Abstract
In solid tumors, the relationship between DNA copy number and global expression over large chromosomal regions has not been systematically explored. We used a 12,626-gene expression array analysis of head and neck squamous cell carcinoma and normal oral mucosa and annotated gene expression levels to specific chromosomal loci. Expression alterations correlated with reported data using comparative genomic hybridization. When genes with significant differences in expression between normal and malignant lesions, as defined by significance analysis of microarrays (SAM), were compared to nonsignificant genes, similar chromosomal patterns of alteration in expression were noted. Individual tumors underwent microsatellite analysis and χ2 analysis of expression at 3p and 22q. Significant 3p underexpression and 22q overexpression were found in all primary tumors with 3p and 22q allelic imbalance, respectively, whereas no tumor without allelic imbalance on these chromosomal arms demonstrated expression differences. Loss and gain of chromosomal material in solid cancers can alter gene expression over large chromosomal regions, including multiple genes unrelated to malignant progression.
Keywords: chromosomal mapping, microarrays, comparative genomic hybridization, allelic imbalance, head and neck squamous cell carcinoma
Gene expression microarray technology has allowed for the rapid screening of several thousand gene transcripts in primary tumors to identify genes responsible for tumor progression or formation. Several studies have looked at gains or losses of DNA copy numbers at different chromosomal loci in tumors; however, to the best of our knowledge, there have not been any systematic methods correlating DNA copy number and global expression alterations over entire chromosomal arms or regions. Using locus-specific annotation of expression alterations in primary head and neck squamous cell carcinoma (HNSC), we examined the relationship of expression alterations and DNA copy alterations for specific chromosomal arms and loci.
Carcinomas of the head and neck represent the sixth most common malignancy in the world, and at least 90% of these are squamous cell carcinomas (1, 2). Chronic exposure of the upper aeorodigestive tract to known carcinogens in alcohol and tobacco ultimately results in the accumulation of multiple genetic aberrations in exposed mucosa (3). Genetic events involved in head and neck carcinogenesis have been characterized by using techniques including microsatellite analysis and comparative genomic hybridization (CGH) (3–8), whereas transcriptional alterations have been characterized by using several techniques, including gene expression profiling based on microarray technology (9, 10).
Initial descriptions of loss of chromosomal arms 3p, 9p, and 17p have been demonstrated by using a variety of the above methods, revealing loss of chromosomal material as a means of inactivation of critical tumor suppressor genes, including p16, p14ARF, and p53 (11–17). Loss of chromosomal arm 3p and chromosomal regions 9p21 and 17p13 have been described as early events in a tumor progression model and have been shown to demonstrate an increased tendency for malignant transformation in premalignant lesions (18).
CGH allows a survey of DNA sequence copy number changes along the length of chromosomes. Regions with increased copy numbers reveal chromosomal sites that may harbor oncogenes or growth-related genes, whereas regions with decreased copy number may contain tumor suppressor gene loci. This technique has been used in the analysis of a variety of tumors, including HNSC. DNA copy number increases have been frequently found on chromosomal arms 3q, 5p, 8q, 16p, 17q, and 19, whereas DNA underrepresentations have been commonly observed on 1p, 3p, 5q, 6q, 8p, 9p, 13q, 18q, and 21q (5, 19, 20).
The use of gene expression profiling allows for rapid genome-wide analysis of tumors in a standardized fashion. Methods of normalizing gene expression that correct for intersample variability and systematic bias inherent in expression array analysis have included Standardization and Normalization of Microarray Data (snomad; www.snomad.org), which provides a standard normalized expression value for each analyzed gene, termed a Z score (21). Additional analysis tools are used to identify differentially expressed genes, including statistical analysis of microarrays (SAM), a tool that uses an adjustable threshold to determine significant genes expressed differentially between two groups (22). Recently, Mao et al. analyzed chromosome 21 gene expression in Down's syndrome by annotating gene expression values to chromosomal loci using Database Referencing of Array Genes On-Line dragon′ (www.dragondb.org), a biological annotation tool that aids in the analysis of differential gene expression data (23, 24). The information provided by dragon, including chromosomal position, can be integrated with other expression data to gain a better understanding of how biological characteristics of specific tumors are related to gene expression patterns. Mao et al. (24) assessed patterns of gene expression in a group of fetal trisomy 21 tissues on a chromosome-wide basis and demonstrated a clear pattern of overexpression localized to chromosome 21. We hypothesized that a similar method could be used to identify more specific areas of increased or decreased expression in solid tumors and could be related to chromosomal copy alterations as well as significant gene expression alterations.
