Abstract
Reducing lignin concentration in lignocellulosic biomass can increase forage digestibility for ruminant livestock and saccharification yields of biomass for bioenergy. In sorghum (Sorghum bicolor (L.) Moench) and several other C4 grasses, brown midrib (bmr) mutants have been shown to reduce lignin concentration. Putative bmr mutants isolated from an EMS-mutagenized population were characterized and classified based on their leaf midrib phenotype and allelism tests with the previously described sorghum bmr mutants bmr2, bmr6, and bmr12. These tests resulted in the identification of additional alleles of bmr2, bmr6, and bmr12, and, in addition, six bmr mutants were identified that were not allelic to these previously described loci. Further allelism testing among these six bmr mutants showed that they represented four novel bmr loci. Based on this study, the number of bmr loci uncovered in sorghum has doubled. The impact of these lines on agronomic traits and lignocellulosic composition was assessed in a 2-yr field study. Overall, most of the identified bmr lines showed reduced lignin concentration of their biomass relative to wild-type (WT). Effects of the six new bmr mutants on enzymatic saccharification of lignocellulosic materials were determined, but the amount of glucose released from the stover was similar to WT in all cases. Like bmr2, bmr6, and bmr12, these mutants may affect monolignol biosynthesis and may be useful for bioenergy and forage improvement when stacked together or in combination with the three previously described bmr alleles.
Keywords: acid detergent lignin (ADL), ethyl methanesulfonate (EMS), bioenergy, forage composition, allelism
Plant cell walls are vast reserves of photosynthetically fixed carbon. Global research efforts are focused on utilizing plant cell walls as renewable resources for the production of energy, chemical precursors, and fuels; however, the traditional uses of plant cell walls as fodder and fiber also remain important. Plant cell walls predominantly consist of the polysaccharides cellulose and hemicellulose and the phenolic polymer lignin. Lignin accounts for approximately 30% of organic carbon in the biosphere (Boerjan et al. 2003), and its environmental abundance is exceeded only by cellulose (Jung and Ni 1998). The lignin polymer cross-links cell wall polysaccharides, thereby stiffening and reinforcing the secondary cell wall structure (Boerjan et al. 2003). The resulting matrix is recalcitrant to both chemical degradation and biological digestion, which impairs hydrolysis of the polysaccharides into their monomeric sugars in ruminant livestock or cellulosic bioenergy systems. Hence, reducing lignin has become an important target for both bioenergy feedstock improvement (Chen and Dixon 2007; Dien et al. 2009; Vermerris et al. 2007) and improving fodder digestibility (Barrière et al. 2003; Guo et al. 2001; Jung and Allen 1995; Vogel and Jung 2001). Although lignin makes liberating sugars from cell walls more difficult, lignin serves critical functions for vascular plants. Lignin is required for vascular elements to transport water under negative pressure, a critical adaptive feature that allowed vascular plants to colonize land during their evolution (Boyce et al. 2004; Sperry 2003). The collapse of vascular elements has been observed in mutants extremely impaired in lignin synthesis (Jones et al. 2001; Piquemal et al. 1998; Ruel et al. 2009). Hence, there is a limit to the extent lignin content may be manipulated without significantly impacting plant fitness.
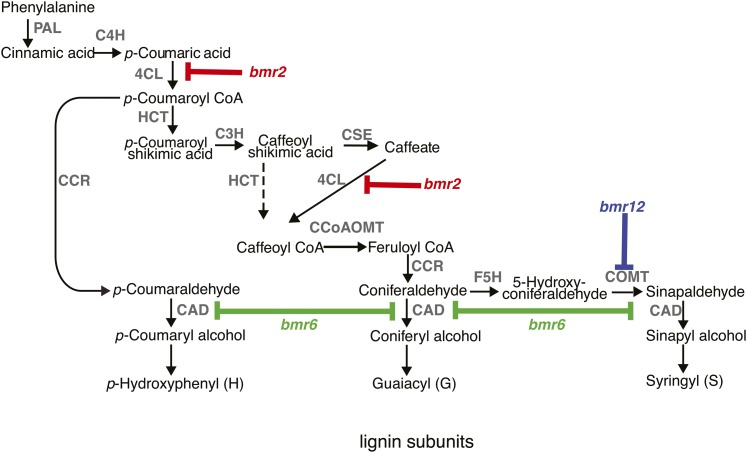
The subunits of lignin are synthesized from the amino acid phenylalanine, and aromatic amino acid synthesis and phenylpropanoid metabolism play central roles in vascular plants. Approximately 30% of the carbon flux passes through phenylalanine in plants (Tohge et al. 2013), which illustrates the importance of this pathway. In angiosperms, there are three main subunits of lignin [p-hydroxyphenol (H), guaiacyl (G), syringyl (S)] that are polymerized within the cell wall from their respective monolignols (p-coumaryl, coniferyl, sinapyl alcohols) via oxidative coupling (Boerjan et al. 2003). A consensus model of the monolignol biosynthetic pathway (Vanholme et al. 2013) has been developed based on the identification and characterization of monolignol biosynthetic enzymes from a range of plant species and the amino acid sequence conservation of these enzymes across plant genomes (Figure 1).
Figure 1.
The monolignol biosynthetic pathway in sorghum based on consensus models from dicot and monocot plants (Vanholme et al. 2013). The enzymatic steps (gray) are as follows: phenylalanine ammonia lyase (PAL); cinnamate 4-hydroxylase (C4H); 4-coumarate-CoA ligase (4CL); hydroxycinnamoyl CoA:shikimate transferase (HCT); p-coumarate 3-hydroxylase (C3H); caffeoyl shikimate esterase (CSE); caffeoyl CoA O-methyltransferase (CCoAOMT); cinnamyl CoA reductase (CRR); ferulate 5-hydroxylase (F5H); caffeic acid O-methyltransferase (COMT); and cinnamyl alcohol dehydrogenase (CAD). The bmr2, bmr6, and bmr12 mutants are impaired in 4CL, COMT, and CAD enzymatic activities, respectively.
The brown midrib phenotype has been useful for identifying mutants impaired in lignin synthesis in C4 grasses (Kuc and Nelson 1964; Gee et al. 1968; Porter et al. 1978; Cherney et al. 1988), because the tan to reddish brown leaf midribs visibly contrast to the white or green midribs observed in wild-type (WT) plants. In maize (Zea mays), spontaneous brown midrib (bm) mutants were first identified in the 1920s (Jorgenson 1931). To date, there are six known brown midrib (bm1-6) loci that have been identified in maize (Ali et al. 2010). In sorghum (Sorghum bicolor), a series of brown midrib (bmr) mutants was isolated from diethyl sulfate (DES) mutagenized populations at Purdue University in the 1970s (Porter et al. 1978; Bittinger et al. 1981). Allelism tests from this series identified four sorghum bmr loci, bmr2, bmr6, bmr12, and bmr19 (Saballos et al. 2008). The bmr19 mutant is not publicly available (effectively reducing the available sorghum brown midrib mutants to a set of three independent loci: bmr2, bmr6, and bmr12). However, bmr19 appears to be of limited value for forage and bioenergy applications, because it did not significantly reduce lignin concentration and did not markedly alter lignin subunit composition (Saballos et al. 2008). Brown midrib mutants have also been isolated in pearl millet (Pennisetum glaucum) (Cherney et al. 1988; Degenhart et al. 1995; Gupta 1995).
