Abstract
Existing transgenic RNA interference (RNAi) methods greatly facilitate functional genome studies via controlled silencing of targeted mRNA in Drosophila. Although the RNAi approach is extremely powerful, concerns still linger about its low efficiency. Here, we developed a CRISPR/Cas9-mediated conditional mutagenesis system by combining tissue-specific expression of Cas9 driven by the Gal4/upstream activating site system with various ubiquitously expressed guide RNA transgenes to effectively inactivate gene expression in a temporally and spatially controlled manner. Furthermore, by including multiple guide RNAs in a transgenic vector to target a single gene, we achieved a high degree of gene mutagenesis in specific tissues. The CRISPR/Cas9-mediated conditional mutagenesis system provides a simple and effective tool for gene function analysis, and complements the existing RNAi approach.
Keywords: CRISPR/Cas9, conditional mutagenesis, gRNA, Gal4, Drosophila
Conditional mutagenesis techniques are required for analyzing the pleiotropic functions of genes at various life stages, especially for genes critical for embryogenesis. Available transgenic RNA interference (RNAi) resources in Drosophila are powerful tools for genomic screens and have been used for many studies of gene function (Dietzl et al. 2007; Ni et al. 2011). This method uses the Gal4-upstream activating site controlled hairpins to trigger sequence-specific mRNA breakdown. Tissue-specific gene silencing is achieved via a cross between UAS-hairpin and Gal4 driver lines; however, the RNAi method suffers from a few drawbacks. For instance, gene expression is rarely completely blocked by RNAi, frequently leading to low efficiency in gene silencing. Furthermore, for reasons unknown, some tissues such as testis and ovary (Raychaudhuri et al. 2012) are less sensitive to RNAi. Although one can always use the traditional approach for making conditional alleles, such as using FLP (fippase)/FRT (fippase recognition target) to generate clones when RNAi fails, the targeting procedures can be time- and labor-consuming (Choi et al. 2009; Gratz et al. 2014; Xue et al. 2014).
Recently, the RNA-guided CRISPR/Cas9 technology has shown potential for highly efficient genome editing in many organisms, including Drosophila (Bassett et al. 2013; Golic 2013; Gratz et al. 2013, 2014; Yu et al. 2013). Stable expression of Cas9 nuclease driven by a germline-specific promoter can induce efficient germline-transmitted mutagenesis in F1 progeny (Kondo and Ueda 2013; Ren et al. 2013; Xue et al. 2014). In addition, the CRISPR/Cas9 system has been successfully applied for conditional genome editing in C. elegans, mouse, and rat (Ma et al. 2013; Yang et al. 2013; Liu et al. 2014). More recently, UAS-driven expression of Cas9 in Drosophila has been shown to work with the CRISPR/Cas9 system (Port et al. 2014). Based on the high efficiency of Cas9/gRNA gene targeting, we reasoned that directly disrupting targeted genes at the DNA level would destroy gene function more efficiently than posttranscriptional breakdown of the targeted mRNA mediated by RNAi. Therefore, we combined the CRISPR/Cas9 and Gal4-UAS systems to develop a CRISPR/Cas9-mediated conditional mutagenesis (CMCM) method and systematically investigated whether it can efficiently inactivate gene activity in most Drosophila tissues. This method contains two steps (Figure 1). First, we generated gene-specific gRNAs directed against a number of genes with known phenotypes and made their 10UAS-Cas9/gRNAs transgenic flies. Second, we induced the expression of 10UAS-Cas9/gRNAs driven by the tissue-specific Gal4 lines and investigated the efficiency of gene disruption and associated phenotypes. Using the CMCM system, we tested six genes (y, notch, bam, nos, cid, and ms(3)k81) and achieved highly effective disruption in wing, eye, ovary, and testis tissues. Moreover, side-by-side comparisons of tissue-specific gene disruption showed the power of the CMCM system. In general, it was more efficient and rapid than RNAi approaches and provided a novel genetic tool for studying gene function in specific tissues in Drosophila (Dietzl et al. 2007; Ni et al. 2011).
