Abstract
Factors affecting splicing of plant U12-dependent introns have been examined by extensive mutational analyses in an in vivo tobacco (Nicotiana tabacum) protoplast system using introns from three different Arabidopsis thaliana genes: CBP20, GSH2, and LD. The results provide evidence that splicing efficiency of plant U12 introns depends on a combination of factors, including UA content, exon bridging interactions between the U12 intron and flanking U2-dependent introns, and exon splicing enhancer sequences (ESEs). Unexpectedly, all three plant U12 introns required an adenosine at the upstream purine position in the branchpoint consensus UCCUURAUY. The exon upstream of the LD U12 intron is a major determinant of its higher level of splicing efficiency and potentially contains two ESE regions. These results suggest that in plants, U12 introns represent a level at which expression of their host genes can be regulated.
INTRODUCTION
Pre-mRNA splicing in eukaryotes is a fundamental step in gene expression and represents an important level at which the expression of protein-coding genes can be regulated (Reed, 2000; Smith and Valcárcel, 2000; Graveley, 2001; Hastings and Krainer, 2001; Cartegni et al., 2002; Black, 2003). In higher eukaryotes, there are two classes of nuclear pre-mRNA introns. The most abundant class consists of U2-dependent introns (U2 introns), whereas the second rarer class (<0.4% of introns) consists of U12-dependent introns (U12 introns). U12 introns have been found in the nuclear genomes of vertebrates, plants, and insects (Hall and Padgett, 1994; Wu et al., 1996; Sharp and Burge, 1997; Tarn and Steitz, 1997; Wu and Krainer, 1998, 1999; Burge et al., 1998; Levine and Durbin, 2001; Patel and Steitz, 2003). Introns belonging to these two distinct classes are spliced by two different spliceosomes: the major U2-type spliceosome and the less abundant U12-type spliceosome (Hall and Padgett, 1996; Tarn and Steitz 1996a, 1996b, 1997). Although the first U12 introns to be described had AT-AC–terminal dinucleotides, the majority of U12-type introns contain GT-AG, and a small number contain other noncanonical terminal dinucleotides, such as AT-AA, AT-AG, AT-AT, GT-AT, or GT-GG (Jackson, 1991; Hall and Padgett, 1994; Dietrich et al., 1997, 2001a; Sharp and Burge, 1997; Burge et al., 1998; Wu and Krainer, 1999; Levine and Durbin, 2001; Zhu and Brendel, 2003). Moreover, functional analyses have shown that AT-AC–terminal dinucleotides are not a defining feature of U12 introns (Dietrich et al., 1997, 2001a). Instead, U12 introns contain highly conserved sequences in the 5′ splice site (exon:G/ATATCCTY) and branchpoint region (TCCTTRAY) (Hall and Padgett, 1994; Sharp and Burge, 1997; Burge et al., 1998), which are both required for prespliceosome complex formation (Frilander and Steitz, 1999). The 3′ splice site consensus sequence of U12 introns (YAC/G:exon) is less informative than their 5′ splice site and branchpoint sequences. U12 introns also lack a polypyrimidine tract and have a short distance between the branchpoint and 3′ splice site (between 10 and 20 nucleotides) (Hall and Padgett, 1994; Burge et al., 1998; Dietrich et al., 2001a; Tarn and Steitz, 1997). In Arabidopsis thaliana, a recent computational search for U12 introns predicted 165 such introns representing ∼0.15% of the predicted total number of introns (Arabidopsis Genome Initiative, 2000; Zhu and Brendel, 2003). Plant U12 introns contain the same splice site and branchpoint consensus sequences as vertebrate U12 introns (Wu et al., 1996; Zhu and Brendel, 2003). Similarly, the first and last nucleotides are variable, with the most common dinucleotide combinations being GT-AG and AT-AC and only two examples that have AT-AA and GT-AT (Zhu and Brendel, 2003). In addition, 153 introns had branchpoint/3′ splice site distances of <21 nucleotides, showing that plant U12 introns in general maintain the core sequence elements required for splicing in vertebrate U12 intron splicing (Zhu and Brendel, 2003).
Both U12- and U2-type spliceosomes contain five small nuclear ribonucleoprotein particles (snRNPs) required for spliceosome assembly and function. The U5snRNP is common to both types of spliceosome (Tarn and Steitz, 1996a). In addition to U5, the U12 spliceosome contains U11, U12, U4atac, and U6atac snRNPs, which are distinct from their major spliceosome counterparts, U1, U2, U4, and U6, but have analogous roles in splice site selection and spliceosome formation (Hall and Padgett, 1996; Tarn and Steitz, 1996a, 1996b, 1997; Kolossova and Padgett, 1997; Yu and Steitz, 1997; Incorvaia and Padgett, 1998; Wu and Krainer, 1999; Patel and Steitz, 2003). In particular, the functions of analogous small nuclear RNAs (snRNAs) are conserved. For example, base-pairing interactions occur between the U11snRNA/5′ splice site and U12snRNA/branchpoint during prespliceosomal complex formation, similar to U1 and U2 in the major spliceosome (Kolossova and Padgett, 1997; Frilander and Steitz, 1999). U4atac and U6atac snRNAs show similar base-pairing interactions and functional domains as U4 and U6 (Shukla and Padgett, 1999, 2001). In addition, both the major and minor spliceosomes share many common spliceosomal proteins (Luo et al., 1999; Will et al., 1999, 2001; Schneider et al., 2002). Plant homologs of U6atac and U12 have been identified in Arabidopsis, and despite large sequence differences, their functional regions are conserved (Shukla and Padgett, 1999). U11 and U4atac remain to be identified in plants but the conservation of intron signals and U12 and U6atac suggest that these snRNAs are likely to exist as part of the plant U12 splicing system. As with U2 introns, U12 intron splicing follows a two-step reaction that involves the formation of a characteristic intron lariat intermediate at an adenosine that can be at one of two possible positions within the stringent branchpoint consensus (Tarn and Steitz, 1996a; McConnell et al., 2002). In addition, exon bridging interactions between complexes forming on splice sites of U12 introns and flanking U2 introns can enhance U12-dependent intron splicing (Wu and Krainer, 1996; Dietrich et al., 2001b; Hastings and Krainer, 2001; Kmieciak et al., 2002), and some U12 introns are involved in alternative splicing (Dietrich et al., 2001b; Zhu and Brendel, 2003).
The distinguishing feature of U2 introns in plants, compared with introns from vertebrates and yeast (Saccharomyces cerevisiae), is their UA richness, which is required for efficient U2 intron splicing and for 5′ and 3′ splice site selection (Goodall and Filipowicz, 1989; Simpson and Filipowicz, 1996; Brown and Simpson, 1998; Lorković et al., 2000; Reddy, 2001). The role of UA-rich elements in plant splicing is still poorly understood, but they may minimize secondary structure of plant introns or bind specific UA binding proteins to recruit splicing factors early in spliceosome formation (Simpson and Filipowicz, 1996; Brown and Simpson, 1998; Lorković et al., 2000; Reddy, 2001). Putative UA-rich binding proteins (UBP1, RBP45, and RBP47) with affinity for U-rich sequences have been isolated and characterized (Gniadkowski et al., 1996; Lambermon et al., 2000; Lorković et al., 2002). Of these, only UBP1 so far has been shown to affect splicing, where overexpression of UBP1 enhanced splicing of otherwise poorly spliced U2-type introns (Lambermon et al., 2000). Putative plant U12 introns are also UA rich in comparison with exonic sequences (Zhu and Brendel, 2003; our unpublished results), but it remains to be seen whether UA richness determines splice site selection or influences splicing efficiency.
To date, the splicing of only one plant U12-dependent intron has been experimentally analyzed in vivo (Kmieciak et al., 2002). Here, we present an analysis of sequence elements that are involved in plant U12 intron splicing efficiency using three different Arabidopsis U12-dependent introns. Differences in splicing efficiency among the introns were investigated by mutational analysis of splice site and putative branchpoint sequences, leading to identification of essential branchpoint nucleotides in vivo. We also demonstrate that UA content influences plant U12 intron splicing efficiency and that two U-rich RNA binding proteins, UBP1 and RBP45, have differential effects on splicing of U2- and U12-type introns. Finally, we show the presence of potential exon splicing enhancer sequences in the 5′ flanking exon of one U12 intron that are required for maximal splicing efficiency.