The brown midrib mutants have been used to identify and characterize the genes that encode the major enzymes for specific steps of monolignol biosynthesis for C4 grasses (Figure 1). The maize Bm3 and sorghum Bmr12 genes both encode orthologous caffeic acid O-methyltransferase (COMT), which catalyzes the penultimate step in monolignol biosynthesis (Vignols et al. 1995; Bout and Vermerris 2003). The maize Bm1 and the sorghum Bmr6 genes both encode orthologous cinnamyl alcohol dehydrogenase (CAD) (Saballos et al. 2009; Sattler et al. 2009; Chen et al. 2012), which catalyzes the last step in monolignol biosynthesis. Bmr2 was shown to encode a 4-coumarate coenzyme A ligase (4CL), which catalyzes an early step in monolignol biosynthesis (Saballos et al. 2012). Recently, the maize Bm2 gene was cloned and shown to encode a methylenetetrahydrofolate reductase (Tang et al. 2014). This enzyme catalyzes the rate-limiting step in one carbon (folate) metabolism and for the synthesis of the methyl donor S-adenosyl-L-methionine (SAM) (Roje et al. 1999, 2002). SAM is a cofactor for two methylation reactions in monolignol biosynthesis catalyzed by the caffeoyl-CoA O-methyltransferase (CCoAOMT) and the caffeic acid O-methyltransferase (COMT), respectively (Figure 1). The maize Bm4, Bm5, and Bm6 and sorghum Bmr19 genes have not yet been cloned and their functions remain to be fully elucidated.
Based on examination of lignin biosynthesis pathways, there is clear potential for major perturbations of monolignol synthesis at key enzymatic steps, any of which could result in reduced lignin concentration and/or altered lignin composition associated with the brown midrib phenotype in sorghum. From an ethyl methane sulfonate (EMS)-induced Targeting Induced Local Lesions in Genomes (TILLING) population (Xin et al. 2009; Xin et al. 2008), 10 new bmr lines were identified in the initial study, and two were shown to have novel mutations in Bmr12 (COMT gene). These other mutants may represent novel loci whose gene products are involved in lignin biosynthesis or in the regulation of this metabolic pathway. To identify new brown midrib lines with mutations in genes other than Bmr2, Bmr6, and Bmr12, a series of putative brown midrib mutant lines isolated from TILLING populations was subjected to allelism testing and subsequent agronomic and chemical characterization. The objectives of these studies were to identify and describe novel brown midrib loci and additional alleles of bmr2, bmr6, and bmr12.
Materials and Methods
Allelism tests
Forty-six putative bmr mutants were isolated from the M2 generation of an EMS mutagenized TILLING population of BTx623 (Xin et al. 2009; Xin et al. 2008), which was derived from approximately 3000 M1 lines. M3 plants shown in Table 1 were crossed to the tester lines A-MP11 (bmr2; USDA-ARS GRIN PI 602898; http://www.ars-grin.gov/), A-N603 (ATx623 bmr6; PI 639713) (Pedersen et al. 2006), and A-N604 (ATx623 bmr12; PI639714) (Pedersen et al. 2006) in the greenhouse at the University of Nebraska–Lincoln during winter 2008–2009. Twelve seeds for each test-cross, mutant line, and check line were planted in the greenhouse in spring 2009; including these check lines not indicated above (A/BMP12, bmr2, PI 602900/602901; A/BMP16, bmr6, PI 602908/602909; A/BMP17, bmr6, PI 602910/602911; A/BMP454, bmr6 and bmr12, PI 602739/602740) (Pedersen et al. 2006). The plants were visually classified as being brown midrib (bmr) or wild-type (WT) phenotype when the plants were approximately 0.5 m in height. Additional test-crosses were performed and evaluated for the complementation tests that yielded inconclusive results in the 2009 evaluation. Digital images were collected to document the leaf midrib phenotype for test-crosses and mutant lines.
Table 1. Midrib genotypes of bmr mutant lines based on results of test-crosses with bmr2, bmr6, and bmr12 tester lines.
| Tester Line |
|||||
|---|---|---|---|---|---|
| AOK11 bmr2 | AN603 (ATx623 bmr6) | AN604 (ATx623 bmr12) | |||
| Line | Mutant Phenotype | Progeny Phenotype | Mutant Locus | ||
| 23 | bmr | WT | bmr | WT | bmr6-23 |
| 29 | bmr | WT | WT | WT | New bmr |
| 30 | bmr | WT | WT | bmr | bmr12-30 |
| 31 | bmr | WT | bmr | WT | bmr6-31 |
| 32 | bmr | WT | bmr | WT | bmr6-32 |
| 34 | bmr | WT | WT | bmr | bmr12-34 |
| 35 | bmr | WT | WT | bmr | bmr12-35 |
| 41 | bmr | WT | WT | WEAK | Inconclusive |
| 45 | bmr | WT | bmr | WT | bmr6-45 |
| 100 | bmr | WT | WT | WT | New bmr |
| 122 | bmr | WT | WT | WT | New bmr |
| 307 | bmr | WT | bmr | WT | bmr6-307 |
| 562 | bmr | bmr | WT | WT | bmr2-2 |
| 741 | bmr | WT | bmr | WT | bmr6-741 |
| 820 | bmr | WT | WT | bmr | bmr12-820 |
| 917 | bmr | WT | bmr | WT | bmr6-971 |
| 1103 | bmr | WT | bmr | WT | bmr6-1103 |
| 1107 | bmr | WT | WT | WT | New bmr |
| 1168 | bmr | WT | WT | WT | New bmr |
| 1277 | bmr | WT | bmr | WT | bmr6-1277 |
| 1937 | bmr | WT | WT | WT | New bmr |
| Tester lines | |||||
| BN603 | bmr | WT | bmr | WT | bmr6 |
| BN604 | bmr | WT | WT | bmr | bmr12 |
| BTx623T | WT | WT | WT | WT | |
| MP11 (PI 602899) | bmr | bmr | WT | WT | bmr2 |
| MP12 (PI 602901) | bmr | bmr | WT | WT | bmr2 |
| MP16 (PI 602909) | bmr | WT | bmr | WT | bmr6 |
| MP17 (PI 602911) | bmr | WT | bmr | WT | bmr6 |
| MP454 (PI 602740) | bmr | WT | bmr | bmr | bmr6 and bmr12 |
The F1 individuals, mutant lines, and check lines were visually classified as being brown midrib (bmr) or wild-type (WT) phenotype (see Materials and Methods). Based on the test-crosses, loci and alleles were designated and appear in the far right column. bmr2-2, bmr12-30, bmr12-34, bmr12-35, and bmr12-820 alleles identified in this study were characterized and reported previously (Saballos et al. 2012; Sattler et al. 2012)
Lines that were confirmed to be brown midrib mutants and produced WT test-cross progeny with all three tester lines were selected as having brown midrib alleles not allelic to bmr2, bmr6, or bmr12. This subset of mutant lines was crossed in all possible combinations using hand emasculation to facilitate crossing in the greenhouse during winter 2009–2010. Progeny were classified by phenotype as described above in the greenhouse during winter 2010–2011.
