Abstract
Objective
Periodontal pathogens in dental plaque are the main causative agents of periodontitis and peri-implantitis. Detection of the presence of such periodontal pathogens early would serve as a useful tool in the diagnosis and treatment of this disease. Therefore, the purpose of this study was to investigate whether the periodontal pathogen levels in saliva were correlated with the periodontal status of patients receiving implant treatment.
Materials and Methods
A total of 291 patients visiting Tokyo Dental College Chiba Hospital were divided into four groups: a no-periodontitis (np) group, a mild-periodontitis (mip) group, a moderate-periodontitis (mop) group, and a severe-periodontitis (sp) group. The levels of the following five periodontal pathogens in saliva were evaluated using real-time polymerase chain reaction: Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Tannerella forsythia, Treponema denticola, and Prevotella intermedia.
Results
The levels of P. gingivalis and T. forsythia were significantly higher in mop group than in np group (P < 0.05). The levels of all periodontal pathogens tested except A. actinomycetemcomitans were significantly higher in sp group than in np group (P < 0.05).
Conclusion
The detection levels of the periodontal pathogens targeted in saliva samples were correlated with the periodontal status. This suggests that using saliva to screen for periodontopathic bacteria offers an easier-to-use clinical tool than the paper point method in the diagnosis and treatment of periodontitis and peri-implantitis.
Keywords: clinical research, diagnosis, microbiology, periodontology
Periodontitis is a multibacterial infection that affects the periodontal tissues and eventually leads to loss of teeth. Periodontitis is a chronic inflammatory disease, and its bacterial etiology has been confirmed by numerous reports (Listgarten & Hellden 1978; Tanner et al. 1979; Moore 1987; Paster et al. 2001). It appears essential, however, to screen for periodontal pathogens that could directly destroy the periodontal tissue. Currently, microbial tests are used to identify pathogenic bacteria, identify patients with high risk of periodontal disease, motivate and educate patients, and provide an indication in the choice of antibiotics. However, whether bacterial tests should be conducted in the treatment of periodontal disease remains open to debate. Although periodontitis is a bacterial infection, there are no established microbiological tests that can be easily applied clinically, and periodontitis is often treated without identifying the responsible bacteria. In recent years, various chair-side microbial tests have been developed aiming at the detection of periodontal disease, including enzyme assay, DNA probe method, and polymerase chain reaction. None of these tests, however, is widely employed due to issues related to cost and convenience (Socransky & Haffajee 1994; Ashimoto et al. 1996; Takahashi et al. 1998; Komiya et al. 2000). At present, it is possible to quantify periodontal pathogens by real-time polymerase chain reaction (RT-PCR), and clinical bacterial tests are being performed much more frequently (Boutaga et al. 2003, 2006, 2007; Kuboniwa et al. 2004; Hyvarinen et al. 2009). Sampling of microbes is usually performed by inserting paper points in periodontal pockets or obtaining samples of saliva, with the latter method being more convenient and suitable for screening of bacteria in the entire oral cavity. Recently, periodontal pathogens have been detected at high frequency in the foci of peri-implantitis, implicating them in the development of this disease (Mombelli & Lang 1998; van Winkelhoff et al. 2000; Quirynen et al. 2002). Periodontal bacteriological examination is expected to be effective as a tool to determine the risk of peri-implantitis.
Therefore, the aim of this study was to determine whether periodontal pathogens in saliva showed a correlation with periodontal status. A periodontal pathogen test that could easily be applied in a clinical setting would serve as an effective tool in identifying the cause of and risk of periodontitis and peri-implantitis.
Material and methods
Patients and clinical evaluation
A total of 291 adult Japanese patients requesting dental implant treatment visiting the Department of Maxillofacial and Oral Implantology at Tokyo Dental College Chiba Hospital between December 2007 and May 2010 were evaluated. The health status and background of each patient were recorded, including age, sex, and probing pocket depth (195 women and 96 men with a mean age ± SD of 53.3 ± 11.3 years). The patients were classified into the following four groups based on probing pocket depth: a no-periodontitis (np) group (all remaining teeth with ≤3 mm probing pocket depth); a mild-periodontitis (mip) group (less than four teeth with ≥4 mm probing pocket depth); a moderate-periodontitis (mop) group (at least four teeth with ≥4 mm probing pocket depth); and a severe-periodontitis (sp) group (at least 30% of the remaining teeth with ≥6 mm probing pocket depth) (Table1). Pocket probing was carried out with a Williams probe and recorded at six sites (mesiofacial, midfacial, distofacial, mesiolingual, midlingual, and distolingual) in each tooth. A modified version of the classification system proposed by the American Academy of Periodontology was used in this study (Armitage 1999). All patients were systemically healthy, had normal salivary flow, and had received no periodontal treatment or antibiotics for at least 6 months prior to participating in this study.
