Abstract
Sir2 enzymes are broadly conserved from bacteria to humans and have been implicated to play roles in gene silencing, DNA repair, genome stability, longevity, metabolism, and cell physiology. These enzymes bind NAD+ and acetyllysine within protein targets and generate lysine, 2′-O-acetyl-ADP-ribose, and nicotinamide products. To provide structural insights into the chemistry catalyzed by Sir2 proteins we report the high-resolution ternary structure of yeast Hst2 (homologue of Sir two 2) with an acetyllysine histone H4 peptide and a nonhydrolyzable NAD+ analogue, carba-NAD+, as well as an analogous ternary complex with a reaction intermediate analog formed immediately after nicotinamide hydrolysis, ADP-ribose. The ternary complex with carba-NAD+ reveals that the nicotinamide group makes stabilizing interactions within a binding pocket harboring conserved Sir2 residues. Moreover, an asparagine residue, N116, strictly conserved within Sir2 proteins and shown to be essential for nicotinamide exchange, is in position to stabilize the oxocarbenium intermediate that has been proposed to proceed the hydrolysis of nicotinamide. A comparison of this structure with the ADP-ribose ternary complex and a previously reported ternary complex with the 2′-O-acetyl-ADP-ribose reaction product reveals that the ribose ring of the cofactor and the highly conserved β1–α2 loop of the protein undergo significant structural rearrangements to facilitate the ordered NAD+ reactions of nicotinamide cleavage and ADP-ribose transfer to acetate. Together, these studies provide insights into the chemistry of NAD+ cleavage and acetylation by Sir2 proteins and have implications for the design of Sir2-specific regulatory molecules.
The Sir2 (silent information regulator 2) or sirtuin family of deacetylases requires NAD+ as a cofactor to hydrolyze the acetyl moiety of an acetyllysine within protein targets to regulate diverse biological functions, including gene silencing, genome stability, longevity, metabolism, and cell physiology (for reviews, see refs. 1 and 2). The mechanism for Sir2 activity has been extensively studied at both structural and enzymatic levels. Structural studies reveal that the Sir2 proteins contain a structurally conserved elongated core domain containing a large Rossmann fold at one end, a structurally more variable zinc-binding motif at the opposite end, and a series of loops connecting these regions and forming a cleft in the central region of the core domain (3–6). The acetyllysine and NAD+ cosubstrates bind to opposite sides of the cleft and highlight the region of the core domain containing the highest degree of sequence conservation within the Sir2 proteins, implying a conserved catalytic mechanism (3, 7). Biochemical and structural studies reveal that the deacetylation of acetyllysine is coupled to the hydrolysis of NAD+ to nicotinamide and 2′-O-acetyl-ADP-ribose (8, 9).
A detailed structural understanding of how the Sir2 proteins mediate nicotinamide cleavage and ADP-ribose transfer to acetate has been hampered by the difficulty in trapping a Sir2 protein bound to a form of NAD+ containing an ordered nicotinamide group (6, 7, 10, 11). To visualize the nicotinamide group of NAD+ bound to a Sir2 protein, we now report the high-resolution crystal structure of the Saccharomyces cerevisiae Sir2 homologue, yHst2, bound to an acetyllysine-containing histone H4 peptide and carbanicotinamide adenine dinucleotide (carba-NAD+). Carba-NAD+ is an inhibitor of NAD+ glycohydrolases and ADP-ribosyltransferases (12, 13), where the β-d-ribonucleotide ring of the nicotinamide ribonucleoside moiety of NAD+ is replaced by a 2,3-dihydroxycyclopentane ring, which significantly disfavors hydrolysis of the pyridinium-N-glycoside bond (14). In this study, we also report on the structure of a corresponding ternary complex with ADP-ribose, representing an intermediate mimic formed after nicotinamide cleavage. A comparison of these structures reveals a conserved nicotinamide-binding site and the mechanistic details underlying nicotinamide cleavage from NAD+. A further comparison of these two structures with a previously reported yHst2 ternary complex with the 2′-O-acetyl-ADP-ribose reaction product reveals how the NAD+ reactions of nicotinamide cleavage and ADP-ribose transfer to acetate are coordinated through significant conformational changes of the ADP-ribose ring of the NAD+ cofactor and the β1–α2 loop of the protein during catalysis. The implications of these studies for the design of compounds for the specific inhibition or activation of Sir2 proteins are also discussed.
