Abstract
Receptor-like kinases (RLKs) belong to the large RLK/Pelle gene family, and it is known that the Arabidopsis thaliana genome contains >600 such members, which play important roles in plant growth, development, and defense responses. Surprisingly, we found that rice (Oryza sativa) has nearly twice as many RLK/Pelle members as Arabidopsis does, and it is not simply a consequence of a larger predicted gene number in rice. From the inferred phylogeny of all Arabidopsis and rice RLK/Pelle members, we estimated that the common ancestor of Arabidopsis and rice had >440 RLK/Pelles and that large-scale expansions of certain RLK/Pelle members and fusions of novel domains have occurred in both the Arabidopsis and rice lineages since their divergence. In addition, the extracellular domains have higher nonsynonymous substitution rates than the intracellular domains, consistent with the role of extracellular domains in sensing diverse signals. The lineage-specific expansions in Arabidopsis can be attributed to both tandem and large-scale duplications, whereas tandem duplication seems to be the major mechanism for recent expansions in rice. Interestingly, although the RLKs that are involved in development seem to have rarely been duplicated after the Arabidopsis–rice split, those that are involved in defense/disease resistance apparently have undergone many duplication events. These findings led us to hypothesize that most of the recent expansions of the RLK/Pelle family have involved defense/resistance-related genes.
INTRODUCTION
The first receptor-like kinase (RLK) was found in maize (Zea mays) 13 years ago (Walker and Zhang, 1990). Subsequently, RLKs have been identified in many other plant species and have been suggested to play diverse roles in plant life history (for review, see Torii, 2000; Shiu and Bleecker, 2001b; Becraft, 2002; Dievart and Clark, 2003). Typically, an RLK contains a signal sequence, a transmembrane region, and a C-terminal domain with eukaryotic protein kinase signatures, resembling the animal receptor tyrosine kinases (van der Geer et al., 1994). Based on our current understanding of receptor tyrosine kinase functions, the plant RLKs likely are transmembrane proteins that perceive signals through their extracellular domains and propagate the signals via their intracellular kinase domains. Consistent with this hypothesis, the ligands for several RLKs have been identified and are found to be integral components of the RLK signaling pathways (Schopfer et al., 1999; Brand et al., 2000; He et al., 2000; Matsubayashi et al., 2002; Scheer and Ryan, 2002). In addition, multiple components downstream to RLKs have been identified (for review, see Shiu and Bleecker, 2001b; Becraft, 2002).
The kinase domains of plant RLKs belong to the same gene family as those of Drosophila melanogaster Pelle and mammalian interleukin receptor–associated kinases (Shiu and Bleecker, 2001a). Drosophila Pelle is involved in the determination of the dorsal–ventral axis (Shelton and Wasserman, 1993), and several other components in this pathway have been implicated in innate immunity (Lemaitre et al., 1996). Mammalian interleukin receptor–associated kinases, on the other hand, are involved in innate immune responses against various pathogens (for review, see O'Neill, 2002). Similar to their animal counterparts, the biological functions of plant RLK/Pelle family members can be classified into two broad categories (Shiu and Bleecker, 2001b). The first category includes RLKs that control plant growth and development (Becraft, 2002), for example, Arabidopsis thaliana ERECTA in determining organ shape (Torii et al., 1996), CLAVATA1 in meristem maintenance (Clark et al., 1997), BRI1 in the regulation of cell growth (Li and Chory, 1997), maize CRINKLY4 in controlling cell morphogenesis and differentiation (Becraft et al., 1996), and carrot (Daucus carota) PSKR in controlling cell proliferation (Matsubayashi et al., 2002). The second category includes RLKs involved in plant–microbe interactions and defense responses. In this category, some RLKs are involved in plant–pathogen interactions, such as rice (Oryza sativa) Xa21 in resistance to bacterial pathogen (Song et al., 1995), Arabidopsis FLS2 in flagellin perception (Gomez-Gomez and Boller, 2000), and tomato (Lycopersicon esculentum) SR160 in systemin signaling (Scheer and Ryan, 2002), whereas the rest are crucial for interactions with plant symbionts, including NORK/SYMRK and HAR1 in fungal and/or bacterial symbiosis (Endre et al., 2002; Krusell et al., 2002; Nishimura et al., 2002; Stracke et al., 2002) and Lys motif–containing RLKs involved in early stages of nodulation and Nod factor perception (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003).
One major difference in this gene family between plants and animals is their relative abundance in the genomes. Plant RLKs belong to a large gene family (e.g., >600 members in Arabidopsis), whereas Plasmodium and animals contain only one to six RLK/Pelle members (Shiu and Bleecker, 2003). Moreover, no RLK/Pelle homolog is found in the fungal genomes sequenced to date. These findings indicate a rather drastic expansion of this gene family in the land plant lineage (Shiu and Bleecker, 2003). Judging from the need for intricate communication networks and the need to respond to a multitude of environmental factors in multicellular organisms, the abundance of plant RLKs represents a plant-specific adaptation for extracellular signal sensing and propagation. Besides its large family size, another important feature of the RLK/Pelle family is the diversity of domain organization (Shiu and Bleecker, 2001a). Approximately 75% of the Arabidopsis RLK/Pelle members have a signal sequence and a transmembrane region. Some, such as proline-rich extensin-like receptor kinase (Silva and Goring, 2002), contain no signal sequence. The rest are likely cytoplasmic kinases. Diverse sequence motifs are present in the putative extracellular regions of RLKs (Shiu and Bleecker, 2001b). These motifs are potentially responsible for interactions with other proteins, carbohydrates, or lipids. Interestingly, some of these motifs are similar to proteins that recognize fungal or bacterial cell wall components, another indication for the involvement of RLKs in plant–microbe interaction.
Although important discoveries have been made in understanding RLK functions, only a small fraction of the members in this gene family have demonstrated biological roles. In addition, the family size differences among eukaryotes raise several questions on the evolution of the RLK/Pelle gene family. The availability of rice genome sequences provides an excellent opportunity to address some of them. In this study, we first identified the kinase superfamily in rice and compared it to that in Arabidopsis. We then estimated the numbers of RLK/Pelles before the divergence of rice and Arabidopsis and the extent of subsequent expansions in each of these two flowering plant lineages. We also evaluated the contribution of different duplication mechanisms. To address questions on the functions of recently expanded RLK/Pelles, we compared the expansion patterns and mechanisms with the plant RLK/Pelle functions. Finally, we analyzed the substitution rates of the extracellular and intracellular domains of RLKs in an attempt to uncover the selection forces that shaped this large and divergent gene family.