RESULTS
Isolation of Three Different Plant U12-Type Introns
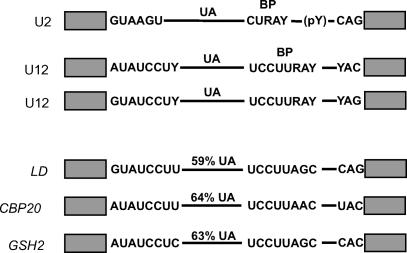
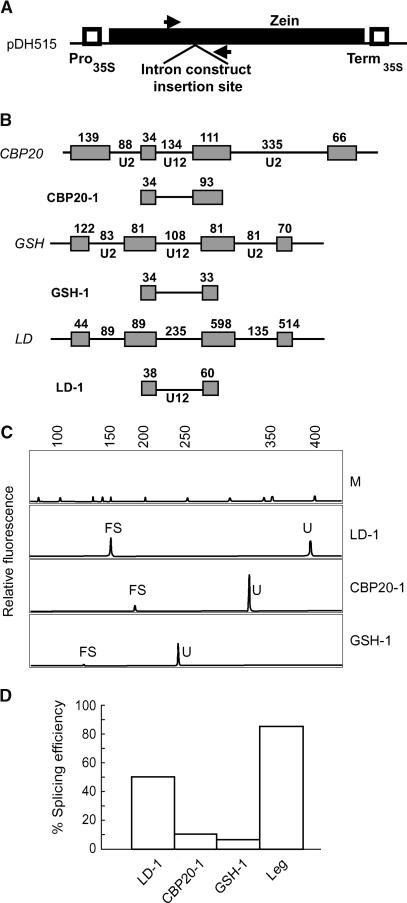
Three different plant U12-dependent introns were selected from three different Arabidopsis genes on the basis of their splice site and branchpoint sequences (Figure 1). The three genes were CBP20, encoding the 20-kD cap binding protein (Kmieciak et al., 2002), GSH2, encoding glutathione synthetase (Wang and Oliver, 1996), and LD, encoding the LUMINIDEPENDENS protein (Lee et al., 1994). The U12 intron from the CBP20 gene is 134 bp long and is the fourth of eight introns in the CBP20 gene. The U12 intron from the GSH2 gene is 108 bp long and is the sixth intron out of 11, and that of LD is 235 bp long and is the tenth intron out of 12. All three introns show sequence features characteristic of U12-dependent introns but have different combinations of splice site and branchpoint sequences. The introns from CBP20 and GSH2 have AU-AC splice site dinucleotides but vary in their branchpoint sequence. CBP20 has the branchpoint sequence UCCUUAAC, whereas GSH2 has UCCUUAGC. The LD U12 intron shows the same branchpoint sequence as GSH2 but has GU-AG termini (Figure 1). The A/G branchpoint variation (underlined) occurs at the branchpoint nucleotide suggested for animal U12-type introns (Tarn and Steitz, 1996a). However, the adenosine directly upstream has also been shown to act as the branchpoint nucleotide in some vertebrate U12 introns (McConnell et al., 2002). The UA content of the U12-type introns from the CBP20, GSH2, and LD genes is 64, 63, and 59%, respectively, which is equal to or just above the minimum UA requirement for accurate and efficient splicing of plant U2-dependent introns (Goodall and Filipowicz, 1989). The introns were isolated from Arabidopsis genomic DNA by PCR and included >30 bp of authentic 5′ and 3′ exon sequences. Amplified fragments were introduced into the plant expression vector pDH515 (Figure 2A; Simpson et al., 1996), which contains the 35S promoter of Cauliflower mosaic virus (CaMV 35S), terminator sequences, and an intronless zein gene, encoding a maize (Zea mays) seed storage protein. Each of the three Arabidopsis U12-dependent introns, together with fragments of authentic flanking exons, was cloned into the unique BamHI site within the zein gene, giving constructs pCBP20.1, pGSH.1, and pLD.1 (Figure 2B).
Figure 1.
Schematic Representation of Splicing Signals of U2- and U12-Dependent Introns.
Consensus 5′ and 3′ splice sites and branchpoints are shown for plant U2 and U12 introns of both the AU-AC and GU-AG types. The splicing signals of the three plant U12 introns studied here (LD, CBP20, and GSH2) are presented. pY, polypyrimidine tract or U-rich sequence; UA, UA richness.
Figure 2.
Schematic Diagrams of Intron Constructs to Examine Splicing of U12 Introns.
(A) Transient expression vector pDH5I5, into which intron constructs are placed for splicing analysis. Closed box, zein gene; open boxes, CaMV 35S promoter and terminator regions.
(B) The wild-type exon-intron structures of part of the CBP20, GSH, and LD genes are shown. Boxes, exons; lines, introns. The sizes of authentic exons and exon fragments in the U12 constructs are given.
(C) GeneScan analysis of splicing of the three U12 introns and a U2 control intron from a pea legumin gene in tobacco protoplasts. FS, fully spliced; U, unspliced; M, DNA size markers.
(D) Histogram of splicing efficiency.
Arabidopsis U12-Dependent Introns Are Spliced Less Efficiently in Tobacco Protoplasts Than U2-Dependent Introns
The U12 intron constructs pCBP20.1, pGSH.1, and pLD.1 were introduced into tobacco (Nicotiana tabacum) protoplasts by polyethylene glycol–mediated transfection. As a control, the splicing efficiency of the U2-dependent intron from a pea (Pisum sativum) legumin gene was monitored in the same experiments. The efficiency of excision of different introns was analyzed by RT-PCR on total RNA isolated from transfected protoplasts, and splicing efficiency expressed as the mean of three independent experiments. The CBP20 and GSH2 introns were spliced with only 10 and 5% efficiency, respectively, whereas the LD intron was spliced with 50% efficiency (Figures 2C and 2D). The control U2-type legumin intron was spliced with much higher efficiency (85 to 90%) as found previously. The accuracy of the splicing reactions was confirmed by sequencing of RT-PCR products. Thus, all three U12 introns were spliced inefficiently in the tobacco protoplast system when compared with the efficient splicing of the U2 control intron (Figures 2C and 2D). Moreover, there were notable differences among the splicing efficiencies of the three U12 introns.
AU-AC and GU-AG U12-Dependent Intron Splice Site Dinucleotides Are Equally Efficient Splice Sites
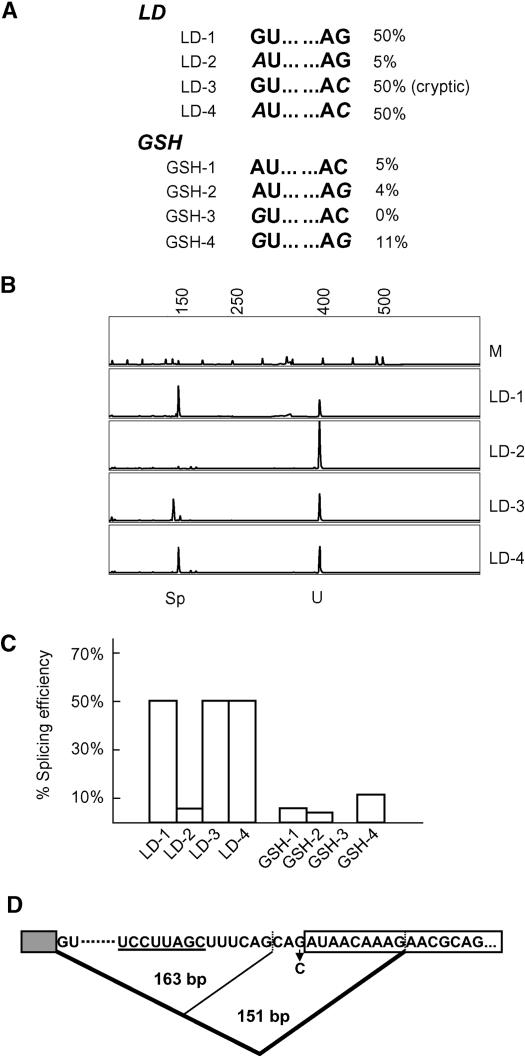
Of the three U12 introns studied in the protoplast system, the LD intron was spliced 5- to 10-fold more efficiently than the CBP20 and the GSH2 introns. Both poorly spliced introns have AU-AC at their termini, whereas the LD intron has GU-AG intron boundaries normally found in the major U2 introns. To investigate whether the increased splicing efficiency of the intron from LD was because of its splice site sequences, the LD and GSH2 U12-dependent introns were mutated from GU-AG to AU-AC and from AU-AC to GU-AG, respectively. Intermediate constructs with only the 5′ or 3′ splice sites mutated were also prepared in both cases (Figure 3A). All mutants were introduced into tobacco protoplasts, and splicing efficiency was measured using RT-PCR. Mutating the GU-AG splice sites of the LD intron to AU-AC (pLD.4) had no effect on splicing efficiency (Figures 3B and 3C). On the other hand, mutation of the AU-AC splice sites to GU-AG in the GSH2 U12 intron (pGSH.4) increased splicing efficiency but remained significantly lower than the splicing efficiency of the LD intron (Figure 3). The constructs containing the intermediate intron mutation with AU-AG termini (pLD.2 and pGSH2.2) showed reduced splicing, which in the case of the LD construct was significant (Figures 3B and 3C). The GU-AC splice site combination abolished splicing of the GSH2 U12 intron (pGSH.3) (Figure 3C). Although the GU-AC combination in the LD U12 intron (pLD.3) also abolished splicing to the mutated 3′ splice site, the overall efficiency of splicing remained at 50% because of activation of two cryptic 3′ splice sites (Figure 3D). Thus, the GU-AC combination of terminal dinucleotides is not competent for splicing. Sequencing of the RT-PCR products confirmed that one cryptic AG- lay three nucleotides upstream of the mutated splice site (used in 5% of transcripts), whereas the second 3′ AG- was located nine nucleotides downstream (used in 95% of the spliced transcripts) (Figure 3D). Thus, AU-AC and GU-AG terminal dinucleotides were equally suitable plant U12 intron splice sites, whereas the intermediate combinations, AU-AG and GU-AC, were unable to support splicing.