Methods
Crystallization and Structure Refinement. The 64-residue C-terminal deletion construct of yHst2 (residues 1–294) was purified as described previously (5, 7). Crystals of yHst2/carba-NAD+/histone H4 and yHst2/ADP-ribose/histone H4 complexes were grown by using the vapor diffusion method at room temperature and were obtained by mixing 2 μl of a complex containing 0.24 mM protein, 1 mM histone peptide, and 1 mM NAD+ analog with 2 μl of reservoir solution containing 2.0 M (NH4)2SO4 and 100 mM [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane (Bis-Tris), pH 5.5/6.5, and equilibrating over 0.5 ml of reservoir solution. Before data collection, crystals were cryoprotected with a five-step gradual transfer into reservoir solution supplemented with higher concentrations of glycerol to a final concentration of 25% (vol/vol). Crystallographic data for the yHst2/carba-NAD+/histone H4 and Hst2/ADP-ribose/histone H4 crystals were collected at the X12B and X25 beamlines at the National Synchrotron Light Source, respectively. All data were processed with HKL 2000 suite (HKL Research, Charlottesville, VA). The structures of both complexes were solved by molecular replacement with the program cns (15), using protein residues 5–293 from the yHst2/2′-O-acetyl-ADP-ribose/histone H4 structure (PDB ID code 1Q1A) as a search model (7). Structures were refined by simulated annealing, torsion angle dynamics, and B factor adjustments in cns with iterative manual adjustments of the models and placement of solvent molecules by using the program o (16). The final models were checked for errors with composite-simulated annealing omit maps, and a final round of refinement resulted in models with excellent refinement statistics and geometry (Table 1, which is published as supporting information on the PNAS web site). The proteins from both structures contain a disordered region from residue 207 to residue 214 that could not be modeled. The two refined structures also showed excellent density for the carba-NAD+ and ADP-ribose, respectively. In both ternary complexes, residues K12 to R19 of the histone H4 peptide (12-KGGAKAcRHRKIL-22) could be modeled, whereas the side chains for K12 and R19 and the three C-terminal side chains (KIL) could not be modeled and are presumed to be disordered.
Isothermal Titration Calorimetry (ITC) Experiment. Full-length yHst2 was prepared as described previously (5). Full-length yHst2 (in the absence or presence of a 3-fold molar excess of K16-acetylated histone H4 peptide was dialyzed against a buffer solution containing 20 mM Hepes at pH 7.5, 100 mM NaCl, and 1.0 mM tris(2-carboxyethyl)phosphine (TCEP, Pierce) before analysis by ITC. ITC measurements were carried out with a Microcal VP-ITC isothermal titration calorimeter (Microcal, Amherst, MA). A 50 μM solution of yHst2 (with or without peptide) diluted in dialysis buffer was added to the sample cell (about 1.4 ml), and a 5.0–5.3 mM solution of ADP-ribose titrant was loaded into the injection syringe. For each titration experiment, a 60-sec delay at the start of the experiment was followed by 35 injections of 2 μl of the titrant solution, spaced apart by 240 sec. The sample cell was stirred at 300 rpm throughout and maintained at a temperature of 10°C. Control titrations were performed by titrating titrant solutions into dialysis buffer. Titration data were analyzed by using the origin 5.0 software supplied by Microcal. Data sets were corrected for baseline heats of dilutions from control runs as appropriate. The corrected data were then fit to a theoretical titration curve describing one binding site per titrant. The area under each peak of the resultant heat profile was integrated and plotted against the molar ratio of yHst2 protein to titrant. A nonlinear best-fit binding isotherm for the data was used to calculate protein/titrant stoichiometry, dissociation constant, and standard change in enthalpy.
Results and Discussion
Overall Structures of yHst2 Complexes with Carba-NAD+ and ADP-Ribose. Crystals of the yHst2/carba-NAD+/histone H4 and yHst2/ADP-ribose/histone H4 complexes were isomorphous and formed in space group P3221 containing one molecule per asymmetric unit. The two structures were refined to high resolution of 1.75 Å (Rfree = 23.9%) and 1.5 Å (Rfree = 23.0%), respectively, and to excellent refinement and geometrical parameters (Table 1). The yHst2/carba-NAD+/histone H4 structure contains both an acetyllysine and an intact NAD+/analog substrate in which a cyclopentane ring replaces the furanose of the nicotinamide-ribonucleotide moiety, and we therefore will refer to this structure as the “substrate analog complex.” We will refer to the yHst2/ADP-ribose/histone H4 structure as an “intermediate analog complex” because it represents a close mimic of a reaction intermediate, immediately after cleavage of the glycosidic bond between nicotinamide and ADP-ribose (it differs only by the addition of a 1′-OH group). The structure of the previously reported yHst2/2′-O-acetyl-ADP-ribose/histone H4 complex, which will be used for comparison in this study, will be referred to as the “product complex” because it contains the Sir2 reaction product of NAD+.
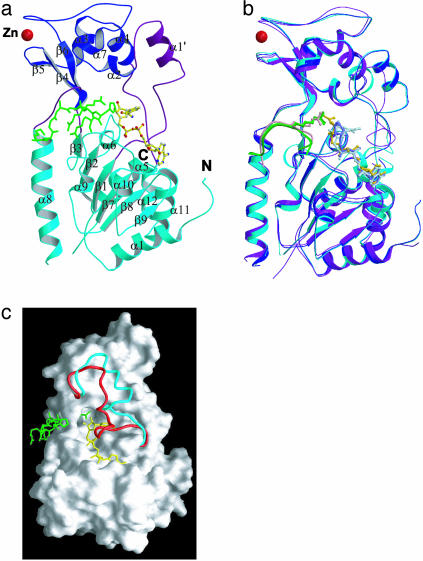
The overall folds of both the yHst2/carba-NAD+/histone H4 and yHst2/ADP-ribose/histone H4 complexes show high similarity to the previously reported structure of the yHst2/2′-O-acetyl-ADP-ribose/histone H4 complex. The protein molecules of the carba-NAD+ substrate and ADP-ribose intermediate analog complexes have an rms deviation of 0.27 Å for Cα atoms, and a comparison of these structures with the ternary complex containing 2′-O-acetyl-ADP-ribose product shows corresponding values of 2.0 and 2.2 Å, respectively (Fig. 1). The most significant differences between the proteins of the three structures are observed in the β1–α2 loop (residues 35–63). A comparison of the β1–α2 loop between the three structures shows an rms deviation of 0.6 Å for all atoms between the carba-NAD+ substrate analog complex and ADP-ribose intermediate analog complex, whereas a significantly larger rms difference of about 3.4 Å is observed when the β1–α2 loop from these structures is compared with the corresponding region of the complex containing the 2′-O-acetyl-ADP-ribose product. Interestingly, in the present structures, the β1–α2 loop is partially formed into a helix (α1′, Fig.1 a and b) and has a more open conformation that is more accessible to NAD+ substrate than the 2′-O-acetyl-ADP-ribose-containing product complex (Fig. 1c). Notably, the β1–α2 loop is partially disordered in the nascent yHst2 structure (5). The β1–α2 loop harbors a high degree of sequence conservation between the Sir2 proteins and mediates multiple cofactor contacts in each of the cofactor-bound structures, implicating its functional importance (6, 7).
