Abstract
The key step in meiosis is synaptonemal complex formation, which mediates homologous chromosome alignment and synapsis. False pairing between homologous chromosomes produces infertility. Here, we present a crystal structure of the mouse meiosis-specific protein SYCE3, which is a component of the synaptonemal complex central element. Our studies show that functional SYCE3 most likely forms a dimer or higher order oligomer in cells. Furthermore, we demonstrate that the SYCE3 N-helix interacts with the SYCE1 C-helix, which is another central element component. Our results suggest that helical packing may mediate intra- or inter-association of each central element protein component, thereby playing an essential role in forming the synaptonemal complex central elements.
Sexual reproduction occurs in eukaryotes through meiosis, a process by which a diploid germ-line cell divides, resulting in the formation of haploid cells. This process involves one round of DNA replication and two rounds of cell division. Errors during meiosis can lead to disorders that result from an abnormal number of chromosomes1,2,3. Prior to the first division, the key step of meiosis is that two homologous chromosomes pair and that a synaptonemal complex (SC) develops between them. Once synapsis is complete, the homologous chromosomes undergo recombination4,5,6,7.
The synaptonemal complex is a ladder-like structure consisting of three components: lateral elements (LEs), central elements (CEs), and transverse filaments (TFs)7. These components are composed of meiosis-specific proteins. When homologous chromosomes begin synapsis, SYCP2 and SYCP3 form the axial elements (AEs)8, and when AEs are associated with a pair of sister chromatids, the mature AEs are called lateral elements7. In the case of SYCP2 lacking its coiled-coil domain, which is required for binding to SYCP3, male mice are sterile because of the failure of SC formation, whereas females are subfertile and have reduced litter size9. However, in mice lacking SYCP3, female mice exhibit aneuploidy because of defective meiotic chromosome segregation10. The TF is a physical linkage between the LE and the CE. SYCP1, a major component of TFs, is a long coiled-coil protein with N- and C-terminal globular domains11,12,13. Mutant SYCP1 with a modified length of the central alpha-helical domain influences the SC width, and deletions of the globular heads result in the failure of SC assembly14.
In the last few years, four proteins of CEs have been identified: SYCE1, SYCE2, SYCE3, and TEX12. All of these proteins localize exclusively to the central element of the SC and contain predicated alpha-helical domains15,16,17. Both SYCE1 and SYCE2 directly interact with SYCP1 in vivo and in vitro. SYCP1 can play a role in recruiting SYCE1 and SYCE2 to the polycomplexes15, and in syce1-/- mutant mice, AEs are formed but homologous chromosomes fail to synapse18. In syce2-/- mutant mice, the SC fails to form, except for small regions, suggesting that SYCE2 could be involved in SC extension19.
SYCE3, a newly identified component of the central element, is reported to co-localize with SYCE1 and SYCP1. SYCE3-knockout mice exhibit both male and female infertility. In syce3-/- spermatocytes, SYCP1 displays a weak pattern at AEs, whereas SYCE1 and SYCE2 are completely absent, indicating that SYCE3 assembles to the CE prior to SYCE1 and SYCE2. SYCE3 is absent from the AEs in sycp1-/- mice. However, SYCE3 localizes to the chromosome axes in mice lacking SYCE1, SYCE2, or TEX12, suggesting that SYCE3 loading requires SYCP116.
The correct formation of the synaptonemal complex is crucial for homologous chromosome recombination and segregation. To further study the molecular basis of central element assembly, we determined the crystal structure of SYCE3 and revealed the extensive interactions involved in its dimer formation. Furthermore, we found that the interaction between SYCE3 and SYCE1 occurs through the N-terminal helix of SYCE3 and the C-terminal helix of SYCE1. Our findings provide evidence that the helical packing of SC proteins contributes to synaptonemal complex assembly.