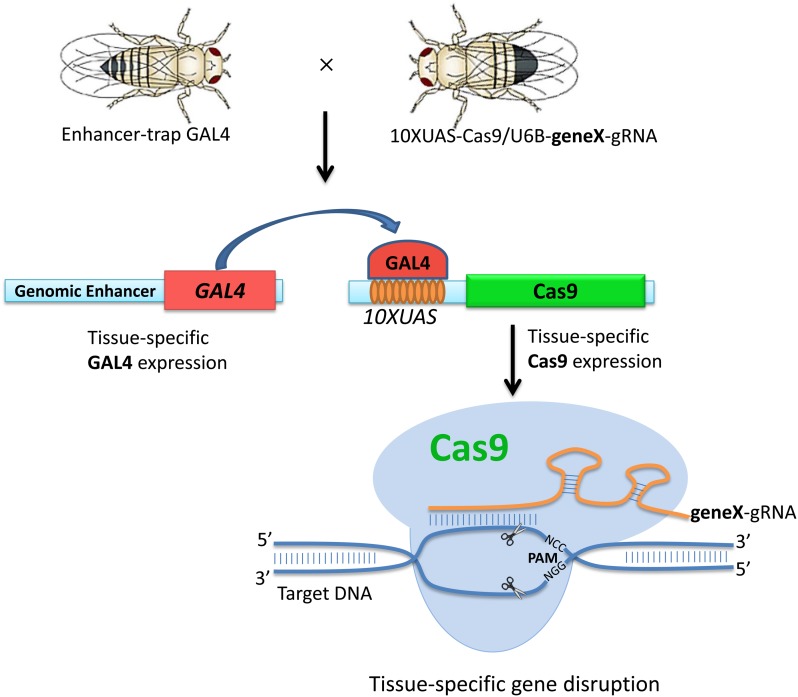
Figure 1.
A schematic overview of the CMCM system. For each gene of interest (gene X), transgenic stocks of U6B-geneX-gRNA, which ubiquitously express gRNA targeting the gene, were established. Transgenic 10ΧUAS-Cas9/U6B-geneX-gRNA male flies were recovered after crossing the 10XUAS-Cas9 stock with the U6B-geneX-gRNA stock. The recovered male flies were crossed with the enhancer-trap Gal4 virgin flies to drive Cas9 expression in specific tissues. Mutations were induced only in the specific tissue determined by the Gal4 stocks.
Materials and Methods
Fly stocks
Two wild-type stocks (w1118 and y) and four Gal4 stocks [tubulin(tub)-Gal4/TM6B,Tb (Ni et al. 2009), C96-Gal4 (Gustafson and Boulianne 1996), GMR-Gal4 (Ni et al. 2009), and Nos-Gal4 (Doren et al. 1998)] were used. Four cid RNAi lines were used, as follows: HMS02160 was provided by the Bloomington Stock Center (Bloomington, IN), and Vienna Drosophila RNAi Center (VRDC) 43856, VRDC43857, and VRDC109020 were provided by the VRDC. All balancers were obtained from the Bloomington Stock Center.
Plasmid construction
PiggyBac-10UAS-Cas9 construction:
PiggyBac-10UAS-Cas9 construction required three steps. First, the 10UAS-HSP70 promoter (Supporting Information, Figure S8) was amplified from the VRDC RNAi flies by polymerase chain reaction (PCR) and was then cloned into an XmaI/NotI double-digested PiggyBac-vasa-cas9 plasmid (Xue et al. 2014) to generate PiggyBac-10UAS-Cas9-vasUTR. Second, an 842-bp αTub84B 3′-UTR-containing sequence (Figure S8) was released from the pM3×P3-RFPattP′ vector (Bischof et al. 2007) by XhoI/SalI and then inserted into the SalI-digested pSP6-2SNL-spcas9 (stored in our lab) to generate the pSP6-2SNL-spcas9-αTub84B3′-UTR. Third, the larger fragment containing cas9-αTub84B3′-UTR was cut from pSP6-2SNL-spcas9-αTub84B3′-UTR by NotI/AscI and cloned into the PiggyBac-10UAS-Cas9-vasUTR plasmid using the same restriction enzymes to yield the final transgenic vector PiggyBac-10UAS-Cas9 (Figure S1A).
