1. Overview
Complete and faithful duplication of the cellular genome is a fundamental life process as the genetic information is passed from one generation to the next. The 4.2 Mb genome of E. coli is duplicated within 40 minutes with a precision of only one misincorporated base per 107 nucleotides. This extremely rapid and highly accurate process requires a dynamic interplay of many different subunits that orchestrate replication in a remarkable way.
Replication of the E. coli circular genome is initiated at a single origin of replication upon which two replisomes assemble to produce replication forks that travel in opposite directions. Each replication fork contains multiple proteins that function in a very dynamic fashion to copy both strands of the parental duplex. Replication is initiated by the action of primase, which synthesizes short RNA primers that are extended by a heterotrimeric DNA polymerase (αεθ), called “Pol III core.” A multiprotein clamp loader complex (γτ2δδ′ψχ) assembles the β sliding clamp on primed sites and tethers Pol III core to DNA for processive synthesis through direct interaction with the α subunit of DNA polymerase. The clamp loader also couples two DNA polymerases through interactions of Pol III core with the two τ subunits. Two Pol III cores associated with one clamp loader forms the large complex called “Pol III*.” The τ subunits of Pol III* also interact with the DnaB helicase that travels ahead of the replicative polymerase and unwinds the parental DNA duplex (Fig. 1).
Fig. 1. Organization of the E. coli replisome.
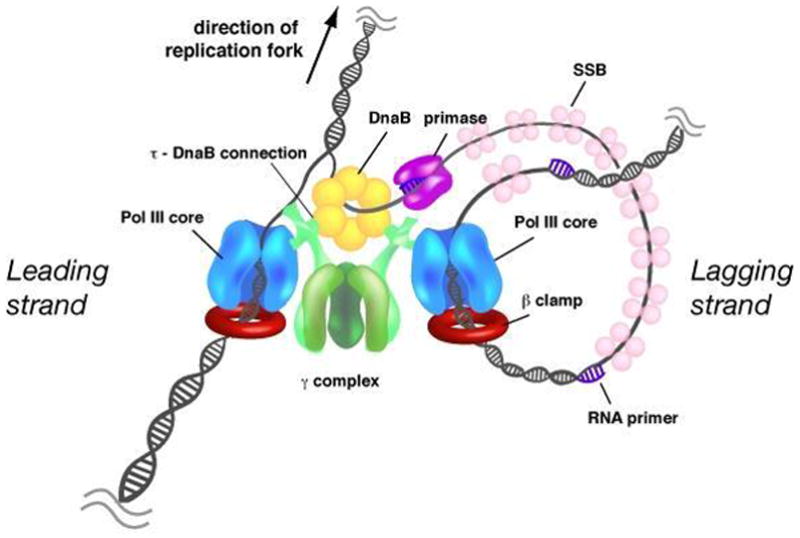
The parental duplex is unwound by the DnaB helicase (yellow) that encircles the lagging strand and travels ahead of the polymerase (blue) in the direction of the moving replication fork. Primase (purple) synthesizes short RNA primers to initiate Okazaki fragment synthesis on the lagging strand. The exposed single strand lagging strand template DNA is covered by SSB (pink). The two DNA polymerases are coupled through the clamp loader (green), which uses the energy of ATP hydrolysis to assemble the β processivity clamp (red) around primed sites on the DNA. For simplicity, the χ and ψ subunits of the clamp loader are omitted from the drawing.
The anti-parallel orientation of the two strands of duplex DNA imposes significant geometric constraints on the mechanism of replication fork progression. This is mainly because all known DNA polymerases synthesize DNA exclusively in the 5′-3′ direction. Therefore, only one strand of the DNA duplex can be synthesized continuously in the direction of the moving replication fork (leading strand), whereas the other strand (lagging strand) must be synthesized in the opposite direction as a discontinuous series of short 1–2 kb Okazaki fragments.
This chapter will describe the components of the E. coli replisome and the dynamic process in which they function and interact under normal conditions. We will also briefly describe the behavior of the replisome during situations in which normal replication fork movement is disturbed, such as when the replication fork collides with sites of DNA damage.
2. The E. coli Pol III holoenzyme
The E. coli DNA polymerase III (Pol III) was first isolated from a mutant E. coli strain (polA−) that lacked the relatively abundant DNA polymerase I (89). Further biochemical studies, and the use of double mutant strains, revealed Pol III to be the replicative DNA polymerase essential to cell viability (48). A large multisubunit form of Pol III, referred to as Pol III holoenzyme (Pol III H.E.) was discovered soon after (109, 170). The multisubunit composition of Pol III H.E. endows it with special properties that distinguish it from other DNA polymerases and transforms Pol III into a unique enzyme, capable of very rapid and processive DNA synthesis needed for replication of the large E. coli genome (108). Studies of the properties of the Pol III H.E. have elucidated principle mechanisms of DNA replication which are conserved in all bacteria as well as in eukaryotes and archaea (65).
Pol III H.E. functions as a large macromolecular machine consisting of 10 distinct subunits that assort into three functional components (Fig. 1): DNA polymerase III core (Pol III core), the clamp loader complex (γ complex) and the β-sliding clamp. Pol III core is a heterotrimer that contains the DNA polymerase (α subunit), the proofreading 3′-5′ exonuclease activity (ε subunit) and the θ subunit. The clamp loader complex (γτ2δδ′χψ) assembles the ring shaped β-sliding clamp onto DNA which then binds to Pol III core and tethers it to DNA for highly processive synthesis. The clamp loader utilizes the energy of ATP hydrolysis to assemble the β sliding clamp onto a primed site. The clamp loader also binds two molecules of Pol III core for simultaneous duplication of both strands of duplex DNA, as described later in this chapter. Overall, Pol III H.E. is a remarkably efficient enzyme that extends DNA at a speed of at least 650 nucleotides (nts)/s with a processivity of several thousand bases and an error rate of only 1 misincorporated base for every 107 incorporated basepair (bp) (88).
The 10 subunit Pol III H.E. can be efficiently reconstituted in vitro using purified components and can function in the context of a replisome with DnaB helicase and primase. The simpler bacteriophage replication machineries (bacteriophages T4 and T7) have also been successfully reconstituted and have taught us an enormous amount of what is known about replisome function (56, 162). Each of these systems display coupled leading and lagging strand synthesis on model replication fork substrates and have elucidated numerous mechanisms that operate at replication forks.
Pol III core
Pol III core is a 1:1:1 heterotrimer consisting of the DNA polymerase α subunit, the ε proofreading 3′-5′ exonuclease subunit, and the small θ subunit (106, 111, 137). The α subunit of Pol III core is a member of the C-family of DNA polymerases, which are found exclusively in eubacteria and do not share sequence similarity with other canonical DNA polymerases. The α subunit is organized into three functional regions (Fig. 2A). The central region harbors the catalytic core, whereas the N- and C-terminal regions contain domains required for interaction with other proteins. The N-terminal region of bacterial α also contains a conserved PHP (polymerase and histidinol phosphatase) domain which has been demonstrated to harbor a 3′–5′ exonuclease activity in a thermophilic α subunit (145). In the E. coli Pol III core, the PHP domain interacts with the ε 3′–5′ exonuclease subunit (171), thereby linking the polymerase with the exonuclease function. The region required for catalysis of DNA synthesis comprises the largest part of the protein and contains the three conserved aspartate residues (Asp401, Asp403, Asp555) that function to coordinate two Mg2+ ions for the two-metal catalyzed reaction of nucleotide incorporation (130), a mechanism observed in all DNA polymerases (148). The C-terminal region of α contains an OB-fold flanked by β binding motifs: an internal β binding motif (residues 920–924) and a C-terminal β binding motif (1154–1160) (31, 34, 102). The internal β binding motif is essential for processive DNA replication, whereas deletion of the C-terminal β binding site reduces β binding and Pol III processivity by approximately 4-fold, indicating that although this β binding motif is not essential, it contributes to polymerase function (34, 92). Genetic studies support these data by indicating a functional role of the C-terminal β binding site in vivo. Interaction of the α subunit with the τ subunit of the clamp loader also occurs within the C terminal 48 amino acids (81), which is important to replisome architecture and function. The OB-domain in the C-terminal region of α is required for processive function of α with the β-sliding clamp (92).
Fig. 2. Domain organization of the replisome components.
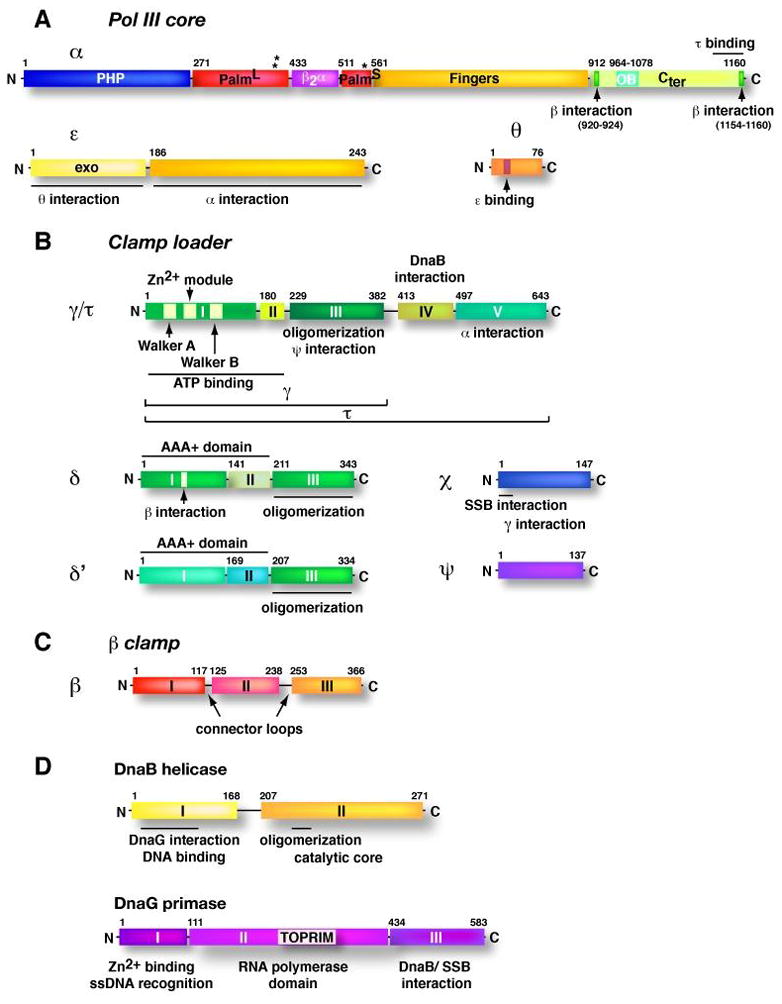
Linear N- to C- terminal drawings of the domain architecture of subunits of the E. coli replisome are shown in scale, relative to their lengths. Distinct domains are numbered with roman letters and the amino acid residues above the drawings indicate the first residue and, if the domains are separated by a linker, the last residue of a particular domain. (A) Subunits of Pol III core. Asterisks indicate the location of the active site residues (Asp401, Asp403, Asp555) in the α subunit. L and S indicate the large and small portions of the palm domain. (B) Subunits of the γ complex clamp loader. Domain architecture of the β clamp monomer is shown in (C) and in (D) for the DnaB helicase and DnaG primase.
The recently solved crystal structures of the α subunits from E. coli (92) and Thermus aquaticus (7) reveal that the catalytic region assumes the shape of a right hand, with fingers, palm and thumb domains, an organization observed for all DNA polymerases (21) (Fig. 3). The three domains form a deep cleft, with the active site located in the palm domain at the bottom of the cleft. Structures of other DNA polymerases show that the fingers domain interacts with the incoming dNTP and the single strand DNA template, while the thumb domain guides the nascent DNA duplex product as it leaves the active site (35, 80, 147). Surprisingly, the detailed structural topology of the palm domain of Pol III of α is strikingly different from members of most other DNA polymerase families and reveals that the Pol III α C family polymerase is structurally related to the Pol β-like nucleotidyltransferase superfamily X. Pol III α also has a much more extensive fingers domain than other DNA polymerases that consists of four distinct sub-domains (i.e., four fingers). A signature β2α structural motif, which is also observed in Pol I, is present within the palm domain suggesting an evolutionary link between Pol III and Pol I. The C-terminal region of α, which contains the two β binding motifs and the OB-domain, extends outward from the fingers domain (see Fig. 3).