Fig. 1.
Overall structure of the yHst2 substrate and intermediate analog complexes. (a) Ribbon diagram of the yHst2/carba-NAD+/H4 complex highlighting the large domain (cyan), small domain (blue), and connecting loops (purple). The NAD+ (yellow), histone H4 peptide (green), and Zn ion (red) are also highlighted. (b) Superimposition of yHst2/carba-NAD+/H4 (cyan), yHst2/ADP-ribose/H4 (blue), and yHst2/2′-O-acetyl-ADP-ribose/H4 (purple). (c) Molecular surface of yHst2, highlighting the conformational difference of the β1–α2 NAD+-binding loops in the yHst2/carba-NAD+/H4 (cyan) and yHst2/2′-O-acetyl-ADP-ribose/H4 (red) complexes.
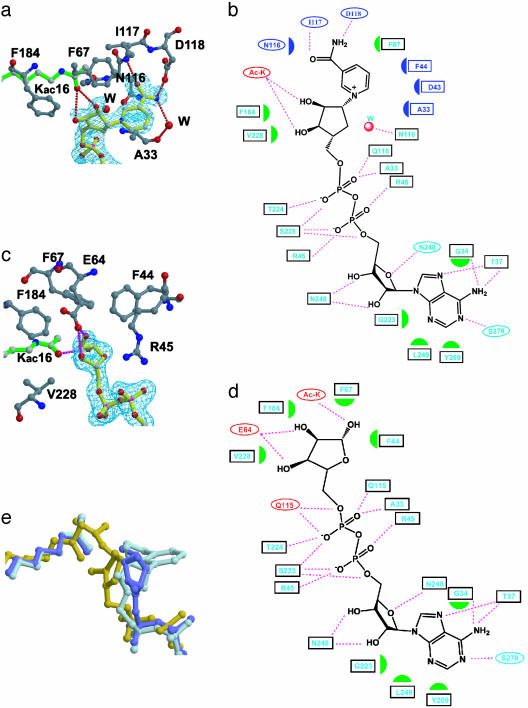
The carba-NAD+ and ADP-ribose have very well refined electron densities, with B factors of 40 Å2 and 29 Å2, respectively, that are close to the average B factor for the protein atoms of the respective structures (Table 1, Fig. 2 a and c). The adenine-ribose and diphosphate groups in the current structures and the 2′-O-acetyl-ADP-ribose product complex are essentially superimposable, with an rms deviation over all atoms of below 0.3 Å. In addition, the interactions that these regions of the NAD+ molecule makes with the protein are essentially maintained in all three structures (Fig. 2 b and d). In contrast, the nicotinamide ribose rings of the NAD+ analogs show significant conformational differences within the three complexes (Fig. 2e). Compared with the 2′-O-acetyl-ADP-ribose product structure, the nicotinamide ribose ring of the carba-NAD+ substrate analog is rotated ≈90° along the ring plane and results in the displacement of the 1′-carbonyl atom by ≈2.5 Å, and the 3′-oxygen by ≈5 Å (Fig. 2e). The ribose ring of the ADP-ribose intermediate analog complex adopts a conformation that is rotated by ≈45° relative to the carba-NAD+ substrate analog, but in a direction opposite to that of the 2′-O-acetyl-ADP-ribose product. An overall comparison of the three structures implicates that the conformational flexibility of the β1–α2 loop of the protein and the nicotinamide ribose ring of the NAD+ cofactor play key roles in catalysis.