pRFP-gRNA construction:
Construction of the standard entry plasmid, pRFP-gRNA, was similar to that reported for the pUAST-RFP-gRNA vector (Xue et al. 2014), with slight modifications (Figure S1B). To generate multiple gRNA insertions against the same target locus in the transgenic vector, pRFP-gRNA, several different subcloning vectors (TA-U6B/CR7T-gRNA-A, TA-U6B/CR7T-gRNA-B, TA-U6B/CR7T-gRNA-C, and TA-U6BCR7T-gRNA-D) harboring different enzyme restriction sites were constructed as previously described (Xue et al. 2014) (Figure S1C). The KOD Mutagenesis Kit (TOYOBO, Osaka, Japan) was used to insert 20-bp target sequences between the promoter and the gRNA scaffold. For single gRNA insertions, the target-specific U6B/CR7T-gRNA was released from the TA-U6B/CR7T-gRNA-A vector by digestion with SpeI/KpnI and then cloned into pRFP-gRNA using the same sites to generate the final transgenic gene-specific vectors, pRFP-U6B/CR7T-1gRNA. For two gRNA insertions, the first gRNA was inserted into the TA-U6B/CR7T-gRNA-A plasmid (between SpeI and AgeI), and the second gRNA was inserted into TA-U6B/CR7T-gRNA-B (between AgeI and KpnI). Next, the second insertion was released by digestion with AgeI/KpnI and inserted into TA-U6B/CR7T-gRNA-A using the same restriction enzymes to generate the TA-U6B/CR7T-2gRNAs vector. Finally, the fragment containing two gRNA insertions was released from the vector TA-U6B/CR7T-2gRNAs by digestion with SpeI/KpnI and then cloned into pRFP-gRNA to generate the final transgenic gene-specific vectors, pRFP-U6B/CR7T-2gRNAs. Using a similar method, we inserted a third and fourth gRNA directly into the pRFP-2gRNAs vector via single enzyme sites (SpeI and KpnI, respectively) to generate transgenic gene-specific vectors pRFP-U6B/CR7T-3gRNAs and pRFP-U6B/CR7T-4gRNAs. In total, 13 transgenic pRFP-gRNAs vectors were generated. The details are presented in Table S1. All primers are listed in Table S2 and Table S3.
Fly transformation and genetics
To obtain transgenic 10UAS-Cas9 flies, the PiggyBac-10UAS-cas9 vector was mixed with helper vector PiggyBac-transposase (Thibault et al. 2004) and injected into w1118 fly embryos. Flies carrying 10UAS-Cas9 were identified by green fluorescent protein (GFP) expression in the eye when viewed under a fluorescence stereomicroscope. Three independent 10UAS-Cas9 lines were generated, mapping to chromosome X, chromosome 2, and chromosome 3. The 10UAS-Cas9 line mapping to chromosome 3 was used for the following experiments. To obtain transgenic gRNA flies, 13 pRFP-U6B/CR7T-gRNA vectors were separately injected into w1118 fly embryos with a hyperactive P-element transposase (Beall et al. 2002), and at least two independent lines per transgenic gRNA were identified by observing the RFP eye marker. After 10UAS-Cas9 flies were crossed with the transgenic gRNA flies, F1 male flies expressing both GFP and RFP fluorescent markers in the eyes were collected and crossed with Gal4 virgin flies (the screen marker was red eyes). In the F2 progeny, flies expressing GFP, RFP fluorescent markers, and the red eye marker were the conditional mutant flies. For most gRNA transgenic vectors, we picked up just one transgene to perform subsequent crossing experiments and phenotype estimation, even if we recovered several gRNA transgenic lines. For some gRNA transgenic vectors such as CR7T-bam1, U6B-nos1nos2, and CR7T-nos1nos2, we picked up two independent gRNA transgenic lines for conditional mutation analyses for each transgenic vector.
Phenotypic and genotypic analysis of conditional mutant flies
The phenotypes and genotypes of conditional mutant flies were assessed. For yellow, the abdomen of the conditional mutant flies was photographed with a Leica Mz10f. For notch, the wings of the conditional mutant flies were scored and photographed with a Leica Mz10f. For bam and nos, each of the conditional mutant female flies was crossed with three w1118 male flies to test the fertility. The ovaries were scored based on their severity of phenotype and photographed with a Leica Mz10f. For ms(3)k81 and cid, each conditional mutant male fly was crossed with three w1118 female virgin flies to test the fertility. The testes were scored according to their fertility and photographed with a Zeiss Imager.Z2. The testes, tips of testes, seminal vesicles, and mature sperm from all conditional mutant flies and control flies were examined under a microscope with light and 4′,6-diamidino-2-phenylindole (i.e., DAPI) staining. The target genomic region from the conditional mutant flies was amplified by PCR using appropriate primers following a previous protocol (Carvalho et al. 2009) (Table S4). For yellow, the whole fly was used for PCR; for notch, the wing, eye, and body were used for PCR; for bam and nos, the ovary was used for PCR; for ms(3)k81 and cid, the testis was used for PCR. The corresponding PCR products were then sequenced.