Fig. 3. Crystal structure of the E. coli Pol III α subunit.
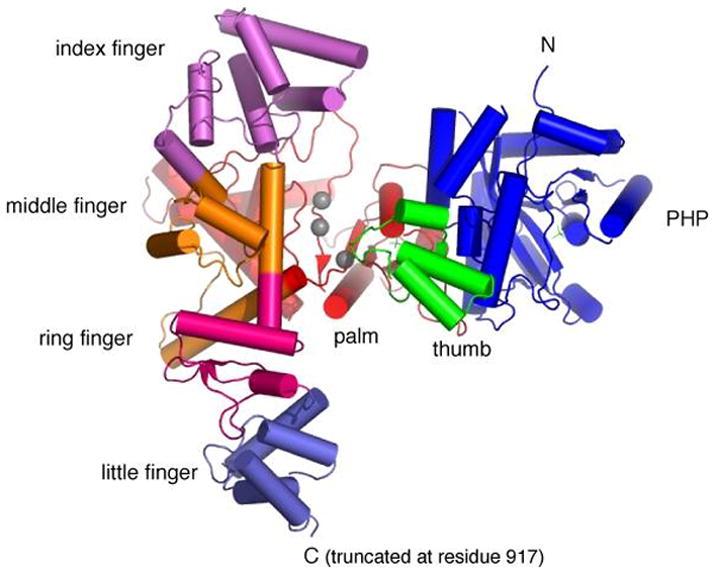
Shown is a top view of the crystal structure of α, lacking the C-terminal region (residues 918–1159) (pdb code, 2hqa). The active site residues in the Palm domain are indicated by grey spheres.
Biochemical characterization of the synthesis rate of the isolated α subunit revealed that it is quite slow (8 nt/s) compared to Pol III H.E. (650 nt/s) (106). The assembly with the ε subunit stimulates the polymerization rate of α (20 nt/s) and increases fidelity 80-fold (104, 105). Interestingly, the ε subunit also greatly stimulates the processivity of Pol III H.E. from approximately 1.5 kb to 50 kb (150), implying that ε contributes to replication speed, fidelity and stability of the moving polymerase.
The ε subunit is comprised of two domains (Fig. 2 A). The 185- residue N-terminal domain of ε contains the exonuclease active site and the θ-binding region, and the C-terminal domain (187–243) interacts with the α subunit (127, 155). The structure of the N-terminal proofreading domain shows a high degree of similarity to other DNA polymerase-associated exonucleases (33, 55). Like other proofreading nucleases, the ε exonuclease activity has a preference for single strand DNA and thus is much more active on a 3′ mismatched primer terminus compared to a fully base-paired primed site (23). As observed with proofreading nucleases of other DNA polymerases, the rate limiting step in the exonuclease reaction is the melting of the duplex DNA to generate a single strand DNA necessary to reach the exonucleolytic site (i.e., about 3 nucleotides) (114). Interestingly, the presence of the α polymerase subunit does not affect the specificity of ε in proofreading, but it stimulates the ε exonuclease activity, most likely by stabilizing the binding of ε to the DNA substrate via interaction of α with DNA (105, 114). In conclusion, cooperative interaction of the α polymerase and the ε exonuclease subunits are essential for efficient and faithful DNA replication.
Most other types of DNA polymerases contain the polymerase and exonuclease active sites on the same polypeptide. It is not known why the 3′–5′ exonuclease of Pol III core is contained on a separate subunit from the DNA polymerase. One may speculate that this organization allows the ε exonuclease to depart from the α DNA polymerase subunit in situations where proofreading may inhibit forward progression of α, for instance to move across a site of DNA damage. Alternatively, the primordial proofreading exonuclease may have been relegated exclusively to the PHP domain, and the recruitment of the more efficient ε exonuclease subunit could be an evolutionary adaptation to enhance speed and fidelity of the Pol III holoenzyme.
The function of the small θ subunit is not yet understood. Deletion of the gene encoding θ (holE) does not affect cell viability (142), but in vitro and in vivo experiments imply a slight stabilization and stimulation of the ε exonuclease activity by θ (151, 155). The solution structure of θ reveals a chain fold that resembles the DNA-interacting domain of eukaryotic DNA polymerase β (78). However, θ has not been demonstrated to bind DNA and does not appear to directly interact with the α subunit (151).
The β sliding clamp
In vivo, the two bidirectional replication forks need to move at a speed of approximately 800 nt/s to completely replicate the 4.2 Mb genome within a 40 min cell cycle. This compares favorably with the value of 650 nt/s obtained from DNA combing studies used to determine the speed of replication forks (22). However, the slow rate of Pol III core (20 nt/s) would require hours for the genome to be replicated. Pol III core therefore depends on additional proteins, which convert the polymerase into a fast and highly processive enzyme. This task is performed by the β sliding clamp, which binds directly to Pol III core (91) and holds the polymerase to DNA for high speed (0.5 – 1kb/s) and processivity (>50 kb) during chain extension (121).
The β sliding clamp is a homodimer that adopts a donut shaped ring structure and encircles duplex DNA (87) (Fig. 4A). The two monomers are arranged in a head-to-tail fashion. Each monomer consists of three globular domains and all three domains have the same chain-folding pattern. Therefore, the circular β dimer exhibits a six-fold pseudosymmetry. The outside perimeter of β is a continuous layer of an antiparallel β sheet structure and the inside cavity is lined with 12 α helices (Fig. 4A). The head-to-tail arrangement of the two protomers produces two structurally distinct faces. The C-terminal face contains the binding site for all proteins that bind β, including the 5 E. coli DNA polymerases, which pull the β sliding clamp behind them during DNA synthesis (Fig. 1).
Fig. 4. Structure of the β sliding clamp.
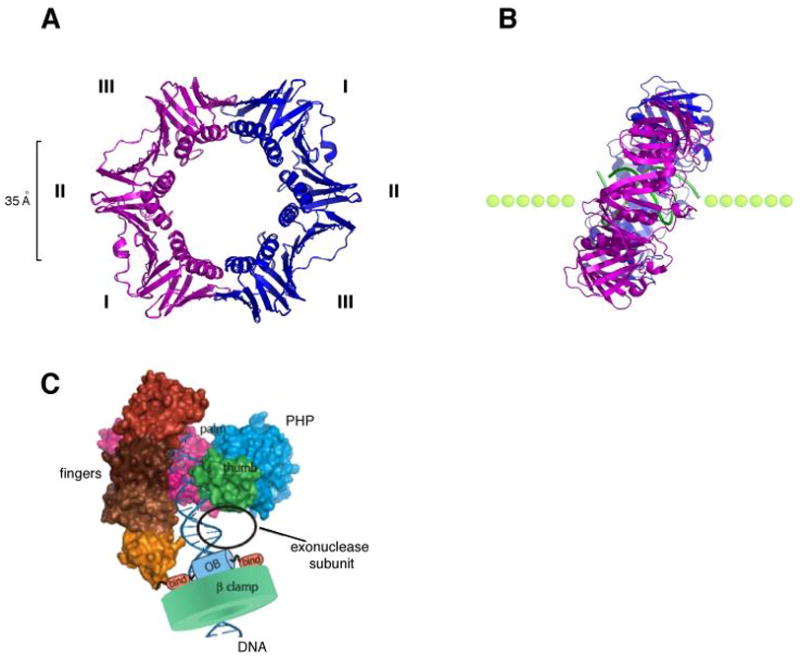
(A) Ribbon representation of the β homodimer (pdb code, 2pol). The two monomers (pink and blue) interact head-to-tail and form a highly symmetrical ring shaped structure that encircles DNA. The three domains (I, II, III) of each subunit have identical chain folding topologies and form an outside perimeter of a continuous antiparallel β sheet. The inside cavity is lined with 12 α helices. (B) Structure of a co-crystal of β with a primed DNA template (green). The side view reveals a tilted conformation of the β clamp on DNA with an angle of approximately 22°. (C) Model of the α subunit of E. coli Pol III bound to the β clamp and DNA (adapted with permission from Fig. 7 in 92).
Structural data reveal that the β ring has an outside diameter of approximately 80 Å, and an inner diameter of about 35 Å, which is sufficiently large to accommodate an A or B form double helix (87). The overall charge of β is negative, but the α-helices that line the central cavity carry a net positive charge. A recent structure of β in complex with a primed DNA template demonstrates that β directly interacts with DNA and is tilted on DNA at a 22° angle (49). This tilt of β on DNA allows direct contacts of β with both strands of duplex DNA (Fig. 4B). The single strand DNA template interacts with a hydrophobic pocket located between domains 2 and 3 of β. This hydrophobic pocket is the protein binding site used by all DNA polymerases (Pol I, II, III, IV, V) and DNA repair factors (MutS, MutL, ligase) that interact with the clamp (31, 101, 103). It seems possible that when the β clamp is assembled at a primed site, the interaction between β and single strand DNA may hold the clamp in place at the 3′ primed template junction until Pol III is recruited to the loaded β clamp.
The β clamp is a homodimer and therefore has two identical protein binding sites. As Pol III α subunit contains two β binding motifs within the C-terminal region of α, one DNA polymerase III may connect to both sites on the β dimer as illustrated in Fig. 4C. Consistent with the α-β model of Fig. 4C is the location of the internal β binding motif of α at the tip of the last finger. In addition, modeling of DNA into the palm domain of α predicts that about two dozen base pairs (bp) exist between the 3′ terminus and the far side of the β clamp, consistent with previous studies indicating that 22–24 bp are required for α to function with β (183). Another possible scenario in which the two protein binding pockets in one β dimer are occupied is one in which two different polymerase molecules occupy the two protomers of the same β dimer. For example, the DNA damage inducible polymerases Pol II, Pol IV and Pol V interact with β at the same site to which Pol III binds. Thus, two different polymerases may interact with one sliding clamp simultaneously. In this case, only one DNA polymerase can be “active” at any given time since there is only one DNA molecule inside the clamp. In situations when Pol III stalls, for example upon encountering a site of DNA damage, a low fidelity DNA polymerase could be present on the same β clamp and take control of the primer/template to facilitate the replication fork advance over a DNA lesion. Once the lesion is passed, the high fidelity Pol III may resume rapid, accurate and processive synthesis with β. Another situation, in which multiple enzymes bound to one β clamp may be useful, could occur during repair of DNA lesions. Various repair enzymes, including Pol I, DNA ligase, MutS and MutL, interact with the sliding clamp independent of replication (101, 103). Sequence comparisons of proteins that bind β reveal a consensus sequence QL[S/D]LF (31, 101, 103, 172). Overall, it has become clear that β is a platform for a variety of proteins involved in several DNA metabolic processes, in addition to serving as a processivity factor during chromosomal DNA replication. A more detailed discussion about different DNA polymerases that interact with β and how they function to reactivate stalled replication forks is presented in section 3 of this chapter.
The β dimer is quite stable on DNA and exhibits a half-life of dissociation from DNA of approximately 100 min at 37°C (184). This high degree of stability may be enabled by the continuous layer of β sheet that extends around the entire ring, including the dimer interfaces (Fig. 4). The dimer interface also involves several electrostatic and hydrophobic interactions (87). During clamp loading of β onto DNA, one of the dimer interfaces is broken for the opened ring to be placed around DNA (164). This process is mediated by the clamp loader, which uses the energy of ATP hydrolysis to assemble β onto DNA as described in the section to follow.