Table 1.
Classification of experimental groups
| Classification | Number (Male/Female) | Age (mean ± SD) | Evaluation basis of periodontal status |
|---|---|---|---|
| No-periodontitis (np) group | 53 (22/31) | 42.6 ± 12.0 | Probing Pocket Depth (PPD) among all residual teeth < 3 mm |
| Mild-periodontitis (mip) group | 103 (49/54) | 51.1 ± 9.6 | Less than four teeth showing PPD 4–6 mm among all residual teeth |
| Moderate-periodontitis (mop) group | 113 (56/57) | 55.3 ± 13.4 | More than four teeth showing PPD 4–6 mm among all residual teeth |
| Severe-periodontitis (sp) group | 22 (10/12) | 54.3 ± 10.3 | More than 9 teeth > PPD 6 mm or Rate of teeth > PPD 6 mm is over 30% among all residual teeth |
Subjects were divided into four groups based on severity of periodontal disease.
Indicate the total number of people, the gender split in each group, mean age.
Informed consent was obtained from each patient. The present study was conducted with approval from the Ethics Review Board of Tokyo Dental College.
Saliva sample
Each patient was required to chew gum for at least 5 min prior to collection of saliva samples. A total of 3–5 ml stimulated whole saliva was then collected in an empty, sterile, 50-ml test tube (Fig.1). The patients were instructed not to brush their teeth or eat for up to 1 h prior to sampling, which was performed between 9:30 and 11:30 AM. The whole saliva was stored at −20°C until processing.
Figure 1.

Sampling of saliva. Patients were required to chew gum for 5 min, after which 3–5 ml saliva was obtained.
Real-time polymerase chain reaction
One hundred microliter diluted saliva samples were used for automated DNA extraction and purification with the Puregene Core kit A (QIAGEN, Tokyo, Japan).
The RT-PCR analysis was performed by the TaqMan® probe method (MIROKU MEDICAL LABORATORY Inc., Nagano, Japan) and the following five periodontopathic bacteria quantified: Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Prevotella intermedia. The primer and probe sets for the five periodontal pathogens targeted and experimental conditions are shown in Fig.2 (Shelburne et al. 2000; Martin et al. 2002; Kuboniwa et al. 2004; Boutaga et al. 2005). The bacterial copy count of each pathogen and proportion of each bacterium to total copy count were determined.
Figure 2.
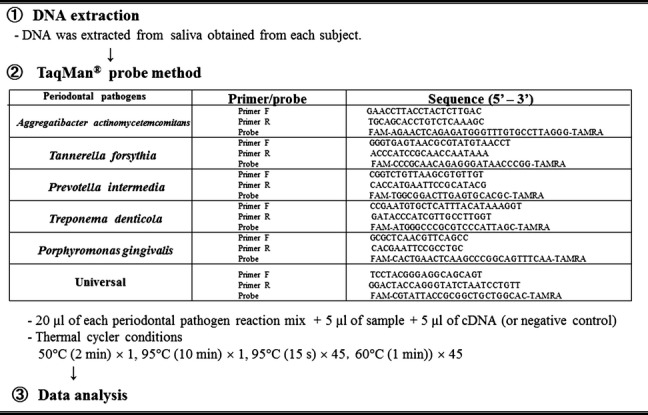
TaqMan® RT-PCR assay was employed to determine count of infectious agents targeted using primers and techniques indicated.
Statistical analysis
The statistical analysis was performed using the SPSS 15.0 statistical package for Windows (SPSS Inc., Chicago, IL, USA). The bacterial copy count of each periodontal pathogen and proportion of each bacterium to the total copy count were compared using the Kruskal–Wallis tests. Tukey’s HSD test was used to assess the difference in each group as post hoc tests. P-values of < 0.05 were considered significant.