RESULTS
A Twofold Larger RLK/Pelle Family in Rice than in Arabidopsis
As the first step for an in-depth comparison between the RLK/Pelle families in Arabidopsis and rice, we identified the protein kinase sequences from the released genome sequences of O. sativa subsp indica and japonica. The indica genome was used for further comparative studies because it was more complete than the japonica genome based on the number of kinase sequences recovered. The indica subspecies is referred to as rice for the rest of the analysis. We found 2210 candidate kinase sequences, 1936 of which contain at least one eukaryotic protein Ser/Thr/Tyr kinase domain based on SMART (Schultz et al., 2000) and Pfam (Sonnhammer et al., 1998). Because the indica genome was sequenced with a whole-genome shotgun strategy, some of these kinases may be redundant even though they were located on distinct scaffolds. To detect these potentially redundant sequences, we examined sequence matches that had >97% identity and found 329 potentially redundant sequences on different scaffolds, most with 100% identity over 600 nucleotides. After eliminating these entries, our data set contains 1607 protein kinase sequences from rice (see Supplement A online for their sequences). To obtain the RLK/Pelle family members from rice, we first aligned the kinase domain sequences of rice kinases and kinase representatives from various families and generated a phylogenetic tree. Within the clade that represents the RLK/Pelle family with 1235 rice sequences, we found that two groups consisted of 77 truncated rice kinase sequences (see Supplement C online, error regions). They were not included in any further analysis. Therefore, >1131 (27 have two kinase domains) RLK/Pelle family members were identified in rice, nearly twice as many as that in Arabidopsis.
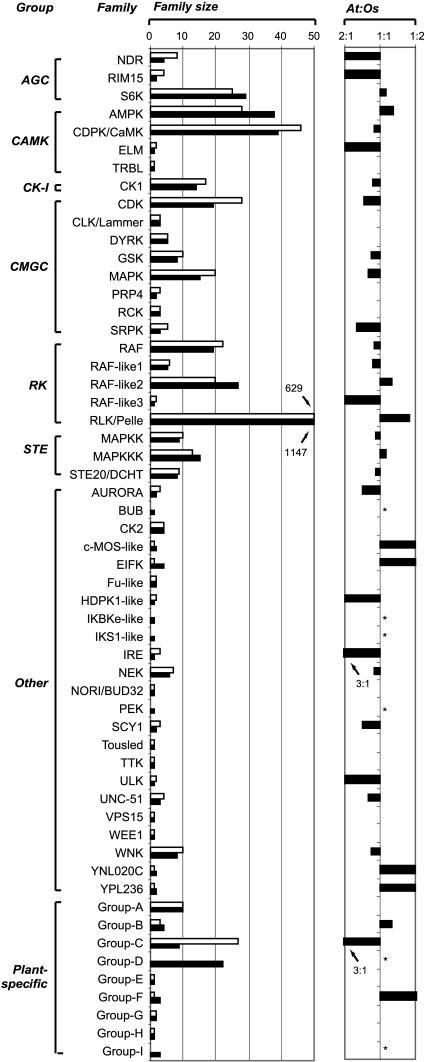
The rice proteome size was estimated to be 32,000 to 55,000 (Goff et al., 2002; Yu et al., 2002), ∼1.24 to 2.15 times larger than that of Arabidopsis (Arabidopsis Genome Initiative, 2000). It is possible that the larger RLK/Pelle family in rice simply reflects a general trend of larger rice gene families than those in Arabidopsis. However, we found that many kinase families in rice are not necessarily larger than those in Arabidopsis (Figure 1; see Supplement A online for the classification and Supplement B online for a phylogeny of Arabidopsis and rice kinases, excluding RLK/Pelle members). These findings suggest that the rice RLK/Pelle family has expanded in a more dramatic fashion than have other kinase families and may represent one of the gene families that have significantly contributed to the gene number differences between rice and Arabidopsis.
Figure 1.
Comparison of Rice and Arabidopsis Protein Kinase Families.
The bars indicate the sizes of kinase families from Arabidopsis (open) and rice (closed). The ratios of sizes of kinase families between these two plants (denoted At and Os) are shown at the right. No consistent size deviation can be seen for either organism. In the cases where the sizes or ratios are larger than the scale used, their values are shown. Asterisks indicate families with only rice members.
Extensive Lineage-Specific Expansions
The RLK/Pelle family in Arabidopsis consists of a large number of subfamilies that differ in both the kinase sequences and domain compositions. The family size difference between Arabidopsis and rice may be the consequence of differential expansion among subfamilies. To infer the patterns of expansion, we aligned the kinase sequences of RLK/Pelle members from both Arabidopsis and rice. The alignments were used to generate the phylogenetic tree shown in Figure 2 (left panel; see Supplement C online for the full tree). Most rice RLK/Pelle members fall into subfamilies defined on the basis of Arabidopsis sequences. Some subfamilies are rice specific (RLCK-OS1-4 and WAKL-OS), but no Arabidopsis-specific subfamily is identified.
Figure 2.
The Kinase Domain Phylogeny and Domain Configurations of RLK/Pelle Members from Rice and Arabidopsis.
(A) Phylogeny of RLK/Pelles from rice and Arabidopsis. The phylogeny was generated using kinase domain amino acid sequences with sequences from Arabidopsis and rice, which are color-coded orange and blue, respectively. Gray branches represent clusters of truncated sequences. The black circles mark the positions where the bifurcating edges were hypothesized to be because of divergence between monocot and dicot (Figure 3A). The subfamily designations are based on a published classification scheme (Shiu and Bleecker, 2003). The full phylogeny is shown in Supplement C online.
(B) Representative domain organizations for RLK/Pelle subfamilies. The RLK/Pelle subfamilies are divided into receptor kinase (RLK) and cytoplasmic kinase (RLCK) categories. Several subfamilies have more than one domain organization shown, representing potential lineage-specific domain fusion events in these subfamilies. Orange and blue arrows represent Arabidopsis- and rice-specific domain organizations, respectively. Arrowheads indicate novel organizations mentioned in the text. The domain organizations of Arabidopsis and rice RLK/Pelles are shown in Supplement C online.
To determine the degrees of expansion in different subfamilies in these two plant lineages, we broke down the phylogeny into ancestral units, which are clades (groups) that were present right before the monocot–dicot (rice–Arabidopsis) split, according to the scheme depicted in Figure 3A. The basal nodes of these ancestral units were labeled with closed circles in Figure 2. Because the RLK/Pelle family members are rather divergent, the phylogeny generated based on the alignments of all members from Arabidopsis and rice may not be entirely correct. To circumvent this problem, we divided the RLK/Pelle family into 17 groups, each with ∼100 genes; each group contained one to several RLK/Pelle subfamilies (see Supplement A online for group designations). The kinase domain sequences from each group were aligned, and the alignments were used for phylogenetic reconstruction. We found that there are 443 ancestral units among 17 trees. Because each tree contains both Arabidopsis and rice RLK/Pelles, no ancestral unit was missed compared with the full tree. Therefore, at least 443 RLK/Pelle members would have been present in the monocot–dicot common ancestor. Because of possible independent gene losses, our estimate of the ancestral gene number should be treated as a lower bound. Taken together, these inferences indicate that the RLK/Pelle family was quite large before the monocot–dicot split and continued to expand in both the rice and Arabidopsis lineages. The expansion may be even more pronounced than what we have observed because gene losses would also have contributed to underestimation of duplication events if both of the duplicated copies were lost.
Figure 3.
Expansions of Different RLK Subfamilies Before and After the Arabidopsis–Rice Split.
(A) Example phylogenies illustrating the rationale for inferring the presence of an ancestral RLK/Pelle. A tree with five taxa is shown at the left, where two genes are from Arabidopsis (At1a and At1b) and three are from rice (Os1, Os2a, and Os2b). According to the parsimony principle, a bifurcating clade with one branch leading to Arabidopsis gene(s) and the other lead to rice gene(s) indicates the presence of one ancestral RLK/Pelle gene, such as clade 1 shown in the tree at the right. Clade 2 is also regarded as an ancestral unit because it has sister group relationship to clade 1. The absence of an Arabidopsis gene in clade 2 is regarded as a gene-loss event (gray line, shaded gene name in the right panel).