The γ complex clamp loader
The E. coli γ complex clamp loader is a multisubunit protein complex (γτ2δδ′ψχ) that also serves as architectural role in the assembly and organization of the replisome (68, 70). The clamp loader binds to Pol III core, DnaB helicase, the β clamp, SSB and DNA. It has become clear that these multiple connections play critical roles during DNA replication, and that the function of the clamp loader extends far beyond the primary function of sliding clamp assembly. As illustrated in Fig. 1, the γ complex physically connects the leading and lagging strand Pol III cores through direct interactions with the two τ subunits of the clamp loader. The τ subunits also interact with the DnaB helicase, thereby coordinating the unwinding activity with DNA synthesis. In addition, the clamp loader binds to SSB (via the χ subunit) and is involved in the recycling of the lagging strand polymerase. This section will describe the biochemical and structural features of the clamp loader, and relate these features to the different functions of the clamp loader during DNA replication.
The two smallest subunits, ψ and χ of the E. coli γ complex are not required for clamp loading, but stabilize the complex through interaction of the χψ complex with γ (45, 124, 178). This occurs most likely through a conserved flexible region within ψ as revealed by the crystal structure of the χ–ψ complex (53). The ψ subunit binds to χ, which directly contacts the single strand DNA binding protein (SSB) that coats the unwound lagging strand and prevents secondary structure formation (1, 141, 178). The χ-SSB interaction mainly contributes to the stability and processivity of the polymerase during elongation (50, 77).
Interestingly, the dnaX gene encodes two proteins, τ and γ (40, 41, 86, 116) (Fig. 2B). The shorter γ subunit (47 kDa) derives from a translational frameshift of the full length τ protein (71 kDa) and therefore lacks the 24 kDa C-terminal residues of τ. The unique 24 kDa region of τ consists of two additional domains, IV and V, which mediate important contacts with the DnaB helicase and Pol III core (30, 46, 47). Domain IV harbors the binding site for the DnaB helicase (46). This interaction is crucial for stimulation of the helicase activity, increasing the rate of unwinding from about 35 bp/s to the rapid rate required for fork movement in vivo (82, 187). The α subunit of Pol III core interacts with domain V of τ (47), and the presence of two τ subunits in one γ complex enables coupling of two molecules of Pol III core, one responsible for leading and the other for lagging strand synthesis (19, 121). In addition to interacting with the helicase and Pol III core, the τ subunit binds single-stranded DNA and is involved in the release of the lagging strand Pol III core from the β clamp when it reaches the end of an Okazaki fragment (96). The C-terminal 24 kDa of τ is not required for clamp loading but is essential for cell viability (15), most probably due to its role in organizing the architecture of Pol III core and DnaB helicase at the replication fork.
The γ subunit shares with τ the first N-terminal three domains that are required for clamp loading activity along with δ and δ′. Different γ complexes containing all the possible ratios of γ versus τ have similar clamp loading activity (112). The τ, γ, δ and δ′ subunits are members of the large family of AAA+ proteins (ATPases Associated with a variety of Activities) (Fig. 2 and 5). AAA+ proteins typically act as circular multimers and use ATP to remodel other proteins (120). The functions of various AAA+ proteins are diverse and widespread. For instance, some AAA+ proteins are involved in protein degradation or vesicular fusion. Not all AAA+ proteins are ATPases, however. For example, δ and δ′ do not bind ATP, only the γ (and τ) subunits are capable of binding and hydrolyzing ATP. The δ subunit of the clamp loader is considered the “wrench” of the γ complex since it is the only subunit that directly interacts with β, and δis capable of opening the β clamp on its own (164).
Fig. 5. Structure of the γ complex clamp loader.
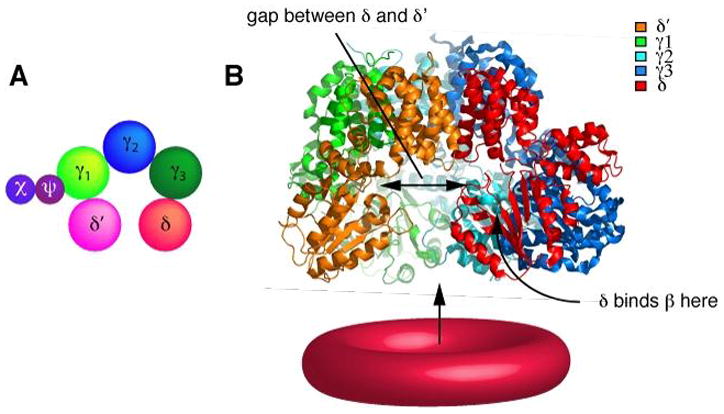
(A) Schematic representation of the arrangement of the clamp loader subunits demonstrating the circular orientation of the five subunits. The pentameric circular assembly is interrupted by a gap between the δ and δ′ subunits, leaving space for the passage of DNA. The χ and ψ subunits are thought to attach to the γ subunit via ψ {Gao, 2001 #816} (B) Ribbon representation of the crystal structure of the minimal γ complex clamp loader γ3δδ′). The Cterminal domains create a tight circular collar. The N-termini containing the two AAA+ domains are suspended downwards and adapt a conformation in which the δ and δ′ subunits create a gap large enough for the DNA to enter. The β clamp interacts with the N terminal domains.
In the absence of ATP, the clamp loader has a very low affinity for the clamp (118). ATP binding induces a conformational change that allows the complex to bind tightly to the β clamp, mediate ring opening, and develop a strong affinity for primed DNA (4, 62) (Fig. 6). Binding of primed DNA stimulates hydrolysis of ATP, allowing the clamp loader to release from β and allows the clamp to close around DNA (11). The crystal structures of the γ3δδ′ clamp loader and the δ-β complex (69, 70) provide important information regarding the organization of the clamp loader and support biochemical studies on the mechanism by which the clamp is opened and closed. The five γ3δδ′ clamp loader subunits are arranged in a circular spiral shape in the order δ′-γ1-γ2-γ3-δ (Fig. 5A) The C-terminal domain of each subunit forms strong intermolecular contacts with one another. These connections result in a tight uninterrupted circular collar from which the N-terminal domains are suspended (see Fig. 5B). The N-terminal domains of the five subunits are arranged in a spiral with a gap between the δ and δ′ subunits. This gap is important for passage of DNA to the inner chamber of the clamp loader, which forms a DNA binding site with specificity for a recessed 3′ terminus. Each of the subunits has the same overall chain fold, including the two N-terminal AAA+ domains and the C-terminal oligomerization domain.
Fig. 6. Mechanism of clamp loading.
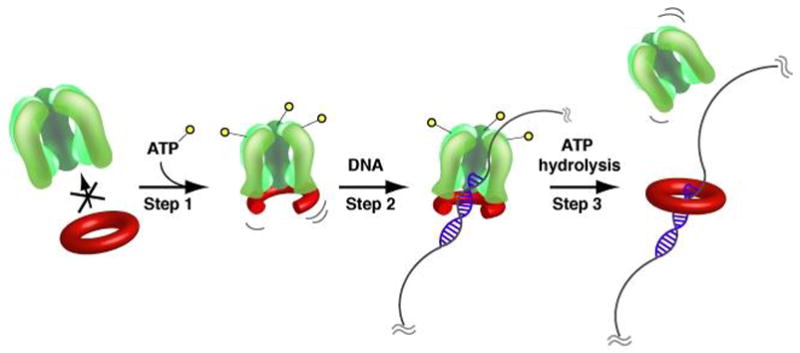
(A) ATP-binding induces a conformational change in the clamp loader that allows β-interaction (Step 1). Binding of the β clamp cracks one β dimer interface open and the β-clamp loader complex gains high affinity for a primer/template junction allowing the clamp to be placed around primed DNA (Step 2). ATP-hydrolysis allows the β dimer to close around primed DNA and ejects the clamp loader (Step 3). For simplicity, the C-terminal extensions of the τ subunits are not shown.
The γ subunits are motor proteins that bind ATP and promote the conformational changes associated with nucleotide binding and hydrolysis needed for ring opening and closing (118, 62). The ATP bound form of the clamp loader is best understood from the structure of the eukaryotic RFC pentameric clamp loader bound to the PCNA sliding clamp (20). Like the E. coli γ complex, the five subunits of RFC are AAA+ subunits and are arranged in a circle. The RFC-PCNA-ATPγS structure shows that the clamp is located directly underneath the AAA+ domains of all 5 subunits (e.g. as indicated in Fig. 5B for γ3δδ′).
The structure of the E. coli δ–β complex (70) reveals details of the clamp opening step and indicates that the β dimer is under spring tension in which the domains of the β monomer form a shallower crescent shape when they are not constrained to form a ring. The interaction domain within the N-terminus of δ is shaped as a triangular wedge, with a tip that is formed by two adjacent β strands and a loop preceding them. Two conserved hydrophobic residues (Leu-73 and Phe-74) that are in the core of the tip fit into the protein binding hydrophobic pocket on the surface of β. The protein binding pocket of β contains highly conserved residues and is located between domains 2 and 3, but does not involve the dimer interface. A second interaction site, which is important for the clamp opening mechanism, exists within the α helix that extends from the triangular wedge in δ. This helix undergoes a large conformational change and interacts with a loop in β, which is connected to an α helix at the dimer interface. The binding of δ distorts the β dimer interface, and opening of the interface allows the domains to relax and the ring to spring open. The interaction domain on β involves a hydrophobic pocket, which is the same pocket that is used for interaction with the DNA polymerase (70). The opening in the ring is positioned below the clamp loader in alignment with the gap between the AAA+ domains of δ and δ′, allowing DNA to pass through the ring and enter the central chamber of the γ complex as illustrated in Fig. 6.
Okazaki fragments in E. coli are about 1–2 kb in length, which requires repeated loading of β onto newly synthesized RNA primers. When polymerase finishes an Okazaki fragment, it rapidly dissociates from the DNA and leaves the clamp behind (152). Considering the stable interaction of β on DNA (t1/2 = 115 min) (184), the pool of 300 molecules of β clamps/cell (26) would be rapidly depleted if there were no active mechanism to disassemble the clamps and make them available for re-loading onto new primers. Clamp unloading is another function of the clamp loader (97, 152). Clamp unloading occurs through a similar mechanism as clamp loading, but only requires binding of ATP and not ATP hydrolysis (164). The δ subunit of the clamp loader also binds to the β dimer and is as efficient in ring opening and clamp unloading as the γ complex (97). The isolated δ subunit is present in 5-fold molar excess over the other components of the clamp loader (97). It is therefore possible that unloading in the cell is mostly accomplished by the free δ subunit, leaving the clamp loader complex available for more critical steps during DNA metabolism that require clamp loading.
The DNA polymerase III holoenzyme is a highly asymmetric structure due to the presence of only one copy of each of several subunits (δ, δ′, χ, ψ) in the clamp loader. Further asymmetry is generated by the replisome architecture due to the presence of DNA helicase, primase and SSB on the lagging strand, which differentiates the environments for the two DNA polymerases within the Pol III H.E. Thus, it has been proposed that the polymerases responsible for leading and lagging strand synthesis are in different environments that impose different behaviors on them, to fit the needs of replicating either one or the other strand (51, 110).
DnaB helicase and DnaG primase
Replicative helicases are circular hexamers that encircle one strand of DNA and use ATP to fuel translocation along it. Unwinding occurs as a consequence, because the DNA strand that is excluded from the inside of the hexamer is forced to part from the DNA strand that resides inside the helicase ring as the helicase moves. The E. coli helicase is called DnaB (2, 94, 133, 175). DnaB is a ring shaped homohexamer that encircles the lagging strand and acts as a wedge to melt the parental duplex as it translocates 5′-3′ along the lagging strand DNA (75, 136). The circular arrangement of the six DnaB subunits requires opening of the ring structure in order to place the DnaB hexamer around the single-strand DNA. At an origin, the helicase loading step is mediated by the activity of the helicase loader, DnaC, which functions with ATP (8) and is discussed in more detail in another chapter in this volume.