Results and Discussion
Overview of the CMCM system
Transgenic Piggybac-10UAS-cas9 (Figure S1A) flies were created to express the Cas9 nuclease under control of the Gal4-driven activating site (UAS). Transgenic gRNA vectors pRFP-U6B-gRNA and pRFP-CR7T-gRNA (Xue et al. 2014) (modified from a pUAST plasmid with RFP replacement of the white gene; and deletion of the UAS site; Figure S1B) were based on two different long noncoding RNA promoters, and corresponding gRNAs were ubiquitously expressed in whole tissues. Using these vectors, we generated gene-specific gRNAs directed against genes with known phenotypes (Table S1). To increase the mutation efficiency, we designed gRNA vectors containing one to three gRNAs directed against different regions of the same gene (Figure S1C). To evaluate tissue bias with respect to the mutation efficiency of gRNA transcripts, we used both promoters to drive gene-specific gRNA transcription in specific tissues (Xue et al. 2014). Furthermore, to test the versatility of the CMCM method, we conducted experiments in body, wing, testis, and ovary tissues at 25° and 28°.
CMCM-mediated conditional mutation of the yellow gene
To test the effectiveness of the CMCM system, we first chose the yellow (y) locus owing to its easily detectable body color phenotype. We generated transgenic fly lines containing pRFP-U6B/CR7T-gRNA constructs that target y. The 10UAS-Cas9 and gRNA transgenic lines were crossed to obtain 10UAS-Cas9/gRNA animals. We then induced 10UAS-Cas9 expression with Tub-Gal4/Tm6b, a line showing ubiquitous high-level expression of the Gal4 activator. All of the lines displayed yellow mosaicism or a completely penetrant yellow phenotype, as expected (Figure 2A), indicating that 10UAS-Cas9/gRNA triggered highly efficient gene disruption in somatic tissues. The U6B and CR7T promoters were equally efficient. The phenotype obtained with CR7T-y1y2-gRNA was indistinguishable from that of y mutant flies, suggesting that targeting a gene with multiple gRNAs improves the efficiency of the conditional mutagenesis. Sequencing of the y locus confirmed cleavage at the targeted site (Figure S2).
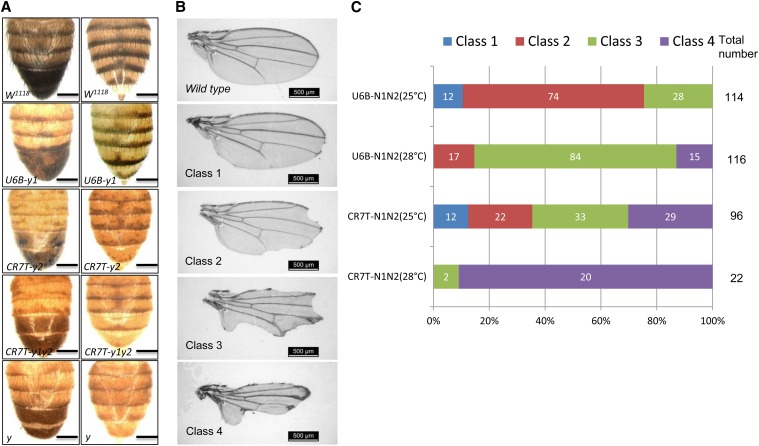
Figure 2.
Conditional mutation of the yellow and notch genes. (A) The yellow mosaic phenotype is shown for the three different gRNA expression vectors, U6B-y1, CR7T-y2, and CR7T-y1y2. Tub-Gal4/Tm6b was used to drive the expression of Cas9. w1118 and y flies are shown as controls. Scale bars: 200 µm. (B) Four classes of wing defects were identified when cas9 was expressed using the wing margin C96-Gal4 driver. Class 1: margin bristles missing but no notches; Class 2: margin bristles missing and moderate wing notching; Class 3: extensive margin bristle loss and notching; and Class4: most of the wing margin missing. Scale bars: 500 µm. (C) Phenotypes of conditional mutant flies for notch in the wing.