(B) Comparison between the numbers of ancestral RLK/Pelle genes (open bar) and those of extant species (closed bar) in different subfamilies. The number of ancestral genes is determined based on the rationale explained in (A). Differential expansion of different subfamilies after the Arabidopsis–rice split is readily detectable in multiple subfamilies, whereas the sizes of others remain relatively constant.
As shown in Figure 3B, the lineage-specific expansions are restricted to certain subfamilies, such as legume lectin (L-LEC), leucine-rich repeat subfamily Ia (LRR-Ia), and wall-asspcoated kinase (WAK), as indicated by their larger sizes in extant plants than in the monocot–dicot common ancestor (Figure 3B). Others, such as the LRR-XI subfamily, are similar in size between the monocot–dicot common ancestor and either rice or Arabidopsis. These subfamilies have not expanded much after the Arabidopsis–rice split.
To further evaluate the extent of expansion in each of the Arabidopsis and rice lineages after their split, we compared the number of rice and Arabidopsis genes in each ancestral unit, and the results are shown in Figure 4A. Interestingly, most data points are clustered around clade sizes ranging from 1 to 5, indicating that most ancestral units underwent limited rounds of duplications or were lost since the divergence between Arabidopsis and rice. However, relatively few but noticeable numbers of ancestral units have expanded rather dramatically. This is especially true in rice, and such expansions account for more than two thirds of the size differences between the rice and Arabidopsis RLK/Pelle families (Figure 4B). Most of the large lineage-specific expansions are not coordinated between rice and Arabidopsis, suggesting differences in RLK/Pelle functions and selection in these two plants.
Figure 4.
Comparison of the Extents of Lineage-Specific Expansion in Arabidopsis and Rice.
(A) The clade sizes indicate the numbers of duplications that occurred after the divergence between Arabidopsis and rice. The relative extent of expansion that occurred after this period is illustrated by comparing the number of Arabidopsis and rice genes in each clade inferred (with the scheme shown in Figure 3A) and are plotted on the x and y axes, respectively. The number of each clade size relationship is counted and plotted on the z axis. Most clades are of similar sizes in Arabidopsis and rice. However, some clades have expanded rather dramatically in a lineage-specific fashion, but very few show similar degrees of expansion in both lineages as the arrow indicates a clade from the LRR-I subfamily.
(B) The contribution of lineage-specific expansions on the sizes of the RLK/Pelle family. Note the gradual separation between Arabidopsis (At) and rice (Os) cumulative total between cluster sizes of 5 and 20. The most significant contribution to the large RLK/Pelle family in rice, however, is because of the presence of lineage-specific expanded clades, accounting for ∼80% of the differences between Arabidopsis and rice.
Contributions of Tandem and Large-Scale Duplications to Family Size
Based on the analysis of the RLK/Pelle family in Arabidopsis, it was suggested that both tandem duplication and large-scale block duplication contributed to its expansion in plants (Shiu and Bleecker, 2001a, 2003). To determine the relative contributions of these two mechanisms to lineage-specific expansions, we examined the physical locations of RLK/Pelle members and estimated the timing of the duplication events. Because the indica genome was sequenced via shotgun sequencing, the sizes of assembled scaffolds were short relative to the average rice gene size. To circumvent this limitation, we first mapped RLK/Pelle members in the indica genome to available japonica BACs. The tandem cluster is defined as a region containing RLK/Pelle members that belong to the same subfamilies with <10 genes apart between neighboring members (see Supplement A online for cluster designation). Among the 646 rice RLK/Pelle members mapped, 270 (42%) are found in tandem clusters. The degree of tandem duplication is substantially higher than that in Arabidopsis, estimated to be 34% with the same criteria (Shiu and Bleecker, 2003).
Large-scale duplication events are defined as simultaneous duplications of genes. Assuming a molecular clock, the synonymous substitution rates (Ks) of these duplicates are expected to be similar over time. There are, however, substantial rate variations among genes. To evaluate the contribution of large-scale duplication and to alleviate the effects of rate variations, we used a relative Ks measure as the proxy for time and examined ancestral units containing both Arabidopsis and rice genes with the scheme shown in Figure 5A. The basal node indicates the divergence point between Arabidopsis and rice. Any node above the base indicates lineage-specific duplication in Arabidopsis or rice. The average synonymous substitution rate (Ks) was calculated for all nodes, and the relative Ks of each duplication event is the ratio of the node average Ks to the basal Ks. The frequency distributions of the relative Ks values are shown in Figures 5B and 5D for Arabidopsis and rice, respectively. The relative Ks distribution peaks at 0.15 to 0.2 in Arabidopsis, suggesting a large-scale event 22.5 to 30 million years ago (based on the divergence time of 150 million years ago between monocot and dicot; Sanderson, 1997; Wikstrom et al., 2001; Chaw et al., 2004). In rice, the results are less conclusive because multiple peaks are present, and the major peak is much wider than that observed in Arabidopsis.
Figure 5.
Large-Scale Duplication in the RLK/Pelle Family.
(A) An example phylogeny showing the rationales for determining the relative ages of duplications. Only ancestral units with both an Arabidopsis branch (A clade) and a rice branch (O clade) were analyzed. The node that represents the species divergence is called basal node (black bar). Four nodes represent duplications occurred in the Arabidopsis lineage (A-I through A-IV, white bars), and two occurred in rice (O-I and O-II, gray bars). The Ks value for each node was calculated by taking the arithmetic mean of all left-right branch combinations. For example, Ks for node A-II was determined by the sum of Ks of sequence pairs A1-A3 and A2-A3 divided by the number of pairs. The node Ks was then divided by the basal Ks to obtain its relative age.
(B) and (C) The frequency distribution of relative ages of Arabidopsis nodes. In (B), the relative ages of all nodes, regardless of their duplication mechanisms, were plotted. In (C), each tandem cluster was treated as one locus, and nodes were determined by excluding all but the first members in tandem clusters.
(D) and (E) The frequency distribution of rice RLK/Pelle relative ages of all nodes or locus-based nodes, respectively.
Because a substantial number of duplicates are located in tandem repeats, we examined the contribution of tandem duplications by treating each tandem cluster as a single locus and taking the first member of each cluster as representative for further analysis. Together with nontandem members of RLK/Pelles, we again determined the ancestral units and relative Ks values. In Arabidopsis, the overall shape of the distribution and location of peaks in the locus-based analysis (Figure 5C) were similar to those using the whole data set (Figure 5B). This finding is consistent with the potential involvement of large-scale events in the expansion of the RLK/Pelle family in the Arabidopsis lineage. In rice, however, the locus-based analysis (Figure 5E) resulted in the removal of the major peaks seen previously using the whole data set (Figure 5D), indicating that tandem duplication, but not large-scale duplication, played a major role in the RLK/Pelle expansion in rice.