Each DnaB monomer is a 50 kDa protein composed of two domains connected by a long flexible linker region (Fig. 2 D). The N-terminal domain contains a DNA binding site and mediates, together with the linker region, interaction of DnaB and DnaG primase (14, 28, 117, 182). The larger C-terminal domain exhibits a RecA-like core fold and contains five conserved sequence motifs (H1, H1a, H2, H3 and H4) that are characteristic of the DnaB helicase family (6). The H1 and H2 motifs are implicated in nucleotide binding and hydrolysis. Furthermore, the C-terminal domain contributes to oligomerization. The C-terminal face of the DnaB hexamer is directed towards the replication fork whereas the N-terminal face is oriented to interact with the DnaG primase. Electron microscopy studies from DnaB homologues of the T4 and T7 phage systems revealed a central channel with a diameter of 25–40 Å, large enough to accommodate single as well as double strand DNA (37, 74). In the absence of a 3′ tail, which normally is excluded from the central channel, DnaB actively translocates over duplex DNA with sufficient force to displace DNA bound proteins (74). In addition, DnaB can drive branch migration of a holliday junction, indicating a role of DnaB during recombination.
In the presence of the primosomal proteins DnaC, DnaG, DnaT, PriA, PriB and PriC, the isolated DnaB helicase exhibits a very slow unwinding rate of approximately 35 nts/sec (82). Connection of Pol III holoenzyme to DnaB through the τ subunit results in increasing the speed of helicase progression to over 500 nts/s (see Fig. 1) (82).
DNA polymerases do not initiate DNA synthesis de novo and therefore depend on a preexisting primed template junction as a substrate for incorporation of new nucleotides. At the origin, and at moving replication forks, primed sites are synthesized by primase (Fig. 1). E. coli DnaG primase is a DNA-dependent RNA polymerase that is capable of synthesizing 60-nt long primers on a single stranded DNA template in vitro. In the context of a replisome however, primer synthesis is restricted to 9–14 nt (188). During lagging strand synthesis, primase synthesizes new ribonucleotide primers every 1–2 kb at a rate of approximately one primer every second or two (135, 165) and the primers are then extended into 1–2-kb-long Okazaki fragments. The length of Okazaki fragments is directly influenced by primase concentration, with shorter Okazaki fragments appearing as primase concentrations are increased (177). Whether this is the result of increased priming frequency or premature release of the lagging strand polymerase (as discussed in the following paragraph on the Okazaki fragment cycle) is not fully understood.
In a replisome, DnaG primase must interact with DnaB for activity, and this constraint ensures that new RNA primers localize to the replication fork (60, 72, 115, 160).
DnaG primase is a 70 kDa protein comprised of three structural domains (Fig. 2D). An Nterminal Zn2+-binding domain, which is required for primase function and mediates recognition of single stranded DNA, a central RNA polymerase domain that catalyses synthesis of ribonucleotide primers and a C-terminal domain that is involved in interaction with the helicase and with SSB (161). The crystal structure of the isolated RNA polymerase core domain revealed a modular, cashew-shaped molecule that is composed of three subdomains (76, 129). The central region shows similarity to unrelated proteins including topoisomerases and is therefore referred to as a TOPRIM (topoisomerase-primase) domain (3). The catalytic core is located within the TOPRIM domain and contains a metalcoordination site and conserved acidic residues that are important for primase function (36, 149). The N-terminal and TOPRIM subdomains form a deep cleft with the catalytic core in the center. In contrast to canonical DNA polymerases that use three conserved aspartate residues for the two-metal catalyzed reaction of nucleotide incorporation, primase appears to use a simple phosphotransferase domain for metal coordination thereby representing a distinct structural class of polymerases. Primases are crucial for multiple steps during DNA replication, including the initiation of DNA synthesis at replication origins, the restart of stalled replication forks and the priming of Okazaki fragments (44, 58, 88). The role of primase during replication initiation and restart is discussed in other chapters in this volume. Here, we focus on the function of primase in the context of a moving replication fork.
Primase acts distributively at a moving replication fork to initiate numerous Okazaki fragments (28). New RNA primers are synthesized every 1–2 kb on the unwound lagging strand (160, 161) and initiate preferably at sites that contain a CTG triplet (79). E. coli primase appears to be slow and highly error prone (154). Primer synthesis occurs in a two-step reaction, in which the initial condensation is slow compared to the extension of the next 10 nucleotides. Hence, the formation of the first phosphodiester bond or a step prior to it is the rate limiting step during primer synthesis (154). Primase has very low affinity for singlestrand DNA templates, especially those coated with SSB. This barrier to substrate binding is removed by transient interaction of primase with DnaB helicase, which is required for primase activity (72, 115, 160). In vitro experiments have shown that DnaB stimulates primer synthesis by increasing the affinity of primase to template DNA and by increasing the catalytic rate (72). Biochemical studies indicate that multiple primase proteins bind to one hexameric helicase molecule, thereby increasing the local concentration of primase for priming to occur more efficiently (115). This functional coordination of primase and helicase activities seems to be conserved throughout species. The Bacillus stearothermophilus helicase, for instance, forms a stable interaction of 2–3 primase molecules/helicase (5). In the bacteriophage T4 system, the helicase (gp41) and primase (gp61) subunits interact strongly to form a primosome complex with the stoichiometry of one helicase hexamer to six primase molecules (71, 181). It is interesting to note that the T7 phage encodes both the primase and helicase activity on one single polypeptide (gp4), thereby covalently connecting the two activities (43, 54). Since T7 gp4 acts as a hexamer, the stoichiometry of helicase and priming activities is 6:6, similar to the T4 phage system (44).
Primase is processive in primer synthesis and remains attached to its product once the RNA primer is complete (146). This stable interaction is mediated through direct interaction of primase with SSB bound to the single-stranded DNA template (186). Primase must be released from the RNA primer for the clamp loader to assemble a β clamp on the primed site prior to recruitment of Pol III core. This step is mediated through the χ subunit of the clamp loader, which competes with DnaG primase for SSB and leads to the displacement of DnaG primase from its RNA product, clearing the way for assembly of a β clamp at the RNA primed site (186). These direct protein-protein interactions during hand-off of the primer to the clamp loader may serve to protect the RNA-DNA hybrid until a β clamp can be assembled onto it.
The lagging strand Okazaki fragment cycle
The leading strand polymerase continually synthesizes DNA in the direction of the replication fork, whereas the lagging strand polymerase synthesizes short discontinuous Okazaki fragments in the opposite direction. Discontinuous lagging strand synthesis requires that the polymerase rapidly dissociates from each new completed Okazaki fragment in order to begin extension of a new RNA primer (Fig. 7). The lagging strand polymerase remains physically attached to the replisome (i.e., via the clamp loader) during the process of polymerase recycling from the end of one Okazaki fragment to the start of the next (83, 176, 187).
Fig. 7. Cycle of lagging strand synthesis.
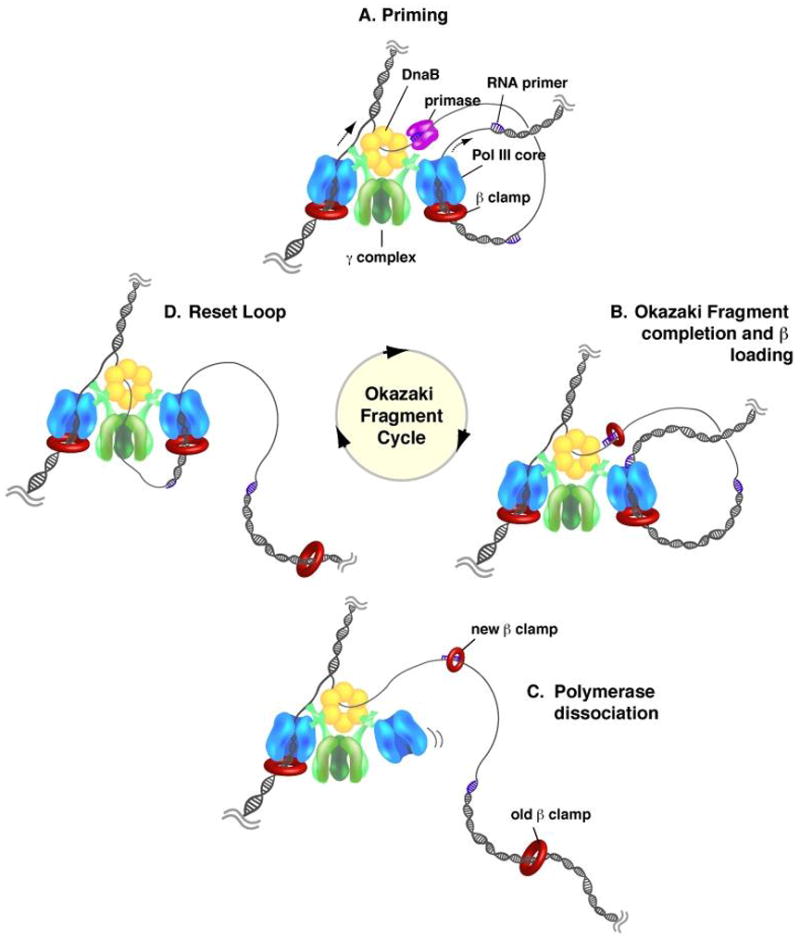
(A) As the replication fork moves, the DnaB helicase recruits DnaG primase, which synthesizes short RNA primers on the unwound lagging strand. (B) While the lagging strand polymerase finishes synthesis of the current Okazaki fragment, the clamp loader displaces primase from the newly synthesized primer and places a β clamp around the primer/template junction. (C) The completion of the Okazaki fragment induces polymerase to dissociate from the β clamp and DNA and allows recruitment to the newly synthesized upstream primer through interaction with the τ subunit of the clamp loader, leaving the β clamp behind. (D) The cycle is complete upon association of the lagging strand polymerase with a new β clamp on an upstream RNA primer to begin synthesis of a new Okazaki fragment.
Pol III H.E. is rapid (>650 nts/s) and highly processive (>50 kb). Such high processivity raises the question of how the lagging strand polymerase can rapidly dissociate from the end of a finished Okazaki fragment? Study of this question has shown the unexpected finding that dissociation of a lagging Pol III from a completed Okazaki fragment is performed by separation of Pol III from β, leaving the β clamp on DNA (see Fig. 7) (122, 152). Studies of replication fork dynamics in vitro demonstrate that the clamp loader repeatedly loads new β clamps on RNA primers as they are formed by primase (Fig. 7B) (186). Model studies show that Pol III core retains a tight grip on β even at a one nucleotide gap, but upon finishing DNA to a nick the Pol III core disengages from the β clamp (Fig. 7B→C) (96). The lagging strand Pol III core reattaches to a new β clamp on an upstream RNA primer to start the next Okazaki fragment (Fig. 7C→D).
Two different processes enable rapid lagging stand polymerase recycling among Okazaki fragments (Fig. 8). Complete synthesis of an Okazaki fragment results in “collision release,” in which the lagging strand polymerase completes the Okazaki fragment and encounters the 5′ terminus of the downstream Okazaki fragment, inducing dissociation of the DNA polymerase from β and DNA (152). Polymerase collision release is facilitated by the τ subunit of the clamp loader, which helps disengage the polymerase from the β clamp only when the single-strand template is completely converted to a duplex (96). The second process is referred to as “premature release” in which the lagging strand polymerase releases from β before it finishes the Okazaki fragment, leaving a single-strand gap to be filled in later (93, 98, 180). The signal that triggers premature release may be either primase, the synthesis of a new upstream RNA primer or the assembly of a β clamp on the new upstream primer. The molecular mechanism that underlies this process, and whether direct protein-protein contacts between primase and the Pol III holoenzyme are involved, has not been elucidated.
Fig. 8. Models of the release of the lagging strand polymerase.