Fig. 2.
yHst2-NAD analog interaction. (a) The carba-NAD+-binding site of yHst2 highlighting the yHst2 residues (carbon, gray; oxygen, red; and nitrogen, blue) that contact the nicotinamide and nicotinamide-ribose (carbon, yellow). A simulated annealing Fo – Fc omit map of carba-NAD+ is counted at 3.0 σ (cyan) and the acetyllysine substrate (green) is also highlighted. (b) Summary of yHst2 carba-NAD+ interactions. Hydrogen bonds are indicated with dashed lines and van der Waals interactions are indicated with half-moon symbols. The residues highlighted in cyan take part in conserved interactions with the yHst2/2′-O-acetyl-ADP-ribose and yHst2/ADP-ribose complexes. The residues colored in red take part in nonconserved interactions and the residues highlighted in blue take part in interactions with the nicotinamide group. (c) The ADP-ribose-binding site of yHst2, highlighting the yHst2 residues that contact the nicotinamide and nicotinamide-ribose. A simulated annealing Fo – Fc omit map of ADP-ribose counted at 3.0 σ and the color coding are as described for a. (d) Summary of yHst2–ADP-ribose interactions. The coding for interactions and color designations are as described for b. (e) Overlay of the carba-NAD+ (light blue), ADP-ribose (dark blue), and 2′-O-acetyl-ADP-ribose (yellow) ligands, using the corresponding proteins for the superpositions. The acetyl group from the acetyllysine substrate of the respective complex is also shown in the same color.
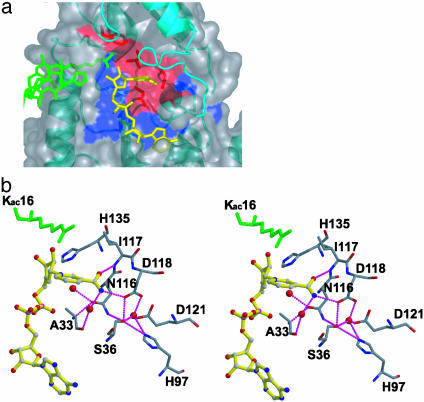
The Nicotinamide-Binding Pocket. The nicotinamide group of the carba-NAD+ substrate analog complex is located in a deep pocket of yHst2 that is adjacent to the ADP-ribose-binding site (Figs. 2a and 3a). This pocket is formed by residues N116, A33, and S36 below the plane of the nicotinamide ring; I117 and D118 on the side of the nicotinamide proximal to the carboxamide group; D43, R45, P42, and I41 on the ribose ring side of nicotinamide; and F67 and F44 on the top of the nicotinamide ring. The NH2 group of nicotinamide is contacted by direct and water-mediated hydrogen bonds from a side-chain carbonyl of D118 and the backbone carbonyl of A33, respectively (Figs. 2a and 3b). The backbone NH of I117 also hydrogen bonds to the carbonyl oxygen of nicotinamide. A33 and N116, as well as D43 and F44 from the β1–α2 loop also take part in van der Waals interactions with the nicotinamide ring (Fig. 2 a and b). Among the residues that contact the nicotinamide, A33, N116, and D118 are strictly conserved and D43 and F44 are highly conserved among the Sir2 proteins, and mutation of N116 and D118 to alanine abolishes nicotinamide exchange and deacetylation, highlighting the functional importance of the observed contacts (4, 10, 17). Of particular interest is N116, in which the side-chain carbonyl is bound to a highly ordered water molecule, which would be in position to hydrogen bond to the ring oxygen of the ribose ring of NAD+. The significance of this interaction is that the side-chain carbonyl of the asparagine would stabilize the oxocarbenium that has been proposed to form upon the hydrolysis of nicotinamide from NAD+ (9, 18, 19). S36 is another residue that is strictly conserved within the Sir2 proteins, and its mutation to alanine has also been shown to abolish the nicotinamide exchange reaction (6). The structure of the carba-NAD+ substrate analog complex reveals that the side-chain hydroxyl of the serine forms direct and water-mediated hydrogen bonds that serve to link N116, D118, H97, and E121 in such a way as to maintain the protein fold and/or protein–nicotinamide interactions mediated by a subset of these residues (Fig. 3b).
Fig. 3.
The yHst2 nicotinamide-binding site. (a) van der Waals surface of yHst2/carba-NAD+/H4 highlighting invariant residues (blue) as well as the subset of these residues that contact nicotinamide (red). I117, which contacts nicotinamide, is also highlighted in red, although this residue has not been subjected to mutagenesis. The surface for the β1–α2 NAD+-binding loop is deleted from this image for clarity. (b) Stereoview of yHst2–nicotinamide interactions, with hydrogen bonds depicted as dotted lines. Protein residues that mediate van der Waals interactions with the nicotinamide are also highlighted. yHst2 and carba-NAD+ color-coding is as in Fig. 2a.
The structure of the ADP-ribose intermediate analog complex reveals that the residues that mediated nicotinamide contacts in the carba-NAD+ product complex superimpose almost perfectly. This observation indicates that there are no significant conformational changes within the nicotinamide-binding site after nicotinamide hydrolysis. In contrast, the 2′-O-acetyl-ADP-ribose product complex reveals that the D43–R45 region shifts by about 5.5 Å toward the nicotinamide-binding pocket and partially occludes the binding site. F44, a phenylalanine or tyrosine within the Sir2 proteins, plays a particularly important role in occluding the nicotinamide-binding site in the 2′-O-acetyl-ADP-ribose product complex. It is notable that in the absence of nicotinamide (in the ADP-ribose and 2′-O-acetyl-ADP-ribose complexes), the nicotinamide-binding pocket is occupied by a series of water molecules that form a water network which mediates the interaction between several residues surrounding the nicotinamide-binding pocket. This water network is conserved in all known Sir2 homologous structures, reflecting the necessity to compensate for the absence of nicotinamide for maintaining the integrity of the core domain of the Sir2 proteins.