Figure 3.
Splicing Analysis of 5′ and 3′ Splice Site Mutations of the LD and GSH U12 Introns.
(A) Mutations to LD and GSH to change splice sites from GU-AG to AU-AC and AU-AC to GU-AG, respectively.
(B) GeneScan analysis of splicing of LD mutants. Sp, spliced product; U, unspliced; M, DNA size markers.
(C) Histogram of splicing efficiencies of LD and GSH2 mutants.
(D) Cryptic splicing of the LD GU-AC mutant (LD-3). The mutation causes selection of either of two cryptic AG dinucleotides, illustrated by lines. The branchpoint sequence is underlined, the G-to-C mutation is indicated, and the sizes of RT-PCR products are given.
Plant U12-Dependent Intron Branchpoint Nucleotides Important for Splicing
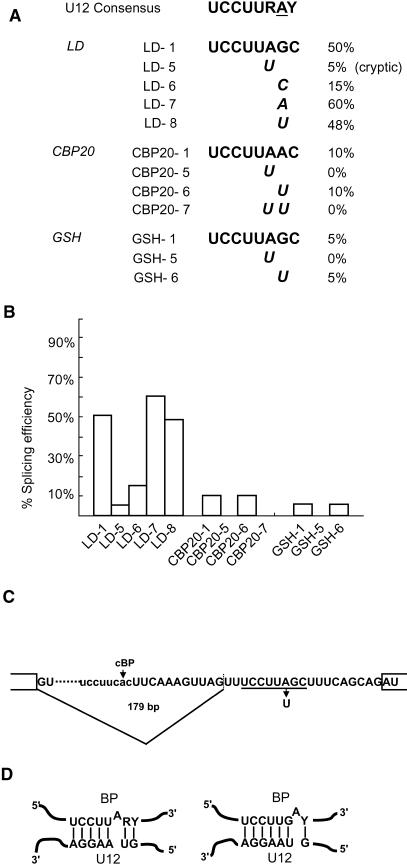
Plant U12-type introns contain the conserved UCCUURAY consensus branchpoint sequence found in vertebrate U12 introns. Adenosines at either of the underlined purine positions have been shown to act as branchpoint nucleotides in vertebrate U12 intron splicing (Tarn and Steitz, 1996a; McConnell et al., 2002). Similarly, some plant U12 introns have an adenosine at only one or other position, suggesting some flexibility in branchpoint selection and utilization in U12-type splicing (Zhu and Brendel, 2003; our unpublished results). Of the three plant U12-type introns tested in these studies, the GSH2 and LD introns have the same branchpoint sequence, UCCUUAGC, whereas CBP20 has the sequence UCCUUAAC (Figures 1 and 4A). The branchpoint sequences are located 11 nucleotides (LD), 12 nucleotides (GSH2), and 13 nucleotides (CBP20) from their associated 3′ splice sites. To investigate whether adenosines at either position could be used in splicing, a series of mutations were made to the branchpoint sequences (Figure 4A). Splicing of the mutated introns was tested in transfected tobacco protoplasts, and the efficiency of splicing was determined (Figure 4A).
Figure 4.
Splicing Analysis of Branchpoint Mutations.
(A) The U12 consensus branchpoint (Zhu and Brendel, 2003) is compared with the branchpoint sequences of LD, CBP20, and GSH2. Single nucleotide mutations are indicated, and efficiencies of splicing are shown next to the mutations.
(B) Histogram of splicing efficiencies of branchpoint mutants.
(C) Cryptic splicing in the branchpoint A-to-U mutation (LD-5) where an upstream cryptic branchpoint and 3′ splice site are activated (lines). The cryptic branchpoint sequence is given in lower case letters, and the putative cryptic branchpoint adenosine (cBP) is indicated by an arrow. The putative authentic branchpoint is underlined, and the A-to-U mutation is indicated.
(D) Models for U12snRNA-branchpoint base-pairing interactions where adenosines at either of two positions act as the branchpoint nucleotide.
In the LD intron, the single nucleotide mutation of A to U at the upstream purine position severely reduced splicing to only 5% (Figure 4B). This residual splicing was because of activation of a cryptic 3′ splice site 19 nucleotides upstream of the mutated 3′ splice site (Figure 4C). Interestingly, a putative U12 branchpoint sequence, UCCUUCAC, was present 13 nucleotides upstream of the cryptic 3′ splice site (Figure 4C), suggesting that the cryptic splicing event was also performed by the U12 spliceosome. Mutation of the G at the downstream purine position in the LD intron did not affect selection of the authentic 3′ splice site, but different nucleotides at this position gave different splicing efficiencies. The G-to-U and G-to-A mutations had little effect, but the G-to-C mutation reduced splicing efficiency from 50 to 15%, perhaps by affecting the base-pairing interactions between U12snRNA and the branchpoint sequence (Figure 4D). Thus, only the upstream adenosine in the LD U12 intron was essential for splicing.
To examine this further and to confirm the results obtained with the LD U12 intron, the nucleotides at both purine positions were also mutated in CBP20 and GSH2. Despite the low splicing efficiency of these introns, mutation of the A to U at the upstream purine position in both introns completely abolished splicing (Figures 4A and 4B, CBP20-5 and GSH-5), whereas mutation of the A at the downstream position in CBP20 and the G in the GSH2 intron had no effect (Figures 4A and 4B, CBP20-6 and GSH-6, respectively). Thus, in all three plant U12 introns tested, the adenosine at the upstream purine position was required for splicing. This nucleotide or the purine in the downstream position can be bulged in putative branchpoint/U12snRNA base-pairing interactions (Figure 4D).
Increasing UA Content of U12 Introns Improves Splicing Efficiency
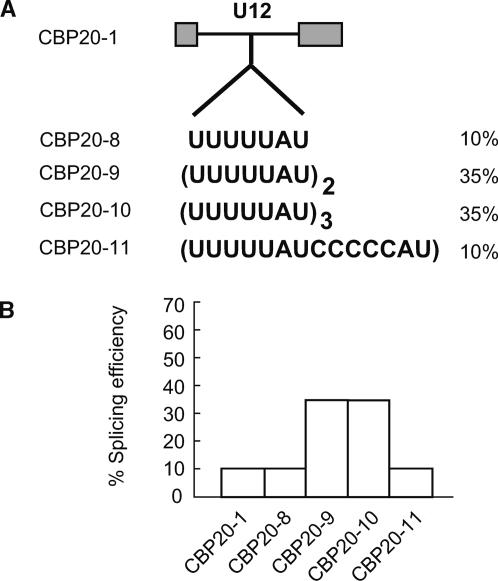
The efficiency of plant U2 intron splicing relies on high intron UA content relative to surrounding exons. Although Arabidopsis U12 introns show a similar UA content range to U2 introns (Zhu and Brendel, 2003; our unpublished results), it is not known whether this UA requirement is important for plant U12 intron splicing. The UA content of the analyzed U12-type introns (59 to 64% UA) lies close to the minimum of 59% UA needed for efficient splicing of U2 introns in tobacco protoplasts (Goodall and Filipowicz, 1989). To examine whether UA-rich elements influence U12 splicing, a series of constructs was made to increase UA content in the poorly spliced CBP20 U12 intron. The constructs consisted of the CBP20 U12 intron, with either one, two, or three copies of a U-rich element (UUUUUAU) introduced 67 nucleotides from the 5′ splice site, giving constructs pCBP20.8, pCBP20.9, and pCBP20.10, respectively (Figure 5A). The site of the insertion was selected to maintain the distance between the branchpoint sequence and the 3′ splice site. Constructs were transfected into tobacco protoplasts, and their splicing was assessed by RT-PCR (Figure 5B).