Agronomic and chemical evaluation of mutants
M3-generation seed received from USDA-ARS in Lubbock, Texas, was increased through self pollinations under pollination bags in the greenhouse during winter 2008–2009 to produce adequate M4-generation seed to plant field studies at the University of Nebraska Agricultural Research and Development Center near Ithaca, Nebraska (Sharpsburg silty clay loam; fine, smectitic, mesic Typic Argiudoll) in 2009 and 2010. Checks BTx623T (line ARS Lubbock, Texas; EMS mutagenized to generate the bmr mutants), BTx623N (line maintained by ARS Lincoln, NE), RTx430, BWheatland, N603, N604, and BMP11 were included in these field studies for comparison. Nitrogen fertilizer was applied at 157 kg ha−1 prior to planting. The experimental design was a randomized complete block with four replications in each year. Plots consisted of 7.6-m rows spaced 0.76 m apart. Single-row plots of each line were planted 21 May 2009 and 25 May 2010. Atrazine [6-chloro-N-ethyl-N′-(1-methylethyl)-1,3,5-triazine-2,4-diamine] was applied at 2.2 kg ha−1 immediately after planting, followed by an application of quinclorac (3,7-dichloro-8-quinolinecarboxylic acid) and atrazine at 0.37 kg ha−1 and 1.1 kg ha−1, respectively, approximately 14 days after emergence. Supplemental irrigation (2.5 cm) was applied on 6 August 2009, 29 August 2009, and 9 August 2010. For plants in each plot, time to 50% anthesis was recorded and height was measured to the top of the mature panicle immediately prior to harvest. Single-plant grain-free forage samples for laboratory analyses were collected by randomly removing three plants from each plot (total of 12 plants per entry), discarding the panicle, and coarsely grinding individual stalks using a commercial silage cutter modified for small plot use (Pedersen and Moore 1995). These individual plant samples were dried in forced-air ovens at 42°, and then stored for subsequent analyses. Total biomass yield was determined by harvesting the remainder of each plot using the same modified commercial silage cutter. Sub-samples were collected and dried in a forced-air oven at 60° to determine dry matter for calculation of plot dry matter yields. Stand counts (number of plants per plot) were made immediately prior to harvest. Plots were harvested 7 October 2009 and 4 October 2010.
The single-plant forage samples were prepared for near-infrared reflectance spectrometry (NIRS) and chemical analyses by grinding in a Wiley mill (2-mm screen; Arthur H. Thomas Co., Philadelphia, PA), followed by grinding to pass a 1-mm screen on a cyclone mill (Udy Corp., Fort Collins, CO). All samples were scanned on a Model 6500 near-infrared reflectance spectrometer (NIRS Systems, Silver Spring, MD3). To cover the expected wide range in spectral diversity, 133 reference samples were selected using cluster analysis of the reflectance data for wet chemistry analysis (Shenk and Westerhaus 1991). Standard wet chemistry methods were used to determine crude protein (CP) (Miller et al. 1998), neutral detergent fiber (NDF), acid detergent fiber (ADF), and acid detergent lignin (ADL) concentration using an ANKOM 200 fiber analyzer of reference samples (ANKOM Tech. Corp., Fairport, NY) (Vogel et al. 1999). Total carbon and ash of reference samples were determined by Ward Labs (Kearney, NE) (Padmore 1990; Leco 2005). The wet chemistry values were then used to develop NIRS prediction equations by partial least squares (Shenk and Westerhaus 1991) and to generate observed values for each plot in each year. The calibration statistics for each trait are shown in Supporting Information, Table S1.
Klason lignin concentration, enzymatic glucose yields, and pyrolysis of new mutant lines
Klason lignin analysis:
A subset of the ground single-plant samples described above containing mutants identified as being non-allelic with bmr2, bmr6, and bmr12 was selected for determination of Klason lignin, and the samples from 2009 were analyzed. Samples were extracted in warm (60°) 50% (v/v) ethanol for 45 min to remove soluble sugars, minerals, and unbound phenolics and oven-dried at 50°. One hundred milligrams of extractive-free sorghum stover was used to determine the Klason lignin concentration. Klason lignin is defined as the ash-corrected residue remaining after the cell wall polysaccharides have been removed via a two-stage hydrolysis in concentrated (12 M) and dilute (0.4 M) sulfuric acid. The procedure was based on the method described Theander and Westerlund (1986), with the modifications of Hatfield et al. (1994).
Enzymatic saccharification:
A volume of 9.7 mL enzyme cocktail containing a 1:1 mix of Trichoderma reesei cellulase and Novozyme 188 (both from SigmaAldrich, St. Louis, MO) in 50 mM sodium citrate buffer pH 4.8 was added to 300 mg of extractive-free stover and incubated at 50° in a shaker-incubator. Novozyme 188 is a β-glucosidase (cellobiase) enzyme preparation from Aspergillus niger that results in the hydrolysis of cellobiose to glucose. The enzyme cocktail had a cellulase activity of 60 FPU/g cellulose based on NREL standard activity measurement LAP006 (Adney and Baker 1996), and the cellobiase activity in the cocktail was 2.0 CBU/mL (based on information from Sigma-Aldrich). The cocktail also contained tetracycline at a final concentration of 20 μg/mL to prevent microbial growth. Hydrolysate samples (100 μL) were collected at 4, 20, and 96 hr, placed in boiling water to inactivate the enzymes, cooled, centrifuged, and used to determine the glucose concentration using a calibrated OneTouch UltraSmart blood glucose meter (Roche Diagnostics, Indianapolis, IN) according to the procedure described by Vermerris et al. (2007). Samples from a minimum of six plants per entry were analyzed, and this analysis included two to three technical replicates per sample for the experiment described above.
Pyrolysis-GC-MS:
Stover samples (∼0.2 mg) from WT and bmr lines were pyrolyzed in a Varian 1079 programmable temperature vaporization (PTV) injector mounted on a Varian 3800 gas chromatograph connected to a Varian 1200 mass spectrometer. The PTV injector was operated at a split ratio of 1:100 at 450°. The pyrolysate was led on a capillary column (25 m, 0.32 mm i.d., fused silica coated with SGE BPX5). Helium (1.2 ml/min) was used as the carrier gas. The GC program started with a hold at 70° for 4 min, followed by a temperature increase to 150° at a rate of 2°/min. The temperature was subsequently increased to 250° at a rate of 4°/min, then to 320° at a rate of 30°/min. The mass spectrometer was operated at 1.0 kV. The mass range included m/z 45 to 350 and was scanned every 0.2 sec. The identification of compounds was based on Ralph and Hatfield (1991). To compare the proportion of the hydroxycinnamic acids p-coumaric acid and ferulic acid in the biomass, stover was subjected to pyrolysis-TMAH-GC-MS as described by Saballos et al. (2012). Samples of approximately 0.2 mg stover from BTx623 and the novel bmr mutants were trans-methylated with 2.5% (v/v) tetramethyl ammonium hydroxide (TMAH; Sigma-Aldrich, St. Louis, MO) in methanol (Mulder et al. 1992) and pyrolyzed in a Varian 1079 PTV injector mounted on a Varian 3800/1200 GC-MS as described above.
Statistical analysis
The data were analyzed using the PROC MIXED procedure of SAS/STAT software4 (SAS, 2002–2008). For analyses of agronomic and chemical evaluations, years and replications were considered random effects (variables). Least squares (LS) means for mutant and check lines were generated and compared using the ADJUST = SIMULATE option in the LSMEANS/DIFF statement to control type I error and to permit multiple comparisons without the requirement for F-test significance (Littell et al. 2006). The rejection level for tests of significance among means was set at P ≤ 0.20 as recommended by FAO (Annicchiarico 2002) to control and achieve a better balance between type I and type II error rates for such experiments (variety trials with large numbers of entries). For analyses of yield data, stand count was treated as a covariate and LS means were adjusted for stand. For analyses of laboratory traits, values from the three individual plant samples per plot were considered repeated measures. Means of all variables were ranked from highest to lowest, and means not significantly different from BTx623T were identified. For Klason lignin and enzymatic saccharification analyses, LS means for mutant and check lines were generated and compared using the LSMEANS/DIFF statement, and significance level for tests among means was set at P ≤ 0.05 because of the smaller number of entries.