Results
Overall SYCE3 structure
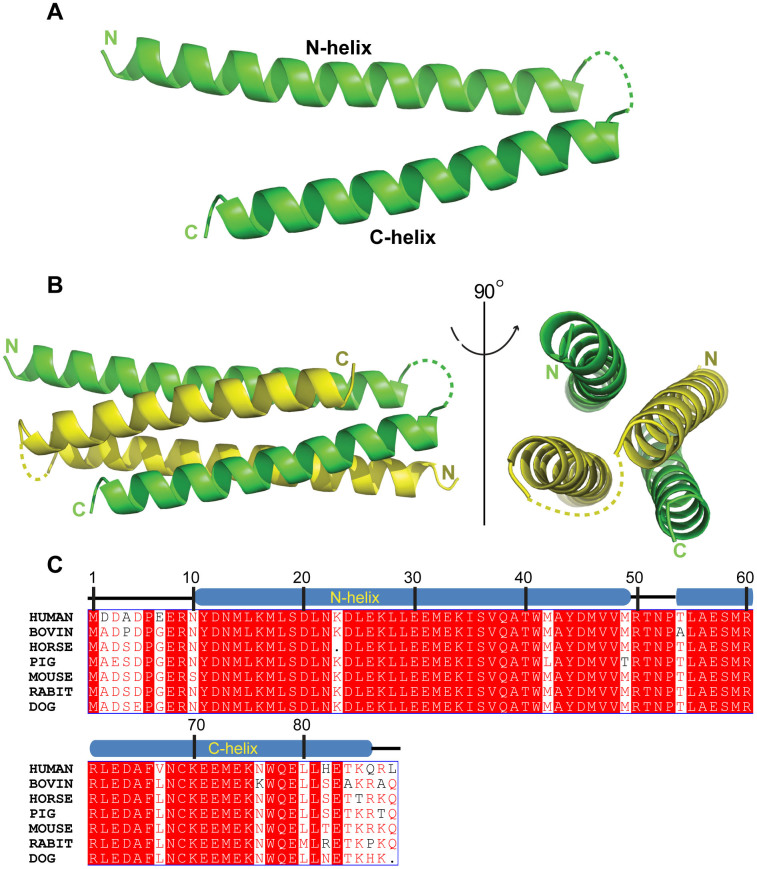
The SYCE3 crystal structure was determined to a 1.9 Å resolution using the single-wavelength anomalous dispersion method. As predicted, the SYCE3 structure consists of two helices: the N-helix (aa 1-48) and C-helix (aa 55-88). Except for the loop connecting the two helices, SYCE3 exhibits a clear electron density (Figure 1A). The protein crystals belong to the primitive rhombohedral space group R3 with two independent molecules in each asymmetric unit that are related by 2-fold non-crystallographic symmetry (Figure 1B). The SYCE3 primary sequence is highly conserved among mammals (Figure 1C), which indicates that both the three-dimensional structure and biological function are conserved.
Figure 1. SYCE3 crystal structure.
(A) A cartoon representation of the SYCE3 molecular structure. (B) A cartoon representation of the SYCE3 dimer structure. The two SYCE3 molecules are shown in yellow and green. The dotted lines represent the region that was not resolved in this structure. (C) A multiple sequence alignment for full-length SYCE3 among various mammals. The SYCE3 primary sequence is highly conserved among mammals.