Figure 5.
Splicing Analysis of U12 Introns Containing Additional UA-Rich Elements.
(A) One, two, or three copies of the UA-rich element UUUUUAU or a control sequence were inserted into the CBP20 intron. Efficiencies of splicing are shown next to the insertions.
(B) Histogram of splicing efficiencies of constructs CBP20-8, CBP20-9, CBP20-10, and CBP20-11 in tobacco protoplasts.
When a single U-rich element was introduced into the CBP20 U12 intron, its splicing efficiency was unaffected, remaining at 10%. Insertion of two or three copies of the UUUUUAU element led to a 3.5-fold increase in splicing efficiency (35%) of the CBP20 U12 intron. To confirm that this effect was because of increasing UA content of the intron and not because of an increase in intron length, a control construct containing a 14-nucleotide-long sequence insertion (UUUUUAUCCCCCAU), consisting of one UA-rich element and seven additional nucleotides, was prepared (pCBP20.11). Splicing of this construct showed only poor splicing (10%), demonstrating that the two or three inserted UA-rich elements were responsible for the increased splicing efficiency. Thus, increased UA content improved the poor splicing of the CBP20 U12-type intron and demonstrated a role for UA sequence elements in plant U12 intron splicing.
Differential Effects of U-Rich Binding Proteins on U12 and U2 Intron Splicing
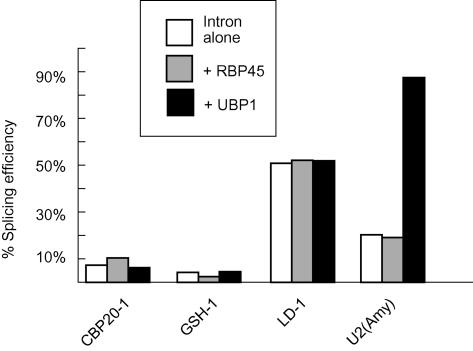
RBP45 and UBP1 are hnRNP-like RNA binding proteins with affinity for U-rich sequences (Gniadkowski et al., 1996; Lambermon et al., 2000; Lorković et al., 2000). UBP1 is able to improve the splicing efficiency of otherwise poorly spliced U2 plant introns (Lambermon et al., 2000). To determine whether overexpression of UBP1 or RBP45 could increase the splicing efficiency of plant U12 introns in tobacco protoplasts, constructs expressing HA-tagged UBP1 or RBP45 were cotransfected with the three U12 introns (Figure 6). As controls, RT-PCR and protein gel blot analysis with antibody to the HA epitope tag were performed to demonstrate the presence of transcripts and protein produced from the transfected U-rich RNA binding protein constructs (data not shown). Overexpression of these proteins did not affect the splicing efficiency of any of the U12 introns (Figure 6). However, overexpression of UBP1 with the wheat (Triticum aestivum) amylase U2 intron, which has a relatively low UA content and splices poorly (5 to 15%) in tobacco protoplasts (Simpson et al., 1996), increased splicing efficiency to 85% (Figure 6). Thus, whereas UBP1 can enhance splicing efficiency of poorly spliced U2 introns, confirming the findings of Lambermon et al. (2000), neither UBP1 nor RBP45 influenced U12 intron splicing directly.
Figure 6.
Effect of Overexpression of U-Rich Binding Proteins RBP45 and UBP1 on Splicing of U2 and U12 Introns.
Splicing efficiencies of U12 introns in the absence and presence of overexpressed RBP47 and UBP1 are shown. As a control, the effect of RBP45 and UBP1 on splicing of a poorly spliced U2 intron (Amy U2) was tested.
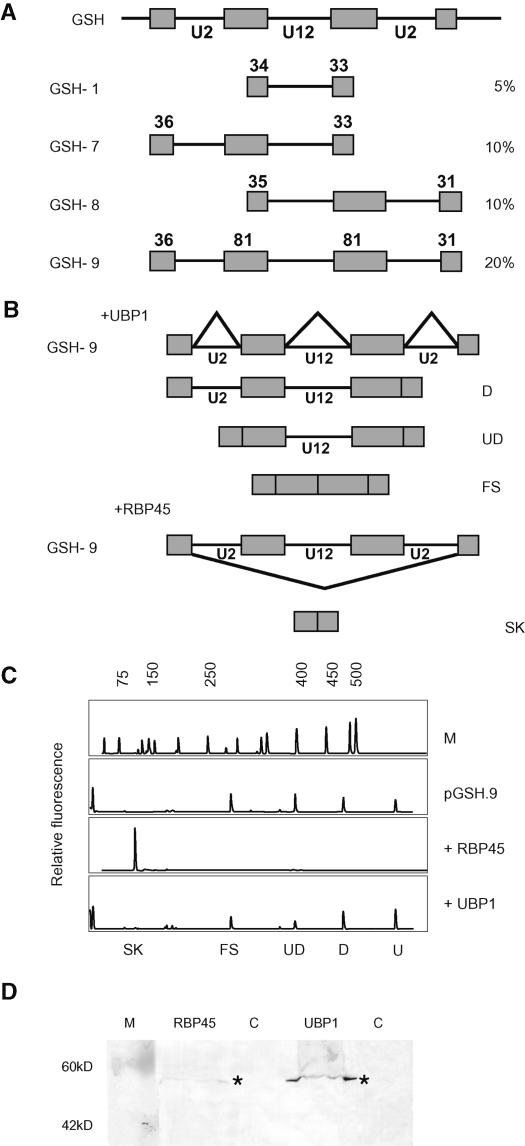
Previously, exon bridging interactions between adjacent U2 introns were shown to enhance splicing of plant pre-mRNAs (McCullough et al., 1996; Simpson et al., 1998, 1999), and flanking U2 introns increased splicing efficiency of the CBP20 U12 intron (Kmieciak et al., 2002). Overexpression of UBP1 or RBP45 did not enhance U12 intron excision in single intron constructs, but may function in intron recognition and recruitment of splicing factors to splice sites via exon bridging interactions. To examine whether UBP1 or RBP45 could enhance U12 intron splicing in such a transcript, a construct with the poorly spliced GSH2 U12 intron and its two authentic flanking U2 introns was made (pGSH.9; Figure 7A). In addition, two intermediate constructs with only the 5′ or the 3′ flanking U2 intron were prepared (pGSH.7 and pGSH.8, respectively). These latter two constructs showed increases in splicing efficiency of the U12 intron from 5 to ∼10%. When the pGSH.9 construct was expressed in tobacco protoplasts, there were four different RT-PCR products, each with similar abundance of ∼20 to 25% (Figures 7B and 7C). These products were characterized by sequencing and represented the unspliced transcript, 564 bp; partially spliced transcripts, 479 bp long, where the downstream U2 intron is removed; a partially spliced transcript, 399 bp long, where both U2 introns are removed; and fully spliced transcripts, where both U2 and the U12 intron are removed (290 bp) (Figures 7B and 7C). There was no evidence of partially spliced transcripts, where only the upstream U2 intron had been spliced, suggesting that the pathway of intron removal is splicing of the downstream U2 intron first, followed by the upstream U2 intron and then the U12 intron. The presence of both U2 introns in the triple intron construct increased splicing efficiency of the U12 intron to 20% compared with the 5% efficiency in the single intron construct. This suggested that the splicing efficiency of the U12 intron was enhanced by exon bridging interactions between flanking U2 introns, as observed previously for the CBP20 U12 intron (Kmieciak et al., 2002).
Figure 7.
RBP45 Causes Exon Skipping in the Complex GSH Intron Construct.
(A) Constructs of the GSH2 U12 intron with flanking U2 introns are shown diagrammatically under the wild-type GSH2 exon-intron structure. GSH.1, U12 intron alone; GSH.7, U12 intron and upstream U2 intron; GSH.8, U12 intron and downstream U2 intron; GSH.9, U12 intron with both upstream and downstream U2 introns.
(B) Splicing of GSH.9 construct gave three different products (in addition to unspliced transcripts) where either the downstream U2 intron was removed (D) or where both U2 introns were removed (UD) and the fully spliced product (FS). Overexpression of UBP1 did not alter the splicing pattern of GSH.9, but overexpression of RBP45 caused exon skipping (SK).
(C) GeneScan analysis of splicing of GSH.9 alone and with overexpressed RBP45 or UBP1 proteins in tobacco protoplasts. M, DNA size markers.