We used annotated expression array data to characterize locus-specific expression alterations in primary HNSC. These findings were correlated with CGH data on chromosomal alteration as reported previously (3–5, 7, 8) by multiple investigators. Chromosomal patterns of gene expression were compared among groups of genes that possess or lack significant expression differences in malignant progression. Finally, patterns of expression in individual tumors were correlated with allelic imbalance (AI) on chromosomal arms 3p and 22q.
Materials and Methods
All microarray data used in this analysis were generated previously as part of efforts to identify the number of significant genes responsible for the progression of head and neck premalignant lesions to invasive cancer and to identify early and late transcriptional alterations in the progression of HNSC (25). Thirteen samples, including oral mucosal samples from six normal control patients and seven patients with invasive carcinoma, were analyzed by hybridization of cRNA to gene expression arrays containing >12,000 genes. Initial normalization using snomad provided local mean normalization and local variance correction that correct for bias and variance, which are nonuniformly distributed across the range of microarray element signal intensities (21). This transformation provides normalization between variations of signal intensity between samples and corrects for systematic bias at extremes of signal intensity, providing a standardized normalized expression value (Z score). Genes were defined as significant when significantly under- or overexpressed in malignant vs. normal samples, as determined by SAM, using a false discovery rate <5% (22), and are listed at www.hopkinsmedicine.org/headneckcancer/physicians.html (25). All genes were subjected to in silico analysis by using dragon. Of the original 12,626 genes, 9,724 were successfully mapped to chromosomal locations. Genes were then sorted based on their respective chromosomal loci by using Microsoft excel software. Average expression Z scores for all genes residing on each chromosomal arm were calculated, and graphs representing the average expression Z scores and 95% confidence interval for each arm in the malignant group were constructed. All significantly over- and underexpressed genes, as determined by SAM, were also annotated. Of the original pool of 2,071 significant genes within the primary HNSC, 1,663 were successfully mapped and used in this study. As with the analysis of all 9,724 genes, graphs were constructed. Average chromosomal arm expressions with 95% confidence intervals are plotted. Intervals that do not overlap 0 are differentially expressed significantly.
Because loss of chromosomal arm 3p has been well characterized in head and neck cancer by using CGH (3–8, 19) and minimal areas of loss further detailed using microsatellite analysis (17, 18, 26), a smaller regional analysis of 3p was performed to identify specific areas of underexpression. A similar analysis was done for chromosomal arm 22q to identify regions with significant overexpression. The expression Z scores for genes on small regions on chromosomal arms 3p and 22q, using the original 9,724 gene group, were pooled and averaged, and graphs were constructed. To demonstrate that the altered expression patterns of genes residing on 3p and 22q extend beyond the significant genes, smaller regional analysis was performed by using only the nonsignificant genes.
To investigate the relationship between expression and AI indicative of chromosomal gain or loss in primary tumors, a panel of 10 microsatellite repeat PCR primers (Research Genetics, Huntsville, AL) was used to amplify loci in the chromosomal 3p21–23, 22q11, 22q11.2, and 22q13-ter regions. Six microsatellite markers (D3S1100, D3S1211, D3S1277, D3S1537, D3S1619, and D3S2405) specific to 3p21–23 and 4 markers (D22S264, D22S343, D22S450, and D22S526) specific for 22q11, 22q13-ter, and 22q11 were selected and used in this study. Before amplification, 50 ng of one primer from each pair was end labeled with [γ-32P]ATP [20 mCi (1 Ci = 37 GBq); Amersham Pharmacia Life Sciences] and T4 kinase (New England Biolabs) in a total volume of 50 μl. PCRs were carried out in a total volume of 12.5 μl containing 10 ng of genomic DNA, 0.2 ng of labeled primer, and 15 ng of each unlabeled primer. The PCR buffer included 16.6 mM ammonium sulfate, 67 mM Tris (pH 8.8), 6.7 mM magnesium chloride, 10 mM 2-mercaptoethanol, and 1% DMSO, to which were added 1.5 mM deoxynucleotide triphosphates and 1.0 unit of TaqDNA polymerase. (Boehringer–Mannheim). PCR amplifications of each primer set were performed for 30–35 cycles consisting of denaturation at 95°C for 30 s, annealing at 50–60°C for 60 s, and extension at 72°C for 60 s. One-third of the PCR products were separated on 8% ureaformamide-polyacrylamide gels and exposed to film from 24 to 48 h. For informative cases, allelic loss was documented if one allele was significantly decreased (>50%) in tumor compared with the same allele in the normal (lymphocyte) DNA. All of the samples were assessed by two observers independently (B.G.M. and J.C.). All of the identified alterations were confirmed by repeating the PCR and electrophoresis. Samples were coded so that the observers were blinded to the expression status of individual samples.