Results
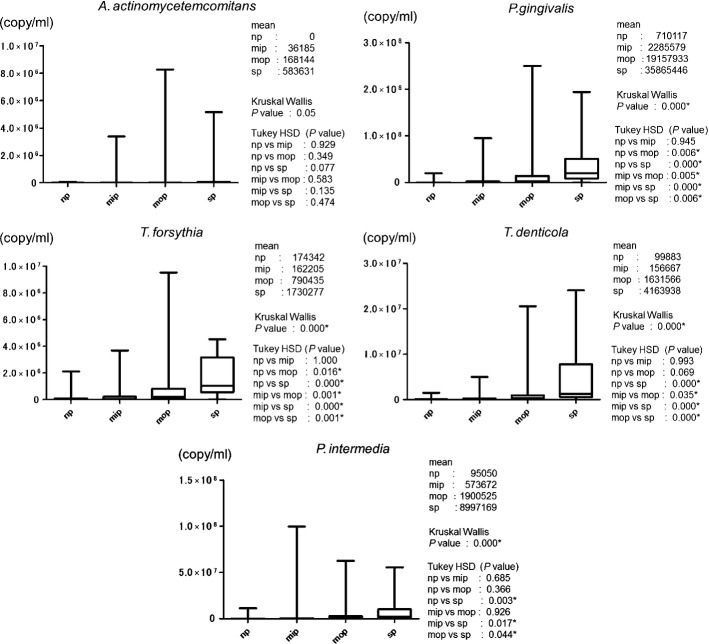
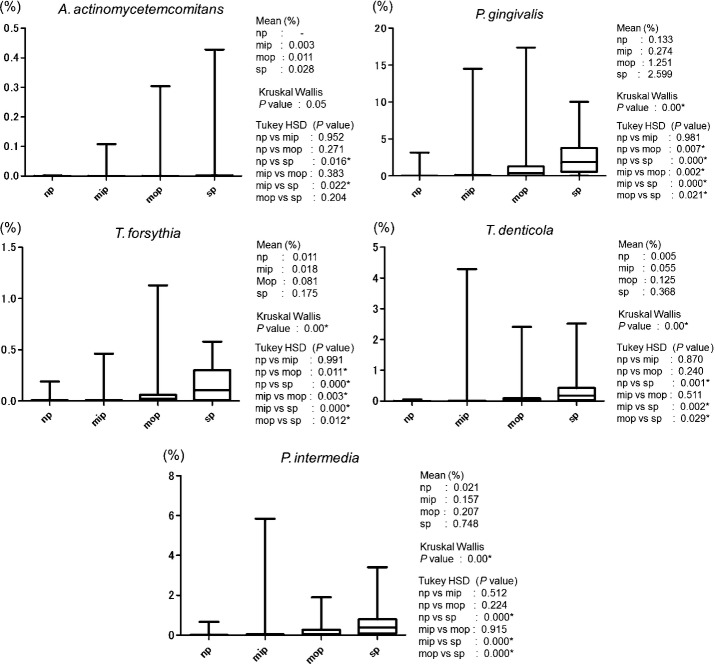
Figure3 show the average bacterial counts of the five periodontal pathogens targeted. No significant difference was detected in the salivary A. actinomycetemcomitans level among the four groups. Salivary levels of P. gingivalis and T. forsythia were significantly higher in both the mop and sp groups (P < 0.05) than in the np group. Salivary levels of T. denticola and P. intermedia were significantly higher in the sp group than in the np group (P < 0.05). Figure4 show the average proportion of each bacterium to total salivary count in each patient. The average proportion for P. gingivalis in the mop and sp groups was markedly high at 1.25% and 2.60%, respectively. In the mop group, the average proportions were 0.08% for T. forsythia, 0.13% for T. denticola, and 0.21% for P. intermedia. In terms of significant differences, these results showed a similar trend to those of previously reported bacterial copy counts of periodontal pathogens (P < 0.05).
Figure 3.
Box plot of copy counts number (copy/ml) of five periodontal pathogens (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Prevotella intermedia) in saliva sample.