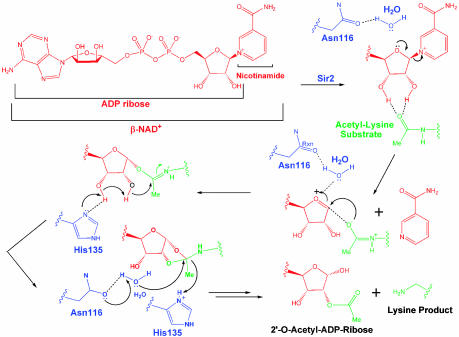
The Nicotinamide Cleavage Reaction. The carba-NAD+ substrate analog complex reveals the stereochemical details of how the binding of acetyllysine is coupled to the nicotinamide cleavage reaction (Figs. 2a and 4). Specifically, the structure shows that the carbonyl oxygen of the acetyllysine forms hydrogen bonds to the 2′- and 3′-hydroxyl groups of the nicotinamide ribose ring. This appears to position what would be the ribose ring oxygen of NAD+ within hydrogen bonding distance of a water-mediated contact to the side-chain carbonyl of N116. This places the asparagine side-chain carbonyl in position to stabilize the oxocarbenium that has been proposed to form upon hydrolysis of the nicotinamide group (9, 18, 19) and is consistent with solution studies showing that this asparagine is essential for the nicotinamide exchange by Sir2 proteins (6). Importantly, the carba-NAD+ substrate analog complex places the ribose ring in a geometry that is not compatible with the acetyllysine deacetylation reaction. In particular, the carbonyl oxygen of the acetyllysine is >4.8 Å away from the 1′-carbon of the ribose ring and is also sterically occluded from attacking this carbon by the 2′-hydroxyl of the ribose ring. In addition, the imidazole nitrogens of H135 are more than 5.4 Å away from either the 2′-or 3′-hydroxyl groups of the ribose ring. It appears that this geometry of the NAD+ substrate helps ensure that nicotinamide cleavage from NAD+ occurs before the subsequent ADP-ribose transfer to acetate from acetyllysine that is initiated by a nucleophilic attack of the 1′-carbon of the ribose ring by the carbonyl group of the acetyllysine and the deprotonation of the 3′-hydroxyl group of the ribose ring by H135 (7) (see Movie 1, which is published as supporting information on the PNAS web site).
Fig. 4.
Proposed catalytic mechanism for yHst2.
In contrast to the situation described above, the ribose ring of the ADP-ribose analog intermediate and the 2′-O-acetyl-ADP-ribose product structures adopt conformations that are incompatible with the nicotinamide cleavage reaction, but favorable for the acetyllysine deacetylation reaction (Figs. 2c and 4). In particular, the ADP-ribose intermediate and 2′-O-acetyl-ADP-ribose product complexes reveal that the carbonyl oxygen of the acetyllysine is 3.6 and 3.3 Å away from the 1′-carbon of the ribose ring, respectively, an appropriate distance for the carbonyl oxygen of the acetyllysine to carry out nucleophilic attack of this carbon. In addition, the 2′-O-acetyl-ADP product complex reveals that an imidazole nitrogen of H135 is within 2.8 Å of the 3′-hydroxyl of the ribose ring and therefore in a suitable position for deprotonation of this hydroxyl group to facilitate the migration of the acetyl group from the 1′ to 2′ position of the ribose ring. The 2′-O-acetyl-ADP-ribose product complex also reveals that a highly ordered water molecule is held in place by N116 to carry out nucleophilic attack of the cyclic-acyldioxalane intermediate that is proposed to form before collapse to the 2′-O-acetyl-ADP-ribose reaction product (7). Together, comparison of the substrate, intermediate, and product complexes of yHst2 reveals that the nicotinamide ribose ring undergoes conformational changes in the enzyme active site to position the cofactor with appropriate geometry and proximity to the appropriate catalytic residues to facilitate, in sequential order, the nicotinamide cleavage and acetyl transfer reactions (Movie 1).