Dimer packing in the SYCE3 structure
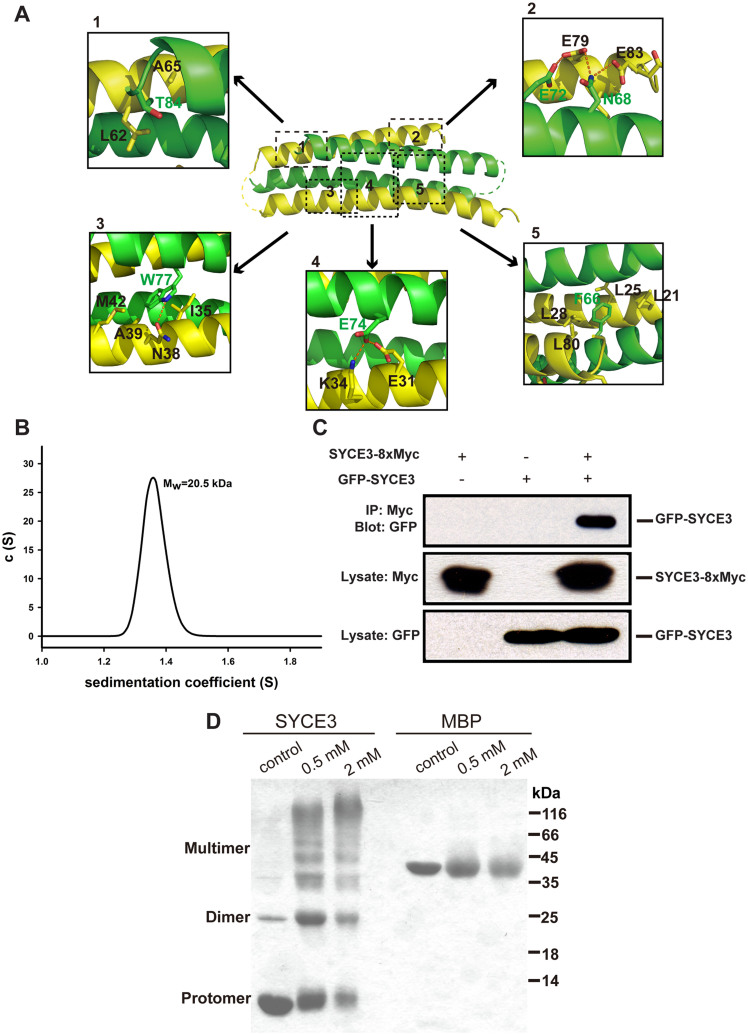
The crystal structure shows that two SYCE3 molecules interact closely (Figure 1B). The interaction between two SYCE3 molecules mainly involves an intensive network of hydrophobic contacts, hydrogen bonds, and salt bridges (Figure 2A). First, the two protomers primarily interact through a salt bridge. Glu74 from one protomer interacts with Lys34 from another protomer and forms a hydrogen bond with the Glu31 OE2 atom from another protomer (Figure 2A-4). Second, the Phe66 side chain from one protomer is buried by the hydrophobic cleft that consists of side chains from another protomer, which is composed of residues Leu21, Leu25, Leu28, and Leu80 (Figure 2A-5). Third, Trp77 is surrounded by a hydrophobic patch consisting of residues Ile35, Ala39, and Met42; Trp77 also forms a hydrogen bond with Asn38 from another protomer (Figure 2A-3). Fourth, Thr84 forms hydrophobic interactions with Leu62 and Ala65 (Figure 2A-1). Fifth, an Asn68 from one protomer forms two hydrogen bonds with another protomer through the side chain Glu79 OE2 atom and Glu83 OE2 atom. A hydrogen bond also forms between the Glu72 OE2 atom and Glu79 OE1 atom from each protomer (Figure 2A-2). These extensive interactions indicate highly stable dimer formation.
Figure 2. SYCE3 dimeric packing.
(A) Detailed interactions between two SYCE3 molecules in the SYCE3 dimer. The residues involved in dimer packing are boxed. The relevant amino acid residues are numbered, and the hydrogen bonds are shown as red dashed lines. (B) An analytical ultracentrifugation sedimentation velocity analysis of the purified SYCE3 protein. The calculated molecular weight is 20.5 kDa. (C) Co-immunoprecipitation assays for SYCE3 homodimerization. COS-7 cells were transfected with EGFP-SYCE3 and SYCE3-8 × Myc. The resulting complexes were analyzed using SDS-PAGE and Western blotting. (D) A cross-linking experiment for the purified SYCE3 protein. The final concentration (0.5 mM and 2 mM) of DSG in the reaction mixture is indicated. Full-length blots/gels are presented in Supplementary Figure 1.