Graphs representing the average expression Z scores for small regions on chromosomes 3p and 22q were created for individual tumor samples. All annotated genes for each tumor sample were divided into two groups. The first group consisted of all genes (n = 9,546), except those mapped to the 3p21–23 region. The second group consisted of those genes assigned to 3p21–23 (n = 177). Similarly, in the analysis of 22q11, all annotated genes were divided into two groups. The first group consisted of all genes (n = 9643) except those mapped to 22q. The second group was comprised of those genes assigned to 22q (n = 217). Genes that were unable to be mapped to a specific chromosomal locus were excluded from the analysis. Average local Z scores for 3p21–23 for individual tumor samples, in standard deviation units, were calculated with local background corrections by using snomad. A total of seven bins containing Z scores were then generated for both groups where bins were defined as follows: ≤ –2, (–2, –1], (–1,0], (0,1], (1,2], (2,3], >3. These cutoffs were chosen empirically to ensure that bins were not sparse. Each gene was assigned to a bin based on its respective Z score. The percentage of genes within each bin was calculated by dividing the number of genes per bin by the total number of genes in that group. P values for the comparisons of these two populations (chromosome 3p21–23 and the rest of the genes) were calculated by using the χ2 test for trend. An independent χ2 trend analysis of 22q was performed as well by using the calculated average Z scores for individual tumors. As with the 3p21–23 analysis, seven bins were used. Each gene was assigned to a bin, and the percentages of genes in each bin were calculated. P values comparing genes assigned to 22q and all other genes were calculated by using χ2 tests for trend.
A review of the literature for CGH profiles for all chromosomes from studies investigating a variety of head and neck tumors was compiled and qualitatively compared to the expression profiles generated from our data. Individual chromosomal arms were recorded as demonstrating either an increase (+) or a decrease (–) in DNA copy number as reported by particular criteria in each study. Those cases where it was impossible to clearly define a chromosomal arm as having a global copy number alteration due to an ambiguous CGH profile for that particular arm were reported as indeterminate. In the setting of equivocal decreases or increases in copy number at regions on a particular chromosomal arm, the decision was made to report that entire arm as equivocal with respect to copy number alteration. Statistical analysis of entire chromosomal arms was performed by using χ2 analysis as described above. Significance was noted for regions on 3p and 22q. To provide an exploratory qualitative comparison of chromosomal arm expression trends and CGH data, we analyzed 95% confidence intervals to identify a chromosomal arm as under- or overexpressing if these intervals excluded no change in expression level, and compared this to CGH data.
Results
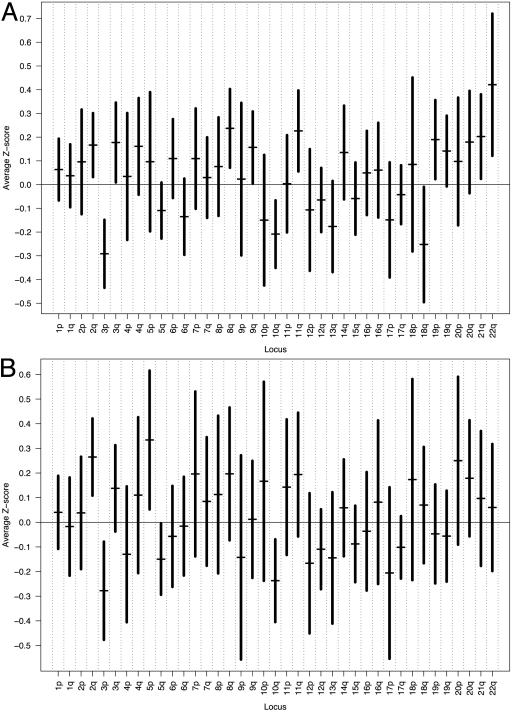
Data from 12,626 gene expression array analysis of malignant tumors and normal oral mucosa were used, and gene expression levels were annotated to specific chromosomal loci to examine chromosomal locus-based expression patterns in HNSC. Expression alterations were then further correlated with previously reported CGH data as well as data from microsatellite analysis of the seven malignant samples. Results of the analysis of the seven primary tumors using all 9,724 annotated genes are shown in Fig. 1A. Using 95% confidence intervals, global underexpression was noted on chromosomal arms 3p, 10q, and 18q. Peaks of