or ECM pathways.
Analysis of tumour subtypes
Splicing mutation analysis in different tumour subtypes revealed between 9–15 mutations per tumour, which generally accounted for 8–9% of all mutations detected (Supplementary Table S20) and are similar levels to those previously reported18. Pathway analyses for each subtype, stratified by lymph node status, indicated higher enrichment of NCAM1-related gene mutations in basal-like and HER2/ERBB2-enriched LN+ tumours (Supplementary Table S21, Supplementary Fig. S5: see word clouds). LN+ basal-like and HER2-enriched tumours were the only tumours found to have significant enrichment in “NCAM signaling for neurite out-growth”, identifying those tumour subtypes and pathways that may play a role in tumour migration. No single gene was significantly mutated within the NCAM1 pathways that were overrepresented in LN+ tumours. This suggests that a general defect in NCAM1-pathway signaling may be associated with lymph node metastasis in breast cancer.
Discussion
Breast carcinoma tumour exomes contain more deleterious mutations than previously recognized. Using Shannon information theory, we have predicted an expanded set of mutations that affect post-transcriptional mRNA processing that either reside in non-coding regions, or overlap known codons. We then employed Veridical24, a high-throughput, genome-scale method, to statistically validate mRNA splicing consequences that result from the predicted variants. This study complements the analyses performed by TCGA5, which comprehensively reported protein-coding mutations, along with gene expression, epigenetic, and copy number changes. Together with known deleterious coding sequence variants, the identification of such splicing mutations can refine and impact our understanding as to which biochemical pathways are dysregulated in these tumours.
Pathway overrepresentation analyses reproduced many of the same pathways identified by TCGA. In our analysis, a number of these attained or increased significance when genes with previously unrecognized splicing mutations were included. Both splicing mutations alone and the complete variant set from all tumours were enriched for genes in pathways known to play a role in tumour development and progression including signaling by growth factors, cell cycle, ECM organization, and cell-to-cell communication. Stratifying the tumours by lymph node status revealed that splicing mutations were enriched for genes within NCAM1 pathways in LN+ tumours, exclusively. Splicing mutations in these pathways were much rarer and sparsely distributed in LN− tumours, with 11 mutations in 92 LN− tumours and 25 mutations in 118 LN+ tumours. Interestingly, this enrichment was not observed when all protein coding substitutions were analyzed, but was significant when assessing all variants that were likely to be deleterious (i.e. validated splicing mutations, stop codon gain or losses and frameshift substitutions). We did not attempt to differentiate loss versus gain of function, however splicing mutations and nonsense codons usually result in loss of function. The percent of tumours with NCAM1-related pathway mutations increased by 6% from lymph node stage N0 to N1 and N3 and by 7% from stage I to III. The lower fraction of tumours with collagen pathway mutations at higher lymph node stages (N3, N4), and with ECM-related mutations in tumour stages III and IV could be related to clonal selection of distinct metastatic phenotypes35, however it is also possible that the decreases may not be significant due to the lower numbers of tumours in these categories.
Our results indicate that NCAM1 pathways are more likely to be dysregulated in tumours that have migrated to lymph nodes. We found the enrichment of NCAM1-related pathway splicing mutations in LN+ tumours was specifically present in HER2-enriched and basal-like tumours. Basal-like, specifically triple-negative, tumours have been associated with poor prognosis and survival36. Early and metastatic HER2 positive tumours were associated with poor prognoses37 until the more recent introduction of HER2-targeted therapies38. In these tumour subtypes, the presence of NCAM1-related pathway mutations may indicate a propensity to migrate and/or form distant metastases.
Dysregulated expression of NCAM1 has been suggested to contribute to tumour migration in other cancers: (i) gene silencing and localization studies have suggested that “NCAM is both necessary and sufficient to promote a migratory and invasive phenotype in EOC cells, with no major effect on cell proliferation”34, (ii) overexpression of NCAM1 has been linked to high ovarian carcinoma tumour grade34 and greater metastatic potential in melanoma cells39; (iii) preserved NCAM1 expression in differentiated thyroid carcinoma has been cited as an indicator for tumours with as increased risk of forming distant metastases40 and (iv) blocking NCAM1 function in murine lung tumour cells led to cell vulnerability to apoptosis. More generally, NCAM1 is known to play a role in apoptotic evasion and matrix degradation, and has potential roles in directional cell migration, cell polarity, extravasation and immunological escape41. NCAM1-mediated stimulation of FGFR activity is causally linked to tumour malignancy, suggesting that this NCAM1-FGFR interaction may be an effective therapeutic target. It is notable that we find mutations in breast tumours that affect the NCAM1-FGFR interaction occur in pathways that are overrepresented in LN+, but not LN− tumour genomes.