Figure 4.
Box plot of ratio (%) of each bacterium to total copy count of five periodontal pathogens (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Prevotella intermedia) in saliva sample.
Discussion
In the present study, patients with periodontitis were classified based on severity of disease (a no-periodontitis group, a mild-periodontitis group, a moderate-periodontitis group, and a severe-periodontitis group), and five periodontal pathogens (A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, and P. intermedia) from saliva samples were quantified and compared. Porphyromonas gingivalis, T. forsythia, and T. denticola are referred to as the red complex bacteria, and along with A. actinomycetemcomitans, the element of exogenous infection is believed to be strong (Socransky & Haffajee 2002). As these four bacteria and P. intermedia are considered to be the main agents of periodontitis, these were the ones targeted in the present study. Both the number and level of P. gingivalis, T. forsythia, T. denticola, and P. intermedia from the saliva samples were significantly higher in the severe-periodontitis group than in the no-periodontitis group. The number and proportions of P. gingivalis were significantly different between the no-periodontitis and moderate-periodontitis groups. Periodontal pathogens are found in increased numbers in severe periodontal disease, and all of the periodontal pathogens targeted in the present study were detected in elevated numbers in the severe-periodontitis group. Large numbers of periodontal pathogens were detected in patients with deep periodontal pockets (Listgarten & Hellden 1978; Edwardsson et al. 1999). Our findings are mostly in agreement with the results of previous papers (Griffen et al. 1998; van Winkelhoff et al. 2002; Sanz et al. 2004). Aggregatibacter actinomycetemcomitans is involved in the pathogenesis of aggressive periodontitis and has been found in younger patients aged between 10 and 30 years (Armitage 1999). The relatively high age of the subjects (mean age, 53.3 ± 9.0 years) may explain the extremely low level of detection of this bacterium in the present study. The bacterial levels observed in the present study were higher than the risk assessment criteria reported in previous studies. Rams et al. (1996) have reported that the possibility of recurrence of periodontitis increased in patients with a threshold proportion of > or = 0.01% for A. actinomycetemcomitans, > or = 0.1% for P. gingivalis, and > or = 2.5% for P. intermedia. Moreover, Brown et al. (1994) noted that a ratio of > or = 2% for P. gingivalis or P. intermedia under adult gingival margin was effective in predicting periodontal progression. The RT-PCR method employed in the present study quantifies bacteria by amplifying the DNA of the sample bacteria. This approach offers greater sensitivity than conventional bacterial tests such as culture and has been used a lot recently for the study of periodontal pathogens (Boutaga et al. 2003, 2006, 2007; Kuboniwa et al. 2004; Hyvarinen et al. 2009). The cutoff point for determining the risk of periodontal disease, however, remains to be established. The present results were little different from the risk assessment criteria employed in conventional culture tests, suggesting that it may be necessary to establish new risk assessment criteria.
The components of saliva are related to those found in blood. Therefore, clinically, saliva is useful in investigating and monitoring the metabolism. It aids not only in the diagnosis of dental caries or periodontitis, but also in diseases affecting other areas of the body, and many studies have been published in this field since 1983 (Ericksson 1983; Rigas & Levine 1983; Vining et al. 1983). The National Institute of Health (NIH) of the United States has shown interest in research on “whether saliva can be used for the medical checkup of the whole body like blood” and earmarked a budget of 5 million dollars for this in 2002 (Wong 2006). All the indications are that saliva testing is destined to become a familiar clinical tool in the future (Ito et al. 2008). Collection of saliva samples is convenient, is painless, and can be performed in a short period of time. Umeda et al. (1998) reported that whole saliva was superior to pooled periodontal pocket samples in the detection of P. gingivalis, P. intermedia, P. nigrescens, and T. denticola in the oral cavity. Studies on periodontal pathogens using saliva are still few (Tamura et al. 2006; Saygun et al. 2011). However, the present results indicate that saliva testing would offer a useful tool in the diagnosis of periodontal disease. With tests in which paper points are placed in periodontal pockets, it may not be possible to identify isolated diseased sites or bacterial flora in the subgingival biofilm. A saliva test, however, reflects the microbial status of the entire oral cavity and may serve as a useful screening technique in the reduction of risk of oral diseases.