(D) Protein gel blot analysis with anti-HA antibody of HA-tagged RBP45 and UBP1 proteins overexpressed in tobacco protoplasts. Protein bands are indicated with asterisks. C, control untransfected protoplasts.
To examine the effect of overexpression of UBP1 and RBP45 on exon-bridging interactions, the three-intron construct, pGSH.9, was cotransfected with HA-tagged UBP1 or RBP45. Overexpression of UBP1 with pGSH.9 in tobacco protoplasts had little effect on splicing pattern or efficiency. However, overexpression of RBP45 completely abolished the normal splicing pattern, and transcripts showed excision of all three introns and both exons in an exon skipping event, giving an RT-PCR product of 126 bp (Figures 7B and 7C). In addition, no unspliced product was visible (Figure 7C). Therefore, RBP45 increased the overall splicing efficiency of the transcript but blocked recognition of the internal GSH2 exon sequences. Thus, neither UBP1 nor RBP45 enhances splicing of U12 introns, but RBP45 can influence splice site selection in pre-mRNAs. This, and the ability of UBP1 to enhance removal of poorly spliced U2 but not U12 introns, demonstrates differential roles for UBP1 and RBP45 in pre-mRNA processing, which may reflect specificity of interaction with different sequences in pre-mRNAs.
The High Splicing Efficiency of the LD Intron Is Because of Exon Splicing Enhancer Activity
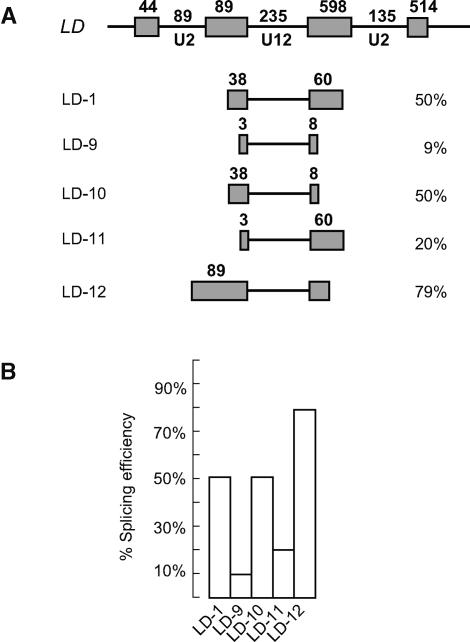
The LD U12 intron is spliced much more efficiently in tobacco protoplasts than the CBP20 or GSH2 introns. As we have already shown, this enhanced splicing efficiency is not because of splice site dinucleotides, branchpoint sequences, or UA content. To examine whether the higher splicing efficiency of this intron was because of sequences in the flanking exons, the length of the 5′, 3′, or both flanking exons was reduced. In pLD.1, the upstream exon contained 38 nucleotides of the authentic 89 nucleotides, whereas the downstream exon consisted of 60 nucleotides from the original 579 nucleotides (Figure 8A). In pLD.9, both upstream and downstream exons were reduced to 3 and 8 bp, respectively. Intermediate constructs with 3 bp upstream and 60 bp downstream exons and 38 bp upstream and 8 bp downstream exons were also generated (pLD.10 and pLD.11). The splicing efficiency of both pLD.9 and pLD.11 dropped significantly to 9 and 20%, respectively, compared with the 50% splicing of the original pLD.1 construct (Figures 8A and 8B). Splicing of pLD.10 remained at 50% efficiency, indicating that sequence(s) between 38 and 3 nucleotides in the upstream exon are important for the higher level of splicing of the LD U12 intron in tobacco protoplasts. Furthermore, when the full-length upstream exon was included (Figures 8A and 8B, pLD.12), splicing efficiency of the LD U12 intron increased to 79%, suggesting that the region between 89 and 38 nucleotides also contained sequences that increased splicing efficiency. Thus, in the case of the LD U12 intron, splicing in tobacco protoplasts required the complete upstream exon (exon 10) sequence for maximal splicing efficiency, and there are potentially two regions that contribute to this enhancement.
Figure 8.
Exon 3 of LD Contains Exon Splicing Enhancer Sequences.
(A) Diagrams of LD U12 intron constructs with differing lengths of flanking exons (LD-1 and LD-9 to LD-12) are shown below the wild-type exon-intron structure of part of the LD gene.
(B) Histogram of splicing efficiencies of the LD constructs in tobacco protoplasts.
DISCUSSION
In the Arabidopsis genome, 165 U12 introns have been predicted by computational analyses, including 113 GT-AG, 50 AT-AC, 1 AT-AA, and 1 GT-AT introns. These introns share similar features with plant U2 introns in terms of length distribution and low GC content (Zhu and Brendel, 2003; our unpublished results). In the absence of a plant in vitro splicing system, we have performed a detailed analysis of the determinants of U12 intron splicing in plants using an in vivo protoplast system. Through extensive mutational analysis, we have examined the influence of splice site and branchpoint signals on splicing efficiency and have identified the likely branchpoint nucleotide used in three plant U12 introns. In particular, we have shown that U12 intron splicing is enhanced by increased UA richness, a characteristic feature of plant U2 and U12 introns, and by the presence of flanking U2 introns, supporting the role of exon bridging interactions between U2 and U12 introns. Finally, exon splicing enhancer sequences may modulate splicing efficiency of U12 introns, raising the possibility of regulation of gene expression of U12 intron–containing genes at the level of pre-mRNA splicing.
Differential Splicing Efficiency of U12 Introns
Two observations were made about splicing efficiency of the plant U12 introns in tobacco protoplasts. First, when introduced as single intron constructs, the efficiency of splicing of all three introns was lower than normally observed for U2 introns. Second, there were significant differences in splicing efficiency among the LD, GSH, and CBP20 introns. In the protoplast system, the U12-containing transcripts were overexpressed from the strong CaMV 35S promoter. This could suggest that transfected protoplasts were not competent to splice the U12 introns efficiently because U12 spliceosomes and/or other factors involved in U12 intron splicing were limiting. In the same protoplast system, U2 intron–containing transcripts expressed from the CaMV 35S promoter were efficiently spliced, possibly reflecting the much higher abundance of U2 spliceosomes. However, the LD U12 intron construct containing the complete 5′ exon (LD-12) was spliced with 79% efficiency, arguing against the simple interpretation that lower splicing efficiency reflects the lower abundance of U12 splicing machinery components. The efficiency of splicing of pLD.12 demonstrates that the protoplast system has the inherent capability to splice U12 introns efficiently; therefore, the observed differences between LD, CBP20, and GSH2 U12 intron–containing pre-mRNAs must reflect variations in splice signal sequences and their ability to interact with splicing factors.
Plant U12 Splice Site Signals
From mutational and computational analyses of vertebrate U12-type introns, it is clear that both AU-AC and GU-AG termini function as U12 intron splice sites (Dietrich et al., 1997). On the basis that the LD U12 intron was spliced with much higher efficiency than those of CBP20 or GSH2 and that LD had GU-AG splice sites, it was possible that the terminal dinucleotides could influence efficiency of plant U12 intron splicing. However, mutational analyses demonstrated that splicing efficiency of plant U12-type introns was unchanged, irrespective of whether the terminal dinucleotides were AU-AC or GU-AG. Moreover, our results indicated that the plant U12-type spliceosome was not able to use the GU-AC–terminal dinucleotide combination, reflecting the lack of plant U12 introns with this sequence (Zhu and Brendel, 2003). The LD AU-AG mutant had reduced splicing, similar to vertebrate mutant U12 introns with AU-AG termini, which splice less efficiently than either AU-AC or AU-AA introns (Wu and Krainer, 1999; Dietrich et al., 2001a).