Correlation between RLK Functions and Expansion Patterns
The RLK/Pelle family expanded rather dramatically in the land plant lineage (Shiu and Bleecker, 2003). An intriguing question is whether RLK/Pelle members with a certain function tend to be duplicated or retained at higher rates than others. We classified RLK/Pelle family members with known functions or relevant expression patterns into three functional categories: developmental control, resistance/defense responses, and symbiotic interactions (Table 1). Interestingly, RLK/Pelles that are involved in defense/disease resistance or their potential orthologs in rice or Arabidopsis belong to subfamilies with high percentage members in clusters and to clades with evidence of lineage-specific duplication events, as indicated by a deviation of the Arabidopsis–rice gene ratios from 1:1. On the other hand, none of the RLK/Pelles or their closest relatives in Arabidopsis or rice that are involved in developmental control are found in tandem clusters. In addition, only one of the RLK/Pelles in this functional category had duplicated after the divergence between rice and Arabidopsis. The degrees of lineage-specific expansions of RLK/Pelles in these two functional categories (developmental versus resistance) are significantly different at 1% level (Fisher's exact test; P = 0.0045). Taken together, these findings suggest that RLK/Pelles that are involved in resistance or defense responses may have been duplicated or retained at higher rates in a lineage-specific fashion.
Table 1.
Functions and Characteristics of Selected RLK/Pelle Family Members
| Synonyma | Source | Subfamilyb | Tandem Clusterb | Gene Namec | Clade Ratiod | Funcate | Functions | Reference |
|---|---|---|---|---|---|---|---|---|
| BRI1/SR160 | Arabidopsis/L. esculentum | LRR-Xb | no | At4g39400 | 1:1 | D/R | Steroid hormone–mediated growth response | Li and Chory (1997); Searle et al. (2003) |
| SERK3/BAK1 | Arabidopsis | LRR-II | At4g33430 | 3:2 | D | Steroid hormone–mediated growth response | Hecht et al. (2001); Li et al. (2002); Nam and Li (2002) | |
| CRINKLY4 | Z. mays | CR4L | no | Osi005386.1 | 1:1 | D | Epidermal cell development | Becraft et al. (1996) |
| EMS1/EXS | Arabidopsis | LRR-Xb | no | At5g07280 | 1:0 | D | Microspore development | Canales et al. (2002); Zhao et al. (2002); |
| ERECTA | Arabidopsis | LRR-XIIIb | no | At2g26330 | 2:2 | D | Organ initiation and elongation | Torii et al. (1996) |
| HAESA | Arabidopsis | LRR-XI | no | At4g28490 | 1:0 | D | Delayed floral organ abscission | Jinn et al. (2000) |
| PRK1 | Petunia inflata | LRR-III | no | Osi009992.1 | 1:1 | D | Microspore development | Lee et al. (1996) |
| PSKR | D. carota | LRR-Xb | no | At2g02220 | 1:1 | D | Phytosulfokine mediated growth response | Matsubayashi et al. (2002) |
| CLAVATA1/HAR1/NARK | Arabidopsis/Lotus japonicus/Glycine max | LRR-XI | no | At1g75820 | 1:1 | D/S | Control of apical meristem proliferation in Arabidopsis, symbiotic interaction in the others | Clark et al. (1997); Krusell et al. (2002); Nishimura et al. (2002); Searle et al. (2003) |
| NORK/SYMRK | Medicago trunculata | LRR-Ic | no | At1g67720 | 1:0 | S | Development of root nodule | Endre et al. (2002); Stracke et al. (2002) |
| NFR1/LYK3 | L. japonicus/M. trunculata | LysM-I | no | Osi009166.2 | 1:2 | S | Perception of Nod factor, infection thread formation | Limpens et al. (2003); Radutoiu et al. (2003) |
| LYK4f | M. trunculata | LysM-I | ND | ND | ND | S | Infection thread formation | Limpens et al. (2003) |
| NFR5/SYM10 | L. japonicus/Pisum sativum | LysM-II | no | Osi015153.1 | 1:2 | S | Perception of Nod factor | Madsen et al. (2003) |
| RKC1 | Arabidopsis | DUF26-Ib | yes | At4g23250 | 37:0 | R | Salicylic acid inducible | Ohtake et al. (2000) |
| RKL1 | Arabidopsis | LRR-III | no | At1g48480 | 2:3 | R | Salicylic acid inducible | Ohtake et al. (2000) |
| RKS1,2 | Arabidopsis | SD-1a | yes | At1g11340-50 | 4:0 | R | Salicylic acid inducible | Ohtake et al. (2000) |
| SFR1,2 | Brassica oleracea | SD-1a | yes | At1g65790, At4g21380 | 4:1 | R | Wound and bacterial inducible expression | Pastuglia et al. (1997) |
| FLS2 | Arabidopsis | LRR-XII | no | At5g46330 | 1:1 | R | Flagellin insensitivity | Gomez-Gomez and Boller (2000) |
| LRK10 | Avena sativa | LRK10-L2 | yes | Osi011907.3 | 0:23 | R | Resistance to fungal pathogen | Feuillet et al. (1997) |
| PBS1 | Arabidopsis | RLCK-VIIa | no | At5g13160 | 1:0 | R | Resistance to bacterial pathogen | Swiderski and Innes (2001) |
| SIRK | Arabidopsis | LRR-Ia | yes | At2g19190 | 44:0 | R | Upregulated during senescence and pathogen challenge | Robatzek and Somssich (2002) |
| WAK1 | Arabidopsis | WAK | yes | At1g21210-50 | 6:4 | R | Defense response | He et al. (1998) |
| Xa21 | O. sativa | LRR-XII | yes | Osi009420.2 | 0:17 | R | Resistance to bacterial pathogen | Song et al. (1995) |
Potentially orthologous sequences from different species are separated by slashes.
Tandem clustering and classification information can be found in Supplements A and C, respectively, online.
The names for closest relatives in Arabidopsis or rice.
The ratio between Arabidopsis and rice genes in the ancestral unit of the gene in question or its closest relative.
Funcat, functional categories: D, developmental control; S, symbiosis; R, resistance or defense responses.
LYK1, LYK4, LYK6, and LYK7 form a sister group to a LysM-I clade that contains both Arabidopsis and rice genes. Therefore, no potential orthologs from either Arabidopsis or rice can be assigned (ND, not determined).
Conservation of Domain Composition and Organization
A wide range of domains and motifs are found in the Arabidopsis RLK/Pelle members. Most of these domains and motifs are not associated with kinases in animals or fungi, suggesting that the domain composition of RLK/Pelle members was established during the course of plant evolution. We examined the predicted domains for RLK/Pelle members in both rice and Arabidopsis in conjunction with their phylogenetic relationships to evaluate the conservation and divergence of protein domain architecture.