overexpression were found on 2q, 3q, 8q, 9q, 11q, 19p, 21q, and 22q. As with the original 9,724 gene data, graphs were constructed by using only those genes previously found to exhibit significant expression alterations as determined by SAM. Results from the analysis of the 1,663 statistically significant annotated genes are shown in Fig. 1B. Significant underexpression was found on 3p, 5q, and 10q; overexpression was seen on 2q and 5p. This demonstrates that multiple chromosomes show similar global expression alteration when significantly differentially expressed genes are compared to all genes, notably chromosomes 2q, 3p, and 10q. Similar trends in average expression scores were seen when chromosomal arms were compared. The discrepancies between these two analyses may be due to differences in the magnitude of effect among groups of significant genes when compared to all genes or to variability introduced by a smaller analyzed number of significant genes. However, in general, graphical comparison demonstrates similar trends. Table 2, which is published as supporting information on the PNAS web site, shows the results of previously reported CGH profiles of all chromosomal arms for a variety of HNSC tumor sites and stages generated from six different studies. Despite the heterogeneity of sample sizes, sites, and stages profiled by these investigators, several chromosomal arms demonstrated consistent copy number aberrations. When comparing the malignant data from our analysis of the 9,724 genes to the CGH profiles from these studies, concordance (defined as complete agreement among all reported CGH profiles in direction of copy number alteration vs. average expression values) was observed on chromosomal arms 1q, 2p, 3q, 5p, 7p, 8q, 9q, 20p, and 20q. Of these 39 chromosomal arms evaluated, discordance between CGH data and expression data, defined as agreement among all reported CGH profiles that is discordant with average expression value, was seen on only three arms, 4q, 9p, and 17q. If only chromosomal arms whose 95% confidence intervals that exclude zero are considered, then concordance is noted for two arms, 3p and 8q, with no arms showing discordance. These discrepancies may be due to the tendency of CGH to oversimplify, in that smaller chromosomal regions with highly variable copy number may tend to become reduced in magnitude by adjacent areas of normal or differing copy number. In addition, review of CGH demonstrates a substantial amount of variability in interpretation on copy number within CGH-based analyses as a group.
Fig. 1.
Graph of average gene expression Z scores for each chromosomal arm in seven primary HNSC using (A) all 9,724 annotated genes and (B) only those 1,663 annotated genes found to demonstrate significant expression alterations as determined by SAM. All annotated genes were subjected to in silico analysis by using dragon. Genes were then sorted based on their respective chromosomal loci. Average expression Z scores for all genes residing on each chromosomal arm (horizontal bars) were calculated, and vertical bars representing the 95% confidence intervals for expression scores are shown.
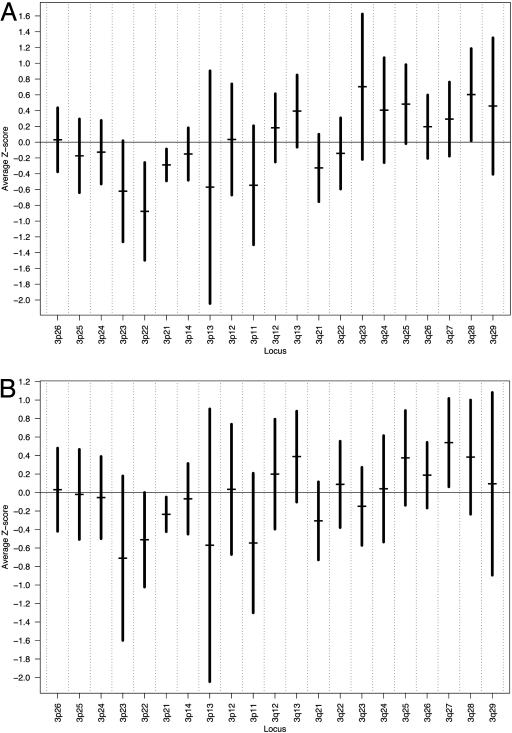
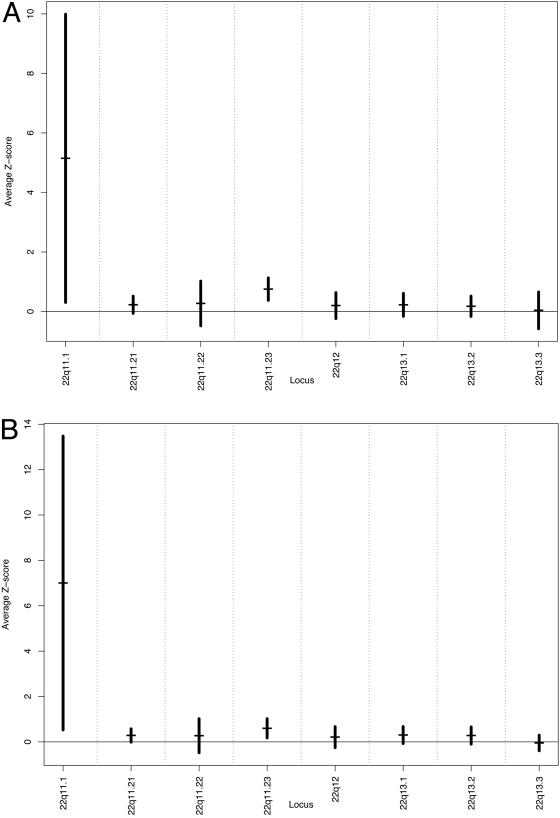