CMCM-mediated conditional mutation of the notch gene in the wing and eye
We also tested the specificity and sensitivity of the CMCM system using the Notch gene, which encodes a signaling receptor essential for wing development (Johnston and Edgar 1998; Presente et al. 2002). We designed two gRNAs against the Notch gene and performed a sensitive wing assay whereby 10UAS-Cas9 was expressed under the control of the C96-Gal4 driver, a line that expresses Gal4 in the blade region of the wing imaginal disc (Gustafson and Boulianne 1996; Presente et al. 2002). Disruption of notch led to marked defects at the wing margin ranging from mild loss of margin bristles to strong wing notching (Figure 2B), consistent with the phenotypes obtained through RNAi (Ni et al. 2008). We consistently observed stronger phenotypes when gRNA expression was driven by the CR7T promoter rather than by the U6B promoter, likely reflecting a difference in promoter activity in the wing (Figure 2C). We further analyzed the notch gene sequence in wing, eye, and body tissues of C96-Gal4 driver-mutated flies. Mutagenesis at the designed site was observed only in the wing, not in the eye or body (Figure S3). In turn, expression of 10UAS-Cas9/gRNAs against notch under the control of an eye-specific GMR-Gal4 driver generated strong defects in eye tissue, but no defects in the wing (Figure S4).
To determine how temperature affects the severity of the notch phenotype induced by 10UAS-Cas9/gRNA, we used the C96-Gal4 driver at both 25° and 28°. Phenotypes obtained were stronger at 28° than at 25° for both promoters. A large fraction of flies (20 of 22) showed extreme defects (class 4) at 28° when using the CR7T promoter, indicating an effect of temperature.
CMCM-mediated conditional mutations of the bam and nos genes in the ovary
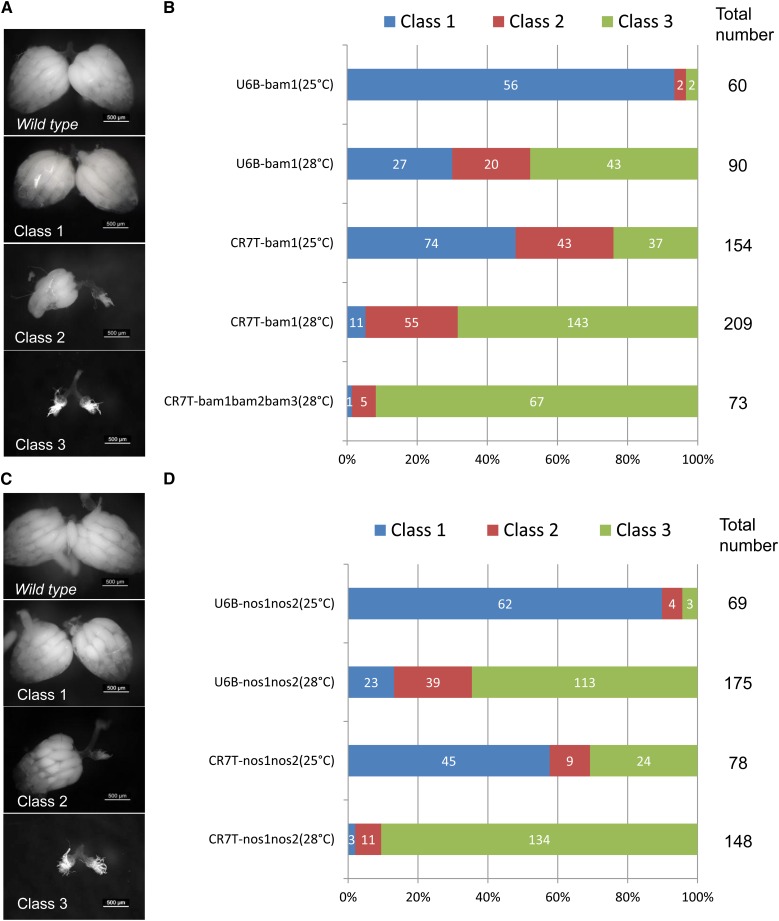
Drosophila oogenesis is an important model system in developmental and evolutionary biology. We next applied CMCM to the ovary using the bam (bag of marbles) gene. Genetic bam-mutants and bam-RNAi flies showed disrupted cyst formation during oogenesis, resulting in ovaries containing an excess number of cells that cannot differentiate into gametes (McKearin and Spradling 1990; Chen and McKearin 2003; Ni et al. 2011). We induced 10UAS-Cas9 expression in the ovary using nos-Gal4, a line expressing Gal4 in all stages of oogenesis (Rørth 1998). We observed phenotypes consistent