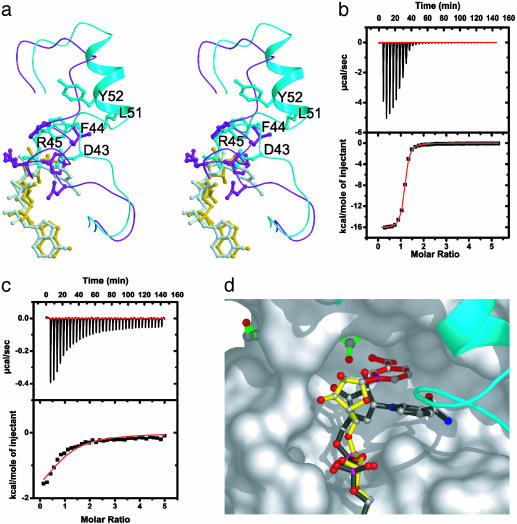
The β1–α2 Loop. The β1–α2 loop of the Sir2 proteins harbors a high degree of sequence conservation, indicating its functional importance, yet a comparison of yHst2 structures shows the greatest structural variability in the β1–α2 loop (Fig. 1 a and b). A comparison of the substrate analog, intermediate analog, and product complexes with acetyllysine-histone H4 reveals that the β1–α2 loop plays at least three important functional roles. First, the carba-NAD+ substrate analog complex, as well as the ADP-ribose intermediate analog complex, suggests that the relatively open conformation of this loop facilitates NAD+ substrate access (Fig. 1 b and c). In particular, residues S43–Y52 of these complexes are shifted away from the NAD+-binding site by about 7 Å relative to the 2′-O-acetyl-ADP-ribose product complex (Fig. 5a). Second, the β1–α2 loop plays an important role in NAD+ substrate interaction, because four conserved residues in this loop (A33, G34, T37, and F44) mediate direct NAD+ interactions in each of the complexes and D43, also conserved in the Sir2 proteins, mediates nicotinamide interactions in the carba-NAD+ substrate analog complex. Third, the more closed conformation of the β1–α2 loop in the 2′-O-acetyl-ADP-ribose structure suggests that the β1–α2 loop may also play a role in nicotinamide release. Consistent with this role, F44 of the β1–α2 loop in the 2′-O-acetyl-ADP-ribose complex partially occupies the binding site for nicotinamide (Fig. 5a). It is also possible that the further burial of the active site, by the more closed conformation of the 2′-O-acetyl-ADP-ribose complex, further facilitates the acetyl-transfer reaction. Taking these observations together, the β1–α2 loop of the Sir2 proteins appears to play a dynamic role in NAD+ association and catalysis.
Fig. 5.
The β1–α2 loop of yHst2 and analog binding. (a) Stereoview of an overlay of the β1–α2 loops from the yHst2/carba-NAD+/H4 (cyan for loop and light blue for carba-NAD+) and yHst2/2′-O-acetyl-ADP-ribose/H4 (purple for loop and yellow for 2′-O-acetyl-ADP-ribose) complexes. Residues D43, F44, R45, and Y52 and the respective NAD+ analogues are also highlighted in each structure. (b and c) Representative data of isothermal titration calorimetry for ADP-ribose titrations into yHst2 (b) and binary yHst2/K16-acetylated histone H4 peptide (c). The area under each injection spike (above) is integrated and fitted by using nonlinear least-squares regression analysis (below). For yHst2 and the binary complex, respectively, the enthalpy of reaction (ΔH = –67.50 ± 0.19 kJ/mol and –17.97 ± 1.20 kJ/mol), dissociation constant (Kd = 0.404 ± 0.001 μM and 29.16 ± 4.50 μM) and stoichiometry (n = 1.114 ± 0.004 and 0.910 ± 0.001) is calculated from the regression analysis. (d) Model for transglycosidation by yHst2. Nicotinamide (red) was modeled onto the ADP-ribose structure (yellow) in a geometry that closely mimics the glycosidic bond geometry between the nicotinamide and ADP-ribose as observed in carba-NAD+ (black, overlaid).
Implications for Modulating the Function of Sir2 Proteins. The p53 tumor suppressor protein has been implicated to be a regulated target for both acetylation and deacetylation in vivo. In particular, several studies have demonstrated that the targeted acetylation of lysine residues at the C terminus of p53 by the transcription coactivators CBP/p300 and P/CAF promotes the stability and transcriptional activation properties of p53 (20–23). More recent studies have shown that the Sir2 homologue from humans, SIRT1, specifically deacetylates p53 in vivo to negatively regulate its activities associated with cellular senescence, apoptosis, and DNA repair (24–27). In light of these recent findings, it may be attractive to develop specific inhibitors to SIRT1 to elevate p53 function in cancer cells. In addition, Sir2-specific inhibitors may be useful as reagents to probe other functions for Sir2 proteins in vivo. Using a cell-based chemical screen, Schreiber and colleagues (28) have identified several putative Sir2 inhibitors, including sirtinol, containing substructures that are similar to the adenine and nicotinamide groups of NAD+. Another small-molecule Sir2 inhibitor, splitomicin, and its derivatives have been identified through library screening (29, 30).
Our studies suggest a more rational approach for the development of Sir2-specific inhibitors. Specifically, our studies suggest that compounds that might mimic the nicotinamide and/or ADP-ribose interactions observed in the substrate or intermediate analog complexes might be particularly effective inhibitors. In addition, compounds that might bind to the β1–α2 loop and reduce its dynamic properties may also serve as effective inhibitors. To further investigate the possibility of developing ADP-ribose as an inhibitor of Sir2 proteins, we determined its binding properties to yHst2 and a binary yHst2/K16-acetylated histone H4 peptide by using isothermal titration calorimetry (Fig. 5 b and c). These studies reveal that ADP-ribose binds to yHst2 and the binary complex with a stoichiometry of 1:1 and dissociation constants of 0.4 μM and 29 μM, respectively. With a dissociation constant of 0.4 μM for the free enzyme, ADP-ribose would appear to be about a 100-fold more potent inhibitor than nicotinamide, splitomicin, or sirtinol (28, 29, 31), and it would certainly provide a suitable scaffold for the further elaboration of Sir2-specific inhibitors.
The possibility of generating Sir2 activating compounds has also generated considerable interest. Specifically, Sir2 has been demonstrated to play a key role in a long established link between calorie-restricted diets and longevity in budding yeast and Caenorhabditis elegans (32–34), a correlation also found in mice, but not yet shown to be Sir2 dependent in this organism or in humans (1, 35). In addition, because it is well documented that as cells age they are more prone to genomic instability, a hallmark of cancer, Sir2-activating molecules may also reduce genomic instability and cancer risk as cells age. Toward developing Sir2-activating compounds, a recent study reported on the identification of a family of polyphenol compounds, several of which are currently being used as chemotherapeutic agents (36, 37). Interestingly, the most potent of these compounds is resveratrol, a plant natural product in high abundance in red wine and correlated with increased lifespan and reduced cancer risk in humans (36, 37). These compounds were shown to increase SIRT1-dependent deacetylation of p53 in vitro and stimulate Sir2-dependant cell survival in yeast. The detailed mechanistic basis for how resveratrol and related compounds stimulates the activity of Sir2 proteins is not well understood, although kinetic data reveal that they stimulate Sir2 activity by lowering the Michaelis constant for both NAD+ and the acetyllysine-bearing substrate (36, 37). Given our results demonstrating that the β1–α2 loop of the Sir2 proteins undergoes significant conformational adjustments to facilitate NAD+ binding and catalysis, we propose that the polyphenol compounds may somehow more optimally reconfigure the β1–α2 loop for NAD+ binding. A comparison between the different yHst2 complexes also reveals that the small zinc-binding domain of the catalytic core also makes adjustments that appear to be important for binding to NAD+ and acetyllysine (Fig. 1b), suggesting that these compounds may also interact with this domain in a way that enhances binding to one or both of the substrates.
Recent studies suggest that nicotinamide, a product formed in the Sir2 reaction, plays an important role in the regulation of Sir2 proteins. In particular, nicotinamide was shown to be an inhibitor of Sir2 enzymes in vitro and also abolishes the silencing phenotype in budding yeast in vivo (31). In addition, increasing expression of the nicotinamidase PNC1, which specifically depletes nicotinamide by hydrolyzing it to nicotinic acid and ammonia, extends the lifespan of Saccharomyces cerevisiae (38, 39). Enzymatic analysis demonstrates that nicotinamide inhibition of Sir2 is noncompetitive with NAD+ and invokes that free nicotinamide intercepts an ADP-ribosyl-enzyme-acetyl peptide intermediate with regeneration of NAD+ (transglycosidation) (18, 19). This mode of nicotinamide inhibition implies that nicotinamide binds to Sir2 at a site distinct from the nicotinamide group of NAD+ but still in a geometry that promotes the regeneration of NAD+. If we assume that the conformation of ADP-ribose in the intermediate analog complex mimics the conformation of the NAD+ intermediate that forms immediately after nicotinamide cleavage, it is particularly striking that an entering nicotinamide group can be modeled onto the 1′-carbon of the ADP-ribose ring with suitable geometry, without stereochemical clash with the protein, and in a binding site that is distinct from the binding site of the nicotinamide group of NAD+ (Fig. 5d). Together, these modeling studies suggest how transglycosylation by Sir2 may occur in a way that is fully consistent with the enzymatic data and further suggest that compounds that block the binding site for free nicotinamide, as suggested from our structural modeling, might serve of potent activators of Sir2 with potential therapeutic applications.
Supplementary Material
Acknowledgments
We thank M. Fitzgerald and W. Ho for useful discussions, the staff at the National Synchrotron Light Source for assistance with Beamlines X12B and X25, and G. Da, K. Li, and M. Hong for assistance with data collection. This work was supported by National Institutes of Health grants to R.M. and by a grant from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health, awarded to the Wistar Institute.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: Coordinates of the Hst2/carba-NAD+ and Hst2/ADP-ribose/histone H4 complexes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1SZC and 1SZD, respectively).
References
- 1.Bitterman, K. J., Medvedik, O. & Sinclair, D. A. (2003) Microbiol. Mol. Biol. Rev. 67, 376–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guarente, L. (2000) Genes Dev. 14, 1021–1026. [PubMed] [Google Scholar]
- 3.Avalos, J. L., Celic, I., Muhammad, S., Cosgrove, M. S., Boeke, J. D. & Wolberger, C. (2002) Mol. Cell 10, 523–535. [DOI] [PubMed] [Google Scholar]