The domain compositions and organizations of RLK/Pelles are similar in general between subfamily members (see Supplement C online for the domain content organizations of Arabidopsis and rice RLK/Pelles; Figure 2, the representative organizations shown to the right). In addition, the domain content and organizations are in most cases very similar between Arabidopsis and rice members in each subfamily. This similarity suggests that the domain organization of most RLK/Pelle subfamilies was established before the Arabidopsis–rice split. Nonetheless, there are several interesting exceptions. In rice, one of the three exceptions is a gene with two PR-1/SCP (for Pathogenesis-Related Protein-1/Sperm-Coating Glycoprotein) domains (Figure 2, arrowhead a). Interestingly, the kinase domain of this PR-1/SCP–containing protein is more closely related to the kinase domains in the rice DUF26 RLKs (see Supplement C online), suggesting that the DUF26 domain was replaced by PR-1/SCP in the rice lineage. In the LRK10-L2 subfamily, there are four nonhomologous extracellular regions (Figure 2, arrowhead b). Among them, only one is shared between rice and Arabidopsis. The extracellular regions that are specific to rice contain epidermal growth factor repeats in one and a glycosyl hydrolase family 18 (chitinase) domain in the other. In both cases, their kinase domain sequences are more closely related to rice LRK10-L2 members than to Arabidopsis ones (see Supplement C online). Meanwhile, the proteins that contain Arabidopsis-specific extracellular regions have kinase domains that are more closely related to Arabidopsis LRK10-L2 members. These findings indicate that the domain organizations of most RLKs are established before the monocot–dicot split. Nonetheless, domain acquisition had continued to occur in both rice and Arabidopsis after the monocot–dicot split.
Different Selection Forces on the Extracellular and Intracellular Domains of RLKs
The modular nature and biological roles of receptor kinases suggest that the extracellular domains (ECDs) and intracellular domains (ICDs; Figure 6A) may be under different functional or selective constraints. To investigate the differences in selective constraints, we first identified reciprocal best matches among RLK/Pelle members from both Arabidopsis and rice and calculated the synonymous and nonsynonymous substitution rates (Ka and Ks) for the ECD, ICD, and kinase domains of each pair (Figures 6B, 6C, and 6D, respectively). Interestingly, the frequency distribution and the mean of the Ka/Ks ratios for ECDs are significantly different from those of ICDs and kinase domains (Table 2), indicating that the ECDs evolved faster than the ICDs. This difference may either be because of relaxed purifying selection or positive selection on the ECDs.
Figure 6.
The Ka/Ks Ratios for Different Domains in RLKs.
(A) A schematic representation of the domains in question. Kinase, kinase domain; signal, signal sequence; TM, transmembrane region.
(B) to (D) The Ka/Ks frequency distribution for ECD, ICD, and kinase, respectively. The average Ka/Ks for each domain is indicated by an arrowhead.
(E) The Ka of ECD plotted against that of ICD of the same protein. The line indicates a one-to-one relationship between Ka of these two domains.
(F) and (G) The relationships between Ka/Ks and Ks of ECDs and ICDs. Regression lines with the highest correlation coefficients are shown. The Ka/Ks values decline sharply as Ks values get larger, a pattern that is more pronounced in ECDs. Because Ks is a proxy for time, this pattern suggests that, for newly duplicated RLKs, the ECDs in general evolved faster than the ICDs of the newly duplicated RLKs.
Table 2.
Statistics of the Ka/Ks Frequency Distribution between Domains
| Welch Two-Sample t Test | F Test | |
|---|---|---|
| ECD versus ICD | 1.35−08 | 4.86−10 |
| ECD versus KINa | 2.25−13 | 3.64−11 |
| ICD versus KIN | 0.008029 | 0.6711 |
KIN, kinase domain.
To further verify the differences in evolutionary rates between these two domains, we compared the Ka values of ECD and ICD of the same protein and found that the numbers of amino acid changing substitutions are in general higher in the ECDs (Figure 6E). Using Ks as a proxy for time, we found that the Ka/Ks ratio declined sharply over time (Figures 6F and 6G). This trend is more pronounced for the ECDs than that for the ICDs, suggesting again positive selection or relaxation of negative selection on the ECDs soon after duplication events. The signatures of positive selection (Ka/Ks > 1) were revealed in several RLK/Pelle pairs when the rate calculation was performed on various size windows (example of one window size is shown in Figure 7). For duplicated pairs that have regions with a Ka/Ks value larger than one, the regions are generally in the ECDs, though few located within the ICD are found as well. Taken together, these findings suggest that parts of the extracellular regions may have experienced positive selection in some of these receptor kinases. It should be noted that the presence of pseudogenes in our data set may have contributed to high Ka values in some gene pairs. However, the comparison of Ka was between ECDs and ICDs of the same protein pairs. If some elevated Ka values were because of pseudogenes, we expect the Ka values to be similar between ECDs and ICDs, which would tend to underestimate the true difference between functional genes. Because we found significant differences between these two domains, the potential bias in Ka values because of pseudogenes will not affect our conclusion.
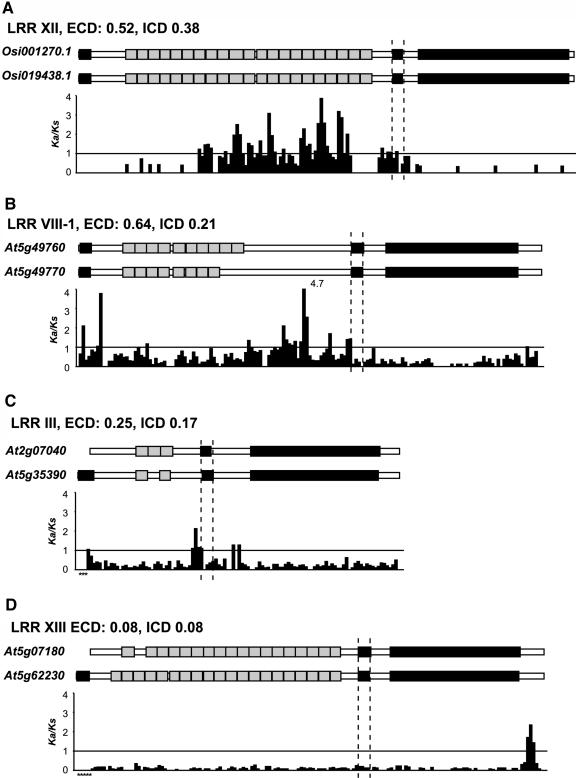
Figure 7.
Detection of Positively Selected Regions in RLKs.
Four LRR RLK pairs differ widely in their Ka/Ks between ECDs and ICDs and are shown as examples. For each pair, the subfamily designation and the Ka/Ks values for ECD and ICD are indicated, followed by the graphical representation of domains and the Ka/Ks values calculated with sliding windows. The signal sequences, if present, are the black boxes at the beginning of each entry. The transmembrane regions are the internal black boxes. Gray boxes indicate motifs in the ECDs; they are LRRs in all examples. The kinase domains are black boxes. Dotted lines define the boundaries between ECDs, transmembrane regions, and ICDs. The window size was 30 amino acids, and the step size was 15. For each window, the Ka value was calculated and divided by the full length Ks value. This was done to reduce false positives because of relatively low Ks values in the windows. The line indicates the Ka/Ks value of 1.
DISCUSSION
History of the RLK/Pelle Family Expansion in Land Plants
The RLK/Pelle family is one of the largest gene families in Arabidopsis, with >600 members (Shiu and Bleecker, 2001b). However, the size of the RLK/Pelle family would likely be small in the common ancestor of animals, plants, and alveolates (Shiu and Bleecker, 2003). In this study, we established that rice contains at least 1132 RLK/Pelle members, providing further support for the land plant expansion hypothesis. We estimated the number of RLK/Pelle members before the divergence of Arabidopsis and rice to be ∼443. Therefore, the RLK/Pelle family was already large in the common ancestor of flowering plants that diverged 150 to 200 million years ago (Wolfe et al., 1989; Goremykin et al., 1997; Sanderson, 1997; Wikstrom et al., 2001; Chaw et al., 2004). After the divergence between the rice and Arabidopsis lineages, further expansions occurred, contributing to their current sizes.