To examine the dimer status of SYCE3 in solution, we performed analytical ultracentrifugation. The results confirm that SYCE3 forms a homogenous dimer in solution with a molecular mass of approximately 20.5 kDa (Figure 2B). Furthermore, to validate SYCE3 homo-dimerization or homo-oligomerization in cells, we performed co-transfection and immunoprecipitation experiments in somatic cells, which do not express meiosis-specific proteins20. Because the Myc-SYCE3 fusion protein did not bind the anti-Myc coupled protein A/G beads, we modified pcDNA4/myc-His A by inserting 7 × Myc tags between the SYCE3 gene and original Myc tag. The SYCE3-8 × Myc and EGFP-SYCE3 fusion constructs were then co-transfected into COS-7 cells. The results show that SYCE3-8 × Myc can successfully pull down GFP-SYCE3 (Figure 2C), which indicates that SYCE3 can form a dimer in cells. To verify whether SYCE3 can form higher order oligomers, we performed a cross-linking experiment using purified SYCE3. The results show that SYCE3 can form a dimer or multimer after treatment with disuccinimidyl glutarate (DSG) (Figure 2D), which suggests that SYCE3 might form higher order oligomers in cells.
The interaction between SYCE3 and SYCE1
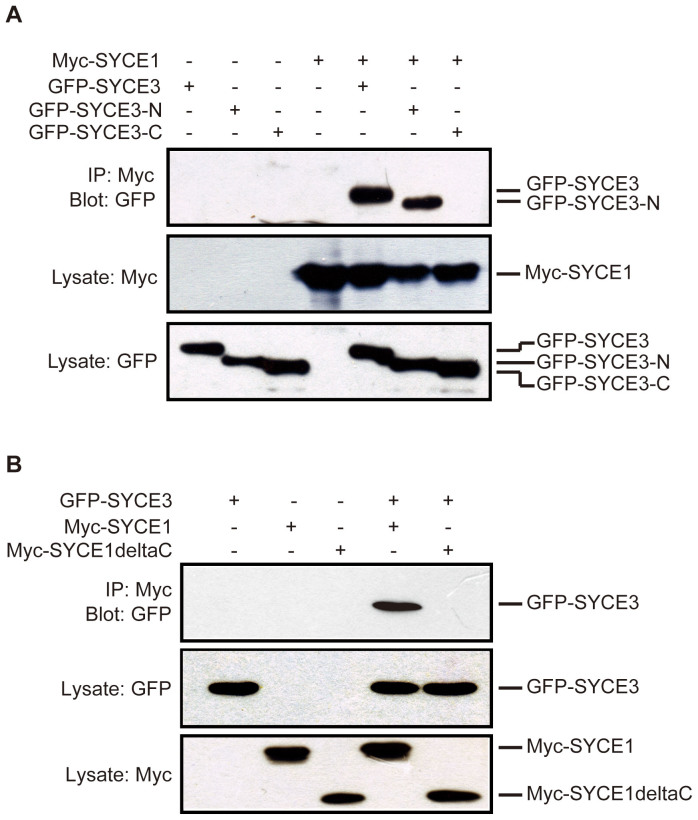
A previous report indicated that SYCE3 co-localizes with SYCE1 and aids in recruiting SYCE1 to SYCP116; thus, we performed co-IP experiments to further study the interaction between SYCE3 and SYCE1. To examine which SYCE3 helix interacts with SYCE1, we co-transfected constructs encoding EGFP-tagged fusion proteins composed of the full-length SYCE3, SYCE3 N-helix (aa1-52), or SYCE3 C-helix (aa 53–88) and a construct encoding Myc-tagged SYCE1 into COS-7 cells. Our data show that both the full-length SYCE3 and SYCE3 N-helix interacts with SYCE1; however, we did not detect an interaction between the SYCE3 C-helix and SYCE1 (Figure 3A).
Figure 3. Interactions between SYCE3 and SYCE1.
(A) COS-7 cells were transfected with Myc-SYCE1 and the respective full-length SYCE3, SYCE3 N-helix, and SYCE3 C-helix EGFP fusion constructs. Myc-SYCE1 can bind the full-length SYCE3 and SYCE3 N-helix. The SYCE3 C-helix does not bind SYCE1. (B) COS-7 cells were transfected with EGFP-SYCE3 and the respective Myc fusion constructs composed of full-length SYCE1 and SYCE1 without C-helix. The results show that SYCE1 without the C-helix does not bind SYCE3. Full-length blots/gels are presented in Supplementary Figure 1.