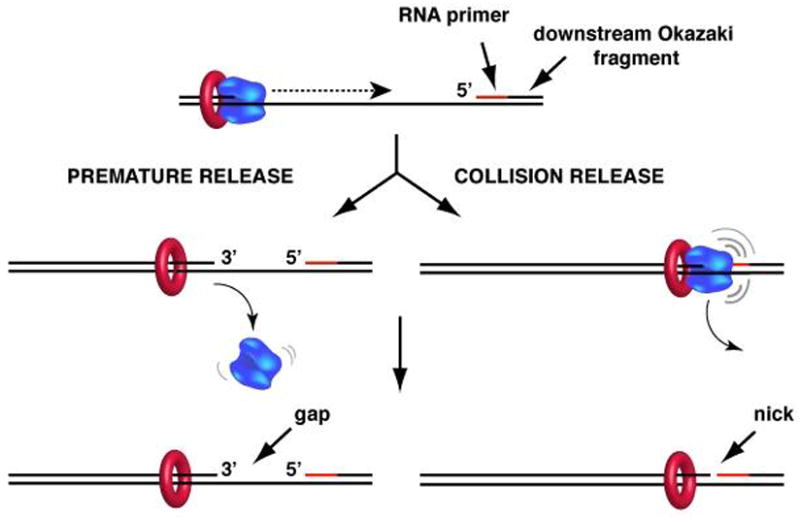
Lagging strand polymerase must be able to dissociate from an Okazaki fragment in order to be recycled to new RNA primers during synthesis of numerous Okazaki fragments. In premature release (left), the polymerase dissociates before finishing the Okazaki fragment, leaving behind a single strand DNA gap. In collision release (right) the lagging strand polymerase completes the Okazaki fragment to a nick and Pol III then disengages from the β clamp. See text for details.
The relative contributions of these two mechanisms of polymerase recycling are not yet understood. There are situations where premature release may be important to keep the fork moving, in particular when the replication fork encounters a damaged nucleotide or DNA structures that lead to stalling of one or both of the polymerases. In section 3 we examine situations that lead to replication fork stalling and discuss alternative DNA polymerases that function with the β clamp and help the replisome to bypass template lesions.
Numerous experiments to study the progression of the two polymerases during DNA replication have shown that in vitro, the leading strand polymerase requires one single priming event to synthesize the daughter strand. This stands in contrast to the lagging strand polymerase, which requires frequent re-priming for the synthesis of short Okazaki fragments. These observations have led to the common view that chromosomal replication is semidiscontinuous, in which leading strand synthesis occurs continuously and lagging strand synthesis is discontinuous. Interestingly, in vivo studies indicate that leading strand synthesis is often interrupted and that discontinuous replication occurs to a significant extent on the leading strand (reviewed in 169). In particular, recent data have shown that a replication fork stalled at a template lesion on the leading strand can be restarted by the action of primase on the leading strand, which re-initiates synthesis downstream of the lesion (57, 59). Discontinuous synthesis on the leading strand in vivo may arise from a number of factors that interfere with normal replication fork progression. These factors may include a variety of types of DNA damage, or proteins that are tightly bound to DNA including repressors, transcription complexes and DNA condensing agents (100). Many of these obstacles can lead to replication fork stalling and/or collapse and result in situations that can lead to premature termination of chain extension and thus form discontinuities in the leading strand. A more detailed discussion of the effects of DNA damage on chromosomal replication is presented in Section 3 in this chapter.
Processing of Okazaki fragments
An important step in generating a complete and intact duplex lagging strand is the removal of RNA primers after Okazaki fragments have been synthesized. This processing step requires exonucleolytic degradation of the RNA followed by fill-in by a DNA polymerase and then the action of DNA ligase to seal the nick, which is performed by DNA ligase I. RNA removal and the gap filling steps are usually performed by Pol I, the first DNA polymerase to be discovered in E. coli (12, 88). Pol I (~90 kDa) is a single subunit protein which harbors a 5′–3′ exonuclease activity in addition to the DNA polymerase and proofreading 3′–5′ exonuclease activities that are normally associated with DNA polymerases. The 5′-3′ exonuclease is actually a Flap endonuclease and functions in concert with the DNA polymerase (179).
Proteolytic cleavage divides Pol I into two active fragments, a small N-terminal (35 kDa) fragment and a large C-terminal fragment (68 kDa, also known as Klenow fragment) (32, 73, 88). The polymerase activity, pyrophosphorolysis, pyrophosphate exchange and 3′–5′ exonuclease proofreading activities are located in the large fragment (32, 90), and the 5′-3′ flap exonuclease activity is located in the smaller N-terminal fragment (42). These activities conspire to provide Pol I with ability to initiate replication at a nick and perform nick translation synthesis (85). Nick translation occurs by strand displacement of duplex DNA, providing 5′ single-strand DNA for the 5′-3′ exonuclease activity of Pol I at the same site as Pol I extends DNA to fill the gap that results from 5′-3′ exonuclease action. This nick translation capability of Pol I efficiently removes RNA primers and simultaneously fills the gap with DNA. Besides its role in RNA primer processing, Pol I is involved in a number of other DNA repair reactions (88).
3. Replication at sites of DNA damage
Cells are constantly exposed to oxidative stress, UV irradiation and reactive chemicals that cause a variety of different types of DNA damage. Some types of damage are easily repaired by nucleotide-repair, mismatch-repair or base-excision repair machineries, while other types of damage are not as efficiently repaired, or are not repaired fast enough to avoid collision with the replication fork. Sites that contain damaged nucleotides generally present a problem for the replication machinery since the high fidelity Pol III H.E. cannot extend DNA across a damaged template base. Several mechanisms exist that allow bypass of lesions and thus promote continued replication fork movement. Interestingly, DNA damage on the lagging strand does not inhibit replication fork movement as illustrated by in vivo and in vitro studies (61, 113). A stalled lagging strand polymerase simply dissociates from β by the premature release mechanism and recycles to a new upstream RNA primer, leaving the lesion behind. A damaged nucleotide on the leading strand presents more of a problem. A damaged template nucleotide on the leading strand induces the polymerase to stall, but the helicase continues to unwind the parental DNA. This produces single strand DNA ahead of the stalled leading strand polymerase (126). Production of single strand DNA is thought to be the primary signal that triggers the induction of a DNA damage response (“SOS-response”), which is initiated by binding of RecA to single strand DNA upon which a RecA filament assembles (reviewed in 139). RecA filament formation activates RecA to function as a coprotease for cleavage of the transcriptional repressor, LexA. Cleavage of LexA results in dissociation of the LexA repressor from DNA, thereby turning on the expression of more than 40 genes involved in the cellular response to damaged DNA. These “SOS-induced” proteins include enzymes required for nucleotide excision repair, base excision repair, DNA recombination, cell division and proteins that are needed to rescue stalled replication forks (29, 39).
There appear to be several mechanisms by which a stalled replication fork may be restarted, and thereby avoid replication fork collapse. In one scenario, referred to as translesion synthesis (TLS), the stalled Pol III is replaced by one of three different specialized damage inducible DNA polymerases that can extend DNA across a damaged template nucleotide. However, this process often results in the insertion of a wrong nucleotide opposite the lesion. These DNA polymerases, and their function with the β clamp, will be described below. Once the lesion is passed, Pol III presumably regains control of the primed site and resumes high fidelity DNA synthesis at the replication fork. The lesion in the template strand may become repaired in a later step through homologous recombination or nucleotide-, mismatch- or base-excision repair machineries. Lesion bypass typically results in an inheritable mutation, but provides a route by which the replication fork continues the essential function of genome duplication. In a second scenario, a leading strand lesion is bypassed by a new priming event downstream of the lesion, leaving the lesion with a gap of single strand DNA (58). This is followed by high fidelity recombination processes that repair the damaged template. These high fidelity recombination based mechanisms are explained in another chapter in this volume. The existence of multiple pathways to resolve a stalled replication fork reflects the importance of recovering from DNA damage and that duplication of the genomic DNA continues to completion. We next describe DNA polymerases that are involved in the process of moving the Pol III H.E. past sites of DNA damage.
Translesion (TLS) polymerases
Lesion bypass can be thought of as a two-step reaction that starts with the incorporation of a nucleotide opposite the lesion followed by extension of the resulting distorted primer terminus. Three different translesion (TLS) DNA polymerases, Pol II, Pol IV and Pol V, are induced during the SOS response (Table 2). Pol II has rather high fidelity as it contains a proofreading 3′–5′ exonuclease and belongs to the B-family of DNA polymerases. Pol IV and Pol V are both members of the error-prone Y-family of DNA polymerases, which lack 3′–5′ proofreading exonuclease activity. These three damage inducible DNA polymerases are regulated somewhat differently during the SOS response and they appear to have distinct preferences for nucleotide insertion opposite certain damaged nucleotide substrates (Table 2) (52, 119). All TLS DNA polymerases may contribute to the increased mutagenesis that is observed after various types of DNA damage (119). The particular DNA polymerase that is chosen to replace Pol III at the replication fork is thought to depend on the timing, the availability of a specific polymerase and the type of DNA damage.
Table 2.
Translesion polymerases
| TLS Polymerase | Subunit | Subunits/ Polymerase | Gene | Mol. wt (kDa) | Molecules/Cell | Molecules/Cell after SOS response | Time after induction | Preferential lesion bypass |
|---|---|---|---|---|---|---|---|---|
| Pol II | PolB | 89.9 | 30–50 | 350 (ref. in 16) | 1–5 min | Abasic sites, N-2-acetylaminofluorene (AAF) guanine adducts | ||
|
| ||||||||
| Pol IV | DinB | 32 | 250 | 2500 (ref. in 29) | 1–5 min | benzo(a)pyrene diol epoxide | ||
|
| ||||||||
| Pol V | - | 15 (ref. in 173) | TT cis-syn photodimers, TT (6–4) photoproduct BaP DE, AAF | |||||
| UmuC | 1 | UmuC | 46 | 30 | 20–60 | 10–45 min | ||
| UmuD | UmuD | 15 | 180 | 400 | 15 min | |||
| UmuD’ | 2 | UmuD | 12 | 350 | 20–25 min | |||
Pol II was originally identified in the 1970s, along with Pol III (89). The 89.9 kDa Pol II protein is encoded by the polB (dinA) gene and is present at 30–50 copies per cell under normal conditions; it is induced approximately 7-fold during the SOS-response (16, 17, 66). Genetic studies have shown that Pol II may be involved in a number of DNA transactions, including the repair of DNA damage upon UV-irradiation (132), repair of inter-strand crosslinks (10), adaptive mutagenesis and long-time survival (38, 185). In vivo and in vitro studies have shown that Pol II is able to bypass AAF (N-2-acetylaminofuorene) and abasic sites, with a preference for incorporating dA opposite the template lesion (17, 159). Interestingly, Pol II may also contribute to fidelity during undisturbed chromosomal replication, since an exonuclease deficient Pol II displays increased levels of mutagenesis (9, 132).
Pol II displays a relatively high fidelity, with a rate of one misincorporated base per 106 nucleotides. This rate is decreased by 1000 fold in an exonuclease deficient mutant of Pol II, which normally very efficiently proofreads replication errors that include single base substitutions, single base additions and deletion errors (27). Pol II, as all the TLS polymerases, interacts with the β clamp and in the case of Pol II the β clamp stimulates polymerase processivity from about 5 to around 1,600 nucleotides (18, 63, 156). Pol II is much slower than Pol III, and extends DNA at a rate of 20–40 nt/s (18).
Pol IV shares high sequence homology to S. cerevisiae Rev1 and E. coli Pol V, both members of the Y-family of DNA polymerases (123). Translesion Y-family polymerases are poorly processive and lack an associated exonuclease activity. They are therefore highly error-prone and have a fidelity of one misincorporated base per 102–103 nt (67). An explanation for the high misincorporation rate of TLS DNA polymerases may be understood by the crystal structures of several members of the Y-family of DNA polymerases (99, 163, 189). Crystal structures of Y-family polymerases reveal a catalytic site architecture that offers sufficient room to accommodate misaligned nucleotides, which may under the observed low fidelity of translesion polymerases. For example, the Pol IV homolog of Sulfolobus solfataricus (Dpo4) shows the basic polymerase structure with the common shape of a right hand consisting of fingers and thumb domains along with the palm domain that contains the conserved key acidic residues in the catalytic site (189). However, the fingers and thumb domains differ significantly from the high fidelity Pol III C-family polymerases. For example, the fingers domain lacks an α helix that is thought to be important in checking the incoming nucleoside triphosphate for a correct base pair to the template. In addition, the binding pocket for the 3′ base pair reveals a relatively open architecture with limited contacts between the protein and the replicating base pair and even contains sufficient space to accommodate an additional template base (99, 189). Overall, the structural data indicate that a much less stringent control of the base to be incorporated, and a catalytic site that offers sufficient space to accommodate misaligned nucleotides, may underlie the observed increase in misincorporation rates observed by TLS polymerases.