In the absence of plant in vitro splicing extracts, branchpoint nucleotides required for splicing were examined by in vivo analysis of branchpoint mutations of the three plant U12 introns, as performed previously for plant U2 introns (Simpson et al., 1996, 2000, 2002). The vertebrate and plant consensus branchpoint sequence is UCCUURAY. In vertebrates, the branchpoint nucleotide of the U12 intron in the human P120 gene was mapped to the adenosine (underlined) (Tarn and Steitz, 1996a). However, some plant and vertebrate U12 introns contain a G or U at this position, with an adenosine immediately upstream, and recently, vertebrate U12 intron branchpoints have been mapped to adenosines in either position (McConnell et al., 2002). For example, the branchpoints of two human U12 introns (from SME and XRP) were mapped by primer extension to the adenosine in the sequence UCCUUAGC, whereas in the U12 introns from XTF and P120, branchpoint adenosines mapped to the downstream adenosine in the sequences UCCUUAAA and UCCUUAAC, respectively. Thus, in vertebrates, there is flexibility in branchpoint nucleotide selection, where an A at either position can function as the branchpoint adenosine (McConnell et al., 2002). Plant U12 introns contain an adenosine at one or both purine positions, suggesting that such flexibility may also occur in plant U12 intron splicing (Zhu and Brendel, 2003; our unpublished results). Alternative base-pairing interactions between the U12snRNA and branchpoint sequence (Figure 4D) could cause the purine at either position to be bulged from the duplex and used as the branchpoint nucleotide, as demonstrated for vertebrate U2 introns (Figure 4D; Query et al., 1994). Nevertheless, mutation of the A to U in the upstream position of all three plant U12 introns inhibited splicing. This suggests that either a uridine in this position is not tolerated, perhaps disrupting U12snRNA–branchpoint interactions, or the upstream adenosine is the preferred branchpoint nucleotide. Finally, in the four human introns (above), the use of the upstream or downstream adenosine correlated with their terminal dinucleotides, GU-AG or AU-AC (Tarn and Steitz, 1996a; McConnell et al., 2002), but this correlation did not hold for the plant introns studied here.
A further significant feature of U12 splicing is the short distance between the branchpoint and 3′ splice site and the lack of a polypyrimidine tract (Dietrich et al., 2001a; Levine and Durbin, 2001). In the LD U12 intron, the authentic 3′ splice site lies 11 nucleotides downstream of the branchpoint. When mutated (GU-AC), two cryptic 3′ splice site -AGs, nine nucleotides downstream or 3 nucleotides upstream of the mutated 3′ splice site, were activated. These 3′ splice sites were positioned 20 and 8 nucleotides from the branchpoint and were selected in 95 and 5% of transcripts, respectively. The frequencies of selection are consistent with the range of branchpoint to 3′ splice site distances between 10 and 20 nucleotides (with an optimum of 12 to 13 nucleotides) proposed for vertebrate U12 introns and with the observation that when this spacing was altered to <10 nucleotides, normal splicing ceased and cryptic splicing was often activated (Dietrich et al., 2001a). In addition, none of the 165 plant U12 introns had branchpoint to 3′ splice distances <11 nucleotides (mean of 12 nucleotides), and only 12 putative plant U12 introns were reported with a branchpoint to 3′ splice site distance >21 nucleotides (Zhu and Brendel, 2003). Therefore, branchpoint/3′ splice site distance is an important determinant of plant U12 intron 3′ splice site selection. In addition, seven examples of alternative splicing involving plant U12 introns were identified underpinning the potential regulation of splicing and expression of genes containing U12 introns (Zhu and Brendel, 2003).
Splice site selection in both plant and vertebrate U2 introns involves exon and/or intron definition. In the exon definition model, splicing factors recognize splice sites at either end of an exon and assemble a complex on the exon; neighboring exons are then brought together to allow the interaction of 5′ and 3′ splice sites and intron removal. In vertebrates, exon definition has been demonstrated between a U12 intron and a downstream U2 intron (Wu and Krainer, 1996). We have shown that both upstream and downstream U2 introns can increase splicing efficiency of two plant U12 introns: GSH2 (this article) and CBP20 (Kmieciak et al., 2002).
UA Richness and U-Rich RNA Binding Proteins
The increased splicing efficiency as a result of inclusion of two or three U-rich elements in the CBP20 U12 intron parallels the stimulatory effect of such sequences on splicing efficiency of U2 introns (Goodall and Filipowicz, 1989; Gniadkowski et al., 1996). However, splicing efficiency did not increase to the levels observed with some U2 introns. For plant U2 introns, UA-rich binding proteins are thought to function early in spliceosome assembly, to bind to U-rich elements within the intron, and to stabilize and recruit snRNPs and other splicing factors. The only partial improvement in splicing of the U12 intron by insertion of the U-rich sequences therefore may be because of (1) a suboptimal arrangement of U-rich elements within the intron, (2) different functional roles for U-rich elements in U12 and U2 intron splicing, or (3) UA-rich binding factors interacting differentially with U12 and U2 splicing machinery components. Thus, both types of intron may depend on U richness in different ways, and/or different U-rich binding proteins may have specific roles in U2 and U12 intron recognition and splicing. These variations may reflect the need to distinguish between U2 and U12 spliceosomal snRNPs and to recruit, for example, the U11-U12 di-snRNP, which interacts with the U12 5′ splice site and branchpoint (Sharp and Burge, 1997).
In this regard, the two highly related plant U-rich RNA binding proteins, UBP1 and RBP45, showed different effects on U2 and U12 intron splicing. Neither of the proteins affected splicing activity of the three plant U12 introns, but overexpression of UBP1 enhanced splicing of the poorly spliced amylase U2 intron, confirming a role for UBP1 in plant U2 splicing (Lambermon et al., 2000; Lorković et al., 2000). These results suggest that the interactions whereby U2 intron splicing was improved by UBP1 were absent from U12 intron splicing and highlight differences between plant U2 and U12 splicing, most likely in early events in intron recognition and spliceosome assembly. Overexpression of RBP45 had little effect on single intron constructs but affected the splicing efficiency and pattern of a more complex transcript containing the U12 intron flanked by U2 introns. The exon-skipping phenotype, suggesting disruption of exon bridging interactions, was specific to RBP45 overexpression because the related protein, UBP1, did not have this effect. It remains to be determined whether the effect of RBP45 overexpression is specific to U12-containing transcripts, but these results clearly demonstrate differential functions for these closely related proteins, probably reflecting different binding affinities of these proteins.
U12 Introns and Regulation of Gene Expression
U12 introns are often found in genes that function in DNA replication and RNA metabolism, and in some cases, the presence and positions of U12 introns are conserved across species, suggesting a role for U12 introns in the regulation of expression of these genes (Burge et al., 1998). In vivo splicing of human U12 introns has been shown to proceed more slowly than splicing of U2 introns in the same transcript, which may reflect the lower abundance of the U12 spliceosome or a slower, less efficient splicing reaction (Patel et al., 2002). Furthermore, it has been postulated that U12 intron removal is a rate-limiting step for complete splicing of U12 intron–containing transcripts, with partially spliced transcripts being targeted for degradation. This would maintain appropriate expression levels that could be regulated by the levels of factors involved in U12 intron splicing (Patel et al., 2002). In in vitro extracts, splicing of U12 introns is also slower than that of U2 introns (Tarn and Steitz, 1996b; Wu and Krainer, 1996). Consistent with this mechanism of gene expression regulation, splicing of plant U12 introns, in vivo in tobacco protoplasts, also occurred more slowly or less efficiently than splicing of U2 introns from transcripts containing both U12 and U2 introns (see Kmieciak et al., 2002). In addition, preliminary results showed different efficiencies of GSH2 U12 intron splicing in different organs of Arabidopsis, being highest in leaves and lowest in roots and flowers (our unpublished results). Thus, it is feasible that plant U12 introns represent one level at which U12 intron–containing genes are regulated posttranscriptionally.
Splicing enhancers play a critical role in the regulation of splicing and in correct splice site recognition of constitutively spliced pre-mRNAs (Hertel et al., 1997; Wang and Manley, 1997; Hertel and Maniatis, 1998; Blencowe, 2000; Smith and Valcárcel, 2000; Graveley, 2001, 2002; Cartegni et al., 2002; Black, 2003). Although they are usually located within the downstream exon, enhancer sequences also have been observed in upstream exons or within the intron itself (Blencowe, 2000). Exon splicing enhancers (ESEs) are often purine rich and are believed to function through the binding of specific protein factors. The best described of these factors are the SR proteins, a large family of polypeptides with one or more RNA binding domains and a variable length domain (the RS domain) containing multiple copies of Arg-Ser dipeptides (Fu, 1995; Valcárcel and Green, 1996; Wang and Manley, 1997; Tacke and Manley, 1999; Black, 2003). Although purine-rich enhancers were identified in many exons flanking U2-dependent introns, it already has been demonstrated, both in vitro and in vivo, that ESEs of this type are also functional in the U12 pre-mRNA splicing pathway (Wu and Krainer, 1998; Graveley, 2000; Dietrich et al., 2001b; Hastings and Krainer, 2001). Many SR proteins have been identified in plants (Lazar et al., 1995; Lopato et al., 1996a, 1996b, 1999a, 1999b, 2002), but to date, only one purine-rich exonic element, which promotes 5′ splice site selection, has been described (McCullough and Schuler, 1997). In this article, we have shown that the exon upstream of the LD U12 intron clearly contains two separate regions that increased the efficiency of splicing. Searching this exon sequence for identified vertebrate ESEs and binding sites of SR proteins (http://www.exon.cshl.org/ESE/) revealed two such sequences that might support SR protein binding. Thus, SR protein (or other similar factors) binding to the exon located upstream of the LD U12-type intron may stimulate recognition of the 5′ splice site. In addition, the LD upstream exon also contained a sequence that has been experimentally demonstrated to act as an ESE in Arabidopsis (S. Mount, personal communication). It is possible that SR (or other) proteins, which recognize ESEs in upstream or downstream exons flanking U12 introns, could contact a component of the U11/U12 di-snRNP, stabilizing its binding to the 5′ splice site (U11) and branchpoint (U12) sequences and promoting U12 spliceosome formation. Interestingly, the LD intron has the lowest UA content of those tested but splices with the highest efficiency and therefore may strongly depend on ESEs for regulation of U12 intron splicing. Further investigations are required to determine whether the ESEs are intron specific or can function in other U12 or U2 splicing pathways. Thus, as with plant U2 introns whose splicing efficiency reflects the strength of various signals, splicing of plant U12 introns depends on both intronic and exonic signals (UA content and ESEs) and exon bridging interactions with flanking U2 introns.
METHODS
Isolation of U12 Introns and Construction of Mutants
The Arabidopsis thaliana U12 introns from CBP20 (intron 4) (Kmieciak et al., 2002), GSH2 (intron 6) (Wang and Oliver, 1996), and LD (intron 10) (Lee et al., 1994) and varying amounts of upstream and downstream intron and exon sequences were isolated from Arabidopsis genomic DNA by PCR (Figures 1 and 2B). Fragments were inserted into the unique BamHI site of expression vector pDH515 (Figure 2A; Simpson et al., 1996). The basic set of constructs consisted of the U12 introns from each of the three genes CBP20, with 34 and 93 nucleotides of upstream and downstream exons (Figure 2B; pCBP20.1), GSH2, with 34 and 33 nucleotides of upstream and downstream exons (Figure 2B; pGSH.1), and LD, with 38 and 60 nucleotides of upstream and downstream exons (Figure 2B; pLD.1). PCR amplification of genomic Arabidopsis DNA was also performed to make GSH2 constructs that contained the upstream-associated U2 intron (pGSH.7; Figure 7A), downstream-associated U2 intron 9 (pGSH.8; Figure 7A), and a construct that contained the U12 intron with both upstream and downstream U2 introns (pGSH.9; Figure 7A). PCR amplification of pLD.1 was performed to reduce the length of both upstream (pLD.9 and pLD.11; Figure 8A) and downstream (pLD.9 and pLD.10; Figure 8A) exon sequences. Genomic Arabidopsis DNA was PCR amplified to extend the length of the upstream exon to its full length of 89 nucleotides (pLD.12; Figure 8A). Site-specific nucleotide substitutions to change the 5′ and 3′ splice site dinucleotides of pLD.1 and pGSH.1 (Figure 3A) and putative branchpoint nucleotides of pLD.1, pCBP20.1, and pGSH.1 (Figure 4A) were performed by site-directed mutagenesis using the Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. One, two, and three copies of the UUUUUAU sequence element (Gniadkowski et al., 1996) were inserted into pCBP20.1, 67 nucleotides downstream from the 5′ splice site and 67 nucleotides upstream of the 3′ splice site, using the Quick-Change site-directed mutagenesis kit (pCBP20.8, pCBP20.9, and pCBP20.10, respectively; Figure 5A). Site-directed mutagenesis was performed on pCBP20.9 to create a pyrimidine-rich control construct (pCBP20.11; Figure 5A). All sequence insertions and mutations were confirmed by sequencing. A list of all the oligonucleotides used for isolation of intron/exon fragments and for mutagenesis is available upon request.
Splicing Analysis by RT-PCR
Intron constructs were transfected into Nicotiana tabacum var Xanthi, and total RNA was isolated as described previously (Simpson et al., 1996). RT-PCR analysis was as described (Simpson et al., 2000) using the PCR primers O8, 5′-CCCAATTGTTCAACCCTAC-3′, labeled with the 5′ fluorescent phosphoramidite 6-FAM, and O9, 5′-GGTAAGATGCCTGTTGCGATTGC-3′. The primer O8 corresponds to the zein sequence 5′ to the site of intron construct insertion, and O9 is complementary to the zein sequence 3′ to the site of intron construct insertion in pDH515 (Simpson et al., 1996). Labeled RT-PCR products were separated on a 4% polyacrylamide denaturing gel on an ABI 377 DNA sequencing machine. Sizes of bands were calculated using GeneScan version 2.1 software by comparison with GeneScan-350 (TAMRA) size standards (Applied Biosystems, Foster City, CA). Quantification of RT-PCR products was by measurement of the fluorescent peak areas of the detected fragments after 24 cycles. Previous quantification established that PCR amplification was linear over a range of 15 to 24 cycles, and Taq polymerase enzyme efficiency was the same for spliced, partially spliced, and unspliced products. Splicing efficiency was calculated from the peak areas for each processed transcript. Each construct was tested at least twice, and standard errors were determined for constructs that were tested three or more times. Standard errors for the poorly spliced GSH2 and CBP20 U12 intron constructs were all <±2%, and for the LD intron, constructs were <±6%. Novel RT-PCR products (e.g., from cryptic splicing) were isolated from a 6% nondenaturing acrylamide gel and eluted from the gel overnight at 4°C in 10 mM Tris-HCl, pH 8, 1 mM EDTA, 0.5 M ammonium acetate, and 0.1% (w/v) SDS overnight at 4°C. The eluted bands were precipitated and resuspended in water before reamplification with oligonucleotides O8 and O9. These PCR products were either cloned into pGEM-T Easy (Promega, Madison, WI) and sequenced using the standard RS and SS primers, or the PCR product was sequenced directly using O8 and O9.
Cotransfection Analysis
HA-tagged U-rich RNA binding protein expression cassettes, pUBP1-HA and pRBP45-HA (Lambermon et al., 2000; Lorković et al., 2002), were mixed separately with an equal molar amount of pCBP20.1, pGSH.1, pGSH.9, pLD.1, and pA, respectively. Protoplasts were transfected with the plasmid mixtures, and splicing analysis was performed as described above. Expression of the tagged proteins was monitored by protein gel blot and RT-PCR analyses. Protein was extracted from protoplasts by boiling in 50 mM Tris-HCl, pH 6.8, 20% (v/v) glycerol, 1 mM EDTA, 1% (w/v) SDS, and 15% (v/v) β-mercaptoethanol and bromophenol blue. Protein gel blot analysis was performed using rabbit anti-HA antibody (Sigma, St. Louis, MO) as primary antibody and anti-rabbit IgG alkaline phosphatase conjugate (Sigma) as secondary antibody to confirm protein expression in protoplasts. RT-PCR analysis was performed using primers specific to the HA-tagged protein transcripts.
Acknowledgments
This research was supported by the Scottish Executive Environment and Rural Affairs Department, the Polish Committee for Scientific Research (Grants 0265/P04/2001/21 and 0045/P04/2002/23), and the Royal Society. We thank Witek Filipowicz (Freidrich Miescher Institute, Basel, Switzerland) for the gift of pUBP1-HA and pRBP45-HA expression cassettes.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Craig G. Simpson (csimps@scri.sari.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020743.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Black, D.L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. (2000). Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25, 106–110. [DOI] [PubMed] [Google Scholar]
- Brown, J.W.S., and Simpson, C.G. (1998). Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 77–95. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Padgett, R.A., and Sharp, P.A. (1998). Evolutionary fates and origins of U12-type introns. Mol. Cell 2, 773–785. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. (2002). Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.C., Incorvaia, R., and Padgett, R.A. (1997). Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell 1, 151–160. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.C., Peris, M.J., Seyboldt, A.S., and Padgett, R.A. (2001. a). Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 21, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.C., Shukla, G.C., Fuller, J.D., and Padgett, R.A. (2001. b). Alternative splicing of U12-dependent introns in vivo responds to purine-rich enhancers. RNA 7, 1378–1388. [PMC free article] [PubMed] [Google Scholar]
- Frilander, M.J., and Steitz, J.A. (1999). Initial recognition of U12-dependent introns requires both U11/5′ splice site and U12/branchpoint interactions. Genes Dev. 13, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.-D. (1995). The superfamily of arginine/serine-rich splicing factors. RNA 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- Gniadkowski, M., Hemmings-Mieszcak, M., Klahre, U., Liu, H.-X., and Filipowicz, W. (1996). Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Res. 24, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall, G.J., and Filipowicz, W. (1989). The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58, 473–483. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. (2001). Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17, 100–107. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. (2002). Sex, agility, and the regulation of alternative splicing. Cell 109, 409–412. [DOI] [PubMed] [Google Scholar]
- Hall, S.L., and Padgett, R.A. (1994). Conserved sequences in a class of rare eukaryotic introns with non-consensus splice sites. J. Mol. Biol. 239, 357–365. [DOI] [PubMed] [Google Scholar]
- Hall, S.L., and Padgett, R.A. (1996). Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- Hastings, M.L., and Krainer, A.R. (2001). Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13, 302–309. [DOI] [PubMed] [Google Scholar]
- Hertel, K.J., Lynch, K.W., and Maniatis, T. (1997). Common themes in the fuction of transcription and splicing enhancers. Curr. Opin. Cell Biol. 9, 350–357. [DOI] [PubMed] [Google Scholar]
- Hertel, K.J., and Maniatis, T. (1998). The function of multisite splicing enhancers. Mol. Cell 1, 449–455. [DOI] [PubMed] [Google Scholar]
- Incorvaia, R., and Padgett, R.A. (1998). Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA 4, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, I.J. (1991). A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmieciak, M., Simpson, C.G., Lewandowska, D., Brown, J.W.S., and Jarmolowski, A. (2002). Cloning and characterization of two subunits of Arabidopsis thaliana nuclear cap-binding complex. Gene 283, 171–183. [DOI] [PubMed] [Google Scholar]
- Kolossova, I., and Padgett, R.A. (1997). U11 interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) introns. RNA 3, 227–233. [PMC free article] [PubMed] [Google Scholar]
- Lambermon, M.H.L., Simpson, G.G., Kirk, D.A., Hemmings-Mieszczak, M., Klahre, U., and Filipowicz, W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19, 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, G., Schaal, T., and Maniatis, T. (1995). Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl. Acad. Sci. USA 92, 7672–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Aukerman, M.J., Gore, S.L., Lohman, K.N., Michaels, S.D., Weaver, L.M., John, M.C., Feldmann, K.A., and Amasino, R.M. (1994). Isolation of LUMINIDEPENDENS: A gene involved in the control of flowering time in Arabidopsis. Plant Cell 6, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A., and Durbin, R. (2001). A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 29, 4006–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Forstner, C., Kalyna, M., Hilscher, J., Langhammer, U., Indrapichate, K., Lorković, Z.J., and Barta, A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277, 39989–39998. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Gattoni, R., Fabini, G., Stevenin, J., and Barta, A. (1999. a). A novel family of plant splicing factors with a Zn knuckle motif: Examination of RNA binding and splicing activities. Plant Mol. Biol. 39, 761–773. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Kalyna, M., Dorner, S., Kobayahshi, R., Krainer, A.R., and Barta, A. (1999. b). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Mayeda, A., Krainer, A.R., and Barta, A. (1996. a). Pre-mRNA splicing in plants: Characterization of Ser/Arg splicing factors. Proc. Natl. Acad. Sci. USA 93, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Waigmann, E., and Barta, A. (1996. b). Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell 8, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković, Z.J., Kirk, D.A.W., Lambermon, M.H.L., and Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5, 160–167. [DOI] [PubMed] [Google Scholar]
- Lorković, Z.J., Wieczorek Kirk, D.A., Lambermon, M.H.L., and Filipowicz, W. (2002). RBP45 and RBP47, two olgouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6, 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H.R., Moreau, G.A., Levin, N., and Moore, M.J. (1999). The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 5, 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, T.S., Cho, S.-J., Frilander, M.J., and Stietz, J.A. (2002). Branchpoint selection in the splicing of U12-dependent introns in vitro. RNA 8, 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, A.J., Baynton, C.E., and Schuler, M.A. (1996). Interactions across exons can influence splice site recognition in plant nuclei. Plant Cell 8, 2295–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, A.J., and Schuler, M.A. (1997). Intronic and exonic sequences modulate 5′ splice site selection in plant nuclei. Nucleic Acids Res. 25, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A., McCarthy, M., and Steitz, J.A. (2002). The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 21, 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A., and Steitz, J.A. (2003). Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 4, 960–970. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Moore, M.J., and Sharp, P.A. (1994). Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes Dev. 8, 587–597. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001). Nuclear pre-mRNA splicing in plants. Crit. Rev. Plant Sci. 20, 523–571. [Google Scholar]
- Reed, R. (2000). Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12, 340–345. [DOI] [PubMed] [Google Scholar]
- Schneider, C., Will, C.L., Makarova, O.V., Makarov, E.M., and Lührmann, R. (2002). Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol. 22, 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.A., and Burge, C.B. (1997). Classification of introns: U2-type or U12-type. Cell 91, 875–879. [DOI] [PubMed] [Google Scholar]
- Shukla, G.C., and Padgett, R.A. (1999). Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA 5, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, G.C., and Padgett, R.A. (2001). The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA 7, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.G., Clark, G.P., Davidson, D., Smith, P., and Brown, J.W.S. (1996). Mutation of putative branchpoint consensus sequences in plant introns reduces splicing efficiency. Plant J. 9, 369–380. [DOI] [PubMed] [Google Scholar]
- Simpson, C.G., Clark, G.P., Lyon, J.M., Watters, J., McQuade, C., and Brown, J.W.S. (1999). Interactions between introns via exon definition in plant pre-mRNA splicing. Plant J. 18, 293–302. [Google Scholar]
- Simpson, G.G., and Filipowicz, W. (1996). Splicing of precursors to messenger RNA in higher plants: Mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32, 1–41. [DOI] [PubMed] [Google Scholar]
- Simpson, C.G., Hedley, P.E., Watters, J.A., Clark, G.P., McQuade, C., Machray, G.C., and Brown, J.W.S. (2000). Requirements for mini-exon inclusion in potato invertase mRNAs provides evidence for exon-scanning in plants. RNA 6, 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.G., McQuade, C., Lyon, J., and Brown, J.W.S. (1998). Characterization of exon skipping mutants of the COP1 gene from Arabidopsis. Plant J. 15, 125–131. [DOI] [PubMed] [Google Scholar]
- Simpson, C.G., Thow, G., Clark, G.P., Jennings, S.N., Watters, J.A., and Brown, J.W.S. (2002). Mutational analysis of a plant branchpoint and polypyrimidine tract required for constitutve splicing of a mini-exon. RNA 8, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.W.J., and Valcárcel, J. (2000). Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Tacke, R., and Manley, J.L. (1999). Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11, 358–362. [DOI] [PubMed] [Google Scholar]
- Tarn, W.-Y., and Steitz, J.A. (1996. a). A novel spliceosome containing U11, U12, and U5snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84, 801–811. [DOI] [PubMed] [Google Scholar]
- Tarn, W.-Y., and Steitz, J.A. (1996. b). Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Tarn, W.-Y., and Steitz, J.A. (1997). Pre-mRNA splicing: The discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 22, 132–137. [DOI] [PubMed] [Google Scholar]
- Valcárel, J., and Green, M.R. (1996). The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 21, 296–301. [PubMed] [Google Scholar]
- Wang, C.L., and Oliver, D.J. (1996). Cloning of the cDNA and genomic clones for glutathione synthetase from Arabidopsis thaliana and complementation of a gsh2 mutant in fission yeast. Plant Mol. Biol. 31, 1093–1104. [DOI] [PubMed] [Google Scholar]
- Wang, J., and Manley, J.L. (1997). Regulation of pre-mRNA splicing in metazoa. Curr. Opin. Genet. Dev. 7, 205–211. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., MacMillan, A.M., Katopodis, N.F., Neubauer, G., Wilm, M., Lührmann, R., and Query, C.C. (2001). A novel U2 and U11/U12 protein that associates with the pre-mRNA branch site. EMBO J. 20, 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., Reed, R., and Lührmann, R. (1999). Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284, 2003–2005. [DOI] [PubMed] [Google Scholar]
- Wu, H.J., Gaubier-Comelia, P., Delseny, M., Grellet, F., Van Montagu, M., and Rouzé, P. (1996). Non-canonical introns are at least 109 years old. Nat. Genet. 14, 383–384. [DOI] [PubMed] [Google Scholar]
- Wu, Q., and Krainer, A.R. (1996). U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science. 274, 1005–1008. [DOI] [PubMed] [Google Scholar]
- Wu, Q., and Krainer, A.R. (1998). Purine-rich enhancers function in the AT-AC pre-mRNA splicing pathway and do so independently of intact U1snRNP. RNA 4, 1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q., and Krainer, A.R. (1999). AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol. 19, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.-T., and Steitz, J.A. (1997). Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc. Natl. Acad. Sci. USA 94, 6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., and Brendel, V. (2003). Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res. 31, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]