We then attempted to map the portion of SYCE1 involved in binding SYCE3. We co-transfected Myc-tagged fusion constructs composed of full-length SYCE1 or SYCE1deltaC (a mutant of full-length SYCE1 with the C-terminal aa 220–329 deleted) and EGFP-tagged fusion constructs composed of full-length SYCE3 into COS-7 cells. We found that deleting the C-terminal portion of SYCE1 significantly weakened the association between SYCE1 and SYCE3 (Figure 3B). Taken together, our results suggest that the association between SYCE3 and SYCE1 is mainly involved in interactions between the SYCE3 N-terminal helix and SYCE1 C-terminal helix.
Discussion
The key step in meiosis is correct synaptonemal complex assembly. Several SC proteins have been identified in recent years; SYCE1, SYCE2, SYCE3, and TEX12 are SC central element proteins. Extensive studies have demonstrated that the CE proteins are crucial for SC assembly15,16,17. Our results, for the first time, provide structural information on the CE proteins and important insights into the central elements' assembly mechanism.
Secondary structure predictions for SYCE1, SYCE2, SYCE3, and TEX12 showed that they form helix-like structural features. In our SYCE3 structure, the extensive interactions between helices of two SYCE3 molecules indicate highly stable dimer formation (Figure 2A). This structure is consistent with our co-immunoprecipitation assay results from transfected COS-7 cells, which show that SYCE3 may function as a dimer or higher-order oligomer in cells during SC assembly. Previous reports indicate that SYCE3 and SYCE1 co-localize along synapsed chromosomes in zygotene and pachytene spermatocytes16. Our co-IP assay results demonstrate that SYCE3 can bind SYCE1. SYCE2 and TEX12 share similar localization patterns with SYCE1 and SYCE3 in the CE17, and previous reports indicate that SYCE2 co-localizes and co-precipitates with TEX12 during meiotic cell division17. Owen et al proposed an SYCE2-TEX12 equimolar hetero-octamer model21; in this model, SYCE2 molecules form a tetramer mediated by the central α-helices, and TEX12 molecules form a dimer mediated by the central α-helices. Further, the SYCE2 tetramer binds two TEX12 dimers through helical packing interactions between SYCE2 and TEX12, which forms a stable higher-order structure.
Based on our study, the SYCE3 N-helix participates in SYCE1 binding; however, the SYCE3 C-helix and SYCE1 do not interact (Figure 3A). We also mapped the SYCE1 C-terminal helix, which is crucial for binding between SYCE3 and SYCE1 (Figure 3B). Based on our structural and biochemical studies, we propose that the SYCE3 dimer binds two or more SYCE1 moieties via interactions between the SYCE3 N-helix and SYCE1 C-terminal helix. In the recently determined SYCP3 structure, four SYCP3 helical chains form a tetramer composed of helix bundles and coiled-coil motifs. The extensive network of hydrophobic contacts and salt bridges in the two helical bundles contributes to the high stability of SYCP3 packing22. These findings support the notion that the helical bundle is important for synaptonemal complex formation. Overall, the helical features of each CE protein and the inter-helix interaction-mediated association between different CE proteins indicate that helical packing mainly drives central element assembly.
Previous studies show that SYCE1 stabilizes the SYCP1 N-terminal interactions in the CE, which was demonstrated through co-IP assays and immunofluorescence analyses15. SYCE1 also co-localizes and co-precipitates with SYCE316. Thus, it is reasonable to assume that SYCE1 functions as a bridge between SYCP1 and SYCE3. However, according to Schramm's research, in Syce3-/- mice, SYCP1 localizes in a weak pattern at AEs, with both SYCE1 and SYCE2 being absent from the axes16. SYCE3 is likely required for initial CE assembly. However, interactions between the SYCP1 N-terminus and SYCE3 remain vague, and co-IP assays do not show that SYCE3 can bind the SYCP1 N-terminal region16. It is possible that additional undiscovered CE proteins may connect SYCP1 and SYCE3.