- 4.Finnin, M. S., Donigian, J. R. & Pavletich, N. P. (2001) Nat. Struct. Biol. 8, 621–625. [DOI] [PubMed] [Google Scholar]
- 5.Zhao, K., Chai, X., Clements, A. & Marmorstein, R. (2003) Nat. Struct. Biol. 10, 864–871. [DOI] [PubMed] [Google Scholar]
- 6.Min, J., Landry, J., Sternglanz, R. & Xu, R. M. (2001) Cell 105, 269–279. [DOI] [PubMed] [Google Scholar]
- 7.Zhao, K., Chai, X. & Marmorstein, R. (2003) Structure (Cambridge, MA) 11, 1403–1411. [DOI] [PubMed] [Google Scholar]
- 8.Jackson, M. D. & Denu, J. M. (2002) J. Biol. Chem. 277, 18535–18544. [DOI] [PubMed] [Google Scholar]
- 9.Sauve, A. A., Celic, I., Avalos, J., Deng, H. T., Boeke, J. D. & Schramm, V. L. (2001) Biochemistry 40, 15456–15463. [DOI] [PubMed] [Google Scholar]
- 10.Chang, J. H., Kim, H. C., Hwang, K. Y., Lee, J. W., Jackson, S. P., Bell, S. D. & Cho, Y. (2002) J. Biol. Chem. 277, 34489–34498. [DOI] [PubMed] [Google Scholar]
- 11.Tanny, J. C., Dowd, G. J., Huang, J., Hilz, H. & Moazed, D. (1999) Cell 99, 735–745. [DOI] [PubMed] [Google Scholar]
- 12.Slama, J. T. & Simmons, A. M. (1989) Biochemistry 28, 7688–7694. [DOI] [PubMed] [Google Scholar]
- 13.Slama, J. T., Aboul-Ela, N., Goli, D. M., Cheesman, B. V., Simmons, A. M. & Jacobson, M. K. (1995) J. Med. Chem. 38, 389–393. [DOI] [PubMed] [Google Scholar]
- 14.Landry, J., Slama, J. T. & Sternglanz, R. (2000) Biochem. Biophys. Res. Commun. 278, 685–690. [DOI] [PubMed] [Google Scholar]
- 15.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong, C. M., Kaeberlein, M., Imai, S. I. & Guarente, L. (2002) Mol. Biol. Cell 13, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauve, A. A. & Schramm, V. L. (2003) Biochemistry 42, 9249–9256. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, M. D., Schmidt, M. T., Oppenheimer, N. J. & Denu, J. M. (2003) J. Biol. Chem. 278, 50985–50998. [DOI] [PubMed] [Google Scholar]
- 20.Barlev, N. A., Liu, L., Chehab, N. H., Mansfield, K., Harris, K. G., Halazonetis, T. D. & Berger, S. L. (2001) Mol. Cell 8, 1243–1254. [DOI] [PubMed] [Google Scholar]
- 21.Liu, L., Scolnick, D. M., Trievel, R. C., Zhang, H. B., Marmorstein, R., Halazonetis, T. D. & Berger, S. L. (1999) Mol. Cell. Biol. 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, A., Lai, C. H., Zhao, X., Saito, S., Hamilton, M. H., Appella, E. & Yao, T. P. (2001) EMBO J. 20, 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakaguchi, K., Herrera, J. E., Saito, S., Miki, T., Bustin, M., Vassilev, A., Anderson, C. W. & Appella, E. (1998) Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, J. Y., Nikolaev, A. Y., Imai, S., Chen, D. L., Su, F., Shiloh, A., Guarente, L. & Gu, W. (2001) Cell 107, 137–148. [DOI] [PubMed] [Google Scholar]
- 25.Vaziri, H., Dessain, S. K., Eagon, E. N., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. & Weinberg, R. A. (2001) Cell 107, 149–159. [DOI] [PubMed] [Google Scholar]
- 26.Langley, E., Pearson, M., Faretta, M., Bauer, U. M., Frye, R. A., Minucci, S., Pelicci, P. G. & Kouzarides, T. (2002) EMBO J. 21, 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng, H. L., Mostoslavsky, R., Saito, S., Manis, J. P., Gu, Y. S., Patel, P., Bronson, R., Appella, E., Alt, F. W. & Chua, K. F. (2003) Proc. Natl. Acad. Sci. USA 100, 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grozinger, C. M., Chao, E. D., Blackwell, H. E., Moazed, D. & Schreiber, S. L. (2001) J. Biol. Chem. 276, 38837–38843. [DOI] [PubMed] [Google Scholar]
- 29.Bedalov, A., Gatbonton, T., Irvine, W. P., Gottschling, D. E. & Simon, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 15113–15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirao, M., Posakony, J., Nelson, M., Hruby, H., Jung, M., Simon, J. A. & Bedalov, A. (2003) J. Biol. Chem. 278, 52773–52782. [DOI] [PubMed] [Google Scholar]
- 31.Bitterman, K. J., Anderson, R. M., Cohen, H. Y., Latorre-Esteves, M. & Sinclair, D. A. (2002) J. Biol. Chem. 277, 45099–45107. [DOI] [PubMed] [Google Scholar]
- 32.Lin, S. J., Defossez, P. A. & Guarente, L. (2000) Science 289, 2126–2128. [DOI] [PubMed] [Google Scholar]
- 33.Lin, S. J., Kaeberlein, M., Andalis, A. A., Sturtz, L. A., Defossez, P. A., Culotta, V. C., Fink, G. R. & Guarente, L. (2002) Nature 418, 344–348. [DOI] [PubMed] [Google Scholar]
- 34.Tissenbaum, H. A. & Guarente, L. (2001) Nature 410, 227–230. [DOI] [PubMed] [Google Scholar]
- 35.Hekimi, S. & Guarente, L. (2003) Science 299, 1351–1354. [DOI] [PubMed] [Google Scholar]
- 36.Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., Zipkin, R. E., Chung, P., Kisielewski, A., Zhang, L. L., et al. (2003) Nature 425, 191–196. [DOI] [PubMed] [Google Scholar]
- 37.Hall, S. S. (2003) Science 301, 1165. [DOI] [PubMed] [Google Scholar]
- 38.Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O. & Sinclair, D. A. (2003) Nature 423, 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallo, C. M., Smith, D. L., Jr., & Smith, J. S. (2004) Mol. Cell. Biol. 24, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.