with genetic mutants and RNAi flies (Figure 3A). A larger fraction of ovaries showed extreme phenotypes (class 3) when gRNA expression was driven by CR7T rather than by the U6B promoter at both 25° and 28° (Figure 3B). Furthermore, disruption of bam driven by multiple gRNAs led to extreme phenotypes in more than 90% of females (class 3) at 28° (Figure 3B). Disruption of nos driven by nos-Gal4 caused a phenotype similar to that resulting from nos-Gal4-driven bam disruption (Figure 3, C and D). Sequencing of dissected mutated ovaries showed many mutations at the designed targeting site (Figure S5 and Figure S6).
Figure 3.
Conditional mutations of the bam and nos genes in the ovary. (A, C) Three classes of ovary defects were distinguished when 10UAS-cas9 was expressed using the nos-Gal4 driver for both bam and nos genes. Class 1: the same as wild-type; Class 2: one side of the ovary was shrunken and the other side was normal; and Class 3: both sides of the ovary were shrunken. (B, D) Phenotypes of the bam and nos genes in the ovaries of conditional mutant flies. High temperature and the use of multiple gRNAs can improve the efficiency of CMCM, and the CR7T promoter works better than the U6B promoter in the ovary.
CMCM-mediated conditional mutations of the cid and ms(3)k81 genes in the testis
We also targeted the cid (cenH3) gene, which encodes a protein essential for somatic centromere assembly and embryogenesis (Blower and Karpen 2001; Blower et al. 2006). We induced 10UAS-Cas9 expression using the nos-Gal4 driver to detect phenotypes in the testis. Testis size was greatly reduced (Figure 4A), probably because nos-Gal4 is expressed very early during spermatogenesis (Rørth 1998). Disruption of the cid gene during early spermatogenesis led to a failure in cell division and to male sterility. We could not detect any mature sperm in the seminal vesicles of the mutated testes (Figure 4A), likely explaining the observed sterility. Low temperature−induced (25°) conditional mutagenesis using both promoters resulted in a small fraction of flies with defects in testis tissue (Figure 4B). However, induced mutagenesis at 28° resulted in a high fraction of flies with defects for both promoters. Notably, all examined mutated male flies under the control of the CR7T promoter (34 of 34) were 100% sterile and showed severe phenotypes in the testes at 28° (Figure 4B).
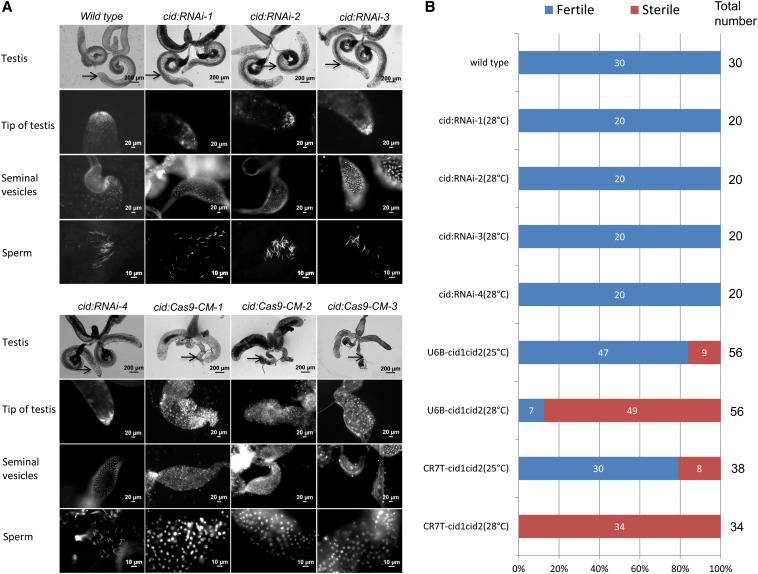
Figure 4.