The CE proteins not only play a structural role in the SC but can also be regulated by other proteins. PLK1, a Polo-like kinase, is required for SYCP1, TEX12, and SYCE1 phosphorylation to promote SC disassembly during the G2/MI transition23. Studies have also predicted phosphorylation sites for SYCE3 that could be regulated by kinase activity16. Further studies will elucidate the structural and regulatory roles of CE proteins in SC assembly.
Methods
Protein expression and purification
Gene fragments encoding SYCE3 were PCR amplified from Mus musculus cDNAs and then cloned into the pET-M vector, a derivative of pET32a (Novagen). The correctness of the constructs was confirmed by DNA sequencing. Escherichia coli BL21 CondonPlus cells harboring the expression plasmid for the fusion protein were grown at 37°C to an OD600 >0.6 and then induced with 0.2 mM isopropyl-β-D-thiogalactoside (IPTG) at 20°C for 16–18 h. The cells were resuspended in T20N300I20 buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 20 mM imidazole) and then lysed by sonication. After centrifugation at 20,000 × g for 40 min, the supernatant was loaded directly onto a Ni-NTA agarose column (Qiagen) equilibrated with T20N300I20 buffer. After washing the Ni-NTA column with 5 column volumes of equilibrating buffer, the His6-tagged protein was eluted with T20N300I300 buffer. Subsequently, the protein elute was subjected to anion-exchange chromatography (Hitrap Q FF; GE Healthcare) using T20N60E1D1 buffer (20 mM Tris-HCl, pH 8.0, 60 mM NaCl, 1 mM EDTA, 1 mM DTT) from 6% to 50% NaCl. The final target protein was loaded onto a HighLoad 26/600 Superdex 200 size-exclusion column (GE Healthcare) and eluted with T20N200D1 buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT). The protein peak was identified by SDS-PAGE, harvested, and concentrated using a Centricon device (Millipore).
Crystallization and data collection
Crystals of wild-type and Se-Met-substituted SYCE3 were grown using sitting-drop vapor diffusion. SYCE3 was crystallized by combining 1 μL of protein solution (8.8 mg/mL in 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT) with an equal volume of well solution containing 0.1 M citric acid, pH 3.5, 7% 2-propanol, and 1% PEG 20,000. The crystals were grown for approximately one week at 4°C and frozen in cryoprotectant consisting of the well solution supplemented with 25% glycerol. Se-Met-substituted SYCE3 was crystallized by the combination of 1 μL of protein solution (3.7 mg/mL in 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT) with an equal volume of well buffer containing 0.1 M citric acid, pH 3.5, 2% 2-propanol, and 5% PEG 20,000. The crystals were grown for approximately one week at 4°C and frozen in cryoprotectant consisting of the well solution supplemented with 25% glycerol.
The data for wild-type were collected at the beamline BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF), and single-wavelength anomalous data were collected for the Se-Met-substituted crystals at the elemental Se peak wavelength at the beamline BL-17A of the Photon Factory (Tsukuba, Japan) and then processed using the HKL2000 software24.
Structure determination and refinement
The SYCE3 crystal structure was determined by SAD methodology. The program HKL2MAP25 was used to search for Se sites, and the initial phases were then calculated using PHENIX software26. The model was built manually using the program COOT27. After the initial main-chain model was built, the wild-type data were applied to carry out an iterative refinement to assign all side chains using the program COOT until the free R factor was converged. The final structure had an Rcrystal value of 21.47% and an Rfree value of 23.45%, with good geometry. The detailed data collection and refinement statistics are summarized in Table 1.