Pol IV preferentially bypasses misaligned substrates with bulges rather than mismatched primer ends (167). Consistent with this, overexpression of Pol IV results in an increase of mutagenesis with a preference for −1 frameshift mutations and single nucleotide substitutions (84, 168). The processivity of Pol IV is greatly stimulated by the presence of the β sliding clamp, reaching 300–400 nucleotides per template binding event in the presence of the β clamp. The increased processivity correlates to a higher affinity of Pol IV to the DNA in the presence of β (166). In addition, binding of Pol IV to β in the presence of the γ complex increases the affinity of Pol IV for dNTPs by 400 fold (157).
Similar to other DNA polymerases and repair factors, Pol IV interacts with β through a conserved motif located at the extreme C-terminus of Pol IV (24, 95, 102). The crystal structure of a C-terminal domain of Pol IV bound to β shows that the C-terminal residues of Pol IV bind to the hydrophobic protein binding pocket of β and also reveals a second interaction site of Pol IV with the edge of the β ring that results in Pol IV angling off the side of the β clamp (24). The authors suggest that the orientation of Pol IV on β may accommodate the binding of two polymerases at the same time. Soon after, it was demonstrated experimentally that the β dimer can indeed bind Pol III and Pol IV simultaneously (64). The latter study went on to show that Pol III controls the primer terminus during uninterrupted chain extension, but upon stalling of Pol III, Pol IV gains control of the primer/template junction (64). Once the lesion has been bypassed, the high fidelity Pol III takes control of the primer terminus and resumes faithful DNA replication. This mechanism, illustrated in Fig. 9, limits the action of the error-prone Pol IV to regions of the template that block Pol III.
Fig. 9. Coordination of two polymerases on one β clamp during bypass of a template lesion.
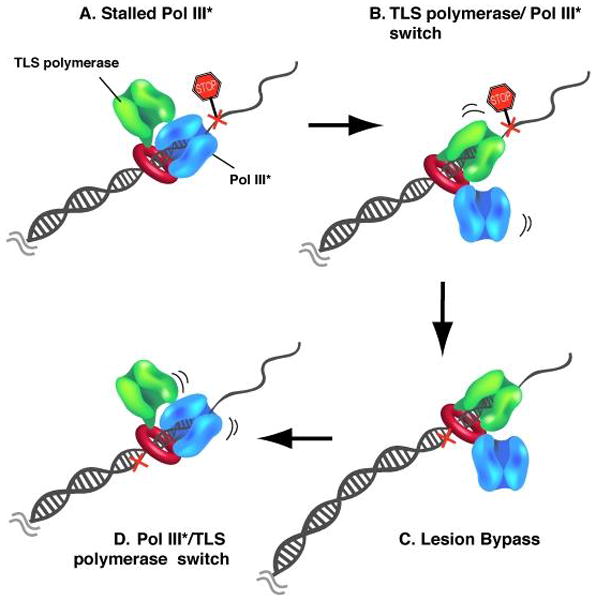
(A) The two protein binding sites on the two protomers of the β clamp homodimer allow interaction with two DNA polymerases simultaneously. Pol III (blue) retains control of the primer/template during replication under undisturbed conditions. Template lesions (cross) ahead of the polymerase induce Pol III to stall. (B) A translesion polymerase (TLS, green) switches places with the stalled Pol III and takes over the primer/template. (C) The TLS polymerase extends the primed site across the lesion. (D) Once the lesion is bypassed, Pol III regains control of the primer/template and continues high fidelity DNA synthesis.
Pol V is the major DNA polymerase responsible for mutagenic bypass of template lesions during the SOS response (134, 158). Pol V is a heterotrimer composed of two UmuD’ subunits (12 kDa each) and one 46 kDa subunit of UmuC which contains the catalytic active site (156, 158, 174). UmuD’ is an N-terminal proteolytic product of full length UmuD and is generated by a self-cleavage reaction mediated by RecA bound to single strand DNA, similar to the RecA mediated auto-cleavage reaction of the LexA repressor (25). It is interesting to note that UmuD is produced within 5 min. after induction of an SOS response. In contrast, the cleaved form, UmuD’, is only detectable after about 25 min. Peak levels of UmuC are only reached after 45 min following SOS induction (173). The early induction of the uncleaved form of UmuD suggests a role for UmuD in addition to formation of Pol V, which requires cleavage of UmuD to UmuD’. In fact, expression of uncleaved UmuD has been shown to delay DNA replication and cell cycle progression, which allows time for accurate repair systems to process the lesion and prevent the replication machinery from hitting damaged nucleotides (125). Thus, cleavage of UmuD to UmuD’ may act to delay assembly of an active translesion polymerase that results in mutagenic bypass. If a blocking lesion cannot be fixed by an error-free process within 45 min., mutations mediated by Pol V are the price to pay for cells to continue replication. It is important to note however, that mutations may also facilitate adaptation by natural selection to evolve an organism that is more fit to a changing environment. In addition, high concentrations of UmuD’ and UmuC appear to inhibit RecAmediated homologous recombination, which suggests that when homologous recombination is not successful, translesion synthesis may become a viable alternative pathway (143).
Pol V lacks a 3′–5′ exonuclease and thus demonstrates low fidelity, with a misincorporation rate of 10−2 to 10−3 nucleotides on damaged and non-damaged templates (156, 157). These characteristics enable Pol V to efficiently bypass TT (6–4) photoproducts, TT cis-syn photodimers and abasic sites (157). Three additional factors facilitate Pol V activity during lesion bypass: RecA, SSB and the β clamp (128). Pol V interacts with the β sliding clamp through a conserved β binding motif (31) located at the extreme C-terminus of UmuC (13, 107). Pol V also binds the β clamp through the UmuD and UmuD’ subunits, with a stronger interaction of UmuD to the β clamp than UmuD’ (153). Pol V activity is greatly stimulated by a RecA filament containing a free 3′ end, in trans (138). Short stretches of RecA filaments are sufficient for stimulation of Pol V, but longer stretches of single-stranded DNA, and higher concentrations of RecA filaments, increase the stimulatory effect (138, 144). The stimulation seems to be mediated through two distinct interactions between Pol V and RecA. First, Pol V directly interacts with RecA in a DNA and ATP independent manner (139). This interaction is required, but is not sufficient for stimulation of Pol V activity. Second, a DNA and ATP dependent interaction between RecA and the UmuD’ subunit of Pol V is required (140).
4. Conclusion
A remarkable property of E. coli, and many other eubacterial organisms, is the speed by which they propagate. Rapid cell division requires the presence of an extremely efficient replication machinery for rapid and faithful duplication of the genome. Characterization of the E. coli chromosomal Pol III holoenzyme shows that it is exceedingly rapid and processive. Compared to a yeast replication fork, which travels at a speed of 48 nt/s (131), the E. coli replication forks move approximately 20 times faster. The molecular basis of this efficient synthesis of DNA is a ring shaped sliding clamp, and a clamp loading machine that together endow the Pol III holoenzyme with highly efficient synthetic capability. It is now apparent that the same strategy, use of a clamp and clamp loader, generalizes to the eukaryotic and archael branches of life as well.
At a functional replication fork, the Pol III machinery is embedded in a complex network of protein interactions with the hexameric DnaB helicase, primase and SSB at a replication fork. Many of the factors and dynamic interactions that are involved in replication fork propagation in E. coli are highly conserved in eubacteria and probably also exist within replication machineries of eukaryotic organisms.
Many fascinating and important questions remain to be addressed in the area of replication fork structure and function. For example, the process that recycles the lagging strand DNA polymerase is still not understood in molecular detail. Nor are the multiple steps in clamp loading action that must underlie coupling of ATP hydrolysis to the opening and closing of the β clamp at a primed template junction. The replisome encounters many different types of blocks, such as DNA bound repressors, RNA polymerases and chromosome condensation factors. How the replisome deals with these various obstacles are important questions for future studies. In addition, the replisome encounters DNA lesions and must interface with DNA repair proteins, recombination machinery and various types of lesion-bypass DNA polymerases. The detailed mechanisms that underlie these processes, and others, will hold the attention of numerous laboratories for many years to come.
Table 1.
E. coli replisome components and associated functions
| Replisome component | Subunit | Subunits/replisome | Molecules/cell | Gene | Mol. wt (kDa) | Function during DNA replication | |
|---|---|---|---|---|---|---|---|
| Pol III H.E. | Pol III* | Pol III Core | 20 (ref. in 109) | DNA synthesis and proofreading | |||
| α | 2 | dnaE | 129.9 | DNA polymerase | |||
| ε | 2 | dnaQ | 27.5 | 3′–5′ exonuclease | |||
| θ | 2 | holE | 8.6 | Stimulates exonuclease activity | |||
| Clamp loader | (ref. in 97) | Clamp loading, stimulates helicase activity, connects leading and lagging strand polymerases, main coordinator of replisome | |||||
| γ/τ | 3 | 140 | dnaX | 47.5/71.1 | ATPase, connects both polymerases, interaction with DnaB | ||
| δ | 1 | 930 | holA | 38.7 | Opens β clamp | ||
| δ′ | 1 | 140 | holB | 36.9 | Stator | ||
| χ | 1 | 1200 | holC | 16.6 | Binds SSB | ||
| ψ | 1 | 340 | holD | 15.2 | Connects clamp loader to SSB | ||
|
| |||||||
| β | 2 | 300 (ref. in 26) | dnaN | 40.6 | Processivity clamp | ||
|
| |||||||
| Primase | 3 | 50–100 (ref. in 135) | dnaG | 65.6 | RNA primer synthesis | ||
| Helicase | 6 | 15–20 (ref. in 133, 175) | dnaB | 52.4 | DNA unwinding | ||
| SSB | 4 | 800 (ref. in 141) | ssb | 18.8 | Binds ssDNA, prevents secondary structure formation, protects against nucleases, interacts with χ and primase | ||
Acknowledgments
We grateful to Chiara Indiani for comments on the manuscript and Roxana E. Georgescu for help with illustrations. This work was supported by NIH grant (GM38839).