Conditional mutation of the cid gene in the testis. (A) The phenotypes in the testis tissue are compared for the RNAi lines and CMCM lines. From top to bottom of each column: whole testis (light), tip of the testis (DAPI), seminal vesicles (DAPI), and sperm detection (DAPI). The arrows indicate testis tissue. cid:RNAi-1, BM40912; cid:RNAi-2, v43856; cid:RNAi-3, v43857; cid:RNAi-4, v109020. cid:Cas9-CM denotes testis from a conditional mutant fly via the CMCM system; nos-Gal4 was used to drive the expression of Cas9, and two vectors (U6B-cid1cid2 and CR7T-cid1cid2) were used to drive the expression of gRNA. cid:cas9-CM-1 is from U6B-cid1cid2 (28°C); cid:cas9-CM-2 is from CR7T-cid1cid2 (25°C); cid:cas9-CM-3 is from CR7T-cid1cid2 (28°C). All examined cid:Cas9-CM testes lacked mature sperm in the seminal vesicles. (B) Fertility tests of flies bearing the transgenic RNAi and the CMCM system of the cid gene. DAPI, 4′,6-diamidino-2-phenylindole.
To directly compare the CMCM system with RNAi, we tested all four cid-RNAi lines from the Vienna Drosophila RNAi Center and Bloomington Stock Center. After crossing each of them to the nos-Gal4 driver, all testes from RNAi flies were indistinguishable from those from wild-type flies (Figure 4A). Indeed, all examined RNAi flies (more than 20 for each line) were fertile (Figure 4B), indicating that RNAi cannot completely block the expression of Cid in testis tissue. A previous study also demonstrated that an RNAi-induced reduction in Cid expression during spermatogenesis has no effect on male fertility (Raychaudhuri et al. 2012).
To rule out any nonspecific effects of the CMCM system in testis, we next tested the male-specific gene ms(3)k81. Mutations in ms(3)k81 cause male sterility without morphologically detectable defects in testis and sperm development (Fuyama 1986; Yu et al. 2013). We then induced 10UAS-Cas9 expression in testis under the nos-Gal4 driver. Testis and sperm development were normal (Figure S7A). Almost 80% of CMCM-induced flies (30 of 38) were completely sterile at 28° (Figure S7B). Sequencing of these sterile flies confirmed disruption of ms(3)K81 in testis tissue (Figure S7, C and D).
In contrast to the RNAi strategy of knocking down gene expression at the mRNA level, the CMCM approach silences a gene at the DNA level in a more complete way. For six well-known genes, we achieved very efficient gene disruption in wing, eye, ovary, and testis tissues using CRISPR/Cas9 combined with the Gal4-UAS system in Drosophila. Detailed examinations of phenotypes resulting from notch gene disruption in the wing and eye using tissue-specific Gal4 drivers revealed that CMCM-mediated gene disruption appears to be strictly restricted to the particular tissue.
However, the efficiency of CMCM-induced phenotypes could be affected by the following parameters: (1) the promoter of gRNA expression, (2) the choice and the number of gRNAs, (3) temperature, (4) incomplete mutation of the target gene, and (5) positional effects of randomly integrated gRNA transgenes. In this report, CR7T promoter-driven gRNAs led to more efficient conditional mutagenesis than U6B promoter-driven gRNAs in most tissues, particularly in the ovary, indicating that the pleiotropic functions of one particular gene in various tissues could be analyzed by crossing the same transgenic 10UAS-Cas9/gRNA under the CR7T promoter with different tissue-specific Gal4 drivers. Nevertheless, RNAi-mediated knockdown efficiency usually is affected by the structure and direction of hairpin RNAs, which can result in unstable mutagenesis in different tissues (Ni et al. 2011). To achieve the strong phenotype of gene silencing, one frequently needs to generate different hairpin RNAi lines to trigger silencing of the same gene in multiple tissue types, especially during oogenesis (Ni et al. 2011). Regarding the choice and the number of gRNAs, the mutation efficiency of the CMCM system depends on the gene locus. To obtain the strongest loss-of-function phenotypes, the use of multiple gRNAs against the same target site could be considered. The presence of both mutant and wild-type alleles from incomplete cleavage by Cas9 may lead to complex phenotypes. The phenotypes generated at 28° were more severe than those generated at 25°, which is consistent with the temperature sensitivity of the Gal4-UAS system. In addition, we did not observe significant efficiency differences in the tissue-specific mutagenesis between two independent gRNA transgenic lines from the same transgenic gRNA. Although the expression of randomly integrated gRNA is known to be influenced by their local genomic environment, position effects of transgenic gRNA in this study did not cause severe effects on the mutant phenotypes. Based on current examples, we propose that a small amount of transgenic gRNA expression may be enough for efficient tissue-specific mutagenesis. However, future CMCM-mediated conditional experiments could be performed at the fitted transgenic gRNA loci via the phiC31-mediated attP/attB system (Ni et al. 2008).