Table 1. Data collection and refinement statistics.
| Crystal name | Wild-type | Se-Met-crystal |
|---|---|---|
| Space group | R3 | R3 |
| Unit cell (Å) | a = b = 75.6 | a = b = 75.7 |
| c = 101.5 | c = 101.7 | |
| Wavelength (Å) | 0.9796 | 0.9796 |
| Resolution range (Å) | 50–1.90 (1.93–1.90) | 50–2.04 (2.11–2.04) |
| No. of unique reflections | 8,742 | 7,272 |
| Redundancy | 3.9(2.8)a | 3.7(3.7)a |
| Rsym (%)b | 5.9(48.6)a | 11.7(60.2)a |
| I/σ | 22.0(1.8)a | 37.2(4.2)a |
| Completeness (%) | 97.1(81.9)a | 99.3(99.6)a |
| FOM | --- | 0.445 |
| Refinement | ||
| Resolution range (Å) | 23.66 ~ 1.90 | |
| Rcrystal (%)c | 21.47 | |
| Rfree (%)d | 23.45 | |
| RMSDbond (Å) | 0.007 | |
| RMSDangle(°) | 0.874 | |
| Number of | ||
| Protein atoms | 1173 | |
| Solvent atoms | 70 | |
| Residues in (%) | ||
| most favored | 99.2 | |
| additional allowed | 0.8 | |
| Generously allowed | 0 | |
| disallowed | 0 | |
| Average B factor (Å2) of | ||
| Chain A | 45.061 | |
| Chain B | 45.154 | |
| Solvent | 54.790 |
athe highest resolution shell.
b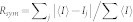 .
.
c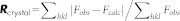 .
.
dRfree, calculated the same as Rcrystal, but from a test set containing 5% of data excluded from the refinement calculation.
Analytical ultracentrifugation
Sedimentation velocity measurements were obtained using a Beckman/Coulter XL-I analytical ultracentrifuge with double-sector and sapphire windows. The SV experiments were conducted at 42,000 rpm and 4°C using interference light detection and double-sector centerpieces loaded with 1 mg/mL protein. The sample was prepared in a buffer containing 20 mM Tris-HCl, pH 8.0, and 200 mM NaCl. The data were analyzed using the SEDFIT program28.
Cell culture and transfection
COS-7 (green monkey kidney) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% v/v fetal bovine serum (FBS). The cells were maintained in a 95% air and 5% CO2 environment at 37°C.
Gene fragments corresponding to full-length SYCE3, SYCE3 N-helix (aa 1–52), and SYCE3 C-helix (aa 53–88) were cloned into the pEGFP-C1 vector (Clontech). The SYCE3 gene was also cloned into the pcDNA4/myc-His A vector (Invitrogen), and 7 Myc tags were inserted after the SYCE3 gene. This modified plasmid was named pcDNA4/8myc-His A. Gene fragments corresponding to full-length SYCE1 and SYCE1deltaC (aa 1–219) were cloned into the pCMV-Myc vector (Clontech).
Co-immunoprecipitation and Western blot analysis
Transfected COS-7 cells were collected in PBS buffer (136 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4). Cells harvested from a 10-cm culture dish were lysed in 500 μL of RIPA buffer (PBS containing 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM β-glycerolphosphate, 1 mM Na3VO4, 1 mM EDTA, 1 mM EGTA) containing Roche protease inhibitor cocktail. After incubation on ice for 1 h, the lysate was centrifuged at 13,000 × g for 30 min. A total of 40 μL of a 1:1 slurry of protein A/G agarose (Pierce) was incubated with 2 μg of anti-Myc antibody for 1 h and then washed twice with cold RIPA buffer. The cell extract supernatant was added to the antibody-coupled protein A/G beads and was incubated for 3 h at 4°C. The beads were washed three times for 10 min with cold RIPA buffer, followed by elution with 40 μL SDS-loading buffer. Subsequently, 5 μL of the elute was loaded onto a 15% SDS-PAGE gel and transferred onto a PVDF membrane for Western blot analysis. The immunoblot analysis was conducted using a primary antibody against GFP. The protein-antibody complexes were detected using enhanced chemiluminescence.
Protein cross-linking
The purified proteins SYCE3 and maltose binding protein (MBP) were prepared in reaction buffer (20 mM HEPES, pH7.5, and 200 mM NaCl), respectively. Disuccinimidyl glutarate was added to the protein sample at the final concentrations 0.5 mM and 2 mM. Next, the reaction mixture was incubated at room temperature for 30 minutes. The samples were loaded onto a 15% SDS-PAGE gel and stained with Coomassie brilliant blue.