NCAM1 homophilic clusters form within lipid rafts on the cell membrane. Spectrin, an NCAM1-binding cytoskeletal protein, colocalizes with NCAM1 and is codistributed within lipid rafts42. Frequent mutations in spectrin (SPTA1) may prevent its association with RPTPα, thereby impeding its subsequent association with the cytoplasmic NCAM1 domain, redistribution of NCAM1 and cluster formation. This could abrogate downstream interactions with FYN and GRB2, ultimately affecting activation of RAS. These findings merit further investigation into how dysregulation in these different partners (i.e. NCAM1, FGFR and the other interacting proteins), acting as an ensemble, may promote tumour metastasis.
The number of aberrant mRNA splicing mutations reported by TCGA5 is <10% of those reported here, and the variants were not functionally validated in the previous study. We predict that 8% of all cis-activating point mutations detected in these tumours will significantly reduce the strength of the corresponding natural splice sites. The 5,206 splicing mutations reported here nearly double the number of mutations that lead to stop-gains or losses (2,587 variants in 1,907 genes), and the number of insertions/deletions leading to frameshift substitutions (2,707 variants in 1,848 genes) in this set of tumours. It is not surprising that these analyses revealed previously unrecognized pathways that may be dysregulated, in addition to those already known in these tumours.
Our analysis of significantly mutated genes based on the protein coding and splicing mutations reproduced many of the genes reported by TCGA, and revealed one additional gene, ARID1A. ARID1A has been implicated in breast cancer in a large-scale genomic study4 and has also been mutated in 57% of ovarian clear-cell carcinoma tumours43. Thirteen genes identified as significantly mutated in breast cancer by the TCGA did not reach statistical significance within our study (Table S4). This can be explained by a number of different factors: differences in variant callers, variant annotation, the number of tumours analyzed and differences in the filtering of variants, once the gene set was derived. In addition, TCGA initially analyzed all variants (SNVs and indels) by tumour subtype, unlike our study, which considered mutations in all tumours, then reanalyzed overrepresented pathways with mutations by subtype. Mutations that lead to a significant level of aberrant splicing can alter or improve genomic signatures, which are important when assessing potential biomarkers, diagnosis and prognosis, and metastatic or treatment-resistant tumour phenotypes.
Methods
This study involved a reanalysis of controlled-access data from The Cancer Genome Atlas Project (NCBI dbGaP Project #988: Predicting common genetic variants that alter the splicing of human gene transcripts, PI: PK Rogan). DNA and RNA breast cancer sequencing data was obtained for 445 tumours from 442 patients (Supplementary Table S1; July, 2012 DNA-Seq download; July, 2013 RNA-Seq Download)5. The tumour-normal pairs used mirrored those published by the TCGA in the Level 2 mutation data. Duplicate mutations in the same patient from two different tumour-normal pairs are reported, but were treated as one tumour for the mutation summaries reported by tumour. Somatic mutations were predicted from the same DNA sequencing data using two different algorithms: Strelka (v1.0.10)6 and SomaticSniper (v1.0.2)44 (See Supplementary Material section I). Realignment was not necessary before running Strelka because of the program's internal realignment capabilities, so Strelka was run on the raw BAM files downloaded from TCGA. Default parameters were used with the provided Burrows-Wheeler Aligner (BWA) configuration file, since BWA was used in the initial exome alignments. Additionally, the isSkipDepthFilters configuration option was changed to true, since such depth filters are designed for use on whole-genome data and would erroneously filter out most data when used with exome sequencing data. Strelka's BWA quality control script was run to remove variants considered low quality. Variants that were found to be common SNPs, defined by those that were annotated with dbSNP135 in over 1% of the population, were filtered out from the variant set before any subsequent analyses.
Somatic mutations, including single-nucleotide variants (SNVs) and insertion/deletions (indels) were used to predict the coding and non-coding genic effects of the variants. Annovar (August 23, 2013 release)8 was used with default parameters to predict which variants are likely to affect amino acid sequence and splicing at the natural splice sites. The Shannon Human Splicing Pipeline Version 2.0 (Shannon Pipeline)18 was used to complete a more in-depth analysis of splicing mutations, which predicts variants that will alter the binding affinity of the natural site or cause cryptic splicing (i.e. extension or truncation of an exon). The Shannon Pipeline results were subsequently filtered to prioritize which variants are most likely to have the greatest effect on mRNA splicing, using the filtering criteria outlined in Supplementary Fig. S6.