In patients requesting implant treatment in the present study, microbiological tests were carried out to screen for periodontal disease before implant treatment. The results of this study indicated that large numbers of periodontal pathogens were detected in more severe periodontitis. Ito et al. (2011) reported that most of the patients who requested dental implant treatment had been suffering from periodontal disease. It has been reported that patients with periodontal disease are more susceptible to peri-implantitis (Hardt et al. 2002; Schou et al. 2006). Moreover, microbiological studies documented a correlation between failed implant therapy and periodontal pathogens (Mombelli & Lang 1998; Quirynen et al. 2002, 2006; Shibli et al. 2008). The onset mechanism for peri-implantitis is believed to be the same as that for periodontitis, as the same bacteria present in periodontal pockets are involved (Takanashi et al. 2004; Aoki et al. 2012). Furthermore, it has been reported that implant therapy in patients with untreated periodontitis increases the risk of peri-implantitis (Renvert & Persson 2009). Because periodontal pathogens are likely to be associated with peri-implantitis, it appears valid to test for periodontal disease-causing bacteria before implant therapy and ascertain the risk of per-implantitis. Finding a correlation between specific oral bacteria and the onset of peri-implantitis would be of considerable clinical use.
The results of the present study confirmed a correlation between the bacterial count of each periodontal pathogen targeted or ratio of each bacterium to total count and periodontitis, suggesting that the present saliva test could be used to assess risk of not only this disease, but also implant failure and peri-implantitis. Further studies are needed, however, to clarify how specific levels of each bacterium correlate with the development of periodontitis and peri-implantitis.
Acknowledgments
We would like to thank Associate Professor Jeremy Williams, Tokyo Dental College, for his assistance with the English of this manuscript. We also would like to thank Mrs. Kiyoko Tamai and Mrs. Toshie Kondou, Miroku Medical Laboratory, for helping us in data analysis of the RT-PCR. This study was supported by Grant-in-Aid for Scientific Research, Young Scientists (B), 18791453, from the Japan Society for the Promotion of Science.
References
- Aoki M, Takanashi K, Matsukubo T, Yajima Y, Okuda K, Sato T, Ishihara K. Transmission of periodontopathic bacteria from natural teeth to implants. Clinical Implant Dentistry and Related Research. 2012;14:406–411. doi: 10.1111/j.1708-8208.2009.00260.x. [DOI] [PubMed] [Google Scholar]
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Annals of Periodontology. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiology and Immunology. 1996;11:266–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- Boutaga K, Savelkoul PH, Winkel EG, Winkelhoff AJ. Comparison of subgingival bacterial sampling with oral lavage for detection and quantification of periodontal pathogens by real-time polymerase chain reaction. Journal of Periodontology. 2007;78:79–86. doi: 10.1902/jop.2007.060078. [DOI] [PubMed] [Google Scholar]
- Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CM, Savelkoul PH. Periodontal pathogens: a quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunology and Medical Microbiology. 2005;45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Boutaga K, van Winkelhoff AJ, Vandenbroucke-Graus CM, Savelkoul PH. Comparison of real-time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque sample. Journal of Clinical Microbiology. 2003;41:4950–4954. doi: 10.1128/JCM.41.11.4950-4954.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaga K, van Winkelhoff AJ, Vandenbroucke-Graus CMJE, Savelkoul PHM. The additional value of real-time PCR in the quantitative detection of periodontal pathogens. Journal of Clinical Periodontology. 2006;33:427–433. doi: 10.1111/j.1600-051X.2006.00925.x. [DOI] [PubMed] [Google Scholar]