Author Contributions
J.L., Y.G. and J.F. did protein purification; J.L. did crystallization, structure refinement, co-IP and biochemical experiments; X.Y. did data collection and structure determination; W.Z. analyzed the data. J.L. and Y.S. designed the study and wrote the paper. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary figure
Acknowledgments
We are grateful to Dr. Zheng Wang for help in analytical ultracentrifugation experiment, to Dr. Zhijie Lin for help in protein expression and purification, to the staff at the beamline BL-17A at Photon Factory (Tsukuba, Japan) and at the beamline BL17U1 of the Shanghai Synchrotron Radiation Facility for excellent technical assistance during data collection. This work was supported by the 973 Program (grants 2012CB917200 and 2013CB910400 to YS; grant 2014CB910201 to XY), the Natural Science Foundation of China (grants 31370826 to YS, 31300628 to XY and 31170684 to WZ), Tianjin Basic Research Program (grant 14JCQNJ09300 to XY) and the Fundamental Research Funds for the Central Universities (grants 65142007 to YS; 65121016 to XY).
References
- Garcia-Cruz R., Roig I. & Caldes M. G. Maternal origin of the human aneuploidies. Are homolog synapsis and recombination to blame? Notes (learned) from the underbelly. Genome Dyn. 5, 128–136 (2009). [DOI] [PubMed] [Google Scholar]
- Handel M. A. & Schimenti J. C. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124–136 (2010). [DOI] [PubMed] [Google Scholar]
- Hassold T. & Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001). [DOI] [PubMed] [Google Scholar]
- Fawcett D. W. The fine structure of chromosomes in the meiotic prophase of vertebrate spermatocytes. J. Biophys. Biochem. Cytol. 2, 403–406 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses M. J. Chromosomal structures in crayfish spermatocytes. J. Biophys. Biochem. Cytol. 2, 215–218 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses M. J. Structure and function of the synaptonemal complex. Genetics. 61, Suppl41–51 (1969). [PubMed] [Google Scholar]
- Page S. L. & Hawley R. S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558 (2004). [DOI] [PubMed] [Google Scholar]
- Eijpe M., Offenberg H., Jessberger R., Revenkova E. & Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J. Cell Biol. 160, 657–670 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F. et al. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 173, 497–507 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L. et al. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296, 1115–1118 (2002). [DOI] [PubMed] [Google Scholar]
- Meuwissen R. L. et al. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11, 5091–5100 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. G. et al. Localization of the N-terminus of SCP1 to the central element of the synaptonemal complex and evidence for direct interactions between the N-termini of SCP1 molecules organized head-to-head. Exp. Cell Res. 226, 11–19 (1996). [DOI] [PubMed] [Google Scholar]
- Schmekel K. et al. Organization of SCP1 protein molecules within synaptonemal complexes of the rat. Exp. Cell Res. 226, 20–30 (1996). [DOI] [PubMed] [Google Scholar]
- Ollinger R., Alsheimer M. & Benavente R. Mammalian protein SCP1 forms synaptonemal complex-like structures in the absence of meiotic chromosomes. Mol. Biol. Cell 16, 212–217 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y. et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118, 2755–2762 (2005). [DOI] [PubMed] [Google Scholar]
- Schramm S. et al. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 7, e1002088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G. et al. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J. Cell Sci. 119, 4025–4032 (2006). [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E. et al. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 5, e1000393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E. et al. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 176, 741–747 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson P. J., Hale C. M., Wirtz D. & Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 314, 1892–1905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies O. R., Maman J. D. & Pellegrini L. Structural analysis of the human SYCE2-TEX12 complex provides molecular insights into synaptonemal complex assembly. Open Biol. 2, 120099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrjanen J. L., Pellegrini L. & Davies O. R. A molecular model for the role of SYCP3 in meiotic chromosome organisation. Elife. 3, e02963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. W., Karppinen J. & Handel M. A. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J. Cell Sci. 125, 5061–5072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z M. W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Pape T. & Schneider T. R. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 37, 843–844 (2004). [Google Scholar]
- Zwart P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008). [DOI] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Brown P. H. & Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90, 4651–4661 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure