Abstract
We have compared the gene expression profiles of inflorescences of the floral homeotic mutants apetala1, apetala2, apetala3, pistillata, and agamous with that of wild-type plants using a flower-specific cDNA microarray and a whole genome oligonucleotide array. By combining the data sets from the individual mutant/wild type comparisons, we were able to identify a large number of genes that are, within flowers, predicted to be specifically or at least predominantly expressed in one type of floral organ. We have analyzed the expression patterns of several of these genes by in situ hybridization and found that they match the predictions that were made based on the microarray experiments. Moreover, genes with known floral organ–specific expression patterns were correctly assigned by our analysis. The vast majority of the identified transcripts are found in stamens or carpels, whereas few genes are predicted to be expressed specifically or predominantly in sepals or petals. These findings indicate that spatially limited expression of a large number of genes is part of flower development and that its extent differs significantly between the reproductive organs and the organs of the perianth.
INTRODUCTION
Much progress has been made in recent years in understanding the initiation of organ formation, in plants as well as in animals, by identifying many of the master regulatory genes that trigger the developmental programs required for organogenesis. However, in most cases, the molecular mechanisms by which the activation of these genes results in organ function are not well understood. One example that illustrates our limited understanding of organogenesis is the development of floral organs. In Arabidopsis thaliana, the floral mutants apetala1 (ap1), ap2, ap3, pistillata (pi), and agamous (ag) show homeotic phenotypes, that is, the replacement of one type of floral organ (namely, sepals, petals, stamens, and carpels) by another (Figure 1). In ap1, sepals are transformed into bract-like organs, and petals are often completely absent. Furthermore, secondary flowers arise from the axils of the outermost whorl organs. In ap2, petals are missing or are transformed into stamens, and sepals are transformed into carpel-like organs. In addition, the number of stamens is reduced. In the mutants ap3 and pi, petals are replaced by sepals, and stamens are replaced by carpels. In ag, stamens are replaced by petals, and carpels are replaced by extra whorls of sepals and petals. Based on the phenotypes of these mutants, the ABC model of floral organ identity determination was proposed (Bowman et al., 1991; Coen and Meyerowitz, 1991). According to this model, the activities of the genes affected in the mutants can be assigned to three different functions, namely A (AP1 and AP2), B (AP3 and PI), and C (AG), with each function required for organ specification in different floral regions. The functions act combinatorially in the specification of floral organ identity: A function alone leads to the formation of sepals, whereas the combination of A and B functions determines petal identity. The combination of B and C functions results in stamens, and C function alone determines carpel formation. The floral organ identity genes were cloned and found to encode transcription factors (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Jofuku et al., 1994). Their expression patterns and (in some cases) molecular interactions (Honma and Goto, 2001) suggest that the combinatorial nature of the ABC model, as indicated by genetics, is reflected at the molecular level.
Figure 1.
Floral Phenotypes of Strains Used in This Study.
Landsberg erecta (A), ap1-1 (B), ap2-2 (C), ap3-3 (D), pi-1 (E), and ag-3 (F) mature flowers are shown. The diagram depicts the expected changes in levels of organ-specific transcripts in the floral mutants compared with wild-type plants. (−), absent; (↑), upregulated; (↓), downregulated; (?), questionable.
However, in spite of their well-characterized roles in floral organ specification, it is unclear to what extent these transcription factors also participate in the continuing formation of floral tissues throughout organogenesis. Furthermore, it is for the most part unknown whether they control the expression of other transcriptional regulators or instead directly affect the transcript levels of genes that encode structural proteins required for cellular differentiation. Thus, for a better understanding of floral organ formation, it is necessary to decipher the regulatory networks that control gene expression during the various stages of flower development. A possible strategy for this would be to identify the components of these networks first and then to determine how they are linked to each other.
So far, the identification of regulatory genes involved in flower development has been primarily accomplished by the characterization of mutants that show severe phenotypic alterations. By comparison, relatively few mutants have been described in which, for example, only a certain floral cell type is affected. One explanation for this is that the inactivation of these genes results in phenotypic alterations that are subtle and are easily missed among thousands of plants in a screening population. It is also possible that functional redundancy prevents the identification of many regulatory genes by conventional mutagenesis.
Early work on tobacco (Nicotiana tabacum) using RNA-excess single-copy DNA hybridization reactions had shown for different plant organs that the composition of their nuclear RNA sequence population is developmentally regulated and contains a set of transcripts that is specific to each organ (Kamalay and Goldberg, 1984). Subsequently, studies have shown that many genes with known or presumed regulatory functions are often expressed in limited domains and/or only during certain stages of flower development and that their ectopic expression results in novel phenotypes (i.e., Yanofsky et al., 1990; Jack et al., 1992; Mizukami and Ma, 1992; Weigel et al., 1992; Weigel and Meyerowitz, 1993; Goto and Meyerowitz, 1994; Gustafson-Brown et al., 1994; Kempin et al., 1995; Mandel and Yanofsky, 1995; Sakai et al., 1995; Weigel and Nilsson, 1995; Krizek and Meyerowitz, 1996; Ferrándiz et al., 2000). These results indicate that the spatial and temporal control of gene expression is crucial for the development of floral organs. Thus, a possible strategy for the identification of components of the regulatory networks that control flower development is to analyze the expression of genes during flower development on a genome-wide level. Microarray technology makes this possible by allowing the simultaneous detection of thousands of transcripts in a single experiment.
In this study, we have analyzed spatial gene expression in Arabidopsis flowers by comparing the gene expression profiles of inflorescences of the floral homeotic mutants with that of wild-type inflorescences. To this end, we have used a cDNA microarray whose composition is strongly enriched for elements representing flower-specific transcripts as well as a whole genome oligonucleotide array. The fact that different organ types are missing in the mutants allowed us to identify transcripts that are, within flowers, specifically expressed, or are at least strongly enriched, in one type of floral organ by combining the data sets from the individual experiments. We have determined the expression patterns of several previously uncharacterized genes by in situ hybridization and found that the predictions based on the microarray data are in agreement with the actual expression patterns. In addition, genes that had previously been shown to have floral organ–specific expression patterns were correctly assigned to the different organ groups by the microarray experiments. Our results indicate that spatially limited expression of several genes is part of Arabidopsis flower development. Furthermore, they suggest that the extent to which spatially limited gene expression is required for floral organ formation differs significantly between the organs of the perianth and of the reproductive organs. Our data also provide a rich source of target genes for reverse genetics approaches and candidates for floral organ–specific or cell type–specific markers.
RESULTS
Analysis of Gene Expression Profiles of Floral Homeotic Mutants
In recent years, gene expression analysis with DNA microarrays has become a powerful tool for the analysis of developmental processes in animals and plants (Reinke and White, 2002). To allow the use of microarray technology for the analysis of flower development, we have constructed a cDNA microarray whose elements are strongly enriched for flower-specific transcripts. The array is comprised of 10,816 elements representing ∼5000 to 6000 genes (see supplemental data online for a detailed description of the strategy used for the construction of the array). We used this array and, in later stages of this study, whole genome microarrays comprised of 26,090 oligonucleotides to analyze the gene expression profiles of inflorescences of the floral homeotic mutants ap1, ap2, ap3, pi, and ag. To this end, tissue samples were collected from the different mutants that contained the inflorescence meristem and floral buds corresponding to developmental stages 1 to 13 (Smyth et al., 1990). Total RNA was isolated from these samples, and the RNA was used to synthesize dye-labeled cDNA (for the cDNA array) or antisense RNA (for the oligonucleotide array) that was cohybridized to the microarrays with control samples derived from inflorescences of wild-type plants (see Methods for further details). In all cases, at least three independent biological samples were used in separate hybridizations. To avoid dye-related artifacts, the dyes used for the labeling of the cohybridized samples were switched in the replicate experiments.
After quantitation of the signal intensities, the data were normalized to compensate for nonlinearity of intensity distributions and differences in probe labeling (Figure 2 and see Methods and supplemental data online for further details). We next identified elements showing significant differences in expression in the replicate experiments using either significance analysis of microarrays (SAM; Tusher et al., 2001) with an estimated false discovery rate of 1% for data from the cDNA array or the Rosetta Resolver analysis platform for data derived from the oligonucleotide array (elements were judged significantly changed when they were assigned a P value < 0.01). In addition, we applied a twofold expression cutoff to make the selection more stringent and to account for the fact that the changes in gene expression for organ-specific transcripts should be relatively large in the mutant/wild type comparisons. Experiments with both types of microarrays led to similar conclusions; therefore, results will be presented together.
Figure 2.
Results of Experiments Using the Whole Genome Oligonucleotide Array.
Expression profiles of ap1-1 (A), ap2-2 (B), ap3-3 (C), pi-1 (D), and ag-3 (E) inflorescences were compared with those of wild-type inflorescences. Plots were generated from experimental data of four replicates with biologically independent samples after normalization and replicate analysis (see Methods for details). log(ratio) = log10 (IMT/IWT); log(intensity) = 0.5 log10 (IMT·IWT), where IMT and IWT are signal intensities for a given element in a mutant or the wild type, respectively.
For ap3, pi, and ag, we observed a large number of elements that were determined significantly changed in the experiments. Most of these elements corresponded to downregulation in the mutants compared with the wild type. In the A function mutants ap2 and especially ap1, a smaller number of elements reporting significant expression changes was detected, and the difference between the number of elements indicating upregulation or downregulation was less pronounced.
Because the B function mutants ap3 and pi have phenotypes that are almost indistinguishable from each other, we calculated the correlation coefficient of the data sets obtained from the microarray experiments and found a high degree of correlation (for the cDNA array, r = 0.98; for the oligonucleotide array, r = 0.91), suggesting that the similar phenotypes coincide with similar gene expression profiles. This result is in agreement with the idea that PI and AP3 are obligate partners in a regulatory complex and that the disruption of one of the genes results in a loss of function of the complex (Riechmann et al., 1996).
Prediction of Organ-Specific Transcripts
For the prediction of organ-specific transcripts, the results of the different mutant/wild type comparisons were combined, and the array elements that were significantly changed in at least one of the experiments were subjected to cluster analysis. Based on the expected changes in gene expression in the different mutants (Figure 1), we were able to identify groups of elements that might represent floral organ–specific transcripts (Figure 3 and supplemental data online). We next searched numerically for patterns in the microarray data sets that could be indicative of organ-specific expression. For this, we focused primarily on elements that represented transcripts significantly downregulated in the mutants. For example, sepals are missing in ap1 and ap2. Sepal-specific transcripts should therefore be absent in the A function mutants. By contrast, flowers of ap3, pi, and ag contain extra sepals so that sepal-specific transcripts should be upregulated. However, it was unclear whether the extent of upregulation is sufficient to allow reliable detection. We therefore assumed that elements representing sepal-specific transcripts are significantly downregulated in ap1 and ap2 and are either unchanged or upregulated in the B and C function mutants. A total of 13 genes were identified that met these criteria.
Figure 3.
Self-Organizing Map Generated from Elements of the Oligonucleotide Array That Showed Significant Expression Changes in at Least One of the Experiments.
Elements that are upregulated in the wild type compared with the mutant are depicted in yellow and downregulated elements in blue. The intensities of the colors increase with increasing expression differences as indicated at the bottom. The diagram was generated with the program Resolver using log10-transformed expression ratios. The clusters predicted to contain organ-specific transcripts are indicated. Ca, carpel; Pe, petal; Se, sepal; St, stamen.
Petals are absent in ap2, ap3, and pi and are strongly reduced in number in ap1, whereas extra petals are found in flowers of ag. In accordance with the criteria used for the prediction of sepal-specific transcripts, we initially assumed that elements representing petal-specific transcripts show statistically significant downregulation in all four A and B function mutants and are either unchanged or upregulated in ag. However, it is possible that elements representing petal-specific transcripts might be missed with these stringent screening criteria. For example, an element representing a petal-specific transcript would be missed if it were not considered significantly downregulated in one of the four experiments perhaps because of variability in the replicate experiments. We therefore slightly loosened the screening criteria by assuming that the downregulation of petal-specific transcripts should be statistically significant in at least three of the four A and B function mutants. This led to the identification of 18 genes with predicted petal-specific expression.
For the prediction of stamen-specific transcripts, we assumed significant downregulation in ap3, pi, and ag because stamens are completely missing in these B and C function mutants. These criteria led to the identification of 1162 genes. Because stamens are reduced in number in ap2, the identified genes should be downregulated, at least to some extent, in this mutant as well. We calculated the log2-transformed median expression ratios for the predicted stamen-specific transcripts and obtained values of −0.7 and −1.1 (for data from the cDNA array and from the oligonucleotide array, respectively), indicating that the corresponding transcripts are indeed generally downregulated in ap2.
The only mutant used in this study that lacks carpels is ag. In the other mutants, the carpel number is either unchanged (ap1), or extra carpelloid tissues are present (ap2, ap3, and pi). We identified several hundred genes that were downregulated only in ag but were upregulated or unchanged in the other mutants. Among these genes, we found several with known carpel-specific expression patterns (see below). However, in contrast with all other types of floral organs, these predictions were based on the significant downregulation of an element in a single mutant; thus, the predictions are potentially less reliable than those for the other organs. To improve the prediction of carpel-specific transcripts, we therefore introduced a second criterion that was based on the assumption that the difference between the expression ratios of ag and the mutants containing extra carpel tissue should be relatively large. For the selection of appropriate cutoff values, we calculated the mean expression ratios and standard deviations for known carpel-specific transcripts (Table 2) for ap2, ap3, and pi. The mean log2-transformed expression ratios minus two standard deviations were used as cutoff values. This led to the prediction of 260 carpel-specific transcripts.
Table 2.
Genes with Known Organ-Specific Expression Patterns in Carpels Were Correctly Assigned to the Organ Groups by the Microarray Experiments
| Gene ID | Name | Expression/Localization | Reference |
|---|---|---|---|
| At1g23420 | INNER NO OUTER | Ovule | Villanueva et al., 1999 |
| At1g69180 | CRABS CLAW | Carpel epidermis; nectaries | Bowman and Smyth, 1999 |
| At2g42830 | SHATTERPROOF2 (AGL5) | Carpel wall, ovule; nectaries | Savidge et al., 1995 |
| At3g23130 | SUPERMAN | Ovule; young floral meristem | Sakai et al., 1995 |
| At3g58780 | SHATTERPROOF1 (AGL1) | Carpel wall, ovule; nectaries | Flanagan et al., 1996 |
| At4g09960 | SEEDSTICK (AGL11) | Ovule | Rounsley et al., 1995 |
| At5g60910 | FRUITFULL (AGL8) | Carpel valves; inflorescence meristem | Gu et al., 1998 |
| At5g67110 | ALCATRAZ | Valve margins | Rajani and Sundaresan, 2001 |
Genes assigned to the carpel group. Gene identifiers and names are listed. The patterns of expression or of protein localization are briefly described, and references are given.
Of the 1453 genes with predicted organ-specific expression (see supplemental data online for a list of all genes), 1380 were identified in experiments with the oligonucleotide array and 247 with the cDNA array (Table 1). A total of 174 genes were detected with both types of arrays. Of the 73 genes that were identified with the cDNA array but not with the oligonucleotide array, 18 are not represented on the oligonucleotide array. Thus, we found a 76% overlap (174 of 229 possible genes) between the data sets derived from the cDNA and oligonucleotide arrays.
Table 1.
Comparison of Organ-Expressed Genes Identified with the Two Types of Microarrays Used in This Study
| Organ | cDNA | Oligonucleotide | Total | Overlap |
|---|---|---|---|---|
| Sepal | 1 | 12 | 13 | 0 |
| Petal | 5 | 15 | 18 | 2 |
| Stamen | 207 | 1106 | 1162 | 151 |
| Carpel | 34 | 247 | 260 | 21 |
The numbers of genes identified with the flower-specific cDNA array or the oligonucleotide array are listed for each organ group. The total number of unique genes in each organ group as well as the number of genes identified with both types of arrays (overlap) are indicated. For the cDNA array, not all elements representing organ-expressed genes were sequenced. These unsequenced elements contain ∼100 additional genes (see Methods for details).
Verification of Predicted Organ-Specific Gene Expression
As mentioned above, several known carpel-specific transcripts were identified in the carpel group (Table 2). The corresponding genes all encode transcription factors that are predominantly expressed in carpel walls (e.g., SHATTERPROOF1, SHATTERPROOF2, and FRUITFULL) or in ovules (SEEDSTICK, INNER NO OUTER, and SUPERMAN) during late stages of flower development. The correct assignment of genes such as INNER NO OUTER, which is expressed only in a small domain of ovules (Villanueva et al., 1999), suggests that expression changes of relatively rare transcripts were reliably detected by the microarray analysis. In the stamen group, several genes with known expression in anthers, especially in the tapetal cell layer or in pollen, were found (Table 3). Among the few transcripts assigned to the petal group, one (At5g45950) previously had been predicted to be petal specific (Zik and Irish, 2003). The sepal group contained the organ identity gene AP1. Although AP1 is indeed strongly expressed in sepals, it is also expressed in petals (Mandel et al., 1992), suggesting that the organ groups might contain some genes whose expression is not strictly organ specific.
Table 3.
Genes with Known Organ-Specific Expression Patterns in Stamens Were Correctly Assigned to the Organ Groups by the Microarray Experiments
| Gene ID | Name | Expression/Localization | Reference |
|---|---|---|---|
| At1g20130 | APG | Various tissues in anthers | Roberts et al., 1993 |
| At1g23240 | EF-hand protein | Pollen coat | Mayfield et al., 2001 |
| At1g24520 | Bcp1 | Tapetum, microspores | Xu et al., 1995 |
| At1g75910 | EXL4 | Pollen coat | Mayfield et al., 2001 |
| At1g75930 | EXL6 | Pollen coat | Mayfield et al., 2001 |
| At1g75940 | ATA27 | Tapetum | Rubinelli et al., 1998 |
| At2g16910 | ABORTED MICROSPORES | Tapetum, microspores, locules | Sorensen et al., 2003 |
| At2g19770 | Profilin 4 | Pollen | Huang et al., 1996 |
| At3g11980 | MALE STERILITY 2 | Tapetum | Aarts et al., 1997 |
| At3g11980 | ATA20 | Tapetum | Rubinelli et al., 1998 |
| At3g42960 | ATA1 | Tapetum | Lebel-Hardenak et al., 1997 |
| At4g14080 | A6 | Tapetum | Hird et al., 1993 |
| At5g07230 | A9 | Tapetum | Paul et al.,1992 |
| At5g07510 | GRP14 | Tapetum; pollen coat | Mayfield et al., 2001; Kim et al., 2002 |
| At5g07520 | GRP18 | Tapetum; pollen coat | Mayfield et al., 2001; Kim et al., 2002 |
| At5g07530 | GRP17 | Tapetum; pollen coat | Alves-Ferreira et al., 1997; Mayfield et al., 2001; Kim et al., 2002 |
| At5g07540 | GRP16 | Tapetum; pollen coat | Alves-Ferreira et al., 1997; Mayfield et al., 2001; Kim et al., 2002 |
| At5g07550 | GRP19 | Tapetum; pollen coat | Mayfield et al., 2001; Kim et al., 2002 |
| At5g07560 | GRP20 | Tapetum; pollen coat | Mayfield et al., 2001; Kim et al., 2002 |
| At5g07600 | Oleosin | Tapetum | Kim et al., 2002 |
Genes assigned to the stamen group. Gene identifiers and names are listed. The patterns of expression or of protein localization are briefly described, and references are given.
We next tested the expression of previously uncharacterized genes. For this, we first confirmed the results of the microarray experiments by real-time PCR for several genes (data not shown). Next, we generated probes for these genes and performed in situ hybridizations. Expression of a NAC-family transcription factor (At1g61110; assigned to the stamen group) was found in stamens in the tapetum at stage 11 of flower development (Figures 4A and 4B). Two of the studied genes were expressed during late stages of pollen development (Figures 4C and 4I): At2g07040, encoding a putative receptor-like kinase, and At2g43230, coding for a putative protein kinase. Both genes were correctly assigned to the stamen group by the microarray analysis. Another gene in the stamen group, At3g26860 (encoding a putative self-incompatibility protein), is expressed during early pollen development in tetrads at floral stage 9 (Figure 4L). For At3g61160, encoding the SHAGGY-like kinase ASKβ, predominant expression in pollen had been shown previously by RNA gel blot analysis and reverse transcriptase PCR (Tichtinsky et al., 1998). In addition to its expression in pollen, we also observed expression in ovules, probably in egg cells (Figures 4J and 4K).
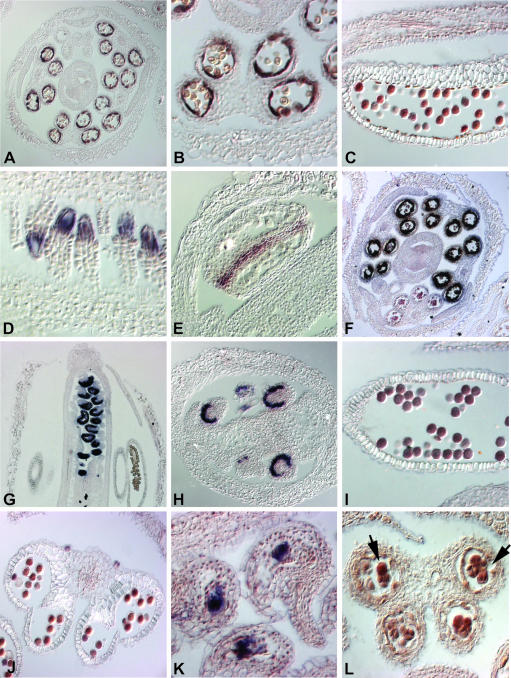
Figure 4.
mRNA Localization of Predicted Floral Organ–Specific Transcripts.
(A) and (B) Transverse sections through a stage 11 flower. Expression of At1g61110 (NAC-family transcription factor) was detected inside anthers in the tapetal cell layer.
(C) Longitudinal section through a stage 11 flower. At2g07040 (receptor-like kinase) is expressed in pollen.
(D) and (E) Longitudinal sections through stage 10 floral buds. Expression of At2g17950 (WUS) is found in developing ovules (D) and between the locules of anthers (E).
(F) Transverse section through a stage 10 floral bud. At1g75030 (thaumatin-like protein) is expressed in the tapetum.
(G) and (H) Longitudinal (G) and transverse (H) sections through stage 12 flower buds. Expression of At4g12960 (expressed protein) was detected in ovules.
(I) Longitudinal section through a stage 12 flower. At2g43230 (kinase) is expressed in pollen.
(J) and (K) Transverse section (J) and longitudinal section (K) through stage 12 flowers. Expression of At3g61160 (ASKβ) is found in pollen (J) and inside ovules, probably in egg cells (K).
(L) Transverse section through a stage 9 flower. mRNA of At3g26860 (putative self-incompatibility protein) was detected in tetrads during pollen development (indicated by arrows). For all in situ hybridizations, the use of sense probes did not result in signals above background (data not shown).
An example of a gene predicted to be expressed in carpels is At4g12960, encoding a protein with unknown function. This gene is expressed starting at floral stage 12 inside of ovules and likely marks the inner integuments (Figures 4G and 4H).
The homeodomain transcription factor WUSCHEL (WUS) is expressed during floral organ formation in the nucellus of developing ovules (Gross-Hardt et al., 2002). Because WUS was assigned to the stamen group by the microarray analysis and WUS expression in stamens had been mentioned but not further characterized in the literature (Gross-Hardt et al., 2002), we tested whether this result could be because of a relatively large expression domain in stamens that masks expression in ovules. In fact, besides the expression in ovules, we detected stripes of WUS expression between the locules of anthers (Figures 4D and 4E). Thus, the assignment of WUS to the stamen group is likely because of the fact that WUS is expressed in more cells and/or at higher levels in anthers than in ovules. This result (as well as the result for ASKβ) confirmed that the organ groups indeed contain some genes whose expression is not strictly organ specific (see above). We therefore concluded that the identified transcripts are, within flowers, either specifically expressed or are at least strongly enriched in one type of floral organ. To account for this fact, genes in the organ groups will hereafter be referred to as organ-expressed genes.
The criteria applied for the analysis of the microarray data were designed to minimize the number of falsely identified organ-expressed transcripts. However, a reduction of false positives coincides with an increase in the number of genes that are organ expressed but were missed in the analysis. Based on the expression ratios, we were able to identity several elements that likely represent false negatives (data not shown). In situ hybridization for one of the corresponding genes, At1g75030, showed that this gene, coding for a thaumatin-like protein, is expressed in the tapetum of anthers and thus was missed by the microarray analysis (Figure 4F).
The results of a recent study allowed us to identify genes of the stamen group expressed most likely specifically in pollen. In this study, the transcriptome of pollen was analyzed by comparing RNA isolated from mature pollen to RNA isolated from whole plants at various stages of development using an Affymetrix 8k Arabidopsis GeneChip array (Honys and Twell, 2003). A total of 992 elements were predicted to represent pollen-expressed genes. Approximately one-third of the identified genes were predicted to be pollen specific based on nondetection of the corresponding transcripts in reference samples. We have compared the list of pollen-expressed genes with the genes identified in this study and found an overlap of 147 genes (see supplemental data online for further information). The vast majority of these genes (83%) were called pollen specific in the previous study and included At2g07040 and At2g43230 that were detected exclusively in pollen by in situ hybridization (Figures 4C and 4I). On the other hand, four of these genes were predicted by our analysis to be expressed in carpels rather than stamens. For one of these genes, At4g12960, the results of in situ hybridizations showed strong expression in ovules starting at floral stage 12, whereas we were not able to detect expression in pollen (Figures 4G and 4H). Furthermore, several other genes predicted to be pollen specific, though present on the microarrays used for this study, were not predicted as stamen expressed by our analysis. One possible explanation for these differences in the data sets is that the corresponding transcripts are found in more than one type of floral organ but were too dilute in the whole plant extracts used as reference samples for the analysis of the pollen transcriptome to be reliably detected.
Functional Assessment of the Identified Genes
For a functional assessment of the identified genes, gene ontology annotations for the Arabidopsis genome were obtained from the Arabidopsis Information Resource. To identify terms that were underrepresented or enriched among the organ-expressed genes, we first determined their distribution in the entire gene ontology data set for the three organizing principles (i.e., molecular function, biological process, and cellular component) separately. Subsequently, the distribution of terms was determined for the organ-expressed genes and compared with the genome-wide distribution (see supplemental data online for results). We found that genes involved in general cellular processes, such as DNA recombination, protein synthesis, protein folding, or photosynthesis, were underrepresented, suggesting that the identified genes might be involved in more specialized processes. Overrepresented categories included genes involved, for example, in cell wall modification, aging, or embryonic development. Enrichment was also detected for certain functional terms (e.g., for genes with pectinesterase or polygalacturonase activity).
We also analyzed the distribution of gene families among the organ-expressed genes and identified several underrepresented or overrepresented families (see supplemental data online for results). For example, members of the class III peroxidase family or the glycoside hydrolase family 28 were enriched, whereas members of the relatively large families of core cell cycle genes and of putative sugar transporters were completely absent in the data set.
We next searched in the data set for pairs of closely related genes that might indicate possible functional redundancy. To this end, we first determined the closest related protein sequence in the Arabidopsis proteome for every organ-expressed gene product using BLAST (Altschul et al., 1990). We then investigated whether any of these proteins were included in the same organ group as the query sequence. For 21 gene products of the carpel group and 278 of the stamen group, the closest related protein sequence was present in the same organ group (see supplemental data online for results). No such pairs were found in the sepal and petal groups. Many of the identified pairs show high levels of sequence similarity and thus are good candidates for functionally redundant proteins.
Among the 1453 organ-expressed genes, 80 or 5.5% encode putative transcription factors (genes are listed in the supplemental data online). This number is close to the percentage of transcription factors in the Arabidopsis proteome (∼5.9%; Riechmann et al., 2000), indicating that they are neither underrepresented nor overrepresented in the data set. We next determined the representation of different transcription factor families among the organ-expressed genes. Eleven of the 82 described members of the MADS box family and 13 of 109 NAC-like proteins were found in the data set. Statistical tests showed that these numbers are consistent with a significant enrichment of the factors among the organ-expressed genes (P value < 0.02; Fisher's exact test). By contrast, only one of 144 AP2/ERF factors in the genome was found in the data set, indicating significant underrepresentation (P value < 0.01; Fisher's exact test).
Because most of the organ identity genes on which this study was based encode MADS box transcription factors and we detected an enrichment of MADS box factors among the organ-expressed genes, we analyzed the distribution of CArG boxes (consensus 5′-CC(A/T)6GG-3′), the putative binding sites of these factors (Pollock and Treisman, 1990; Shiraishi et al., 1993; Pellegrini et al., 1995; Riechmann et al., 1996), among the organ-expressed genes. To this end, we first determined the genome-wide distribution of CArG boxes in the regions 500, 1000, or 3000 bp upstream of the transcription (or translation) start site, 500 or 1000 bp downstream of the 3′ end of a transcription unit (or the stop codon), and in introns. Compared with the genome-wide distribution, no significant enrichment or differences in the distribution of CArG boxes were observed for the organ-expressed genes (see supplemental data online for details). We repeated this analysis with the putative AG binding site (5′-TT(A/T/G)CC(A/T)4NNGG(A/T/C)(A/T)2-3′; consensus derived from Shiraishi et al., 1993; Huang et al., 1993) that contains a CArG box–like sequence (underlined). However, no enrichment of this site was observed among the genes of the stamen and carpel groups that could be directly regulated by AG (see supplemental data online for details).
The identification of organ-expressed genes allowed the search for overrepresented motifs in their putative promoters that might mediate organ-specific expression. Five different prediction programs were used to analyze the regions 1000 bp upstream of the translation start site (see supplemental data online for details) of genes assigned to the sepal, petal, and carpel groups as well as the genes with predicted pollen-specific expression (see above). The latter group was chosen as an example for genes with potentially cell type–specific expression. Although some overrepresented motifs were identified by the promoter analysis (data not shown), these are present in only a small portion of the genes in an analysis group, suggesting that spatially limited gene expression in flowers might not be mediated by a small number of conserved cis-regulatory elements.
DISCUSSION
In this study, we have analyzed spatial gene expression in flowers by comparing the gene expression profiles of floral homeotic mutants. We have identified a large number of genes that are, within flowers, specifically or predominantly expressed in one type of floral organ and are probable components of the gene networks involved in floral organ development. Several of these genes encode proteins with presumed regulatory function, most of which have not yet been characterized in detail.
In Arabidopsis, the targeted inactivation of genes has become a very powerful approach for functional analysis. RNA interference can be used to induce loss-of-function phenotypes (Chuang and Meyerowitz, 2000), and T-DNA insertion lines are available for many genes (Alonso et al., 2003). Thus, the function of the identified genes can now be systematically studied by reverse genetics.
In large microarray data sets, such as the one presented in this study, not every observation can be confirmed by independent methods. Therefore, we chose selection criteria that aimed to minimize the number of transcripts that are falsely predicted to be floral organ specific. For the analysis of the individual mutant/wild type comparisons, we have used parameters that led to a false discovery rate for significantly changed elements of ∼1%. We then combined the results of the significance analyses for the individual experiments to increase the level of confidence for the predictions of organ-expressed transcripts. For all organs (with the exception of carpels), these predictions were based on a reported downregulation of an element in more than one of the experiments. The application of stringent screening criteria for the identification of organ-expressed genes has led inevitably to a relative increase in the number of organ-expressed genes that have been missed in the analysis. However, the number of these false negatives cannot be reliably estimated based on the data.
Among the genes in the different organ groups, we found several with known floral organ–specific expression that were correctly assigned (Tables 2 and 3). This result indicates that the selection criteria applied here were suitable for the identification of organ-expressed transcripts. The analysis of expression patterns of previously uncharacterized genes further supported this conclusion (Figure 4). For all genes tested, the actual expression patterns matched the predictions that were based on the microarray data. However, the results of in situ hybridizations also showed that some genes that were assigned to one of the organ groups are also expressed in other floral organs either at low levels or in relatively small domains. Examples of such genes are WUS and ASKβ. Here, the strong expression of the genes in anthers or in pollen, respectively, masks their expression in small domains of ovules.
We observed a large overlap between the data sets from the two different types of microarrays used in this study. Seventy-six percent of the genes identified with the flower-specific cDNA array were also identified with the oligonucleotide array. The differences between the data sets are likely because of differences in the properties of the two array platforms that affect critical parameters such as sensitivity, probe specificity, or reproducibility (Kothapalli et al., 2002; Barczak et al., 2003; Tan et al., 2003). It is noteworthy that the presence of multiple probes for many genes on the flower-specific cDNA array results in a significant increase in statistical power compared with the data from the oligonucleotide array that were in general derived from a single probe per gene. On the other hand, data from the cDNA array are more likely to be affected by cross-hybridization than the oligonucleotides of the whole genome array that were designed to have optimal specificity.