- Brown LF, Beck JD, Rozier RG. Incidence of attachment loss in community-dwelling older adults. Journal of Periodontology. 1994;65:316–323. doi: 10.1902/jop.1994.65.4.316. [DOI] [PubMed] [Google Scholar]
- Edwardsson S, Bing M, Lindberg B, Soderfeldt B, Attstrom R. The microbiota of periodontal pockets with different depths in therapy – resistant periodontitis. Journal of Clinical Periodontology. 1999;26:143–152. doi: 10.1034/j.1600-051x.1999.260303.x. [DOI] [PubMed] [Google Scholar]
- Ericksson Y. Monofluorophosphate physiology: general considerations. Caries Research. 1983;17:46–55. doi: 10.1159/000260728. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. Journal of Clinical Microbiology. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt CR, Gröndahl K, Lekholm U, Wennström JL. Outcome of implant therapy in relation to experienced loss of periodontal bone support: a retrospective 5-year study. Clinical Oral Implants Research. 2002;13:488–494. doi: 10.1034/j.1600-0501.2002.130507.x. [DOI] [PubMed] [Google Scholar]
- Hyvarinen K, Laitinen S, Paju S, Hakala A, Suominen-Taipale L, Skurnik M, Kononen E, Pussinen PJ. Detection and quantification of five major periodontal pathogens by single copy gene-based real-time PCR. Innate Immunity. 2009;15:195–204. doi: 10.1177/1753425908101920. [DOI] [PubMed] [Google Scholar]
- Ito T, Komiya-Ito A, Arataki T, Furuya Y, Yajima Y, Yamada S, Okuda K, Kato T. Relationship between antimicrobial protein levels in whole saliva and periodontitis. Journal of Periodontology. 2008;79:316–322. doi: 10.1902/jop.2008.070348. [DOI] [PubMed] [Google Scholar]
- Ito T, Yasuda M, Norizuki Y, Sasaki H, Honma S, Furuya Y, Kato T, Yajima Y. Periodontal conditions in patients requesting dental implant treatment. The Bulletin of Tokyo Dental College. 2011;52:53–57. doi: 10.2209/tdcpublication.52.53. [DOI] [PubMed] [Google Scholar]
- Komiya A, Kato T, Nakagawa T, Saito A, Takahashi J, Yamada S, Okuda K. A rapid DNA probe method for detection of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Journal of Periodontology. 2000;71:760–767. doi: 10.1902/jop.2000.71.5.760. [DOI] [PubMed] [Google Scholar]
- Kuboniwa M, Amano A, Kimura KR, Sekine S, Kato S, Yamamoto Y, Okahashi N, Iida T, Shizukuishi S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiology and Immunology. 2004;19:168–176. doi: 10.1111/j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- Listgarten MA, Hellden L. Relative distribution of bacteria at clinically healthy and periodontally diseased sites in humans. Journal of Clinical Periodontology. 1978;5:115–132. doi: 10.1111/j.1600-051x.1978.tb01913.x. [DOI] [PubMed] [Google Scholar]
- Martin FE, Nadkarni MA, Jacques NA, Hunter N. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. Journal of Clinical Microbiology. 2002;40:1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombelli A, Lang NP. The diagnosis and treatment of peri-implantitis. Periodontology 2000. 1998;17:63–76. doi: 10.1111/j.1600-0757.1998.tb00124.x. [DOI] [PubMed] [Google Scholar]
- Moore WE. Microbiology of periodontal disease. Journal of Periodontal Research. 1987;22:335–341. doi: 10.1111/j.1600-0765.1987.tb01595.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Erickson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. Journal of Bacteriology. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirynen M, De Soete M, van Steenberghe D. Infectious risks for oral implants: a review of the literature. Clinical Oral Implants Research. 2002;13:1–19. doi: 10.1034/j.1600-0501.2002.130101.x. [DOI] [PubMed] [Google Scholar]
- Quirynen M, Vogels R, Peeters W, van Steenberghe D, Naert I, Haffajee A. Dynamics of initial subgingival colonization of ‘pristine’ peri-implant pockets. Clinical Oral Implants Research. 2006;17:25–37. doi: 10.1111/j.1600-0501.2005.01194.x. [DOI] [PubMed] [Google Scholar]
- Rams TE, Listgarten MA, Slots J. Utility of 5 major putative periodontal pathogens and selected clinical parameters to predict periodontal breakdown in patients on maintenance care. Journal of Clinical Periodontology. 1996;23:346–354. doi: 10.1111/j.1600-051x.1996.tb00556.x. [DOI] [PubMed] [Google Scholar]