References
- 1.Anderson SG, Williams CR, O’Donnell M, Bloom LB. A function for the psi subunit in loading the Escherichia coli DNA polymerase sliding clamp. J Biol Chem. 2007;282:7035–45. doi: 10.1074/jbc.M610136200. [DOI] [PubMed] [Google Scholar]
- 2.Arai K, Yasuda S, Kornberg A. Mechanism of dnaB protein action. I. Crystallization and properties of dnaB protein, an essential replication protein in Escherichia coli. J Biol Chem. 1981;256:5247–52. [PubMed] [Google Scholar]
- 3.Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–13. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ason B, Handayani R, Williams CR, Bertram JG, Hingorani MM, O’Donnell M, Goodman MF, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III beta sliding clamp on DNA. Bona fide primer/templates preferentially trigger the gamma complex to hydrolyze ATP and load the clamp. J Biol Chem. 2003;278:10033–40. doi: 10.1074/jbc.M211741200. [DOI] [PubMed] [Google Scholar]
- 5.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–63. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 6.Bailey S, Eliason WK, Steitz TA. The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 2007;35:4728–36. doi: 10.1093/nar/gkm507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 8.Baker TA, Sekimizu K, Funnell BE, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45:53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- 9.Banach-Orlowska M, I, Fijalkowska J, Schaaper RM, Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- 10.Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181:2878–82. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram JG, Bloom LB, Hingorani MM, Beechem JM, O’Donnell M, Goodman MF. Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when gamma complex loads beta on DNA. J Biol Chem. 2000;275:28413–20. doi: 10.1074/jbc.M910441199. [DOI] [PubMed] [Google Scholar]
- 12.Bessman MJ, Kornberg A, Lehman IR, Simms ES. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–8. doi: 10.1016/0006-3002(56)90127-5. [DOI] [PubMed] [Google Scholar]
- 13.Beuning PJ, Sawicka D, Barsky D, Walker GC. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol Microbiol. 2006;59:460–74. doi: 10.1111/j.1365-2958.2005.04959.x. [DOI] [PubMed] [Google Scholar]
- 14.Biswas EE, Biswas SB. Mechanism of DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding, and oligomerization. Biochemistry. 1999;38:10919–28. doi: 10.1021/bi990048t. [DOI] [PubMed] [Google Scholar]
- 15.Blinkova A, Hervas C, Stukenberg PT, Onrust R, O’Donnell ME, Walker JR. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J Bacteriol. 1993;175:6018–27. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonner CA, Hays S, McEntee K, Goodman MF. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:7663–7. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonner CA, Randall SK, Rayssiguier C, Radman M, Eritja R, Kaplan BE, McEntee K, Goodman MF. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988;263:18946–52. [PubMed] [Google Scholar]
- 18.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O’Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–8. [PubMed] [Google Scholar]
- 19.Bowman GD, Goedken ER, Kazmirski SL, O’Donnell M, Kuriyan J. DNA polymerase clamp loaders and DNA recognition. FEBS Lett. 2005;579:863–7. doi: 10.1016/j.febslet.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 21.Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 22.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci U S A. 2005;102:3942–7. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenowitz S, Kwack S, Goodman MF, O’Donnell M, Echols H. Specificity and enzymatic mechanism of the editing exonuclease of Escherichia coli DNA polymerase III. J Biol Chem. 1991;266:7888–92. [PubMed] [Google Scholar]
- 24.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003;22:5883–92. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burckhardt SE, Woodgate R, Scheuermann RH, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci U S A. 1988;85:1811–5. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgers PM, Kornberg A, Sakakibara Y. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of escherichia coli. Proc Natl Acad Sci U S A. 1981;78:5391–5. doi: 10.1073/pnas.78.9.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai H, Yu H, McEntee K, Kunkel TA, Goodman MF. Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J Biol Chem. 1995;270:15327–35. doi: 10.1074/jbc.270.25.15327. [DOI] [PubMed] [Google Scholar]
- 28.Chang P, Marians KJ. Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J Biol Chem. 2000;275:26187–95. doi: 10.1074/jbc.M001800200. [DOI] [PubMed] [Google Scholar]
- 29.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dallmann HG, Kim S, Pritchard AE, Marians KJ, McHenry CS. Characterization of the unique C terminus of the Escherichia coli tau DnaX protein. Monomeric C-tau binds alpha AND DnaB and can partially replace tau in reconstituted replication forks. J Biol Chem. 2000;275:15512–9. doi: 10.1074/jbc.M909257199. [DOI] [PubMed] [Google Scholar]
- 31.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–32. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derbyshire V, Freemont PS, Sanderson MR, Beese L, Friedman JM, Joyce CM, Steitz TA. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 33.DeRose EF, Darden T, Harvey S, Gabel S, Perrino FW, Schaaper RM, London RE. Elucidation of the epsilon-theta subunit interface of Escherichia coli DNA polymerase III by NMR spectroscopy. Biochemistry. 2003;42:3635–44. doi: 10.1021/bi0205451. [DOI] [PubMed] [Google Scholar]
- 34.Dohrmann PR, McHenry CS. A bipartite polymerase-processivity factor interaction: only the internal beta binding site of the alpha subunit is required for processive replication by the DNA polymerase III holoenzyme. J Mol Biol. 2005;350:228–39. doi: 10.1016/j.jmb.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 35.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–8. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 36.Dracheva S, Koonin EV, Crute JJ. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J Biol Chem. 1995;270:14148–53. doi: 10.1074/jbc.270.23.14148. [DOI] [PubMed] [Google Scholar]
- 37.Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci U S A. 1995;92:3869–73. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escarceller M, Hicks J, Gudmundsson G, Trump G, Touati D, Lovett S, Foster PL, McEntee K, Goodman MF. Involvement of Escherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation. J Bacteriol. 1994;176:6221–8. doi: 10.1128/jb.176.20.6221-6228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–72. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 40.Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990;87:3713–7. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flower AM, McHenry CS. Transcriptional organization of the Escherichia coli dnaX gene. J Mol Biol. 1991;220:649–58. doi: 10.1016/0022-2836(91)90107-h. [DOI] [PubMed] [Google Scholar]
- 42.Freemont PS, Ollis DL, Steitz TA, Joyce CM. A domain of the Klenow fragment of Escherichia coli DNA polymerase I has polymerase but no exonuclease activity. Proteins. 1986;1:66–73. doi: 10.1002/prot.340010111. [DOI] [PubMed] [Google Scholar]
- 43.Frick DN, Baradaran K, Richardson CC. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc Natl Acad Sci U S A. 1998;95:7957–62. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 45.Gao D, McHenry CS. Tau binds and organizes Escherichia coli replication proteins through distinct domains. Domain III, shared by gamma and tau, binds delta delta ‘ and chi psi. J Biol Chem. 2001;276:4447–53. doi: 10.1074/jbc.M009827200. [DOI] [PubMed] [Google Scholar]
- 46.Gao D, McHenry CS. tau binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of tau, binds the replication fork, helicase, DnaB. J Biol Chem. 2001;276:4441–6. doi: 10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- 47.Gao D, McHenry CS. tau binds and organizes Escherichia coli replication through distinct domains. Partial proteolysis of terminally tagged tau to determine candidate domains and to assign domain V as the alpha binding domain. J Biol Chem. 2001;276:4433–40. doi: 10.1074/jbc.M009828200. [DOI] [PubMed] [Google Scholar]
- 48.Gefter ML, Hirota Y, Kornberg T, Wechsler JA, Barnoux C. Analysis of DNA polymerases II and 3 in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc Natl Acad Sci U S A. 1971;68:3150–3. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glover BP, McHenry CS. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem. 1998;273:23476–84. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 51.Glover BP, McHenry CS. The DNA polymerase III holoenzyme: an asymmetric dimeric replicative complex with leading and lagging strand polymerases. Cell. 2001;105:925–34. doi: 10.1016/s0092-8674(01)00400-7. [DOI] [PubMed] [Google Scholar]
- 52.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 53.Gulbis JM, Kazmirski SL, Finkelstein J, Kelman Z, O’Donnell M, Kuriyan J. Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur J Biochem. 2004;271:439–49. doi: 10.1046/j.1432-1033.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 54.Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J Biol Chem. 1999;274:30303–9. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 55.Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–46. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 56.Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–49. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 57.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–62. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 58.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–43. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 59.Heller RC, Marians KJ. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol Cell. 2005;17:733–43. doi: 10.1016/j.molcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Hiasa H, Marians KJ. Initiation of bidirectional replication at the chromosomal origin is directed by the interaction between helicase and primase. J Biol Chem. 1999;274:27244–8. doi: 10.1074/jbc.274.38.27244. [DOI] [PubMed] [Google Scholar]
- 61.Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003;8:437–49. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 62.Hingorani MM, O’Donnell M. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J Biol Chem. 1998;273:24550–63. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 63.Hughes AJ, Jr, Bryan SK, Chen H, Moses RE, McHenry CS. Escherichia coli DNA polymerase II is stimulated by DNA polymerase III holoenzyme auxiliary subunits. J Biol Chem. 1991;266:4568–73. [PubMed] [Google Scholar]
- 64.Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–15. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 65.Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7:751–61. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 66.Iwasaki H, Nakata A, Walker GC, Shinagawa H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990;172:6268–73. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–7. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 68.Jeruzalmi D, O’Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr Opin Struct Biol. 2002;12:217–24. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 69.Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–41. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 70.Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O’Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–28. [PubMed] [Google Scholar]
- 71.Jing DH, Dong F, Latham GJ, von Hippel PH. Interactions of bacteriophage T4-coded primase (gp61) with the T4 replication helicase (gp41) and DNA in primosome formation. J Biol Chem. 1999;274:27287–98. doi: 10.1074/jbc.274.38.27287. [DOI] [PubMed] [Google Scholar]
- 72.Johnson SK, Bhattacharyya S, Griep MA. DnaB helicase stimulates primer synthesis activity on short oligonucleotide templates. Biochemistry. 2000;39:736–44. doi: 10.1021/bi991554l. [DOI] [PubMed] [Google Scholar]
- 73.Joyce CM, Kelley WS, Grindley ND. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982;257:1958–64. [PubMed] [Google Scholar]
- 74.Kaplan DL, O’Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–57. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 75.Kaplan DL, O’Donnell M. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol Cell. 2004;15:453–65. doi: 10.1016/j.molcel.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 76.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–6. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 77.Kelman Z, Yuzhakov A, Andjelkovic J, O’Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. Embo J. 1998;17:2436–49. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keniry MA, Berthon HA, Yang JY, Miles CS, Dixon NE. NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli. Protein Sci. 2000;9:721–33. doi: 10.1110/ps.9.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khopde S, Biswas EE, Biswas SB. Affinity and sequence specificity of DNA binding and site selection for primer synthesis by Escherichia coli primase. Biochemistry. 2002;41:14820–30. doi: 10.1021/bi026711m. [DOI] [PubMed] [Google Scholar]
- 80.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–7. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 81.Kim DR, McHenry CS. Biotin tagging deletion analysis of domain limits involved in protein-macromolecular interactions. Mapping the tau binding domain of the DNA polymerase III alpha subunit. J Biol Chem. 1996;271:20690–8. doi: 10.1074/jbc.271.34.20690. [DOI] [PubMed] [Google Scholar]
- 82.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–50. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 83.Kim S, Dallmann HG, McHenry CS, Marians KJ. tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J Biol Chem. 1996;271:21406–12. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 84.Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci U S A. 1997;94:13792–7. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klett RP, Cerami A, Reich E. Exonuclease VI, a new nuclease activity associated with E. coli DNA polymerase. Proc Natl Acad Sci U S A. 1968;60:943–50. doi: 10.1073/pnas.60.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kodaira M, Biswas SB, Kornberg A. The dnaX gene encodes the DNA polymerase III holoenzyme tau subunit, precursor of the gamma subunit, the dnaZ gene product. Mol Gen Genet. 1983;192:80–6. doi: 10.1007/BF00327650. [DOI] [PubMed] [Google Scholar]
- 87.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–37. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 88.Kornberg A, Baker TA. DNA replication. 2. W.H. Freeman; New York: 1992. [Google Scholar]
- 89.Kornberg T, Gefter ML. Purification and DNA synthesis in cell-free extracts: properties of DNA polymerase II. Proc Natl Acad Sci U S A. 1971;68:761–4. doi: 10.1073/pnas.68.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–7. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 91.LaDuca RJ, Crute JJ, McHenry CS, Bambara RA. The beta subunit of the Escherichia coli DNA polymerase III holoenzyme interacts functionally with the catalytic core in the absence of other subunits. J Biol Chem. 1986;261:7550–7. [PubMed] [Google Scholar]
- 92.Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–92. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 93.Langston LD, O’Donnell M. DNA replication: keep moving and don’t mind the gap. Mol Cell. 2006;23:155–60. doi: 10.1016/j.molcel.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 94.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–48. [PubMed] [Google Scholar]