CMCM system has two important applications. First, the CR7T promoter can be used to drive multiple gRNAs to target a single gene to achieve high degree of inactivation in specific tissues; all gRNAs could easily be integrated into one simple transgenic vector using our method. Second, it provides an independent method for validation of the loss-of-function phenotypes after RNAi genetic screening. In summary, our CMCM system provides a simple and efficient strategy for conditional gene disruption, making it possible to investigate the different functions of a single gene in various tissues during development.
Supplementary Material
Acknowledgments
We thank J. C. Pastor-Pareja, J. Dai, and T. Xie for their critical comments on our manuscript. This work was supported by grants from the National Natural Science Foundation of China (31171278, 31271542).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014159/-/DC1
Communicating editor: M. Boutros
Literature Cited
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E. L., Mahoney M. B., Rio D. C., 2002. Identification and analysis of a hyperactive mutant form of Drosophila P-element transposase. Genetics 162: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H., 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Daigle T., Kaufman T., Karpen G. H., 2006. Drosophila CENP-A mutations cause a BubR1-dependent early mitotic delay without normal localization of kinetochore components. PLoS Genet. 2: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho G. B., William W. J., Benzer S., 2009. Non-lethal genotyping of single Drosophila. Biotechniques 46: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., McKearin D. M., 2003. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130: 1159–1170. [DOI] [PubMed] [Google Scholar]
- Choi C. M., Vilain S., Langen M., Van Kelst S., De Geest N., et al. , 2009. Conditional mutagenesis in Drosophila. Science 324: 54. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Doren M. V., Williamson A. L., Lehmann R., 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–246. [DOI] [PubMed] [Google Scholar]
- Fuyama Y., 1986. Genetics of parthenogenesis in Drosophila melanogaster. I. The modes of diploidization in the gynogenesis induced by a male-sterile mutant, ms(3)K81. Genetics 112: 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K. G., 2013. RNA-guided nucleases: a new era for engineering the genomes of model and nonmodel organisms. Genetics 195: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 194: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson K., Boulianne G., 1996. Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome 39: 174–182. [DOI] [PubMed] [Google Scholar]
- Johnston L. A., Edgar B. A., 1998. Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature 394: 82–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 915: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Long L., Xiong K., Yu B., Chang N., et al. , 2014. Heritable/conditional genome editing in C. elegans using a CRISPR-Cas9 feeding system. Cell Res. 24: 886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang X., Shen B., Lu Y., Chen W., et al. , 2013. Generating rats with conditional alleles using CRISPR/Cas9. Cell Res. 24: 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D. M., Spradling A. C., 1990. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 4: 2242–2251. [DOI] [PubMed] [Google Scholar]
- Ni J.-Q., Markstein M., Binari R., Pfeiffer B., Liu L.-P., et al. , 2008. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods 5: 49–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Liu L. P., Binari R., Hardy R., Shim H. S., et al. , 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presente A., Shaw S., Nye J. S., Andres A. J., 2002. Transgene-mediated RNA interference defines a novel role for notch in chemosensory startle behavior. Genesis 34: 165–169. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri N., Dubruille R., Orsi G. A., Bagheri H. C., Loppin B., et al. , 2012. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 10: e1001434–e1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C., et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118. [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Xue Z., Ren M., Wu M., Dai J., Rong Y. S., et al. , 2014. Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3 (Bethesda) 4: 925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., et al. , 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.