Abstract
The straight-chain C10 to C18 unsaturated aliphatic compounds containing an oxygenated functional group (aldehyde, alcohol, or acetate ester) derived from saturated C16 or C18 fatty acids are a major class of sex pheromone components produced by female moths. In the biosynthesis of these pheromone components, various combinations of limited chain-shortening and regio- and stereospecific desaturation reactions significantly contribute to the production of a vast number of the species-specific pheromone components in Lepidoptera. Biosynthesis of the silkmoth sex pheromone bombykol, (E,Z)-10,12-hexadecadien-1-ol, involves two consecutive desaturation steps, the second of which is unique in that it generates a conjugated diene system from the Δ11-monoene C16 intermediate. In experiments designed to characterize the acyl-CoA desaturases responsible for bombykol biosynthesis, we have cloned three cDNAs encoding desaturase family members from the pheromone gland of the inbred strain of the silkmoth, Bombyx mori. Transcript analyses by RT-PCR and subsequent functional assays using a Bac-to-Bac baculovirus expression system revealed that desat1 is the only desaturase gene prominently expressed during pheromonogenesis and that its gene product, B. mori Desat1, possesses both Z11 desaturation and Δ10,12-desaturation activities. Consequently, we have concluded that B. mori Desat1 is not only a bifunctional desaturase involved in bombykol biosynthesis but that it is also the enzyme responsible for both desaturation steps.
Many species of moths (Insecta: Lepidoptera) release sex pheromones, which are species-specific multicomponent blends of semiochemicals, to attract conspecific mates (1). A major class of sex pheromone components produced by these female moths has been characterized by the presence of straight-chain C10-C18 unsaturated aliphatic compounds containing an oxygenated functional group (aldehyde, alcohol, or acetate ester) derived from saturated C16 or C18 fatty acid (FA) intermediates that are synthesized de novo in the abdominal pheromone gland cells (2).
Studies over the past 2 decades have established a general scheme regarding the biosynthetic pathway of this class of pheromone components in which the FA intermediates are stepwise converted to the final components through a combination of desaturation and chain-shortening reactions followed by reductive modifications of the carbonyl carbon (3). In this biosynthetic pathway, various combinations of limited chain-shortening and regio- and stereospecific desaturation steps significantly contribute to the production of a large number of the species-specific pheromone components in Lepidoptera. Although the enzymes used in the limited chain-shortening steps have yet to be characterized, intensive research on the desaturases expressed in the pheromone gland of various moth species has revealed that diverse desaturases with different regio- and stereospecificities participate in generating the pheromone components (4).
Bombykol, (E,Z)-10,12-hexadecadien-1-ol, was the first chemically identified pheromone from organisms and is the major sex pheromone component produced by the domesticated silkworm, Bombyx mori (5). The biosynthesis of bombykol from the intermediate palmitate (C16:acyl) is a rather simple procedure that eschews the common chain-shortening and hydroxyl group modification steps for three consecutive steps, i.e., Z11 desaturation, (E,Z)-10,12 desaturation, and fatty-acyl reduction (2, 6). Among these steps, we have recently characterized the enzyme responsible for the fatty-acyl reduction, which we named pheromone gland fatty-acyl reductase (pgFAR) (7). On the other hand, the desaturases responsible for the two desaturation steps have yet to be identified and characterized. The first desaturation step seems to be a less species-specific step that is presumably catalyzed by the Z11-desaturase commonly used in the pheromone biosyntheses of numerous moth species. In contrast, the second desaturation step is less common in that it generates a conjugated diene system through 1,4-elimination of two hydrogen atoms at the allylic positions of the double bond in the Z11-monoene C16 intermediate (2). Similar 1,4-desaturation reactions involving monoene acyl precursors have only been observed in a handful of sex pheromone biosynthetic pathways (8–10). In this paper, we describe the functional characterization of a cDNA from the B. mori pheromone gland that encodes a bifunctional desaturase responsible for both of the desaturation steps in bombykol biosynthesis.
Materials and Methods
Insects. B. mori were reared at 25°C under 16:8 (light:dark) on an artificial diet as described in ref. 11.
cDNA Cloning. To characterize the B. mori desaturase genes that are expressed in the pheromone gland during pheromonogenesis, pheromone glands from the inbred strain p50 of B. mori were dissected immediately after adult eclosion (12). For the most abundant desaturase gene, desat1, the p50 pheromonegland cDNA library described in ref. 7 was screened with a 519-bp DNA fragment of desat1 as a probe that was prepared by using the direct nucleic acid labeling and detection systems (Amersham Pharmacia). The 519-bp DNA fragment used as a probewasamplifiedbyusingtheprimers5′-CTAAATTACTAAAATGCCTCC-3′ and 5′-AAATATCTGACATGTCAAGGC-3′, which were designed from the 2,029-bp B. mori desat1 cDNA previously cloned from the hybrid strain Shuko × Ryuhaku (12). After subcloning into pBluescript vectors, the inserts of the positive clones were sequenced with an ABI PRISM 377XL DNA Sequencer (Applied Biosystems) by using the Big Dye Terminator cycle sequencing kit v1.1 (Applied Biosystems) according to the manufacturer's instruction.
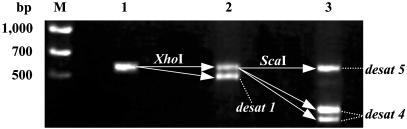
For other desaturase genes expressed in the pheromone gland, poly(A)+ RNA (300 ng) was prepared with the microfast-track 2.0 mRNA isolation kit (Invitrogen) and reverse-transcribed with Oligo(dT)20 by using the Thermoscript RT-PCR system (Invitrogen). To amplify DNA fragments of all of the desaturase genes present, a set of degenerate primers, DsF (5′-ATHACNGCNGGNGCNCAYMG-3′) and DsR (5′-AANRYRTGRTGRTARTTRTG-3′), were designed from the conserved sequences encoding the first and third histidine-cluster regions of known insect desaturases (4). Amplification was performed with KOD-Plus-(Toyobo, Osaka), and conditions for the PCR used were 94°C for 2 min followed by 35 cycles at 94°C for 15 s, 50°C for 30 s, and 68°C for 40 s. The 566-bp PCR products containing the various desaturase gene fragments were fractionated on 2% agarose gel by means of sequential digestions with XhoI and ScaI (Fig. 1) and then cloned with the ZeroBlunt TOPO PCR cloning kit for sequencing (Invitrogen).
Fig. 1.
Fractionation of the 566-bp PCR products containing pheromone gland desaturase gene fragments after sequential digestions with restriction enzymes. Lane 1, undigested PCR products; lane 2, XhoI digests separating desat1 fragments; lane 3, ScaI digests after XhoI digestion; M, size markers.
RT-PCR. Total RNAs (1 μg of each) from various tissues taken from the newly emerged female moths of B. mori were prepared with the TRIzol reagent (Invitrogen) and reverse-transcribed with the RNA PCR kit Ver. 2.1 (Takara Bio, Otsu, Japan). RT-PCR was performed with LA Taq polymerase (Takara Bio), the conditions for which were 94°C for 3 min followed by 20 cycles at 94°C for 1 min, 55°C for 1 min, and 68°C for 30 s. The gene-specific primer sets used were 5′-CGAGATCCTGTTGATGGTC-3′ and 5′-ATGCATTTGCCATGCCAC-3′ for desat1, 5′-GGCCTCTACGTGTTATCC-3′ and 5′-ACACGGAGATGTTCTCGG-3′ for desat4, and 5′-GGCCACTGCAGATCATAC-3′ and 5′-GCAGGGATACGATCAGG-3′ for desat5. PCR products were analyzed on a 2% agarose gel and visualized with ethidium bromide.
Construction of Recombinant Baculoviruses. A plasmid, designated as pFASTBac-desat1, containing the 330-aa ORF of p50 desat1 behind the polyhedrin promoter was generated with the pFast-Bac1 vector (Invitrogen). For construction of pFASTBac-desat1, a pair of gene-specific primers was designed: 5′-AAAGAATTCAAAAAATGCCTCCTAATTCAG-3′ (an EcoRI site is included) and 5′-AAAGGTACCTCAATTCATTCCATTTGGTCG-3′ (a KpnI site is included). PCR was performed by using KOD-Plus-, the conditions of which included 94°C for 2 min followed by 25 cycles at 94°C for 15 s, 45°C for 30 s, and 68°C for 72 s. The PCR product was double digested with KpnI and EcoRI (Takara Bio), separated by agarose-gel electrophoresis, and purified from the gel with a QIAquick gel extraction kit (Qiagen, Valencia, CA). The purified DNA was ligated into a linearized pFastBac 1 vector that had been predigested with KpnI and EcoRI, and then sequenced. The expression cassette of pFASTBac-desat1 was incorporated into the baculovirus genome (bacmid DNA) in DH10Bac competent cells (Invitrogen) by using Tn7 site-specific transposition according to the manufacturer's instructions for the Bac-to-Bac expression system (Invitrogen). The resulting recombinant bacmid DNA was purified with a Wizard Plus SV Minipreps DNA purification system (Promega) and transfected into Sf9 cells (Invitrogen) by using Lipofectin reagent (Invitrogen). After incubation at 27°C for 4 days, the resultant recombinant baculovirus, named Desat1-AcNPV, was plaque-purified and propagated in Sf9 cells. Bac1-AcNPV, a control recombinant baculovirus, was prepared in the same way by using pFastBac 1. Viral titers were determined with a conventional plaque assay.
Functional Assay. Sf9 cells (1.4 × 107 cells) seeded in a cell culture dish (150 × 25 mm) were infected with Desat1-AcNPV or Bac1-AcNPV at a multiplicity of infection of 10 and incubated in 20 ml of IPL41 medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Sigma). After incubation at 27°C for 4 days, the cell pellet was extracted with 2 ml of n-hexane and was then reextracted with 2 ml of chloroform/methanol (2:1). After evaporation, the combined extracts were base-methanolyzed in 0.2 ml of 0.5 N KOH/methanol at room temperature for 2 h and was then neutralized with 1 N HCl. The resulting FA methyl esters (FAMEs) were analyzed by GC/MS by using a Hewlett Packard 6890 gas chromatograph equipped with an HP-5 capillary column (0.25 mm i.d. × 30 m, Agilent, Palo Alto, CA) coupled to an HP5973 series mass selective detector. The oven temperature gradient was programmed at 50°C for 1 min, then 10°C/min to 160°C, and finally 4°C/min to 220°C and held for 5 min.
Free FAs used as substrates were D3-palmitic acid (hexadecanoic-16,16,16,-D3 acid, D3-C16:acid) (Isotec) and (Z)-11-hexadecenoic acid (Z11–16:acid) (7). These substrates, dissolved in n-hexane, were added to the incubation vials, and the solvent was allowed to evaporate before incubation. The double bond position of unsaturated FAMEs was verified by MS analysis of the dimethyl disulfide adducts for monoene acids (13) or the 4-methyl-1,2,4-triazoline-3,5-dione adducts for conjugated diene acids (14). The oven temperature gradient was programmed at 50°C for 1 min, then 10°C/min to 240°C, and finally 4°C/min to 320°C and held for 5 min. Stereoisomers were assigned based on retention times with synthetic standards. E10,E12–16:Me and E10,Z12–16:Me were prepared from their corresponding acids (7). E12,Z14–18:Me and E12,E14–18:Me were synthesized from 12-[(tert-butyldimethylsilyl)oxy]-1-dodecanol (15), which was oxidized under Swern's conditions (16) and then subjected to the Wittig–Horner–Emmons reaction (17). Diisobutylaluminum hydride reduction of the resulting product, ethyl (E)-14-[(tert-butyldimethylsilyl)oxy]-2-tetradecenoate, followed by MnO2 oxidation yielded the corresponding aldehyde, which was converted to E12,Z14–18:acid by the same method as used for the preparation of E10,Z12–16:acid (7). Methylation of the acid products yielded a mixture of E12,Z14–18:Me and E12,E14–18:Me (100:14 ratio, by 1H-NMR analysis).
Results
cDNA Clones Encoding Pheromone Gland Fatty-Acyl Desaturases in B. mori. We previously obtained genes encoding acyl-CoA desaturase homologues (desat1–3) from an EST database of B. mori (12). Although the genes desat1 and desat2 (GenBank accession nos. AF157627 and AF182405) were shown to be expressed specifically in the pheromone gland, they were considered to be alleles derived from the heterozygote of B. mori (a hybrid strain, Shuko × Ryuhaku). To eliminate this kind of ambiguity in the present experiments, we used p50, an inbred strain of B. mori, to clone and characterize the acyl-CoA desaturase genes that are expressed in the pheromone gland during pheromonogenesis. We first cloned desat1 cDNA from a p50 pheromone-gland cDNA library by using a 519-bp fragment as a probe that was prepared from the 2,029-bp B. mori desat1 cDNA of the hybrid strain Shuko × Ryuhaku (12). The full-length cDNA of the cloned p50 desat1 spans 2,019 nt with an ORF encoding a 330-aa protein. Sequence analysis indicated a conserved amino acid substitution (Leu for Ile) at position 105 when compared with the Shuko × Ryuhaku Desat1 protein as well as 70% and 64% identities to the known Z11 desaturases from Helicoverpa zea (GenBank accession no. AF272342) and Trichoplusia ni (AF035375), respectively. Genomic Southern blot analysis confirmed that desat1 is a single copy gene in the p50 genome (data not shown).
To obtain other desaturase cDNAs that might be less abundantly expressed in the pheromone gland, we prepared poly(A)+ RNA from the p50 pheromone glands immediately after adult eclosion and performed RT-PCR to amplify portions of the desaturase cDNAs included. For RT-PCR, we designed a set of degenerate primers based on the conserved sequences corresponding to the first and third histidine-cluster regions of known insect desaturases, and the ensuing amplification yielded the expected 566-bp PCR products (Fig. 1, lane 1). We hypothesized that the PCR products should be a mixture of DNA fragments derived from various desaturase cDNAs; consequently, the products were digested with XhoIto remove the desat1-derived fragment (Fig. 1, lane 2). Subsequent cloning and sequencing of products that failed to be digested revealed the existence of a desaturase gene designated desat4. Furthermore, because the desat4 fragment contained a ScaI digestion site, the XhoI-resistant 566-bp PCR products were further digested with ScaI (Fig. 1, lane 3). Cloning of the resulting ScaI unsusceptible products afforded an additional desaturase-derived fragment that we have designated desat5. The full-length cDNAs of these desaturases were subsequently cloned from the p50 pheromone-gland cDNA library. The desat4 ORF encodes a 352-aa protein with 86% identities to both the Ostrinia furnacalis Z9-desaturase (GenBank accession no. AY057862) and the Ostrinia nubilalis Z9-desaturase (AF243047). In contrast, the desat5 ORF encodes a 339-aa protein with 69% identities to both the Spodoptera littoralis Z,E11-desaturase (AY362879) and the H. zea Z11-desaturase (AF272342). The amino acid sequences deduced from the ORFs of desat4 and desat5 are aligned in Fig. 2 with that of the p50 desat1.
Fig. 2.
Multiple alignment of B. mori pheromone gland fatty-acyl desaturases. The consensus residues are highlighted in black, and the residues that exhibit similarity are shaded in gray. The three conserved histidine clusters are overlined. The regions corresponding to the primer sequence used for RT-PCR are indicated by arrows. Alignment was made by using the clustalw program (http://hypernig.nig.ac.jp/homology/clustalw.shtml) and boxshade 3.21 file (www.ch.embnet.org/software/BOX_form.html).
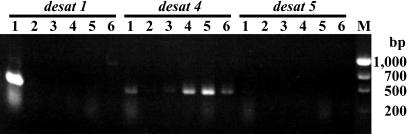
Detection of Desaturase Transcripts by Using RT-PCR. To examine the levels of the desaturase-gene transcripts, we prepared B. mori pheromone glands from newly emerged female moths and analyzed transcript levels by RT-PCR using gene-specific primers (Fig. 3). The most abundant of the transcripts found in the pheromone gland was desat1. In contrast, the levels of the other two desaturase-encoding transcripts were quite low. We also analyzed the expression of these transcripts in other tissues at the same stage, and the results showed specific expression of the 2-kb desat1 transcript within the pheromone gland with no detectable signal observed in the other tissues examined, which included whole head, flight muscle, Malpighian tubule, testis, and ovary (Fig. 3). Although the desat4 transcript was detected in some tissues, i.e., the Malpighian tubule, testis, and ovary, no detectable signal was observed for the desat5 transcript in the tissues examined.
Fig. 3.
RT-PCR analysis of desat transcripts from B. mori tissues. Tissues were dissected from newly emerged female moths (day 0). Testes were dissected from newly emerged male moths. 1, pheromone gland; 2, head; 3, flight muscle; 4, Malpighian tubule; 5, testis; 6, ovary; M, size markers.
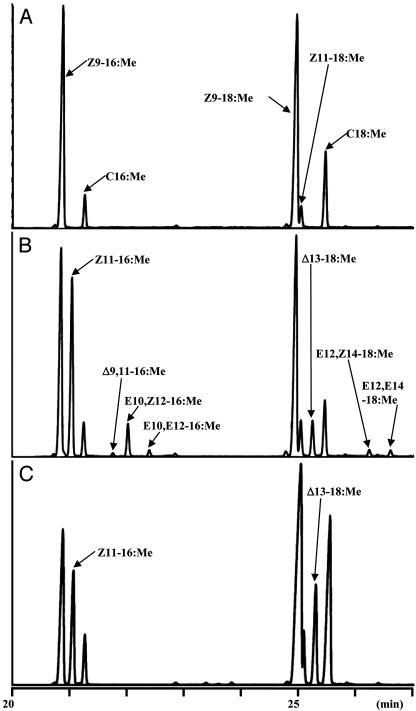
Functional Assay of B. mori Desat1 in a Bac-to-Bac Baculovirus Expression System. Analysis of the desaturase gene transcripts by RT-PCR indicated that desat1 is the only desaturase gene actively expressed in the pheromone gland during pheromonogenesis. To investigate the desaturase activity of B. mori Desat1, we constructed the recombinant baculoviruses Desat1-AcNPV and Bac1-AcNPV. Desat1-AcNPV, but not Bac1-AcNPV, contains the 330-aa ORF of desat1 and expresses the B. mori Desat1 protein under the control of the polyhedrin promoter. After infection with the recombinant viruses and incubation at 27°C for 4 days, infected Sf9 cells were subjected to extraction of fatty-acyl compounds, which were then converted to FAMEs by base methanolysis and analyzed by GC/MS. In control experiments with Bac1-AcNPV, the infected cells exhibited several background FAMEs (Z9–16:Me, C16:Me, Z9–18:Me, Z11–18:Me, and C18:Me; Fig. 4A). In contrast, cells infected with Desat1-AcNPV yielded additional peaks in the regions typical of C16- and C18-unsaturated FAMEs (Fig. 4B), indicating that the B. mori desat1 gene was functionally expressed. In these experiments, Z11–16:Me was the most predominantly produced of the newly generated FAMEs, indicating that B. mori Desat1 has a Z11-desaturase activity and is, therefore, responsible for the first desaturation step from C16:acyl in bombykol biosynthesis. Interestingly, we also found that the cells infected with Desat1-AcNPV concomitantly produced three distinct C16 dienes in addition to the monoene Z11–16:Me (Fig. 4B). Structure elucidation of these dienes by MS for their 4-methyl-1,2,4-triazoline-3,5-dione adducts and comparison with synthetic compounds confirmed them to be conjugated diene isomers; the major product was E10,Z12–16:Me, whereas the minor products were E10,E12–16:Me and Δ9,11–16:Me. With respect to the production of these conjugated dienes, the Δ9,11–16:Me likely results from the conversion of the highly abundant endogenous Z9–16:acyl by the overexpressed recombinant Z11-desaturase activity of B. mori Desat1. In contrast, we found the production of E10,Z12–16:Me and E10,E12–16:Me (5:1 ratio, Fig. 4B) to be quite intriguing; it has been demonstrated that E10,Z12–16:acyl is the immediate precursor for bombykol and is the product derived from Z11–16:acyl in the second desaturation step of bombykol biosynthesis (6). Importantly, in addition to the predominant bombykol (=E10,Z12–16:OH), female moths of B. mori usually produce a minor pheromone component, E10,E12–16:OH, in a ratio of ≈1:5 with bombykol (18). Thus, the newly generated diene E10,E12–16:acyl is also the likely immediate precursor for the minor component, E10,E12–16:OH derived from Z11–16:acyl. Because these two diene isomers were produced by the Sf9 cells infected with Desat1-AcNPV, but not with Bac1-AcNPV, it is, therefore, quite possible that these two diene isomers are produced by a 1,4-desaturase activity of B. mori Desat1 to form the Δ10,12-conjugated diene from Z11–16:acyl.
Fig. 4.
GC/MS analysis by total ion current for the FAMEs prepared from Sf9 cells infected with the recombinant Desat1-AcNPV or the recombinant Bac1-AcNPV (control). (A) Sf9 cells infected with Bac1-AcNPV. (B) Sf9 cells infected with Desat1-AcNPV. (C) Sf9 cells infected with Bac1-AcNPV and incubated in the presence of Z11–16:acid.
It has been reported that chain-elongation reactions occur in Sf21 insect cells (19). This also seems to be the case with Sf9 cells. In control Sf9 cells, we found production of the unusual Z11–18:Me (Fig. 4A), which is likely the result of an elongation reaction involving endogenous Z9–16:acyl. Furthermore, if we added Z11–16:acid to the medium, the control cells produced a large amount of Δ13–18:Me (Fig. 4C). This observation, in conjunction with the following experiment using D3-C16:acid, confirms that our Sf9 system does contain a strong chain-elongation activity, thereby explaining why Δ13–18:Me and other C18-FAME dienes (identified as E12,Z14–18:Me and E12,E14–18:Me) were produced in Sf9 cells infected with Desat1-AcNPV (Fig. 4B).
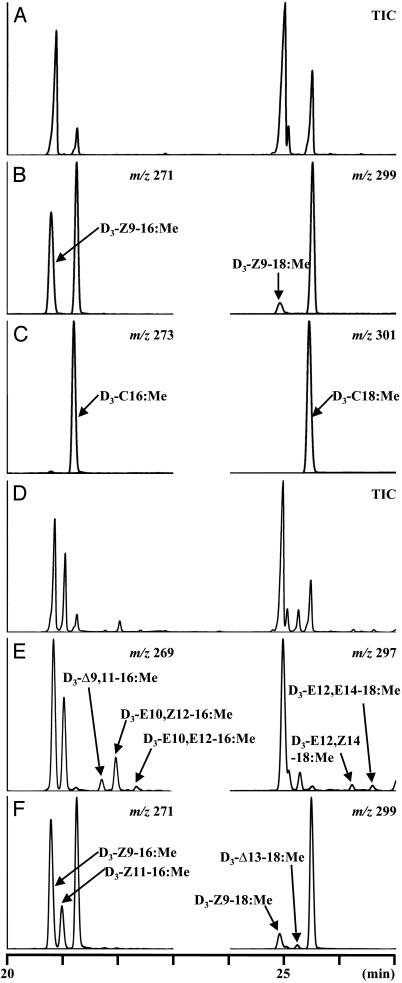
To further examine the production of C16-FAME mono- and dienes by B. mori Desat1, we analyzed the conversion of a deuterium-labeled FA in Sf9 cells infected with Desat1-AcNPV. When we added D3-C16:acid to the incubation medium, the control cells infected with Bac1-AcNPV produced deuteriated products, D3-Z9–16:Me, D3-Z9–18:Me, and D3-C18:Me (Fig. 5 A–C), indicating that Sf9 cells possess a Z9-desaturase activity in addition to the chain-elongation activity, as reported in Sf21 cells (19). In contrast, the cells infected with Desat1-AcNPV produced the deuterium-labeled B. mori pheromone precursors, i.e., D3-Z11–16:Me, D3-E10,Z12–16:Me, and D3-E10,E12–16:Me (Fig. 5 D–F). Because these precursors were not produced by the Sf9 cells infected with Bac1-AcNPV even in the presence of Z11–16:acid (Fig. 4C), they indeed must be produced by the action of B. mori Desat1 from the incorporated D3-C16:acid. We can, therefore, conclude that B. mori Desat1 is a desaturase with dual activities of Z11 desaturation and Δ10,12 desaturation, and that, consequently, it is responsible for the two consecutive desaturation steps in bombykol biosynthesis. With respect to the other unsaturated FAMEs produced by cells infected with Desat1-AcNPV, the diene D3-Δ9,11–16:Me can be attributed to the conversion of D3-Z9–16:Me by the overexpressed recombinant Z11-desaturase activity of the B. mori Desat1, whereas the unsaturated C18-FAMEs, i.e., D3-Δ13–18:Me, D3-E12,Z14–18:Me, and D3-E12,E14–18:Me, are the products derived from the corresponding C16-FAMEs after chain-elongation reactions.
Fig. 5.
GC/MS analysis by total ion current or mass chromatograms for the FAMEs prepared from Sf9 cells. Sf9 cells were infected with Bac1-AcNPV (A–C) or Desat1-AcNPV (D–F) and incubated in the presence of 0.1 mM D3-C16:acid; m/z 269, [M]+ of D3-diene C16:Me; m/z 271, [M]+ of D3-monoene C16:Me; m/z 273, [M]+ of D3-C16:Me; m/z 297, [M]+ of D3-diene C18:Me; m/z 299, [M]+ of D3-monoene C18:Me; m/z 301, [M]+ of D3-C18:Me.
Discussion
Over the past 20 years, numerous studies have established a general scheme for the biosynthetic pathway of a major class of Lepidopteran sex pheromone components in which the common C16 or C18 FA intermediate is stepwise converted to the final components through various combinations of chain-shortening and desaturation reactions that are then followed by reductive modifications of the carbonyl carbon (3). For the biosynthesis of such pheromone components, various pheromone gland-specific desaturases with diverse regio- and stereospecificities have been shown to contribute significantly to the generation of species-specific pheromone components (4).
In the silkmoth, B. mori, bombykol biosynthesis involves two consecutive desaturation steps, i.e., Z11 desaturation and (E,Z)-10,12 desaturation (2, 6). The first Z11-desaturation step is a prevalent one that has been described in many other moth species, whereas the second (E,Z)-10,12-desaturation step generates a conjugated diene system by means of 1,4-elimination of two allylic hydrogen atoms (2). In the present experiments designed to characterize the enzymes responsible for these two desaturation steps, we assumed that there would be two distinct desaturases acting sequentially. Consequently, we designed a pair of degenerate primers based on two conserved sequences corresponding to the first and third histidine-cluster regions of known desaturases in Lepidoptera (4) and tried to amplify all desaturase gene fragments present in the pheromone gland during pheromonogenesis. Through sequential fractionation by digesting the PCR products with XhoI and ScaI (Fig. 1), we have cloned three cDNAs (desat1, desat4, and desat5) that encode members of the desaturase family (Fig. 2). Because subsequent analysis of the three desaturase gene transcripts indicated that desat1 is the only desaturase gene actively expressed in the pheromone gland (Fig. 3), only desat1 was selected for functional analysis by means of a Bac-to-Bac baculovirus expression system. As a result, we uncovered functional aspects of its gene product, B. mori Desat1 (Figs. 4 and 5). Although the Z11-desaturase activity of B. mori Desat1 was initially anticipated based on the sequence homology to other desaturases, we unexpectedly found that Sf9 cells infected with Desat1-AcNPV produced the pheromone precursor dienes, E10,Z12–16:Me and E10,E12–16:Me, in addition to the expected intermediate Z11–16:Me (Fig. 4B). Further experiments by using the deuterium-labeled C16:acid confirmed that B. mori Desat1 is a desaturase with activities of Z11 desaturation and Δ10,12 desaturation. Consequently, we have concluded that B. mori Desat1 is the sole desaturase involved in bombykol biosynthesis and that it catalyzes both desaturation steps in the biosynthetic pathway.
In addition to the above-mentioned findings, we found that production of the pheromone component precursors, E10,Z12– 16:Me and E10,E12–16:Me, by B. mori Desat1 occurred in a ratio of 5:1 (Fig. 4B). Interestingly, this ratio coincides with the proportion of the final components in the pheromone blend of this species; B. mori produces the pheromone components E10,Z12–16:OH (=bombykol) and E10,E12–16:OH in a ratio of 84:16 with a trace of E10,Z12–16:AL (18). Although the regulatory mechanisms to determine the proportion of species-specific pheromone blend components in Lepidoptera have yet to be understood, our results indicate that the ability of B. mori Desat1 to produce the pheromone precursor dienes at a defined ratio seems to significantly contribute to the determination of the final pheromone blend proportion in this species.
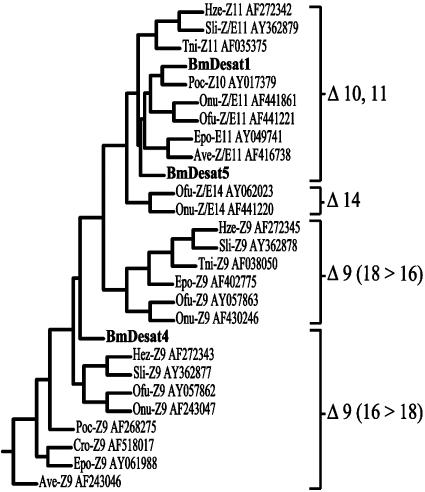
Although the pheromone gland of B. mori contains three different desaturase gene transcripts, only desat1 generates a functional protein product used for pheromone biosynthesis. Phylogenetic analysis of desaturases in Lepidoptera indicate that the B. mori Desat1 can be grouped within the Δ10,11 branch, albeit on the periphery (Fig. 6) (20, 21). This selective partitioning is undoubtedly the result of the enzyme's bifunctionality and clearly demonstrates the divergence of the B. mori Desat1 from the other Δ11 desaturases. Analysis of the other pheromone gland-derived desaturases indicates that desat4 can be clustered with Δ9 desaturases, whereas desat5 can be grouped within the Δ10,11 branch of desaturase genes (20).
Fig. 6.
Phylogeny of various lepidopteran desaturases. Tree was reconstructed by using the phylogeny inference package (phylip) by Felsenstein (21) (http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). The accession numbers for the sequences are listed to the right of the species abbreviation. The B. mori desaturase genes are listed in bold.
It is known that some higher plants accumulate conjugated linolenic acids as major components of seed oils (22). In the biosynthesis of conjugated linolenic acids, enzymes termed conjugases catalyze the formation of conjugated double bonds; the Z12 double bond of linoleic acid, for instance, is converted into the two E11 and E13 double bonds found in α-eleostearic acid (23). Recently, several plant seed cDNAs encoding conjugases have been characterized (22–25). Interestingly, one such conjugase associated with the formation of the E11,Z13-diene in punicic acid (22) has been reported to exhibit both Z12 desaturation and E11,Z13-conjugation activities. This enzyme, however, shares only 28% amino acid identity with B. mori Desat1.
In this paper, we have demonstrated that B. mori Desat1 is a bifunctional enzyme that catalyzes both a Z11 desaturation and a Δ10,12 desaturation and that it converts palmitate to the conjugated dienes, Δ10,12–16:acyls. In the tobacco hornworm, Manduca sexta, female moths produce a pheromone blend composed of eight C16 aldehydes, of which E10,Z12–16:AL and E10,E12,Z14–16:AL have proven to be essential for biological activity (9). From studies on the M. sexta pheromone biosynthetic pathway, it has been suggested that the intermediate Z11–16:acyl is first converted to Δ10,12-conjugated dienes and then to Δ10,12,14-conjugated trienes (9). Because the procedure to form the Δ10,12-conjugated dienes through Δ11 desaturation is similar to that of B. mori, it is most likely that an enzyme similar to B. mori Desat1 participates in the biosynthesis of the M. sexta pheromone components. This enzymatic process has also been observed in other moth sex pheromone biosynthetic pathways; in S. littoralis, Z11–14:acyl is transformed into E10,E12–14:acyl (10), whereas in Cydia pomonella E9–12:acyl is transformed into E8,E10–12:acyl (8). These findings indicate that the 1,4-desaturation reaction is not exclusive of C16 substrates, as it also occurs with other monounsaturated acyl substrates, although the desaturases responsible for these reactions have yet to be characterized.
Acknowledgments
We thank Shinji Atsusawa and Masaaki Kurihara for their technical assistance. This work was supported by the Bioarchitect Research Program and the Chemical Biology Research Program from RIKEN, and Ministry of Education, Science, Sports, and Culture of Japan Grant-in-Aid (A) 12306003 for Scientific Research and Grant-in-Aid 14656022 for Exploratory Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FA, fatty acid; FAME, fatty acid methyl ester; C16:acyl, palmitate; Z11–16:acid, (Z)-11-hexadecenoic acid; Z9–18:Me, (Z)-9-octadecenoic acid methyl ester; E10,Z12–16:Me, (E,Z)-10,12-hexadecadienoic acid methyl ester; E10,E12–16:OH, (E,E)-10,12-hexadecadien-1-ol; E10,E12,Z14–16:AL, (E,E,Z)-10,12,14-hexadecatrien-1-al (fatty acyls, FAs, FAMEs, fatty alcohols, and fatty aldehydes of other configurations, chain length, and positions of unsaturation are named similarly).
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB166851, AB166852, and AB166853).
References
- 1.Tamaki, Y. (1985) in Comprehensive Insect Physiology, Biochemistry, and Pharmacology, eds. Kerkut, G. A. & Gilbert, L. I. (Pergamon, New York), Vol. 9, pp. 145–191. [Google Scholar]
- 2.Bjostad, L. B., Wolf, W. A. & Roelofs, W. L. (1987) in Pheromone Biochemistry, eds. Prestwich, G. D. & Blomquist, G. J. (Academic, Orlando), pp. 77–120.
- 3.Tillman, J. A., Seybold, S. J., Jurenka, R. A. & Blomquist, G. J. (1999) Insect Biochem. Mol. Biol. 29, 481–514. [DOI] [PubMed] [Google Scholar]
- 4.Roelofs, W. L., Liu, W., Hao, G., Jiao, H., Rooney, A. P. & Linn, C. E., Jr. (2002) Proc. Natl. Acad. Sci. USA 99, 13621–13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butenandt, A., Beckman, R., Stamm, D. & Hecker, E. (1959) Z. Naturforsch. B14, 283–284. [Google Scholar]
- 6.Ando, T., Hase, R., Arima, R. & Uchiyama, M. (1988) Agric. Biol. Chem. 52, 473–478. [Google Scholar]
- 7.Moto, K., Yoshiga, T., Yamamoto, M., Takahashi, S., Okano, K., Ando, T., Nakata, T. & Matsumoto, S. (2003) Proc. Natl. Acad. Sci. USA 100, 9156–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lofstedt, C. & Bengtsson, M. (1988) J. Chem. Ecol. 14, 903–915. [DOI] [PubMed] [Google Scholar]
- 9.Tumlinson, J. H., Teal, P. E. A. & Fang, N. (1996) Bioorg. Med. Chem. 4, 451–460. [DOI] [PubMed] [Google Scholar]
- 10.Navarro, I., Mas, E., Fabrias, G. & Camps, F. (1997) Bioorg. Med. Chem. 5, 1267–1274. [DOI] [PubMed] [Google Scholar]
- 11.Fónagy, A., Matsumoto, S., Uchiumi, K., Orikasa, C. & Mitsui, T. (1992) J. Pesticide Sci. 17, 47–54. [Google Scholar]
- 12.Yoshiga, T., Okano, K., Mita, K., Shimada, T. & Matsumoto, S. (2000) Gene 246, 339–345. [DOI] [PubMed] [Google Scholar]
- 13.Buser, H. R., Arn, H., Guerin, P. & Rauscher, S. (1983) Anal. Chem. 55, 818–822. [Google Scholar]
- 14.McElfresh, J. S. & Millar, J. G. (1999) J. Chem. Ecol. 25, 687–709. [Google Scholar]
- 15.Krafft, M. E., Crooks, W. J., III, Zorc, B. & Milczanowski, S. E. (1988) J. Org. Chem. 53, 3158–3163. [Google Scholar]
- 16.Omura, K. & Swern, D. (1978) Tetrahedron 34, 1651–1660. [Google Scholar]
- 17.Fieser, L. F. & Fieser, M. (1967) in Reagents for Organic Synthesis, eds. Fieser, L. F. & Fieser, M. (Wiley, New York), Vol. 1, pp. 1216–1217. [Google Scholar]
- 18.Kasang, G. & Schneider, D. (1978) Naturwissenschaften 65, 337–338. [Google Scholar]
- 19.Liu, W., Jiao, H., Murray, N. C., O'Connor, M. & Roelofs, W. L. (2002) Proc. Natl. Acad. Sci. USA 99, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roelofs, W. L. & Rooney, A. P. (2003) Proc. Natl. Acad. Sci. USA 100, 9179–9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein, J. (1993) phylip, Phylogeny Inference Package (University of Washington, Seattle), Version 3.5c.
- 22.Iwabuchi, M., Kohno-Murase, J. & Imamura, J. (2003) J. Biol. Chem. 278, 4603–4610. [DOI] [PubMed] [Google Scholar]
- 23.Cahoon, E. B., Carlson, T. J., Ripp, K. G., Schweiger, B. J., Cook, G. A., Hall, S. E. & Kinney, A. J. (1999) Proc. Natl. Acad. Sci. USA 96, 12935–12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahoon, E. B., Ripp, K. G., Hall, S. E. & Kinney, A. J. (2001) J. Biol. Chem. 276, 2637–2643. [DOI] [PubMed] [Google Scholar]
- 25.Qui, X., Reed, D. W., Hong, H., MacKenzie, S. L. & Covello, P. S. (2001) Plant Physiol. 125, 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]