Because the overall predicted gene number in rice is 1.5- to 2.2-fold higher than that in Arabidopsis (Goff et al., 2002; Yu et al., 2002), the nearly doubled rice RLK/Pelle family may simply be the consequence of a larger predicted gene set in rice. However, rice gene families are in general similar in size to those in Arabidopsis, such as the various kinase families shown in Figure 1 and other gene families, such as HAK potassium transporters (Banuelos et al., 2002), LRR extensins (Baumberger et al., 2003), P-type ATPases (Baxter et al., 2003), Dof transcription factors (Lijavetzky et al., 2003), and ATP binding cassette transporters (Jasinski et al., 2003). Therefore, the RLK/Pelle family may represent one of the gene families differentially expanded in rice. In addition, lineage-specific expansions of the RLK/Pelle family occurred in Arabidopsis and rice since their last common ancestor. These findings indicate that biological innovations through this versatile family of proteins are still favored, and many duplicates have been retained over the past 150 million years. One may argue that the RLK/Pelle family is larger in rice because of the presence of more rice pseudogenes. In our analysis, we found that the Ka/Ks values of pairwise comparison were generally significantly smaller than 0.5 (data not shown; Figures 6B to 6D). The number of pseudogenes will be very limited in our data set because most of the RLK/Pelles show signs of purifying selection (with Ka/Ks < 1) rather than neutral evolution (with Ka/Ks ≈ 1, expected for pseudogenes). In addition, rice and Arabidopsis have similar proportions of RLK/Pelles with ESTs and/or cDNAs (61.4 and 67%, respectively; see Supplement D online). These findings indicate that pseudogenes are not likely the major contributing factor in the size differences between rice and Arabidopsis.
Owing to the recent history of rice domestication (∼10,000 years ago; Huke and Huke, 1990) and the involvement of some RLK/Pelles in defense/resistance, the differential expansion of this gene family in rice may be the consequence of artificial selection for disease-resistant varieties. The recent domestication of rice implies that duplicates arisen through artificial selection will be young with very low sequence divergence. Most rice RLK/Pelle duplicates have rather large relative Ks values (as shown in Figure 5D), indicating their ancient origins and arguing against their retention because of rice domestication. However, we excluded ∼400 sequences that are nearly identical to the sequences we analyzed in their kinase domains. Until a better assembly of the rice genome is available, we cannot rule out the possibility that they are distinct RLK/Pelles that were retained via artificial selection.
Lineage-Specific Expansions and Mechanisms of Duplications
The RLK/Pelle family can be divided into multiple subfamilies with distinct kinases and domain organizations (Shiu and Bleecker, 2001a, 2001b). We showed that subfamilies differ widely in the degree of expansion after the Arabidopsis–rice split. Most ancestral units underwent limited rounds of duplications or were not duplicated since the divergence between these two plants. However, the few ancestral units that have expanded contribute profoundly to the family size differences between rice and Arabidopsis. This is because of larger lineage-specific expansions in the rice lineage. Several lines of evidence indicate that Arabidopsis is a paleopolyploid (Arabidopsis Genome Initiative, 2000; Vision et al., 2000; Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003). The most recent large-scale duplications that likely involved the whole genome occurred before the Arabidopsis–Brassica split and after the Malvaceae–Brassiceae split 14.5 to 40 million years ago (Blanc et al., 2003; Bowers et al., 2003). The peak of relative Ks distribution of Arabidopsis RLK/Pelle duplicates falls into this period. In addition, 33.6% of the Arabidopsis RLK/Pelle family members are located within tandem clusters (Shiu and Bleecker, 2003). Both mechanisms likely have contributed to the net gain of Arabidopsis RLK/Pelles after the rice–Arabidopsis split.
In rice, however, tandem duplication seems to be the major mechanism contributing to the RLK/Pelle family expansion. The contribution of tandem duplication in rice may be even higher because BAC sequences rather than chromosome assemblies were used to determine tandem duplicates. Although it was reported that rice underwent large-scale duplication 40 to 50 million years ago (Goff et al., 2002), the substitution rate distribution was generated using all possible paralogous protein pairs, which may result in a biased representation of rates. Vandepoele et al. (2003) reported that rice is likely an ancient aneuploid rather than a polyploid. Consistent with this finding, we do not see evidence for the involvement of large-scale duplications within a relative short time period in the expansion of rice RLK/Pelles.
Functional Correlations
Most RLK/Pelles implicated in defense/resistance responses are located in tandem clusters and tend to have biased Arabidopsis-to-rice ratios in the ancestral units (Table 1). These findings suggest that recent expansions of the RLK/Pelle family involved defense/resistance-related genes. Among plant resistance (R) genes, the nucleotide binding site (NBS)/LRR-containing family is also quite large compared with most gene families in Arabidopsis (Michelmore and Meyers, 1998; Mondragon-Palomino et al., 2002). Similar to the RLK/Pelle family, both tandem duplications and large-scale duplications are implicated in the expansion of the NBS-LRR gene family (Meyers et al., 2003). It will be interesting to determine if this gene family shows similar patterns of expansion after the Arabidopsis–rice split. In mammals, it has been shown that large gene families, such as mammalian olfactory receptors (Young et al., 2002) and taste receptors (Shi et al., 2003), have differentially expanded in mouse and human. The interesting parallel between these two gene families and at least the receptor kinases in the RLK/Pelle family is that they are likely involved in the perception of diverse extracellular signals. A need for perceiving these signals may allow the retention of duplicates.
In rice, one of the RLK subfamilies that differentially expanded the most is the LRR-XII subfamily with >150 rice and only six Arabidopsis members. Defense/resistance genes in this subfamily include Xa21, involved in resistance to bacterial pathogens (Song et al., 1995), and FLS2, involved in the perception of bacterial flagellin (Gomez-Gomez and Boller, 2000). Interestingly, the Xa21 clade but not FLS2 has expanded in rice, suggesting that some defense/resistance RLK/Pelles did not follow the general trend we hypothesized. It is possible that these RLK/Pelles are involved in the perception of conserved components from pathogens, such as pathogen-associated molecular patterns (Nurnberger and Brunner, 2002). Alternatively, they may not interact directly with pathogen-derived signals but are downstream intermediates in defense/resistance responses.
By contrast, only one of the RLK/Pelles with known functions in developmental control has evidence of duplication after the Arabidopsis–rice split. Furthermore, none of these Arabidopsis or rice genes is located in tandem clusters. One explanation is that these genes were not duplicated. Although this may be true in rice, evidence points to the involvement of whole-genome duplication in Arabidopsis, indicating that, instead of the absence of duplication, gene losses likely have led to the absence of duplicates. This is consistent with the notion that transcription factors, instead of signaling molecules, are the major innovative factors in development (Doebley and Lukens, 1998). The lack of lineage-specific expansion in genes with development functions also is seen in the animal tyrosine kinase family where strict one-to-one relationships were found between mouse and human members (Shiu and Li, 2004). However, these findings by no means rule out the possibility that developmental innovation can be accomplished by duplicated receptor kinases. The tyrosine kinase family has experienced a period of net gain early in the evolution of chordates and very likely contributed to the complicated body plan in extant vertebrates (Shiu and Li, 2004). One potential example of developmental innovation via plant RLK/Pelles is ERECTA and its close relatives, which were duplicated both in rice and Arabidopsis after their divergence. A factor functionally redundant to Arabidopsis ERECTA has been identified recently (Shpak et al., 2003). We speculate that the ERECTA paralog identified in this study may represent the functionally redundant factor.