- Renvert S, Persson GR. Periodontitis as a potential risk factor for peri-implantitis. Journal of Clinical Periodontology. 2009;36:9–14. doi: 10.1111/j.1600-051X.2009.01416.x. [DOI] [PubMed] [Google Scholar]
- Rigas B, Levine L. Human salivary eicosanoids: circadian variation. Biochemical and Biophysical Research Communications. 1983;115:201–205. doi: 10.1016/0006-291x(83)90989-0. [DOI] [PubMed] [Google Scholar]
- Sanz M, Lau L, Herrera D, Morillo JM, Silva A. Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: a review. Journal of Clinical Periodontology. 2004;31:1034–1047. doi: 10.1111/j.1600-051X.2004.00609.x. [DOI] [PubMed] [Google Scholar]
- Saygun I, Nizam N, Keskiner I, Bal V, Kubar A, Acikel C, Serdar M, Slots J. Salivary infectious agents and periodontal disease status. Journal of Periodontal Research. 2011;46:235–239. doi: 10.1111/j.1600-0765.2010.01335.x. [DOI] [PubMed] [Google Scholar]
- Schou S, Holmstrup P, Worthington HV, Esposito M. Outcome of implant therapy in patients with previous tooth loss due to periodontitis. Clinical Oral Implants Research. 2006;17:104–123. doi: 10.1111/j.1600-0501.2006.01347.x. [DOI] [PubMed] [Google Scholar]
- Shelburne CE, Prabhu A, Gleason RM, Mullally BH, Coulter WA. Quantitationof Bacteroides forsythus in subgingival plaque: comparison of immunoassay and quantitative polymerase chain reaction. Journal of Microbiology Methods. 2000;39:97–107. doi: 10.1016/s0167-7012(99)00106-2. [DOI] [PubMed] [Google Scholar]
- Shibli JA, Melo L, Ferrari DS, Figueiredo LC, Faveri M, Feres M. Composition of supra- and subgingival biofilm of subjects with healthy and diseased implants. Clinical Oral Implants Research. 2008;19:975–982. doi: 10.1111/j.1600-0501.2008.01566.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Evidence of bacterial etiology: a historical perspective. Periodontology 2000. 1994;5:7–25. doi: 10.1111/j.1600-0757.1994.tb00016.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Dental biofilms: difficult therapeutic targets. Periodontology 2000. 2002;28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- Takahashi J, Saito A, Nakagawa T, Yamada S, Ishihara K, Okuda K. Dynamics of serum Immunoglobulin G avidity for Porphyromonas gingivalis in adult periodontitis. Journal of Periodontology. 1998;69:367–373. doi: 10.1902/jop.1998.69.3.367. [DOI] [PubMed] [Google Scholar]
- Takanashi K, Kishi M, Okuda K, Ishihara K. Colonization by Porphyromonas gingivalis and Prevotella intermedia from teeth to osseointegrated implant regions. The Bulletin of Tokyo Dental College. 2004;45:77–85. doi: 10.2209/tdcpublication.45.77. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nakano K, Hayashibara T, Nomura R, Fujita K, Shintani S, Ooshima T. Distribution of 10 periodontal bacteria in saliva samples from Japanese children and their mothers. Archives of Oral Biology. 2006;51:371–377. doi: 10.1016/j.archoralbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. Journal of Clinical Periodontology. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- Umeda M, Contreras A, Chen C, Bakker I, Slots J. The utility of whole saliva to detect the oral presence of periodontopathic bacteria. Journal of Periodontology. 1998;69:828–833. doi: 10.1902/jop.1998.69.7.828. [DOI] [PubMed] [Google Scholar]
- Vining RF, McGinley RA, Maksvytis JJ, Ho KY. Salivary cortisol: a better measure of adrenal cortical function than serum cortisol. Annals of Clinical Biochemistry. 1983;20:329–335. doi: 10.1177/000456328302000601. [DOI] [PubMed] [Google Scholar]
- van Winkelhoff AJ, Goene RJ, Benschop C, Folmer T. Early colonization of dental implants by putative periodontal pathogens in partially edentulous patients. Clinical Oral Implants Research. 2000;11:511–520. doi: 10.1034/j.1600-0501.2000.011006511.x. [DOI] [PubMed] [Google Scholar]
- van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. Journal of Clinical Periodontology. 2002;29:1023–1028. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. Journal of American Dental Association. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]