- 95.Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–9. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leu FP, Georgescu R, O’Donnell M. Mechanism of the E. coli tau processivity switch during lagging-strand synthesis. Mol Cell. 2003;11:315–27. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 97.Leu FP, Hingorani MM, Turner J, O’Donnell M. The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem. 2000;275:34609–18. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- 98.Li X, Marians KJ. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. J Biol Chem. 2000;275:34757–65. doi: 10.1074/jbc.M006556200. [DOI] [PubMed] [Google Scholar]
- 99.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 100.Liu B, Alberts BM. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science. 1995;267:1131–7. doi: 10.1126/science.7855590. [DOI] [PubMed] [Google Scholar]
- 101.Lopez de Saro F, Georgescu RE, Leu F, O’Donnell M. Protein trafficking on sliding clamps. Philos Trans R Soc Lond B Biol Sci. 2004;359:25–30. doi: 10.1098/rstb.2003.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lopez de Saro FJ, Georgescu RE, Goodman MF, O’Donnell M. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. Embo J. 2003;22:6408–18. doi: 10.1093/emboj/cdg603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lopez de Saro FJ, O’Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A. 2001;98:8376–80. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maki H, Horiuchi T, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. I. Amplification of the dnaE gene product and polymerase activity of the alpha subunit. J Biol Chem. 1985;260:12982–6. [PubMed] [Google Scholar]
- 105.Maki H, Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc Natl Acad Sci U S A. 1987;84:4389–92. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maki H, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985;260:12987–92. [PubMed] [Google Scholar]
- 107.Maor-Shoshani A, Livneh Z. Analysis of the stimulation of DNA polymerase V of Escherichia coli by processivity proteins. Biochemistry. 2002;41:14438–46. doi: 10.1021/bi0262909. [DOI] [PubMed] [Google Scholar]
- 108.Marians KJ, Hiasa H, Kim DR, McHenry CS. Role of the core DNA polymerase III subunits at the replication fork. Alpha is the only subunit required for processive replication. J Biol Chem. 1998;273:2452–7. doi: 10.1074/jbc.273.4.2452. [DOI] [PubMed] [Google Scholar]
- 109.McHenry C, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem. 1977;252:6478–84. [PubMed] [Google Scholar]
- 110.McHenry CS. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–65. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- 111.McHenry CS, Crow W. DNA polymerase III of Escherichia coli. Purification and identification of subunits. J Biol Chem. 1979;254:1748–53. [PubMed] [Google Scholar]
- 112.McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–38. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 113.McInerney P, O’Donnell M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279:21543–51. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 114.Miller H, Perrino FW. Kinetic mechanism of the 3′-->5′ proofreading exonuclease of DNA polymerase III. Analysis by steady state and pre-steady state methods. Biochemistry. 1996;35:12919–25. doi: 10.1021/bi960326d. [DOI] [PubMed] [Google Scholar]
- 115.Mitkova AV, Khopde SM, Biswas SB. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J Biol Chem. 2003;278:52253–61. doi: 10.1074/jbc.M308956200. [DOI] [PubMed] [Google Scholar]
- 116.Mullin DA, Woldringh CL, Henson JM, Walker JR. Cloning of the Escherichia coli dnaZX region and identification of its products. Mol Gen Genet. 1983;192:73–9. doi: 10.1007/BF00327649. [DOI] [PubMed] [Google Scholar]
- 117.Nakayama N, Arai N, Kaziro Y, Arai K. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem. 1984;259:88–96. [PubMed] [Google Scholar]
- 118.Naktinis V, Onrust R, Fang L, O’Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. II. Intermediate complex between the clamp loader and its clamp. J Biol Chem. 1995;270:13358–65. [PubMed] [Google Scholar]
- 119.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–65. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 121.O’Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–6. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 122.O’Donnell ME. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1987;262:16558–65. [PubMed] [Google Scholar]
- 123.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 124.Olson MW, Dallmann HG, McHenry CS. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. The chi psi complex functions by increasing the affinity of tau and gamma for delta.delta′ to a physiologically relevant range. J Biol Chem. 1995;270:29570–7. [PubMed] [Google Scholar]
- 125.Opperman T, Murli S, Smith BT, Walker GC. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci U S A. 1999;96:9218–23. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pages V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300:1300–3. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 127.Perrino FW, Harvey S, McNeill SM. Two functional domains of the epsilon subunit of DNA polymerase III. Biochemistry. 1999;38:16001–9. doi: 10.1021/bi991429+. [DOI] [PubMed] [Google Scholar]
- 128.Pham P, Bertram JG, O’Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–70. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 129.Podobnik M, McInerney P, O’Donnell M, Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J Mol Biol. 2000;300:353–62. doi: 10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- 130.Pritchard AE, McHenry CS. Identification of the acidic residues in the active site of DNA polymerase III. J Mol Biol. 1999;285:1067–80. doi: 10.1006/jmbi.1998.2352. [DOI] [PubMed] [Google Scholar]
- 131.Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–21. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 132.Rangarajan S, Woodgate R, Goodman MF. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci USA. 1999;96:9224–9. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reha-Krantz LJ, Hurwitz J. The dnaB gene product of Escherichia coli. I. Purification, homogeneity, and physical properties. J Biol Chem. 1978;253:4043–50. [PubMed] [Google Scholar]
- 134.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–6. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 135.Rowen L, Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Biol Chem. 1978;253:758–64. [PubMed] [Google Scholar]
- 136.San Martin MC, Stamford NP, Dammerova N, Dixon NE, Carazo JM. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995;114:167–76. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 137.Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A. 1983;80:7085–9. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–7. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 139.Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol. 2007;8:587–94. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- 140.Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell. 2005;17:561–72. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 141.Sigal N, Delius H, Kornberg T, Gefter ML, Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972;69:3537–41. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Slater SC, Lifsics MR, O’Donnell M, Maurer R. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J Bacteriol. 1994;176:815–21. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sommer S, Bailone A, Devoret R. The appearance of the UmuD’C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol. 1993;10:963–71. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 144.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD’C interaction. Mol Microbiol. 1998;28:281–91. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 145.Stano NM, Chen J, McHenry CS. A coproofreading Zn(2+)-dependent exonuclease within a bacterial replicase. Nat Struct Mol Biol. 2006;13:458–9. doi: 10.1038/nsmb1078. [DOI] [PubMed] [Google Scholar]
- 146.Stayton MM, Kornberg A. Complexes of Escherichia coli primase with the replication origin of G4 phage DNA. J Biol Chem. 1983;258:13205–12. [PubMed] [Google Scholar]
- 147.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–2. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 148.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–8. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 149.Strack B, Lessl M, Calendar R, Lanka E. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J Biol Chem. 1992;267:13062–72. [PubMed] [Google Scholar]
- 150.Studwell PS, O’Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990;265:1171–8. [PubMed] [Google Scholar]
- 151.Studwell-Vaughan PS, O’Donnell M. DNA polymerase III accessory proteins. V. Theta encoded by holE. J Biol Chem. 1993;268:11785–91. [PubMed] [Google Scholar]
- 152.Stukenberg PT, Turner J, O’Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–87. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 153.Sutton MD, Narumi I, Walker GC. Posttranslational modification of the umuD-encoded subunit of Escherichia coli DNA polymerase V regulates its interactions with the beta processivity clamp. Proc Natl Acad Sci USA. 2002;99:5307–12. doi: 10.1073/pnas.082322099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Swart JR, Griep MA. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 1995;34:16097–106. doi: 10.1021/bi00049a025. [DOI] [PubMed] [Google Scholar]
- 155.Taft-Benz SA, Schaaper RM. The theta subunit of Escherichia coli DNA polymerase III: a role in stabilizing the epsilon proofreading subunit. J Bacteriol. 2004;186:2774–80. doi: 10.1128/JB.186.9.2774-2780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O’Donnell M, Goodman MF. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD’2C mutagenic complex and RecA protein. Proc Natl Acad Sci USA. 1998;95:9755–60. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tang M, Pham P, Shen X, Taylor JS, O’Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–8. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 158.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–24. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tessman I, Kennedy MA. DNA polymerase II of Escherichia coli in the bypass of abasic sites in vivo. Genetics. 1994;136:439–48. doi: 10.1093/genetics/136.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996;271:21398–405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 161.Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–82. [PubMed] [Google Scholar]
- 162.Trakselis MA, Mayer MU, Ishmael FT, Roccasecca RM, Benkovic SJ. Dynamic protein interactions in the bacteriophage T4 replisome. Trends Biochem Sci. 2001;26:566–72. doi: 10.1016/s0968-0004(01)01929-6. [DOI] [PubMed] [Google Scholar]
- 163.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–26. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 164.Turner J, Hingorani MM, Kelman Z, O’Donnell M. The internal workings of a DNA polymerase clamp-loading machine. Embo J. 1999;18:771–83. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van der Ende A, Baker TA, Ogawa T, Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc Natl Acad Sci USA. 1985;82:3954–8. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 2000;1:484–8. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–6. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 168.Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–95. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang TC. Discontinuous or semi-discontinuous DNA replication in Escherichia coli? Bioessays. 2005;27:633–6. doi: 10.1002/bies.20233. [DOI] [PubMed] [Google Scholar]
- 170.Wickner W, Kornberg A. A holoenzyme form of deoxyribonucleic acid polymerase III. Isolation and properties. J Biol Chem. 1974;249:6244–9. [PubMed] [Google Scholar]
- 171.Wieczorek A, McHenry CS. The NH2-terminal php domain of the alpha subunit of the Escherichia coli replicase binds the epsilon proofreading subunit. J Biol Chem. 2006;281:12561–7. doi: 10.1074/jbc.M513844200. [DOI] [PubMed] [Google Scholar]
- 172.Wijffels G, Dalrymple BP, Prosselkov P, Kongsuwan K, Epa VC, Lilley PE, Jergic S, Buchardt J, Brown SE, Alewood PF, Jennings PA, Dixon NE. Inhibition of protein interactions with the beta 2 sliding clamp of Escherichia coli DNA polymerase III by peptides from beta 2-binding proteins. Biochemistry. 2004;43:5661–71. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- 173.Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229:10–6. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 174.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD’. Proc Natl Acad Sci U S A. 1989;86:7301–5. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wright M, Wickner S, Hurwitz J. Studies on in vitro DNA synthesis. Isolation of DNA B gene product from Escherichia coli. Proc Natl Acad Sci USA. 1973;70:3120–4. doi: 10.1073/pnas.70.11.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wu CA, Zechner EL, Hughes AJ, Jr, Franden MA, McHenry CS, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. IV. Reconstitution of an asymmetric, dimeric DNA polymerase III holoenzyme. J Biol Chem. 1992;267:4064–73. [PubMed] [Google Scholar]
- 177.Wu CA, Zechner EL, Reems JA, McHenry CS, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. V. Primase action regulates the cycle of Okazaki fragment synthesis. J Biol Chem. 1992;267:4074–83. [PubMed] [Google Scholar]
- 178.Xiao H, Dong Z, O’Donnell M. DNA polymerase III accessory proteins. IV. Characterization of chi and psi. J Biol Chem. 1993;268:11779–84. [PubMed] [Google Scholar]
- 179.Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. Contacts between the 5′ nuclease of DNA polymerase I and its DNA substrate. J Biol Chem. 2001;276:30167–77. doi: 10.1074/jbc.M100985200. [DOI] [PubMed] [Google Scholar]
- 180.Yang J, Nelson SW, Benkovic SJ. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol Cell. 2006;21:153–64. doi: 10.1016/j.molcel.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 181.Yang J, Xi J, Zhuang Z, Benkovic SJ. The oligomeric T4 primase is the functional form during replication. J Biol Chem. 2005;280:25416–23. doi: 10.1074/jbc.M501847200. [DOI] [PubMed] [Google Scholar]
- 182.Yang S, Yu X, VanLoock MS, Jezewska MJ, Bujalowski W, Egelman EH. Flexibility of the rings: structural asymmetry in the DnaB hexameric helicase. J Mol Biol. 2002;321:839–49. doi: 10.1016/s0022-2836(02)00711-8. [DOI] [PubMed] [Google Scholar]
- 183.Yao N, Leu FP, Anjelkovic J, Turner J, O’Donnell M. DNA structure requirements for the Escherichia coli gamma complex clamp loader and DNA polymerase III holoenzyme. J Biol Chem. 2000;275:11440–50. doi: 10.1074/jbc.275.15.11440. [DOI] [PubMed] [Google Scholar]
- 184.Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, Pan ZQ, Hurwitz J, O’Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–13. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 185.Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci USA. 2002;99:8737–41. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yuzhakov A, Kelman Z, O’Donnell M. Trading places on DNA--a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–63. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 187.Yuzhakov A, Turner J, O’Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–86. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 188.Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase-primase interaction governs primer size. J Biol Chem. 1992;267:4054–63. [PubMed] [Google Scholar]
- 189.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–37. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]