Evolution of Domains in RLKs
The domain architectures of RLKs are mostly conserved between rice and Arabidopsis, indicating that domain organizations were mostly established before their divergence. Nevertheless, we identified some novel domain configurations that are specific to Arabidopsis or rice. Interestingly, the ECDs in two novel rice receptors, PR-1/SCP and chitinase RLKs, are both pathogenesis-related proteins implicated in defense responses. In tobacco (Nicotiana tabacum), the expression of PR-1a is tightly correlated with the onset of systemic acquired resistance, and its overexpression results in higher tolerance to fungal pathogens (Alexander et al., 1993). Our findings indicate that new receptors were made and retained through the process of selection in these two plant lineages independently. In line with the prediction that a large number of genes in this family may be involved in defense/resistance, these fusions may provide novel ways to recognize extracellular biotic agents, particularly pathogens, and were selected for retention.
The preferential expansion of defense/resistance-related RLK/Pelles could be the consequence of strong selection pressure for recognizing pathogens. In plant R genes, the NBS-LRR family (Michelmore and Meyers, 1998; Mondragon-Palomino et al., 2002) and the Cladosporium fulvum resistance gene family (Meyers et al., 1998) both show signatures of positive selection. Similarly, the RLK/Pelles also show signatures of positive selection but in a domain-specific manner. We found that ECDs in general have higher nonsynonymous changes compared with those of ICDs. However, few pairs have Ka/Ks > 1 unless a sliding window analysis was conducted (Figure 7). It is possible that the long evolutionary history of the duplicated pairs reduce the resolution of our analysis. It also highlights the possibility that only certain residues of the ECDs were under positive selection, as demonstrated in NBS-LRRs (Mondragon-Palomino et al., 2002). In any case, these findings are consistent with the notion that the ECDs of RLKs may have experienced weaker purifying selection or stronger diversifying selection to recognize various extracellular signals. By contrast, the ICDs may have been under stronger purifying selection because of functional constraints in transducing signals to downstream components faithfully.
Although substantial progress has been made in the functional dissection of several founding members of this gene family, the functions of most RLK/Pelles remain elusive. The extensive expansion of this gene family only in the land plants suggests their functional importance to land plant evolution. Based on the limited number of RLK/Pelles with known functions, we hypothesize that, after the divergence between rice and Arabidopsis, the majority of RLK/Pelles duplicated are involved in defense/resistance function, whereas those involved in developmental control have rarely been duplicated. Further studies will be required to verify these proposals.
METHODS
Sequence Retrieval and Annotation
The eukaryotic protein Ser/Thr/Tyr kinases in rice (Oryza sativa) were identified with the procedures detailed below. Following Hanks and Hunter (1995) and Hardie (1999), we used 52 plant and animal sequences from different eukaryotic Ser/Thr/Tyr kinase families to conduct batch BLAST searches (Altschul et al., 1997) against the predicted genes of O. sativa subsp indica with a permissive E value cutoff of 1. The rice genes from the indica subspecies was predicted using the whole genome shotgun assembly with FGENESH (Solovyev, 2002) as described (Yu et al., 2002). The japonica BACs and corresponding annotation were extracted from the MOsDB database (Karlowski et al., 2003). The presence of kinase domains was verified with the eukaryotic kinase Hidden Markov Model from Pfam (Sonnhammer et al., 1998). Upon further examination of the 1946 confirmed kinase sequences, we found that 17% of them have a nearly perfect (≥99%) match for >600 nucleotides. Because of the potential problems in shotgun assemblies, we regarded these sequences as potentially redundant, and they were not included in any further analysis. For the sequences of rice kinases, see Supplement A online. They are also deposited in the PlantsP database (http://plantsp.sdsc.edu/). The Arabidopsis thaliana kinases analyzed were as defined previously (Shiu and Bleecker, 2003). The same procedures were used to recover kinase sequences from O. sativa subsp japonica. As of May 2003, ∼700 kinase sequences were identified. Structural domains of all sequences were annotated according to the SMART (Schultz et al., 2000) and Pfam databases using a batch sequence submission and parsing script.
Alignments and Phylogenetic Reconstruction
For the identification of RLK/Pelle family members, the rice kinase sequences were aligned with kinase family representatives as described previously (Shiu and Bleecker, 2001a). All sequence alignments were conducted using Clustal (Higgins et al., 1996). The RLK/Pelle family is defined as the clade that contains RLK/Pelle representative sequences and forms sister group relationships to Raf kinases and receptor tyrosine kinases. The other kinase families were defined based on the kinase classification schemes for budding yeast (Hunter and Plowman, 1997), worm (Plowman et al., 1999), and human kinases (Manning et al., 2002). Two to three representative sequences from each kinase family in each organism were chosen to generate alignments with Arabidopsis or rice kinases. These alignments were used to generate phylogenetic trees with the neighbor-joining method (Saitou and Nei, 1987). The multiple substitutions were corrected using the Poisson distance. The alignment gaps were treated as missing characters. Sequences in the same clades as known kinase families were classified as such. Otherwise, they were identified as plant-specific kinases. All except one phylogenetic tree (Figure 2) in this analysis were generated with 500 bootstrap replicates.
Inference of Duplication Time
For inferring the dates of gene duplications, only the kinase domain coding sequences were used. The large phylogeny shown in Figure 2 was partitioned into 17 sections with similar numbers of genes. The genes in each section were aligned for generating a phylogenetic tree. Ancestral units (clades) as defined in Figure 3A were identified from these phylogenetic trees. The clades that contain both Arabidopsis and rice sequences were analyzed further. For each clade, the synonymous substitution rates (Ks) of all pairwise combinations were estimated using the yn00 program in PAML (Yang, 1997). The duplication time was determined by dividing the node average Ks with that of the basal node (Figure 5A).
Substitution Rate Estimates for RLK Domains
To compare the evolutionary rates of ICDs and ECDs of RLKs, we first identified the reciprocal best matches of Arabidopsis and rice RLK/Pelle family members. An alignment was generated for each pair. Any pair with an aligned area <80% of the longer protein was excluded from further analysis. Any pair with a Ks value >1.5 over the full length also was excluded. For each pair, at least one predicted protein had to have a predicted transmembrane region. For proteins with more than one predicted transmembrane region, we examined the alignments and defined the transmembrane region as the one that overlapped between two proteins. In the data set, no other type of permutation in terms of transmembrane region arrangement was found. ECDs and ICDs were defined as sequences N-terminal and C-terminal to the predicted transmembrane regions, respectively. The signal sequences were deleted from the ECDs. The nonsynonymous substitution rate (Ka) and Ks were calculated for ECD, ICD, and kinase domains using the yn00 program in PAML. For detecting positive selection in regions of gene pairs, the Ka and Ks were determined on windows of aligned sequences taken from full-length alignments.