Results
Determining allelism among the bmr mutants
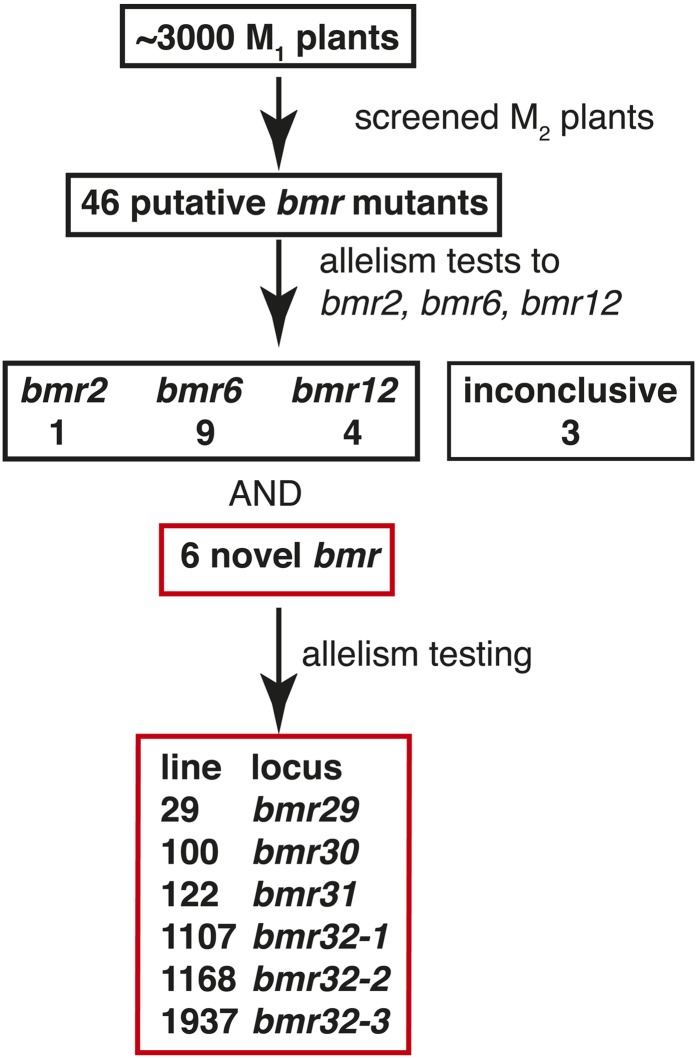
In the greenhouse, putative bmr mutant lines were crossed to cytoplasmic male-sterile lines representing the three previously described bmr loci, bmr2, bmr6, and bmr12, and the leaf midrib phenotype of F1 progeny and mutant lines were visually scored when the plants were approximately 0.5 m tall (Table 1). Through this visual examination, the midribs of 23 of the lines were determined to fall within the naturally observed range of midrib color, and to not have a brown midrib phenotype (Table S2). For three lines, the results of these test-crosses were inconclusive (Table S2). In addition, one allele of bmr2, nine alleles of bmr6, and four alleles of bmr12 were identified based on leaf phenotype of the F1 progeny (Figure 2 and Table 1). The line number was incorporated into the allelic designation for these previously described bmr loci—bmr2, bmr6, or bmr12. The mutations responsible for the bmr2-2, bmr12-30, bmr12-34, bmr12-35, and bmr12-820 alleles isolated were characterized and reported previously (Saballos et al. 2012; Sattler et al. 2012). In addition, there were six mutant lines with bmr leaf phenotypes that were not allelic to bmr2, bmr6, or bmr12. The leaf midrib phenotypes of these six mutant lines were similar to the previously described loci bmr2, bmr6, or bmr12, although brown coloration was not dark as bmr6 midrib (Figure 3). Additional crosses between these six lines were performed and the F1 progeny were scored for the bmr leaf midrib phenotype to determine allelism among them (Table 2). The results of these test-crosses indicated that the six bmr mutant lines (29, 100, 122, 1107, 1168, and 1937) represent four novel loci (Figure 2), which we named bmr29 through bmr32 to avoid overlap with the designators bmr1 through bmr28 for bmr mutants previously isolated at Purdue University in the 1970s (Porter et al. 1978; Saballos et al. 2008). Mutant line 41 was excluded from this group because its leaf phenotype and the results of the allelism tests were inconsistent (Table 2). In summary, 20 lines appear to carry mutations responsible for the bmr leaf midrib phenotype.
Figure 2.
This diagram summarizes the results of the screening process that identified the bmr mutants in the three characterized loci and the four newly identified loci.
Figure 3.
The leaf midrib phenotype of the wild-type BTx623T (WT), B OK11 bmr2, BTx623 bmr6, BTx623 bmr12, bmr29, bmr30, bmr31, bmr32-1, bmr32-2, and bmr32-3. The sixth leaf was photographed from 6-wk-old plants.
Table 2. Midrib phenotype of progeny from test-crosses among seven mutant lines not allelic to bmr2, bmr6 or bmr12.
| ♀ Parent | ♂ Parent |
||||||
|---|---|---|---|---|---|---|---|
| 29 | 41 | 100 | 122 | 1107 | 1168 | 1937 | |
| 29 | – | WT | WT | WT | WT | WT | WT |
| 41 | WT | – | WT | bmr | WT | WT | WT |
| 100 | WT | WT | – | WT | WT | WT | – |
| 122 | WT | ? | WT | – | WT | WT | WT |
| 1107 | WT | WT | WT | WT | – | bmr | bmr |
| 1168 | WT | WT | WT | WT | bmr | – | bmr |
| 1937 | WT | WT | WT | WT | bmr | bmr | – |
The F1 individuals, mutant lines, and check lines were visually classified as being brown midrib (bmr), wild-type (WT), or inconclusive (?) (see Materials and Methods).
Field evaluation of bmr mutant lines
Concurrently, the bmr mutant lines were grown in the field to evaluate agronomic characteristics over two seasons (Table S3). At full maturity, biomass samples were collected for chemical analysis. Time to 50% anthesis was recorded, and the putative bmr mutant lines did not show statistically significant differences (analysis of lsmeans; P < 0.2) compared with BTx623T, from which these lines were derived through EMS mutagenesis (Table S3). Three lines (bmr6-307, bmr30, and bmr32-3) had statistically significantly reduced stand counts (plants/m) relative to BTx623T (Table S3). Eight of the 20 bmr lines had statistically significantly reduced plant heights (m) at maturity relative to BTx623T. Significant reductions in dry matter yield (T/ha) of the above-ground biomass (stover) were observed for all 20 lines relative to BTx623T (Table S3). Statistically significant differences in stover yield relative to BTx623T were not observed for any of the control lines, including the BTx623 near-isogenic lines containing bmr6-ref or bmr12-ref.
Analysis of stover chemical composition of bmr mutant lines
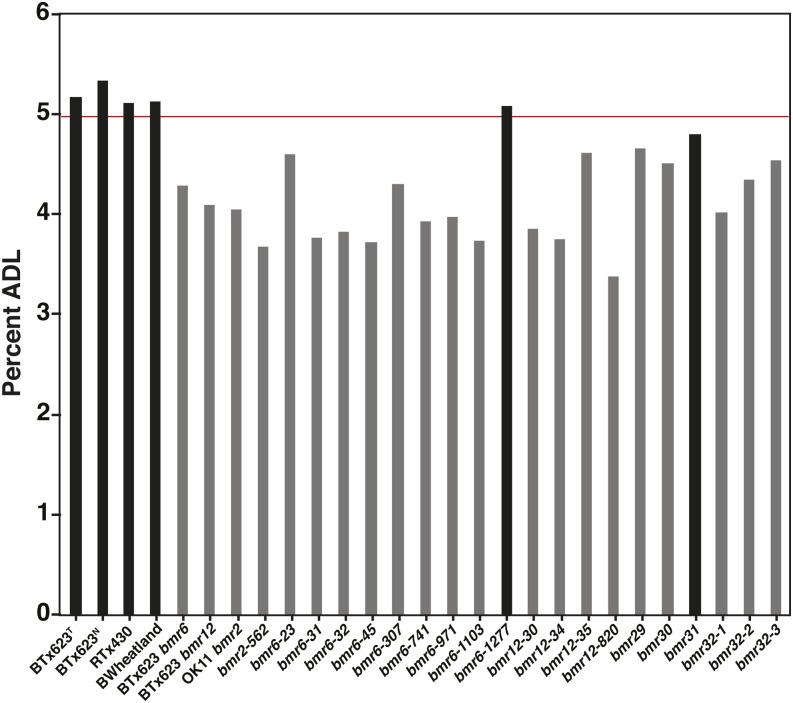
The stover chemical composition was determined through NIRS to determine the impact these 20 bmr mutants had on lignin concentrations (Table S4). Three lines (OK11 bmr2, bmr6-45, bmr6-1277) had statistically significant differences for neutral detergent fiber (NDF) relative to BTx623T (Table S4), which reflects the amount of cellulose, hemicellulose, and lignin in the stover. Fourteen lines showed statistically significant differences for acid detergent fiber (ADF) relative to BTx623T (Table S4), which reflects the combined amount of cellulose and lignin in the stover. Twelve of the bmr lines had reduced ADF levels, which was likely due to reduced lignin concentration of the stover (Table S4). However, two lines (bmr6-1277 and bmr29) had significantly higher ADF levels than WT, which was unexpected. Except for bmr6-1277 and bmr31, all bmr mutant lines (Table 1) had significantly reduced acid detergent lignin (ADL) levels relative to WT BTx623T (Figure 4 and Table S4). The lowest ADL level was observed in bmr12-820 (lsmeans 3.37), which represents an approximate 35% reduction in ADL relative to BTx623T (lsmeans 5.18). Hence, these results are consistent with previously reported reductions in ADL levels of the stover associated with bmr6 and bmr12 phenotypes (Oliver et al. 2005a, b; Sattler et al. 2010).
Figure 4.
Acid detergent lignin (ADL) concentrations were determined by NIRS prediction based on the calibration equation (Table S1). Additional details are found in the Materials and Methods section. The values presented are least squares means (lsmean) for bmr mutant and check lines (Table S4). Gray bars indicate values that were statistically significantly different (P ≤ 0.20) from the values of BTx623T, the line used for mutagenesis. The red line indicates lower limit of all ranked variables for WT (BTx623T). Additional details of the statistical analysis are described in the Materials and Methods section.
In addition, the total ash concentration, crude protein concentration, and total carbon concentration of the stover were determined through NIRS (Table S5). Eight lines had significantly reduced ash concentration relative to WT BTx623T, whereas four lines (bmr2 and bmr6 checks, bmr6-45, bmr6-307) had significantly increased ash concentration (Table S5). Four lines (bmr6-307, bmr12-30, bmr31, bmr32-3) showed significant increases in crude protein concentration of stover (Table S5), which has been associated with abiotic and biotic stresses (Buxton 1996; Wilkins and Humphreys 2003). Total carbon concentration of the stover was significantly increased in eight bmr lines. Total carbon concentration was significantly decreased for three lines, bmr6-307, and the bmr2 and bmr6 checks, all of which had significantly increased ash concentration of the stover. There were significant differences in total carbon or ash concentration of the stover for bmr lines confirmed through complementation tests, but these results did not follow a trend except for lines allelic to bmr12 and bmr32 (Table S5). Stover of all three bmr32 lines had a significant decrease in ash concentration and a significant increase in total carbon concentration. Similarly, stover from three of the four bmr12 lines, but not the check BTx623 bmr12-ref, had a significant decrease in ash concentration, and three of the four bmr12 lines had a significant increase in total carbon concentration.
Characterizing the chemical composition of four novel bmr mutants
The effect of the four novel bmr mutations on Klason lignin concentration was determined, and bmr29 stover had a statistically significant reduction (of 14.3%) in Klason lignin relative to WT BTx623T at P ≤ 0.05 (Table 3). The stover from bmr31 and bmr32-1 through bmr32-3 had lsmeans for Klason lignin that were lower than lsmeans for WT stover, but at the 0.2 level rather than the 0.05 level of probability. Overall, the lower ranges of Klason lignin concentration for stover from these mutants (Table 3) combined with the ADL data (Table S4) suggest that these mutations reduce lignin concentration, but that the effect is, overall, smaller than in the bmr2, bmr6, and bmr12 mutants. Other EMS-induced mutations in these lines may contribute to the observed variation among the samples (see Discussion).
Table 3. Klason lignin concentration and saccharification efficiency of stover from four newly identified bmr loci.
| Glucose Release (mg/g Stover) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Klason Lignin (%) |
Hydrolysis 4 hr |
Hydrolysis 20 hr |
Hydrolysis 96 hr |
|||||||||
| Line | lsmean | Lower | Upper | lsmean | Lower | Upper | lsmean | Lower | Upper | lsmean | Lower | Upper |
| BTx623T | 21.2 | 20.5 | 22.0 | 39.9 | 28.9 | 50.8 | 57.7 | 47.1 | 68.2 | 68.8 | 60.5 | 77.0 |
| bmr29 | 18.2 | 16.7 | 19.7 | 39.2 | 30.2 | 48.1 | 49.5 | 38.9 | 60.0 | 57.3 | 49.1 | 65.5 |
| bmr30 | 20.2 | 18.3 | 22.1 | 51.4 | 40.5 | 62.4 | 73.9 | 63.3 | 84.4 | 79.5 | 71.2 | 87.7 |
| bmr31 | 19.7 | 18.1 | 21.2 | 54.7 | 43.8 | 65.7 | 61.1 | 50.6 | 71.7 | 74.3 | 66.1 | 82.5 |
| bmr32-1 | 19.2 | 17.2 | 21.1 | 41.6 | 28.2 | 55.0 | 55.3 | 42.4 | 68.2 | 65.0 | 54.9 | 75.2 |
| bmr32-2 | ND | ND | ND | 43.0 | 29.6 | 56.5 | 58.9 | 46.0 | 71.8 | 59.7 | 49.2 | 70.2 |
| bmr32-3 | 19.2 | 17.5 | 21.0 | 44.5 | 36.4 | 52.6 | 58.0 | 50.2 | 65.8 | 65.5 | 59.1 | 71.8 |
Klason lignin concentration was determined as described in the Materials and Methods section. Glucose yields were obtained after enzymatic saccharification with cellulase of native (unpretreated) stover after 4, 20, and 96 hr. Least squares means (lsmean) for mutant and check lines were generated, and means of all variables were ranked from highest (Upper) to lowest (Lower). Bold text indicates values that were statistically significantly different (P ≤ 0.05) from the values of BTx623T, the line used for mutagenesis. Additional details of the statistical analysis are described in the Materials and Methods section.
To assess the potential of these novel bmr mutations for improving the biomass conversion efficiency of the stover, enzymatic saccharification of the cellulose was performed. Ground native (unpretreated) stover samples were treated with cellulases, and the amount of glucose released was monitored at fixed time points. The stover from two of the mutants, bmr30 and bmr31, yielded more released glucose than WT at P ≤ 0.2 at 4 hr for bmr30, and at 20 hr and 96 hr for bmr31 (Table 3). Interestingly, bmr29 stover, which had the lowest mean Klason lignin concentration among the novel bmr mutants, did not release more glucose than the WT control following enzymatic saccharification (Table 3). Hence, a clear relationship between glucose release following saccharification and lignin concentration, determined as either Klason lignin or ADL, was not observed.
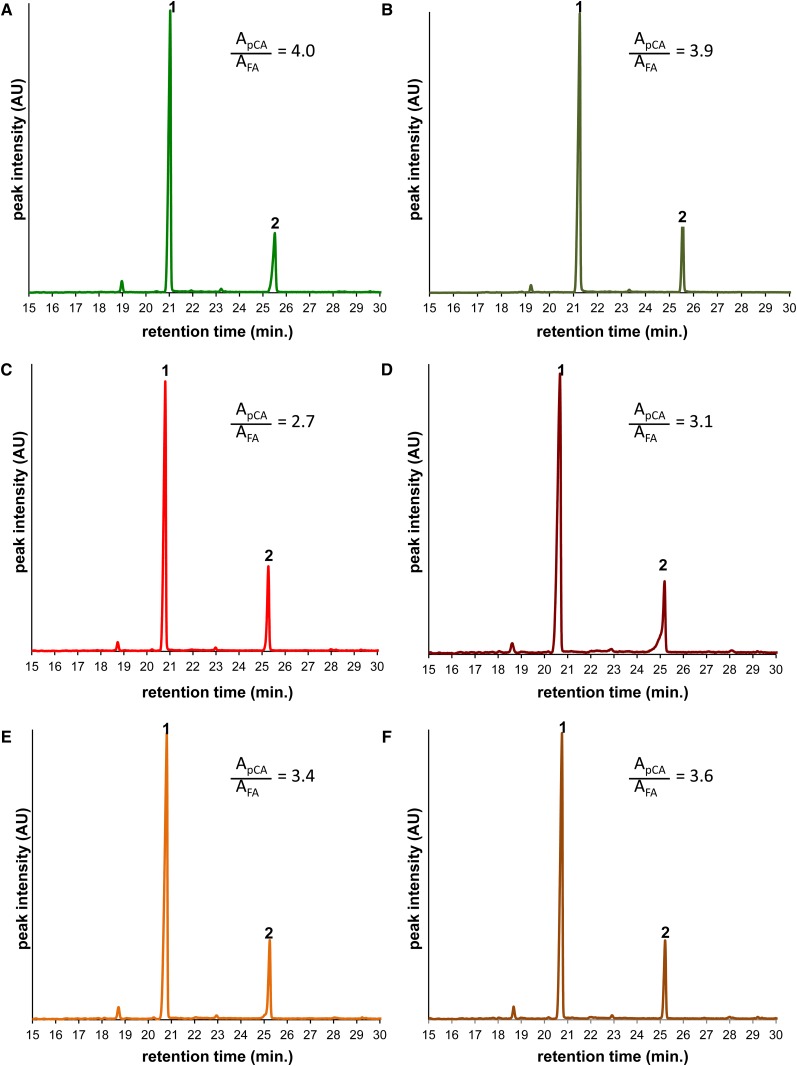
Lignin subunit composition resulting from the novel bmr mutations was analyzed using pyrolysis-GC-MS. Unlike what has been observed in the bmr2 (Saballos et al. 2012), bmr6 (Saballos et al. 2009; Sattler et al. 2009), and bmr12 mutants (Bout and Vermerris 2003; Palmer et al. 2008), no apparent changes in phenolic compounds derived from either guaiacyl or syringyl residues were identified (Figure S1). Stover from three of the four novel bmr mutants (bmr30, bmr31, bmr32) did, however, contain more ferulic acid based on analysis of the pyrograms obtained from TMAH-treated stover (Figure 5). Ferulic acid is primarily esterified to the hemicellulosic polysaccharide glucuronoarabinoxylan (GAX), where it serves as a nucleation site for lignification. The higher content of ferulic acid identified in the pyrograms is consistent with the higher hemicellulose content observed in the bmr30, bmr31, and bm32 mutants, as calculated by subtracting ADF from NDF (Table S4) (27% for BTx623 vs. 29% for the three bmr mutants). The increased ferulate levels in these bmr mutants may reflect cell wall compositional and/or architectural changes that may influence the rate of glucose release during enzymatic saccharification from bmr30 and bmr31 stover.
Figure 5.
Normalized total ion current chromatograms displaying the content of esterified p-coumaric (pCA) and ferulic acid (FA) in the cell walls of wild-type and bmr stover as determined by pyrolysis-TMAH-GC-MS. (A) BTx623 (wild-type). (B) bmr29. (C) bmr30. (D) bmr31. (E) bmr-32-1. (F) bmr32-2. The peaks representing the double methylesters of (1) trans-p-coumaric acid (m/z 192) and (2) trans-ferulic acid (m/z 222) have been selectively displayed. The ratio of the peak areas (pCA/FA) is displayed. This ratio is not reflective of the actual molar ratio of pCA and FA in the cell wall due to the differences in sensitivity of detection.
Discussion
In this study, 20 novel bmr mutant lines were identified and characterized based on allelism tests. Among this set of 20, six bmr mutants were not allelic to bmr2, bmr6, or bmr12. Further complementation testing showed these mutants represent four novel bmr loci. Hence, this study has doubled the number of bmr loci identified and characterized in sorghum from the original four loci bmr2, bmr6, bmr12, and bmr19 isolated and described at Purdue University (Porter et al. 1978; Saballos et al. 2008), although it should be noted that bmr19 was not available to test whether it is allelic to any of the six new mutants (bmr29 through 32). Pyrolysis-GC-MS analysis of bmr19 stover showed a reduced amount of G-derived pyrolysis products relative to WT (Saballos et al. 2008), and this change was not observed in the any six novel mutants described here, which suggests that none of the bmr mutants described here represent alleles of bmr19. The bmr mutants obtained from this TILLING population provide both novel bmr sources for plant breeding and new tools to study lignin synthesis and bioenergy conversion processes. However, the relative ease in which bmr mutants were isolated from this mutagenized population also has an obvious drawback for their subsequent uses. Previously, it was estimated that there was an EMS-induced mutation every 516 kb within the population (Xin et al. 2008). These heavy mutation loads affect overall plant fitness based on evaluation of agronomic traits (Table S3). Although most lines were not statistically different from the WT line BTx623T for stand count, time to 50% anthesis, and plant height, dry matter (stover) yield was significantly reduced in all 20 bmr lines. In contrast, the near-isogenic bmr6 and bmr12 lines previously introgressed into BTx623 (Pedersen et al. 2006) had stover yields that were not significantly different from WT (Table S3). Clearly, these new bmr lines will require several generations of backcrossing to reduce the number of deleterious mutations each line is carrying before they can be used in plant breeding programs.
The brown midrib mutations have been previously shown to reduce the lignin concentration of biomass in maize, sorghum, and pearl millet (Pennisetum glaucum L.) (Kuc and Nelson 1964; Gee et al. 1968; Porter et al. 1978; Cherney et al. 1988). Likewise, stover from all bmr lines had significantly reduced ADL levels except for bmr6-1277 and bmr31, which were not significantly different from WT (Figure 4). The ash and total carbon concentration of the stover were significantly different than the WT BTx623T for some of the bmr lines (Table S5). However, there were also significant differences for these two traits between BTx623T stover and the stover from four of the control lines (RTx430, BTx623 bmr6, OK11 bmr2). These results may indicate that both ash and total carbon concentration are more variable components of stover as compared with NDF, ADF, and ADL. The inverse relationship between significant differences in ash concentration and total carbon concentration that was observed for nine of the bmr stover samples may reflect the influence of ash concentration on measurement of total carbon concentration.
One of the goals of these experiments was to identify novel bmr loci and to determine whether the impact these mutations have on lignin biosynthesis translated into increased saccharification of the stover. Despite the commonly accepted view that lignin impedes enzymatic saccharification (Chen and Dixon 2007; Grabber 2005), the data presented here do not indicate a correlation between Klason lignin or ADL concentration and glucose yield following enzymatic saccharification (Table 3 and Table S4). For example, among the mutants examined, the bmr30 mutant yields the highest amount of glucose while its Klason lignin concentration is not different from WT, and the bmr29 mutant has the lowest amount of Klason lignin but does not yield more glucose. Prior studies have indicated that lignin subunit composition influences glucose release. In the bmr6 and bmr12 mutants of sorghum, the yield of fermentable sugars was shown to be not only higher (Dien et al. 2009; Saballos et al. 2008) but also additive in nature when both mutations were combined (Dien et al. 2009). In both of these mutants, the S/G ratio was lower (Bout and Vermerris 2003; Palmer et al. 2008; Sattler et al. 2009), largely due to the reduction in S-residues, resulting in an almost complete lack of these residues in the double mutant. Similar observations have been made in maize mutants in which the brown midrib1 (bm1) and bm3 mutations were combined (Vermerris et al. 2007). Other factors that have been shown to play a role in biomass conversion efficiency in maize and sorghum are the crystallinity index of the cellulose (Vandenbrink et al. 2012) and the concentration of hemicellulose in the stover (Torres et al. 2014). The latter factor may contribute to the enhanced release of glucose from the stover of bmr30 and bmr31. Finally, given that the novel bmr mutants were identified in a TILLING population, it is possible that there are other mutations with a negative impact on enzymatic saccharification. However, it is unlikely that this would have occurred in all six of the novel bmr mutants.
In conclusion, the newly identified bmr alleles of bmr2, bmr6, and bmr12 and four novel bmr loci represent tools to manipulate lignin concentration and biomass conversion. The four novel bmr loci may represent genes involved in lignin biosynthesis and could be used to further our understanding of this pathway in sorghum, even though they did not significantly increase saccharification yield compared with WT. Because the bmr2, bmr6, and bmr12 mutations significantly decreased lignin concentration and increased enzymatic saccharification yields relative to WT (Bout and Vermerris 2003; Palmer et al. 2008; Saballos et al. 2008, 2012), the most practical use of the novel bmr mutants may be to combine them with the three previously described mutants or with each other, similar to “stacking” bmr6 and bmr12, which resulted in a greater reduction in lignin concentration and larger increase in biomass conversion efficiency than achieved with either mutation alone (Sattler et al. 2010; Dien et al. 2009).
Supplementary Material
Acknowledgments
We thank Tammy Gries, Steve Masterson, Pat O’Neill, and John Toy for their technical assistance with experimental data presented in this manuscript, and Dr. Heather Van Buskirk for critically reviewing the manuscript. This research was supported by the Office of Science (BER), U.S. Department of Energy grant DE-FG02-07ER64458 (Wilfred Vermerris, Scott E. Sattler, Jeffrey F. Pedersen), and additional funding from USDA-ARS, CRIS project 5440-21220-032-00D (S.E.S, Deanna L. Funnell-Harris.), USDA AFRI grant number 2011-67009-30026 (S.E.S, D.L.F.H., J.F.P), and USDA Biomass Research and Development Initiative grant number 2011-10006-30358 (W.V.).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014001/-/DC1
The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, age, disability, and where applicable, sex, marital status, familial status, parental status, religion, sexual orientation, genetic information, political beliefs, reprisal, or because all or part of an individual’s income is derived from any public assistance program. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA’s TARGET Center at (202) 720-2600 (voice and TDD). To file a complaint of discrimination, write to USDA, Director, Office of Civil Rights, 1400 Independence Avenue, S.W., Washington, D.C. 20250-9410, or call (800) 795-3272 (voice) or (202) 720-6382 (TDD). USDA is an equal opportunity provider and employer.
Communicating editor: J. D. Faris
Literature Cited
- Adney B., Baker J., 1996. Measurement of Cellulase Activities LAP-006 NREL Analytical Procedure, National Renewable Energy Laboratory, Golden, CO. [Google Scholar]
- Ali F., Scott P., Bakht J., Chen Y., Lübberstedt T., 2010. Identification of novel brown midrib genes in maize by tests of allelism. Plant Breed. 129: 724–726. [Google Scholar]
- Annicchiarico P., 2002. Estimation of individual effects and comparison of means, FAO Plant Production and Protection, Rome. [Google Scholar]
- Barrière Y., Guillet C., Goffner D., Pichon M., 2003. Genetic variation and breeding strategies for improved cell wall digestibility in annual forage crops. A review. Anim. Res. 52: 193–228. [Google Scholar]
- Bittinger T. S., Cantrell R. P., Axtell J. D., 1981. Allelism tests of the brown-midrib mutants of sorghum. J. Hered. 72: 147–148. [Google Scholar]
- Boerjan W., Ralph J., Baucher M., 2003. Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Bout S., Vermerris W., 2003. A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 269: 205–214. [DOI] [PubMed] [Google Scholar]
- Boyce C. K., Zwieniecki M. A., Cody G. D., Jacobsen C., Wirick S., et al. , 2004. Evolution of xylem lignification and hydrogel transport regulation. Proc. Natl. Acad. Sci. USA 101: 17555–17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton, D.R., 1996 Quality-related characteristics of forages as influenced by plant environment and agronomic factors. Animal Feed Sci. Technol. 59 (1–3 Spec. Iss.):37–49.
- Chen F., Dixon R. A., 2007. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25: 759–761. [DOI] [PubMed] [Google Scholar]
- Chen W., VanOpdorp N., Fitzl D., Tewari J., Friedemann P., et al. , 2012. Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol. Biol. 80: 289–297. [DOI] [PubMed] [Google Scholar]
- Cherney J. H., Axtell J. D., Hassen M. M., Anliker K. S., 1988. Forage quality characterization of a chemically-induced brown-midrib mutant in pearl-millet. Crop Sci. 28: 783–787. [Google Scholar]
- Degenhart N. R., Werner B. K., Burton G. W., 1995. Forage yield and quality of a brown mid-rib mutant in pearl-millet. Crop Sci. 35: 986–988. [Google Scholar]
- Dien B. S., Sarath G., Pedersen J. F., Sattler S. E., Chen H., et al. , 2009. Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. Bioenerg. Res. 2: 153–164. [Google Scholar]
- Gee M. S., Nelson O. E., Kuc J., 1968. Abnormal lignins produced by the brown-midrib mutants of maize. II. Comparative studies on normal and brown-midrib-1 dimethylformamide lignins. Arch. Biochem. Biophys. 123: 403–408. [DOI] [PubMed] [Google Scholar]
- Grabber J. H., 2005. How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci. 45: 820–831. [Google Scholar]
- Guo D., Chen F., Wheeler J., Winder J., Selman S., et al. , 2001. Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res. 10: 457–464. [DOI] [PubMed] [Google Scholar]
- Gupta S. C., 1995. Inheritance and allelic study of brown midrib trait in pearl-millet. J. Hered. 86(4): 301–303. [Google Scholar]
- Hatfield R. D., Jung H. J. G., Ralph J., Buxton D. R., Weimer P. J., 1994. A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J. Sci. Food Agric. 65: 51–58. [Google Scholar]
- Jones L., Ennos A. R., Turner S. R., 2001. Cloning and characterization of irregular xylem4 (irx4) (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26: 205–216. [DOI] [PubMed] [Google Scholar]
- Jorgenson L. R., 1931. Brown midrib in maize and its linkage relations. J. Am. Soc. Agron. 23: 549–557. [Google Scholar]
- Jung H. G., Allen M. S., 1995. Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J. Anim. Sci. 73: 2774–2790. [DOI] [PubMed] [Google Scholar]
- Jung H. J. G., Ni W., 1998. Lignification of plant cell walls: Impact of genetic manipulation. Proc. Natl. Acad. Sci. USA 95: 12742–12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuc J., Nelson O. E., 1964. The abnormal lignins produced by the brown-midrib mutants of maize. I. The brown midrib-1 mutant. Arch. Biochem. Biophys. 105: 103–113. [DOI] [PubMed] [Google Scholar]
- LECO, 2005 Organic Application Note, Carbon, Hydrogen, and Nitrogen in Flour and Plant Tissue–Form No. 203–281–273. LECO, St. Joseph, MI. [Google Scholar]
- Littell R. C., Milliken G. A., Stroup W. W., Wolfinger R. D., Schabenberger O., 2006. SAS for Mixed Models, SAS Institute Inc., Cary, NC. [Google Scholar]
- Miller, R. O., J. Kotuby-Amacher, and J. Rodriguez, 1998 Total Nitrogen in Botanical Materials—Automated Combustion Method, pp. 116–117 in Western States Laboratory Proficiency Testing Program. Colorado State University, Ft. Collins, CO. [Google Scholar]
- Mulder M. M., Van der Hage E. R. E., Boon J.J., 1992. Analytical in source pyrolytic methylation electron impact mass spectrometry of phenolic acids in biological matrices. Phytochem. Anal. 3: 165–172. [Google Scholar]
- Oliver A. L., Pedersen J. F., Grant R. J., Klopfenstein T. J., 2005a Comparative effects of the sorghum bmr-6 and bmr-12 genes: I. Forage sorghum yield and quality. Crop Sci. 45: 2234–2239. [Google Scholar]
- Oliver A. L., Pedersen J. F., Grant R. J., Klopfenstein T. J., Jose H. D., 2005b Comparative effects of the sorghum bmr-6 and bmr-12 genes: II. Grain yield, stover yield, and stover quality in grain sorghum. Crop Sci. 45: 2240–2245. [Google Scholar]
- Padmore J. M., 1990. Ash of Animal Feed; Method No 942.05, p. 70 in Official Methods of Analysis of the Association of Official Analytical Chemists, edited by K. Herlich AOAC, Arlington, VA. [Google Scholar]
- Palmer N. A., Sattler S. E., Saathoff A. J., Funnell D., Pedersen J. F., et al. , 2008. Genetic background impacts soluble and cell wall-bound aromatics in brown midrib mutants of sorghum. Planta 229: 115–127. [DOI] [PubMed] [Google Scholar]
- Pedersen J. F., Moore K. J., 1995. An automated plot harvest system for use with a commercial forage harvester. Agron. J. 87: 605–607. [Google Scholar]
- Pedersen J. F., Funnell D. L., Toy J. J., Oliver A. L., Grant R. J., 2006. Registration of twelve grain sorghum genetic stocks near-isogenic for the brown midrib genes bmr-6 and bmr-12. Crop Sci. 46: 491–492. [Google Scholar]
- Piquemal J., Lapierre C., Myton K., O’Connell A., Schuch W., et al. , 1998. Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13: 71–83. [Google Scholar]
- Porter K. S., Axtell J. D., Lechtenberg V. L., Colenbrander V. F., 1978. Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci. 18: 205–208. [Google Scholar]
- Ralph J., Hatfield R. D., 1991. Pyrolysis-GC-MS characterization of forage materials. J. Agric. Food Chem. 39: 1426–1437. [Google Scholar]
- Roje S., Wang H., McNeil S. D., Raymond R. K., Appling D. R., et al. , 1999. Isolation, characterization, and functional expression of cDNAS encoding NADH-dependent methylenetetrahydrofolate reductase from higher plants. J. Biol. Chem. 274: 36089–36096. [DOI] [PubMed] [Google Scholar]
- Roje S., Chan S. Y., Kaplan F., Raymond R. K., Horne D. W., et al. , 2002. Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the methylenetetrahydrofolate reductase reaction in vivo. J. Biol. Chem. 277: 4056–4061. [DOI] [PubMed] [Google Scholar]
- Ruel K., Berrio-Sierra J., Derikvand M. M., Pollet B., Thévenin J., et al. , 2009. Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 184: 99–113. [DOI] [PubMed] [Google Scholar]
- Saballos A., Vermerris W., Rivera L., Ejeta G., 2008. Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenerg. Res. 1: 193–204. [Google Scholar]
- Saballos A., Ejeta G., Sanchez E., Kang C., Vermerris W., 2009. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the Brown midrib6 gene. Genetics 181: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A., Sattler S. E., Sanchez E., Foster T. P., Xin Z., et al. , 2012. Brown midrib2 (Bmr2) encodes the major 4-coumarate: Coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 70: 818–830. [DOI] [PubMed] [Google Scholar]
- Sattler S. E., Funnell-Harris D. L., Pedersen J. F., 2010. Efficacy of singular and stacked brown midrib 6 and 12 in the modification of lignocellulose and grain chemistry. J. Agric. Food Chem. 58: 3611–3616. [DOI] [PubMed] [Google Scholar]
- Sattler S. E., Saathoff A. J., Haas E. J., Palmer N. A., Funnell-Harris D. L., et al. , 2009. A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiol. 150: 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler S. E., Palmer N. A., Saballos A., Greene A. M., Xin Z. G., et al. , 2012. Identification and characterization of four missense mutations in Brown midrib 12 (Bmr12), the caffeic acid O-methyltransferase (COMT) of sorghum. Bioenerg. Res. 5: 855–865. [Google Scholar]
- Shenk J. S., Westerhaus M. O., 1991. Population definition, sample selection, and calibration procedures for near-infrared reflectance spectroscopy. Crop Sci. 31: 469–474. [Google Scholar]
- Sperry J. S., 2003. Evolution of water transport and xylem structure. Int. J. Plant Sci. 164(Suppl. 3): S115–S127. [Google Scholar]
- Tang H. M., Liu S., Hill-Skinner S., Wu W., Reed D., et al. , 2014. The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 77: 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theander O., Westerlund E. A., 1986. Studies on dietary fiber. 3. Improved procedures for analysis of dietary fiber. J. Agric. Food Chem. 34: 330–336. [Google Scholar]
- Tohge T., Watanabe M., Hoefgen R., Fernie A. R., 2013. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. F., Noordam-Boot C. M. M., Dolstra O., van der Weijde T., Combes E., et al. , 2014. Cell wall diversity in forage maize: genetic complexity and bioenergy potential. Bioenerg. Res. 10.1007/s12155-014-9507-8 [Google Scholar]
- Vandenbrink J. P., Hilten R. N., Das K. C., Paterson A. H., Feltus F. A., 2012. Analysis of crystallinity index and hydrolysis rates in the bioenergy crop Sorghum bicolor. Bioenerg. Res. 5: 387–397. [Google Scholar]
- Vanholme R., Cesarino I., Rataj K., Xiao Y., Sundin L., et al. , 2013. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in arabidopsis. Science 341: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Vermerris W., Saballos A., Ejeta G., Mosier N.S., Ladisch M.R., et al. , 2007. Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Science 47 (Suppl. Dec.):S147–S153. [Google Scholar]
- Vignols F., Rigau J., Torres M. A., Capellades M., Puigdomenech P., 1995. The brown midrib3 (Bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K. P., Jung H. J. G., 2001. Genetic modification of herbaceous plants for feed and fuel. Crit. Rev. Plant Sci. 20: 15–49. [Google Scholar]
- Vogel K. P., Pedersen J. F., Masterson S. D., Toy J. J., 1999. Evaluation of a filter bag system for NDF, ADF, and IVDMD forage analysis. Crop Sci. 39: 276–279. [Google Scholar]
- Wilkins P. W., Humphreys M. O., 2003. Progress in breeding perennial forage grasses for temperate agriculture. J. Agric. Sci. 140: 129–150. [Google Scholar]
- Xin Z., Wang M., Burow G., Burke J., 2009. An induced sorghum mutant population suitable for bioenergy research. Bioenerg. Res. 2: 10–16. [Google Scholar]
- Xin Z., Wang M. L., Barkley N. A., Burow G., Franks C., et al. , 2008. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.