A limitation of our analysis is that the cell type in which a gene is expressed in a floral organ cannot be predicted based on the microarray data. To further characterize the localization of the organ-expressed transcripts, several approaches could be taken. First, genes of interest could be studied by in situ hybridizations as demonstrated in this study. A second possibility would be to analyze the expression profiles of mutants or transgenic lines whose phenotypic defects are limited to certain floral tissues or cell types. Examples for such mutants are extra sporogenous cells/excess microsporocytes1 that do not form the tapetal cell layer in anthers (Canales et al., 2002; Zhao et al., 2002) or nozzle/sporocyteless that fail to form both male and female sporocytes (Schiefthaler et al., 1999; Yang et al., 1999). Lastly, the expression profiles of certain floral tissues could be determined by microdissection of these tissues followed by RNA extraction and microarray analysis. In any case, a comparison of the results of these experiments with the organ-expressed transcripts presented here would introduce an additional level of confidence to the predictions. For example, the comparison between the organ-expressed genes described here and the genes predicted to be expressed in pollen by a previous study (Honys and Twell, 2003) revealed an overlap of 147 genes. Most of these genes were predicted to be expressed specifically in pollen. Because the microarray used in the previous study contained only approximately one-quarter of all Arabidopsis genes, it can be estimated that the total number of transcripts in the stamen group that are pollen specific is about four times higher than the overlap between the two data sets (i.e., ∼600). Thus, specific gene expression in pollen likely accounts for about half of the stamen-expressed transcripts.
The results of in situ hybridizations as well as the expression patterns of the previously characterized organ-specific transcripts suggest that the majority of transcripts are likely expressed at late stages of flower development. The earliest expression found by in situ hybridization for an organ-expressed transcript was detected for At3g26860 in tetrads during pollen development at floral stage 9 (Figure 4L). This observation can be explained by the fact that the RNA preparations used for the experiments were, because of the differences in size of young and old floral buds, strongly enriched for RNA from older buds. Therefore, genes that are expressed in one of the floral organs during early stages of flower development might have been too dilute in the RNA samples to be detected in these experiments.
The majority of the genes that were predicted to be organ expressed were assigned to the stamen and carpel groups, whereas only very few were assigned to the organs of the perianth. This difference in number is likely because of key developmental events, such as the formation of pollen and ovules that occur during late stages of flower development in the reproductive organs. In addition, the reproductive organs contain many different tissues and cell types, whereas the anatomy of sepals and petals appears to be less complex.
In a previous study, the expression profiles of inflorescences of different strains, including ap3 and pi, were compared, leading to the identification of 47 genes that were predicted to be expressed in petals and/or stamens (Zik and Irish, 2003). Based on the total number of genes represented on the cDNA array used for their experiments, the authors estimated that the total number of genes whose expression depends on AP3/PI activity is ∼200. This number is about sixfold below the number of genes with expression in stamens or petals that is reported here. In addition, not all genes regulated by AP3/PI have to be expressed specifically or predominantly in petals or stamens but could be expressed simultaneously in both types of floral organs or could have more complex expression patterns. Thus, the actual number of genes whose expression is affected by AP3/PI activity is likely markedly higher than what has been suggested.
The elements of the cDNA microarray used in the previous study were derived from ESTs from all stages of the Arabidopsis life cycle (Zik and Irish, 2003). In total, this array contained probes representing approximately the same number of genes as the flower-specific cDNA array described here. Because of the different strategies used to design the cDNA arrays, the results of the previous study allow an assessment of the relative enrichment of elements for flower organ–specific transcripts on our array. The total number of genes in the stamen or petal groups that were identified with the flower-specific cDNA array is 212 (Table 1). We estimate that ∼100 additional genes with petal or stamen expression are included in the elements that were not sequenced (see Methods). Thus, the strategy used for the construction of the flower-specific cDNA array has led to a significant increase in elements representing genes with specific expression in floral organs. This result further suggests that cDNA arrays that are based on clones derived from various tissues are of limited use for the analysis of developmental processes in a specific organ or tissue type. However, even the use of clones derived exclusively from floral tissues did not lead to a nearly complete coverage of organ-expressed genes because the number of genes identified with the whole genome array was about four times as high as the number identified with the flower-specific cDNA array. This result is likely because of underrepresentation of low abundance transcripts in the libraries used for the construction of the array.
Underrepresentation of low abundance transcripts on cDNA arrays could also explain why only few transcription factors were found downstream of AP3/PI (Zik and Irish, 2003). Based on this observation, it was suggested that the AP3/PI transcription factor complex acts relatively directly on most genes required for cellular differentiation in petals and stamens. However, we identified a relatively large number of transcription factors among the organ-expressed genes. Thus, the gene regulatory networks mediating floral organ development are likely more complex than previously suggested. In agreement with this idea, we did not observe an enrichment of CArG boxes, the putative binding sites for most of the organ identity factors, nor did we identify a small set of conserved motifs in the putative promoters of the organ-expressed genes that would suggest regulation by a limited number of transcription factors.
METHODS
Strains and Plant Growth
The mutant strains used in this study were ap1-1, ap2-2, ap3-3, pi-1, and ag-3 (Bowman et al., 1989, 1991, 1993; Jack et al., 1992). Plants of the accession Landsberg erecta were used as wild-type control. Plants were grown on a soil:vermiculite:perlite mixture under constant illumination at 21°C.
Microarray Setup
The production of the two types of microarrays used in this study is described in detail in the supplemental data online. In brief, elements for the flower-specific cDNA microarray were obtained from three different sources. First, for those genes that are known or suspected to be involved in flower development, we either amplified fragments of their transcribed regions from cDNA or, if available, obtained cDNA clones from EST clone collections. Second, we generated cDNA libraries using RNA extracted from different floral tissues (see supplemental data online for details). For all libraries, the RNA preparations were subtracted with leaf RNA to reduce the abundance of clones representing ubiquitously expressed genes and, thus, to enrich for flower-specific transcripts. The identity of most of the library clones was not determined before their use for preparing the array. Lastly, we added a set of 2632 nonredundant clones from a nonsubtracted flower-derived library (Asamizu et al., 2000).
The oligonucleotide array was based on the Arabidopsis Genome Oligo Set version 1.0 (Qiagen Operon, Alameda, CA). This set consists of 26,090 oligonucleotides that correspond to 23,542 annotated genes in release 4.0 of the Institute for Genomic Research Arabidopsis thaliana Genome Annotation Database.
Tissue Collection and RNA Isolation for Microarray Experiments
Four sets of tissue samples from each of the homeotic mutants and from wild-type plants were prepared. Tissue collection for the different sets was done on different days but at the same time of day. For each sample, floral buds (corresponding to floral stages 1 to 13) from ∼50 plants were collected. RNA was isolated from 100 mg of tissue with the RNeasy RNA isolation kit (Qiagen, Valencia, CA). RNA quality was assessed by gel electrophoresis.