Supplementary Material
Acknowledgments
We thank Melissa D. Lehti-Shiu for reading the manuscript and the PlantsP database for hosting the rice kinase sequences identified in this study. The work was funded by the National Institutes of Health National Research Service Award (Grant 5F32GM066554-02) to S.-H.S., the Genomanalyse im Biologischen System Pflanze project of the German Ministry for Education and Research to W.M.K. and K.F.X.M., and National Institutes of Health grants to W.-H.L.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shin-Han Shiu (shiu@uchicago.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020834.
References
- Alexander, D., Goodman, R.M., Gut-Rella, M., Glascock, C., Weymann, K., Friedrich, L., Maddox, D., Ahl-Goy, P., Luntz, T., Ward, E., and Ryal, J. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 90, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generate of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Banuelos, M.A., Garciadeblas, B., Cubero, B., and Rodriguez-Navarro, A. (2002). Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130, 784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N., Doesseger, B., Guyot, R., Diet, A., Parsons, R.L., Clark, M.A., Simmons, M.P., Bedinger, P., Goff, S.A., Ringli, C., and Keller, B. (2003). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, I., Tchieu, J., Sussman, M.R., Boutry, M., Palmgren, M.G., Gribskov, M., Harper, J.F., and Axelsen, K.B. (2003). Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 132, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft, P.W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18, 163–192. [DOI] [PubMed] [Google Scholar]
- Becraft, P.W., Stinard, P.S., and McCarty, D.R. (1996). CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273, 1406–1409. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12, 1718–1727. [DOI] [PubMed] [Google Scholar]
- Chaw, S.-M., Chang, C.-C., Chen, H.-L., and Li, W.-H. (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol., in press. [DOI] [PubMed]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Dievart, A., and Clark, S.E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6, 507–516. [DOI] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., Schachermayr, G., and Keller, B. (1997). Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 11, 45–52. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goremykin, V.V., Hansmann, S., and Martin, W.F. (1997). Evolutionary analysis of 58 proteins encoded in six completely sequenced chloroplast genomes: Revised molecular estimates of two seed plant divergence times. Plant Syst. Evol. 206, 337–351. [Google Scholar]
- Hanks, S.K., and Hunter, T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596. [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131. [DOI] [PubMed] [Google Scholar]
- He, Z., Wang, Z.Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- He, Z.-H., He, D., and Kohorn, B.D. (1998). Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–63. [DOI] [PubMed] [Google Scholar]
- Hecht, V., Vielle-Calzada, J.P., Hartog, M.V., Schmidt, E.D., Boutilier, K., Grossniklaus, U., and de Vries, S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. [PMC free article] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gibson, T.J. (1996). Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383–402. [DOI] [PubMed] [Google Scholar]
- Huke, R.E., and Huke, E.H. (1990). Rice: Then and Now. (Manila, Philippines: International Rice Research Institute).
- Hunter, T., and Plowman, G.D. (1997). The protein kinases of budding yeast: Six score and more. Trends Biochem. Sci. 22, 18–22. [DOI] [PubMed] [Google Scholar]
- Jasinski, M., Ducos, E., Martinoia, E., and Boutry, M. (2003). The ATP-binding cassette transporters: Structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol. 131, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., and Walker, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. [PMC free article] [PubMed] [Google Scholar]
- Karlowski, W.M., Schoof, H., Janakiraman, V., Stuempflen, V., and Mayer, K.F. (2003). MOsDB: An integrated information resource for rice genomics. Nucleic Acids Res. 31, 190–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell, L., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., Karunanandaa, B., McCubbin, A., Gilroy, S., and Kao, T.-H. (1996). PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J. 9, 613–624. [Google Scholar]
- Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J.M., and Hoffmann, J.A. (1996). The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. [DOI] [PubMed] [Google Scholar]
- Manning, G., Whyte, D.B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002). The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y., Ogawa, M., Morita, A., and Sakagami, Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., Meyers, B.C., Michelmore, R.W., and Gaut, B.S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Nishimura, R., Hayashi, M., Wu, G.J., Kouchi, H., Imaizumi-Anraku, H., Murakami, Y., Kawasaki, S., Akao, S., Ohmori, M., Nagasawa, M., Harada, K., and Kawaguchi, M. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. [DOI] [PubMed] [Google Scholar]
- Ohtake, Y., Takahashi, T., and Komeda, Y. (2000). Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 41, 1038–1044. [DOI] [PubMed] [Google Scholar]
- O'Neill, L.A. (2002). Signal transduction pathways activated by the IL-1 receptor/toll-like receptor superfamily. Curr. Top. Microbiol. Immunol. 270, 47–61. [PubMed] [Google Scholar]
- Pastuglia, M., Roby, D., Dumas, C., and Cock, J.M. (1997). Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman, G.D., Sudarsanam, S., Bingham, J., Whyte, D., and Hunter, T. (1999). The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. USA 96, 13603–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sanderson, M.J. (1997). A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 14, 1218–1231. [Google Scholar]
- Scheer, J.M., and Ryan, C.A., Jr. (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C., Nasrallah, M., and Nasrallah, J. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. (2000). SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, I.R., Men, A.E., Laniya, T.S., Buzas, D.M., Iturbe-Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112. [DOI] [PubMed] [Google Scholar]
- Shelton, C.A., and Wasserman, S.A. (1993). Pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell 72, 515–525. [DOI] [PubMed] [Google Scholar]
- Shi, P., Zhang, J., Yang, H., and Zhang, Y.P. (2003). Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol. Biol. Evol. 20, 805–814. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. a). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. b). Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, RE22. [DOI] [PubMed]
- Shiu, S.H., and Bleecker, A.B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., and Li, W.H. (2004). Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol. Biol. Evol. 21, 828–840. [DOI] [PubMed] [Google Scholar]
- Shpak, E.D., Lakeman, M.B., and Torii, K.U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, N.F., and Goring, D.R. (2002). The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 50, 667–685. [DOI] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M.C., Zabeau, M., and Van De Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99, 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, V.V. (2002). Finding genes by computer: Probabilistic and discriminative approaches. In Current Topics in Computational Biology, T. Jiang, T. Smith, Y. Xu, and M. Zhang, eds (Cambridge, MA: MIT Press), pp. 365–401.
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gadner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P.C. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., Eddy, S.R., Birney, E., Bateman, A., and Durbin, R. (1998). Pfam: Multiple seuqnce alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26, 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. [DOI] [PubMed] [Google Scholar]
- Swiderski, M.R., and Innes, R.W. (2001). The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26, 101–112. [DOI] [PubMed] [Google Scholar]
- Torii, K.U. (2000). Receptor kinase activation and signal transduction in plants: An emerging picture. Curr. Opin. Plant Biol. 3, 361–367. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Geer, P., Hunter, T., and Lindberg, R.A. (1994). Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 10, 251–337. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Simillion, C., and Van de Peer, Y. (2003). Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15, 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Walker, J.C., and Zhang, R. (1990). Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 345, 743–746. [DOI] [PubMed] [Google Scholar]
- Wikstrom, N., Savolainen, V., and Chase, M.W. (2001). Evolution of the angiosperms: Calibrating the family tree. Proc. R. Soc. Lond. B Biol. Sci. 268, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.W., Sharp, P.M., and Li, W.H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- Young, J.M., Friedman, C., Williams, E.M., Ross, J.A., Tonnes-Priddy, L., and Trask, B.J. (2002). Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11, 535–546. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Zhao, D.Z., Wang, G.F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.