Multiple factor analyses used the R package FactoMineR (version 1.25)45. Clinical parameters were obtained from the TCGA including AJCC tumour staging (metastasis stage code, neoplasm disease lymph node stage, and neoplasm disease stage), receptor statuses (estrogen, progesterone, and HER2/neu immunohistochemistry receptor statuses) as well as patient status (neoplasm cancer status and vital status). These clinical parameters were input into FactoMineR as qualitative groups, as listed above, along with the number of NCAM1 pathway mutations. Within the program, options were set to perform clustering after MFA, and to automatically determine the choice of the number of clusters. A second MFA was performed based on the number of NCAM1 pathway mutations per tumour in genes present only in the NCAM1 related pathways that were also not present in the collagen or extracellular matrix pathways.
Word Clouds were generated to portray the overrepresentation analysis of mutated pathway results generated with Reactome29,30 and, in particular, the differences between lymph node-positive and -negative tumour samples. The primary input data for these graphics was the overrepresented pathways from Reactome, partitioned according to subtype and lymph status. Additional sets were composed of all subtypes and all subtypes with only pathways not found within both lymph status partitions. However, this direct data was not suitable for plotting, as many pathways were vastly too specific and varied to portray any broader trends. Pathway abstraction was undertaken to mitigate these difficulties and allow for visual perception of trends in the data. The full Reactome human pathway hierarchy was downloaded, using the provided RESTful API46. A query to abstract pathways was performed using the BaseX XML database engine47. The abstraction was designed to generalize the pathways, while still maintaining sufficient specificity to confer biological meaning in this context. To accomplish this, corresponding pathways of specific depths were retrieved and abstracted by taking instead higher-order pathways in the hierarchy. Reactions or black box events that were four or five levels deep, as well as pathways that were four levels deep, were abstracted by taking the corresponding element of depth three (i.e. their parent or grandparent). Pathways one level higher in the hierarchy (i.e. the parent pathway) of all other pathways, reactions, or black box events (i.e. those not at the aforementioned depths) were retrieved. The resulting abstracted pathways were then used as input for the word clouds. They were generated using R (v3.0.2) with the RColorBrewer (v1.0.5 tm, and wordcloud packages (v2.4)48. Parameters used to generate the word clouds were as follows: scale = c(wordFit,0.3), min.freq = 2, random.order = F, colors = brewer.pal(6, “Dark2”)[−1])), vfont = c(“serif”,“plain”).
The Mutational Significance in Cancer (v0.4) (MuSiC)25 suite of tools was employed to identify genes significantly mutated in the breast cancer samples analyzed with the variant set derived in this study. Three tools from genome MuSiC were used with all default parameters: bmr calc-bmr, bmr calc-covg, and smg. NCBI Reference Sequence Genes release 62 (RefSeq)49 were used as the regions of interest (ROI) file with the Human Feb. 2009 (GRCh37/hg19) assembly reference sequence for bmr calc-bmr and bmr calc-covg. All FDRs that we report pertaining to the MuSiC analysis used the Fisher's combined P-value (FCPT), convolution (CT) and likelihood ratio (LRT) statistical tests.
The software program Veridical24 was used for in silico validation of all predicted splicing mutations using its default settings. At the time the program was run, Veridical rounded p-values to 2 decimal places. Validated results reported were filtered for cryptic variants using reads demonstrating junction-spanning cryptic sites, junction-spanning exon skipping, or read-abundance intron inclusion, whereas reads for predicted natural splice site mutations variants were filtered for all of the above evidence types, except for cryptic splice site-activating, junction-spanning reads. Variants were considered validated if at least one of the above categories for the indicated variant type were excluded from normal controls, but present in the transcriptome containing the predicted mutation (p ≤ 0.05, after transformation of both sample and control read counts to a normal distribution and use of a parametric Z test). Validation was not always possible in instances where predicted mutations occurred in genes or exons with minimal cDNA coverage, resulting from either low expression in the breast tumours carrying the mutation50, tissue-specificity of gene expression, or transcript instability from nonsense-mediated decay. Although Veridical provided experimental validation of predicted splicing mutations, the impact of these and protein coding mutations on tumour progression and biology could not be determined from the present analyses. Further laboratory studies with the original tumour tissues (which were not available), cell line or model organism studies would be required to prove biological significance.