Probe Labeling and Microarray Hybridization Protocols
Probe labeling and hybridization protocols are described in detail in the supplemental data online. In brief, first and second strand cDNA was synthesized from 3 μg of total RNA using a poly(A)-primer with a T7 promoter sequence. Then, in vitro transcription was performed using the Megascript T7 kit (Ambion, Austin, TX). For the cDNA array, dye-labeled cDNA was generated from the amplified RNA using the Atlas glass fluorescent labeling kit (Clontech, Palo Alto, CA) and hybridized to the array. For the oligonucleotide array, dye molecules were coupled to the amplified RNA, and the dye-labeled RNA was fragmented before hybridization.
Data Acquisition, Normalization, and Statistical Analysis
Hybridized arrays were scanned with an Axon 4000A scanner using the GenePix 3.1 analysis software (Axon Instruments, Foster City, CA). The photomultiplier tube voltages for the two channels were adjusted so that the ratio of the mean signal intensities was ∼1 and the percentage of spots with saturated pixels was <0.25% but >0%. Two lines of average were used for the scanning process. The spot intensities as well as the local background intensities were quantified using the GenePix 3.1 analysis software.
For experiments with the cDNA array, data were analyzed as follows. Before normalization, the data points were removed if a spot was flagged during data acquisition (e.g., as a bad spot, absent spot, etc.) or if a spot intensity was calculated based on >5% of saturated pixels. In addition, low intensity elements were removed if the sum of the median signal intensities of a spot was below the sum of the median background intensities plus two standard deviations of background.
For the remaining elements, the median background intensities were subtracted from the median spot intensities. Subsequently, the data sets for the replicate experiments were combined. Elements for which less than three data points were available were excluded from further analysis. In addition, all the elements whose amplification during the generation of the array did not result in high-quality PCR products were removed from the data sets.
After quantitation of the signal intensities, the data were normalized to compensate for nonlinearity of intensity distributions and differences in probe labeling. Commonly used normalization methods include global normalization or linear regression. These methods are based on the assumption that the percentage of the elements reporting significant expression changes is small and that the number of upregulated and downregulated genes is similar (Causton et al., 2003). However, when we plotted the experimental data, we noticed that the percentage of elements reporting downregulation relative to the wild type in the data sets for ap3, pi, and ag was ∼20%, whereas a much smaller proportion corresponded to upregulated transcripts (see supplemental data online). Therefore, the use of the standard normalization methods in these cases would lead to a misinterpretation of the experimental results. To allow data normalization without the introduction of a bias, we applied an alternative normalization method (Kepler et al., 2002) that performs local regression based on an invariant subset of elements that are identified in the replicate experiments (see supplemental data online). For this, the background-corrected spot intensities were loaded into the program NoSe-CoLoR version 1.0 (Kepler et al., 2002). For every experiment, the normalization procedure was repeated with different values for the assumed percentile of invariant elements (r). For all experiments, the number of elements with significant expression changes after normalization was nearly constant below a certain value of r, suggesting that it reflected a good estimate of the invariant number of elements. The following values for r were used: 0.9 (ap1), 0.9 (ap2), 0.75 (ap3), 0.75 (pi), and 0.75 (ag).
After normalization, ratios of signal intensities were calculated for each experiment followed by the coefficient of variation of the ratios for the replicate experiments. To assess the overall reproducibility of the experiments, the median coefficient of variation was calculated for each mutant/wild type comparison. The following values were obtained: 16% (ap1), 19% (ap2), 15% (ap3), 16% (pi), and 16% (ag).
Next, we performed SAM (Tusher et al., 2001) using the normalized signal intensities to identify differentially expressed genes. The parameters for SAM were adjusted so that the false discovery rate for every experiment was ∼1%. A twofold expression cutoff was applied to make the analysis more stringent.
The log2-transformed mean ratios of the elements that were judged significant in SAM in at least one of the experiments were loaded into the program CLUSTER (Eisen et al., 1998), and hierarchical clustering was performed. In addition, a self-organizing map was generated (see supplemental data online). Clustering results were visualized with the program TREEVIEW (Eisen et al., 1998). Several elements of each cluster that presumably contained organ-specific genes were sequenced, and the sequences were matched to the Arabidopsis genome. In general, we found that elements corresponding to the same gene clustered together. From the remaining elements, we sequenced several hundred that were picked at random. We found that only ∼10% of these elements represent novel genes that had not been identified by the sample sequencing of the clusters. Because we had at this point already sequenced approximately half of the ∼2000 array elements predicted to represent organ-specific transcripts, we concluded that the remaining elements contain ∼100 genes that had not been identified previously.
For experiments with the oligonucleotide array, data were analyzed using the Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware, Kirkland, WA). In Resolver, spots that are flagged during data acquisition by the Genepix software are not included in the analysis. In addition, if for a given spot intensities in both channels are below zero after background subtraction, the data are excluded from further analysis. Additional data processing in this system consists of error correction and calculation of a P value of differential expression using the intensity-error estimation from the .gpr file. Error correction consists of a simplified version of the algorithm described by Schadt et al. (2001), in which a piecewise linear function replaces smoothing splines to fit and correct intensity nonlinearity. Calculation of P values consists of a statistic that combines additive and multiplicative error components in both channels of a two-colored experiment. The resultant ratio profiles (that is, two-channel, error processed microarray scans) were combined into ratio experiments in the Resolver system as described in Stoughton and Dai (2002). Within the Resolver system, we performed the analysis at the so-called sequence level (i.e., when there are multiple data points for the same gene in the same hybridization such data are combined). When combining data, it is assumed that the ratio measurement with the lowest overall error is closest to the true value for that sequence, and weights are constructed such that the feature or reporter with the lowest error is given the greatest weight (Stoughton and Dai, 2002).
In Situ Hybridizations
In situ hybridizations were performed as previously described (Long and Barton, 1998) using inflorescences of wild-type plants. Primers used for the generation of probes are listed in the supplemental data online. The probe used for detection of WUS transcript was described previously (Brand et al., 2000). For ASKβ, a cDNA fragment produced by RACE-PCR was used as a probe.
Microarray data from this article have been deposited with the NCBI Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE1265 through GSE1275.
Supplementary Material
Acknowledgments
We are very grateful to Vijaya Rao and Ali Mortazavi for help with microarray data analysis, to Arnavaz Garda for technical assistance, and to members of the Meyerowitz laboratory for discussion and for comments on the manuscript. We thank S. Tabata (Kazusa DNA Research Institute, Japan) for the generous gift of the flower-derived EST collection. This work was supported by National Institutes of Health Grant GM45697 to E.M.M. and by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology. F.W. was supported by an Emmy Noether fellowship of the Deutsche Forschungsgemeinschaft. M.A.-F. is indebted to the Instituut voor Plantenbiotechnologie voor Ontwikkelingslanden and Marc Van Montagu for a fellowship sponsored by Aventis Crop Sciences.

Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the Instructions for Authors (www.plantcell.org) is: Elliot M. Meyerowitz (meyerow@caltech.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021741.
References
- Aarts, M.G., Hodge, R., Kalantidis, K., Florack, D., Wilson, Z.A., Mulligan, B.J., Stiekema, W.J., Scott, R., and Pereira, A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12, 615–623. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Alves-Ferreira, M., Engler, J.A., Miguens, F.C., Van Montagu, M., Engler, G., and Oliveira, D.E. (1997). Oleosin gene expression in Arabidopsis thaliana coincides with accumulation of lipid in plastids and in cytoplasmatic bodies. Plant Physiol. Biochem. 35, 729–739. [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7, 175–180. [DOI] [PubMed] [Google Scholar]
- Barczak, A., Rodriguez, M.W., Hanspers, K., Koth, L.L., Tai, Y.C., Bolstad, B.M., Speed, T.P., and Erle, D.J. (2003). Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res. 13, 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Alvarez, J., Weigel, D., Meyerowitz, E.M., and Smyth, D. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743. [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 15, 1718–1727. [DOI] [PubMed] [Google Scholar]
- Causton, H.C., Quackenbush, J., and Brazma, A. (2003). Microarray Gene Expression Data Analysis. (Oxford, UK: Blackwell Publishing).
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz, C., Liljegren, S.J., and Yanofsky, M.F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438. [DOI] [PubMed] [Google Scholar]
- Flanagan, C.A., Hu, Y., and Ma, H. (1996). Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 10, 343–353. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gross-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Q., Ferrandiz, C., Yanofsky, M.F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1994). Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76, 131–143. [DOI] [PubMed] [Google Scholar]
- Hird, D.L., Worrall, D., Hodge, R., Smartt, S., Paul, W., and Scott, R. (1993). The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to beta-1,3-glucanases. Plant J. 4, 1023–1033. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132, 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Mizukami, Y., Hu, Y., and Ma, H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., McDowell, J.M., Weise, M.J., and Meagher, R.B. (1996). The Arabidopsis profilin gene family. Evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol. 111, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G., Van Montagu, M., and Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalay, J.C., and Goldberg, R.B. (1984). Organ-specific nuclear RNAs in tobacco. Proc. Natl. Acad. Sci. USA 81, 2801–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin, S.A., Savidge, B., and Yanofsky, M.F. (1995). Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267, 522–525. [DOI] [PubMed] [Google Scholar]
- Kepler, T.B., Crosby, L., and Morgan, K.T. (2002). Normalization and analysis of DNA microarray data by self-consistency and local regression. Genome Biol. 3, 0037.1–0037.12. [DOI] [PMC free article] [PubMed]
- Kim, H.U., Hsieh, K., Ratnayake, C., and Huang, A.H. (2002). A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 21, 22677–22684. [DOI] [PubMed] [Google Scholar]
- Kothapalli, R., Yoder, S.J., Mane, S., and Loughran, T.P., Jr. (2002). Microarray results: How accurate are they? BMC Bioinformatics 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Lebel-Hardenack, S., Ye, D., Koutnikova, H., Saedler, H., and Grant, S.R. (1997). Conserved expression of a TASSELSEED2 homolog in the tapetum of the dioecious Silene latifolia and Arabidopsis thaliana. Plant J. 12, 515–526. [DOI] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., Fiebig, A., Johnstone, S.E., and Preuss, D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292, 2482–2485. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. [DOI] [PubMed] [Google Scholar]
- Paul, W., Hodge, R., Smartt, S., Draper, J., and Scott, R. (1992). The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol. Biol. 19, 611–622. [DOI] [PubMed] [Google Scholar]
- Pellegrini, L., Tan, S., and Richmond, T.J. (1995). Structure of serum response factor core bound to DNA. Nature 376, 490–498. [DOI] [PubMed] [Google Scholar]
- Pollock, R., and Treisman, R. (1990). A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 18, 6197–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11, 1914–1922. [DOI] [PubMed] [Google Scholar]
- Reinke, V., and White, K.P. (2002). Developmental genomic approaches in model organisms. Annu. Rev. Genomics Hum. Genet. 3, 153–178. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Krizek, B.A., and Meyerowitz, E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93, 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, M.R., Foster, G.D., Blundell, R.P., Robinson, S.W., Kumar, A., Draper, J., and Scott, R. (1993). Gametophytic and sporophytic expression of an anther-specific Arabidopsis thaliana gene. Plant J. 3, 111–120. [PubMed] [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinelli, P., Hu, Y., and Ma, H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37, 607–619. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Medrano, L.J., and Meyerowitz, E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203. [DOI] [PubMed] [Google Scholar]
- Savidge, B., Rounsley, S.D., and Yanofsky, M.F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E.E., Li, C., Ellis, B., and Wong, W.H. (2001). Feature extraction and normalization algorithm for high-density oligonucleotide gene expression array data. J. Cell. Biochem. 84 (suppl.), 120–125. [DOI] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi, H., Okada, K., and Shimura, Y. (1993). Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4, 385–398. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A.M., Krober, S., Unte, U.S., Huijser, P., Dekker, K., and Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423. [DOI] [PubMed] [Google Scholar]
- Stoughton, R.S., and Dai, H. (2002). Statistical combining of cell expression profiles. U.S. Patent 6,351,712.
- Tan, P.K., Downey, T.J., Spitznagel Jr, E.L., Jr., Xu, P., Fu, D., Dimitrov, D.S., Lempicki, R.A., Raaka, B.M., and Cam, M.C. (2003). Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 31, 5676–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichtinsky, G., Tavares, R., Takvorian, A., Schwebel-Dugue, N., Twell, D., and Kreis, M. (1998). An evolutionary conserved group of plant GSK-3/shaggy-like protein kinase genes preferentially expressed in developing pollen. Biochim. Biophys. Acta 8, 261–273. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Meyerowitz, E.M. (1993). Activation of floral homeotic genes in Arabidopsis. Science 261, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500. [DOI] [PubMed] [Google Scholar]
- Xu, H., Knox, R.B., Taylor, P.E., and Singh, M.B. (1995). Bcp1, a gene required for male fertility in Arabidopsis. Proc. Natl. Acad. Sci. USA 92, 2106–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Zhao, D.-Z., Wang, G.-F., Speal, B., and Ma, H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-repeat receptor protein kinase that controls somatic and reproductive cell fates in Arabidopsis. Genes Dev. 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik, M., and Irish, V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15, 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.