RSeQC's (v2.3.7) ReadDist51 script was used to generate the genome-wide intron inclusion data with the RefSeq gene annotation file to determine intronic genomic sequences. We ran BedTools multicov (v2.17.0)52 upon the RefSeq49 exome annotation BED file retrieved from the UCSC table browser53 with a minimum map quality of 1. The returned coverage values were multiplied by the read length, and divided by the number of exonic bases. In cases of genes with more than one transcript, the shortest transcript was used such that the coverage values per exonic base were maximized, which is the most conservative assumption to adopt when excluding variants due to low coverage. The heat map, provided in Supplementary Fig. S1, was generated by breast cancer subtype for this data using the R packages Hmisc (v3.14.3) and gplots (v2.12.1).
Author Contributions
P.K.R. and S.N.D. conceived of the study. P.K.R. directed the project, and S.N.D. and C.V. performed all analyses. S.N.D. and P.K.R. wrote and revised the paper, with input from C.V. All authors have approved the final manuscript.
Supplementary Material
Supplementary Materials
Tables S1 - S21
Acknowledgments
We would also like to thank Ben Shirley of Cytognomix, Inc., who worked on the Shannon Human Splicing Pipeline software and provided support for the splicing variant prediction. This work was made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET) and Compute/Calcul Canada. PKR is supported by the Canadian Breast Cancer Foundation, Canadian Foundation for Innovation, Canada Research Chairs Secretariat and the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant 371758-2009). SND received fellowships from the Ontario Graduate Scholarship Program, the Pamela Greenaway-Kohlmeier Translational Breast Cancer Research Unit, and the CIHR Strategic Training Program in Cancer Research and Technology Transfer, and the University of Western Ontario.
Footnotes
PKR is the inventor of US Patent 5,867,402 and other patents pending, which underlie the prediction and validation of mutations. He is one of the founders of Cytognomix, Inc., which is developing software based on this technology for complete genome or exome splicing mutation analysis. This software is available from Cytognomix (www.cytognomix.com).
References
- Banerji S. et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. J. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C. et al. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C. T. et al. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- Ng P. C. & Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I., Jordan D. M. & Sunyaev S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 7, 7.20.1-41; 10.1002/0471142905.hg0720s76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang J. & Chen L. Whole-exome sequencing reveals recurrent somatic mutation networks in cancer. Cancer Lett. 340, 270–276 (2013). [DOI] [PubMed] [Google Scholar]
- Ali M. A. & Sjöblom T. Molecular pathways in tumor progression: From discovery to functional understanding. Mol. BioSyst. 5, 902–908 (2009). [DOI] [PubMed] [Google Scholar]
- Mucaki E. J., Ainsworth P. & Rogan P. K. Comprehensive prediction of mRNA splicing effects of BRCA1 and BRCA2 variants. Hum. Mutat. 32, 735–742 (2011). [DOI] [PubMed] [Google Scholar]
- Menéndez M. et al. Assessing the RNA effect of 26 DNA variants in the BRCA1 and BRCA2 genes. Breast Cancer Res. Treat. 132, 979–992 (2012). [DOI] [PubMed] [Google Scholar]
- Krawczak M., Reiss J. & Cooper D. N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum. Genet. 90, 41–54 (1992). [DOI] [PubMed] [Google Scholar]
- Mucaki E. J., Shirley B. C. & Rogan P. K. Prediction of Mutant mRNA Splice Isoforms by Information Theory-Based Exon Definition. Hum. Mutat. 34, 557–565 (2013). [DOI] [PubMed] [Google Scholar]
- Pertea M., Lin X. & Salzberg S. L. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 29, 1185–1190 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churbanov A., Vorechovský I. & Hicks C. A method of predicting changes in human gene splicing induced by genetic variants in context of cis-acting elements. BMC Bioinformatics 11, 1–12; 10.1186/1471-2105-11-22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. C. et al. Interpretation, Stratification and Evidence for Sequence Variants Affecting mRNA Splicing in Complete Human Genome Sequences. Genomics Proteomics Bioinformatics 11, 77–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bigas N., Audit B., Ouzounis C., Parra G. & Guigó R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 579, 1900–1903 (2005). [DOI] [PubMed] [Google Scholar]
- Venables J. P. Aberrant and alternative splicing in cancer. Cancer Res. 64, 7647–7654 (2004). [DOI] [PubMed] [Google Scholar]
- Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int. J. Cell Biol. 2013, 463786; 10.1155/2013/463786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J., Lam K. C. L., Dias C. & Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 5(12), e1000766 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K. et al. Identification of a novel protein isoform derived from cancer-related splicing variants using combined analysis of transcriptome and proteome. Proteomics 11, 2275–2282 (2011). [DOI] [PubMed] [Google Scholar]
- Viner C., Dorman S. N., Shirley B. C. & Rogan P. K. Validation of predicted mRNA splicing mutations using high-throughput transcriptome data. F1000Res. 3, 8; 10.12688/f1000research.3-8.v2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees N. D. et al. MuSiC: Identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J. M. et al. Frequent somatic mutations of GATA3 in non-BRCA1/BRCA2 familial breast tumors, but not in BRCA1-, BRCA2- or sporadic breast tumors. Breast Cancer Res. Treat. 119, 491–496 (2010). [DOI] [PubMed] [Google Scholar]
- Rahman N. et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39, 165–167 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D. et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–D697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J. et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald J., Holden P. & Hansen U. The expanded collagen VI family: New chains and new questions. Connect. Tissue Res. 54, 345–350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Håkansson J.
et al.
Neural cell adhesion molecule-deficient
 tumorigenesis results in diminished extracellular matrix molecule expression and tumour cell-matrix adhesion. Tumor Biol.
26, 103–112 (2005). [DOI] [PubMed] [Google Scholar]
tumorigenesis results in diminished extracellular matrix molecule expression and tumour cell-matrix adhesion. Tumor Biol.
26, 103–112 (2005). [DOI] [PubMed] [Google Scholar] - Zecchini S. et al. The adhesion molecule NCAM promotes ovarian cancer progression via FGFR signalling. EMBO Mol. Med. 3, 480–494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry M. & Yardley D. A. Updates in the treatment of basal/triple-negative breast cancer. Curr. Opin. Obstet. Gynecol. 25, 40–48 (2013). [DOI] [PubMed] [Google Scholar]
- Slamon D. J. et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987). [DOI] [PubMed] [Google Scholar]
- Jelovac D. & Emens L. A. HER2-directed therapy for metastatic breast cancer. Oncology (Huntington, N. Y.) 27, 166–175 (2013). [PubMed] [Google Scholar]
- Osborne J. K. et al. NeuroD1 regulation of migration accompanies the differential sensitivity of neuroendocrine carcinomas to TrkB inhibition. Oncogenesis 2, e63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. H., Chen J. Y., Lee C. H. & Chen J. Y. Expression of NCAM and OCIAD1 in well-differentiated thyroid carcinoma: Correlation with the risk of distant metastasis. J. Clin. Pathol. 65, 206–212 (2012). [DOI] [PubMed] [Google Scholar]
- Wai Wong C., Dye D. E. & Coombe D. R. The role of immunoglobulin superfamily cell adhesion molecules in cancer metastasis. Int. J. Cell Biol. 2012, 340296; 10.1155/2012/340296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Leshchyns'ka I., Sytnyk V., Morrow J. S. & Schachner M.
Neural cell adhesion molecule (NCAM) association with
 via
via  spectrin is implicated in NCAM-mediated neurite outgrowth. J. Cell Biol.
161, 625–639 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
spectrin is implicated in NCAM-mediated neurite outgrowth. J. Cell Biol.
161, 625–639 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar] - Jones S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D. E. et al. Somaticsniper: Identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S., Josse J. & Husson F. FactoMineR: An R package for multivariate analysis. J. Stat. Software 25, 1–18 (2008). [Google Scholar]
- Milacic M. et al. Annotating cancer variants and anti-cancer therapeutics in Reactome. Cancers 4, 1180–1211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün C., Gath S., Holupirek A. & Scholl M. H. XQuery full text implementation in BaseX. Lect. Notes Comput. Sci. 569, 114–128 (2009). [Google Scholar]
- Feinerer I., Hornik K. & Meyer D. Text Mining Infrastructure in R. J. Stat. Software 25, 1–54 (2008). [Google Scholar]
- Pruitt K. D., Tatusova T. & Maglott D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang S. & Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- Quinlan A. R. & Hall I. M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D. et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Tables S1 - S21