Because loss of chromosomal arm 3p has been frequently found in head and neck cancer, and highly significant overexpression of 22q was noted, a smaller regional analysis was performed to identify specific areas of under- and overexpression in our grouped malignant samples. Fig. 2A shows the results of in silico analysis of chromosome 3 using the 9,724-gene data. Nadirs of expression were found on chromosomal regions 3p21 and 3p22. Fig. 3A shows the results of the analysis of chromosomal arm 22q. Peaks of expression were noted on chromosomal regions 22q11.1and 22q11.23. Overexpression in this region was found to be statistically significant (P < 0.001). To separate out effects on global expression from outliers that include genes with significant expression differences, we removed those genes with significant expression differences as determined by SAM from the analysis. Again nadirs of expression were observed on 3p21 and 3p23 (Fig. 2B). Similarly, statistically significant (P < 0.001) peaks of expression were preserved in the 22q11.1 and 22q11.23 region (Fig. 3B).
Fig. 2.
Graph of average gene expression Z scores in small regions of chromosome 3 in HNSC using all 9,724 annotated genes (A) and average expression Z scores of only nonsignificant chromosome 3 genes (B). All annotated genes localized to chromosome 3 were identified and sorted based on their chromosomal localization. Average gene expression Z scores for small regions on chromosome 3 were calculated (horizontal bars) with 95% confidence intervals (vertical bars).
Fig. 3.
Graph of average gene expression Z scores in small regions of chromosomal arm 22q in primary HNSC using all 9,724 annotated genes (A) and average expression Z scores of only nonsignificant 22q genes (B). All annotated genes that could be localized to chromosomal arm 22q were identified and sorted based on their respective chromosomal localization. Average gene expression Z scores for small regions on chromosome 22q were calculated (horizontal bars) with 95% confidence intervals (vertical bars).
The CGH data for chromosomal arm 3p from previous studies were compiled, and areas of decreased copy numbers were identified to determine whether copy number aberrations corresponded with expression alterations at specific regions on 3p. Bockmuhl et al. (5) reported nadirs of copy numbers on 3p11-p14, 3p21-p22, and 3p25. Several areas of overlap with our data are shown in Fig. 2, specifically, 3p11, 3p13, and 3p21–22. Bergamo et al. (4) reported significantly decreased copy numbers on 3p24–26. We similarly found decreased expression on 3p24-p25. Data generated from other investigators unfortunately did not specifically identify subregions of decreased copy numbers on 3p. Even after subtracting the significant chromosome 3 genes as determined by SAM analysis, these regions of decreased expression were clearly preserved.
We performed microsatellite analysis by using a panel of 10 markers to identify AI in single primary tumors. Six microsatellite markers (D3S1100, -1211, -1277, -1537, -1619, and -2405) specific to 3p21–23 and 4 markers (D22S264, -343, -450, and -526) specific for 22q.11, 22q13-ter, and 22q11 were used. Four of the seven tumors (samples 2291, 2313, 2397, and 2717) demonstrated AI on 3p21–23 in at least three of the six microsatellite markers chosen (Table 1). The remaining three samples (samples 2304, 2674, and 2700) exhibited retentions at all six markers specific for 3p21–23 (Table 1). To establish the relationship between expression and AI on 3p21–23, the average expression scores of all genes in this region were calculated for individual tumors and compared to the results of the microsatellite analysis. Statistical significance of expression scores was determined by using χ2 tests for trend. Table 1 summarizes our results and demonstrates a clear average expression Z score demarcation between those samples with and those without AI. Samples 2291, 2313, 2397, and 2717, all with documented AI, demonstrated significant underexpression with average Z scores ranging from –0.4 to –0.68 (P < 0.0001–0.00026). Of the three samples with documented retention in this region, however, only one demonstrated significant underexpression, with average expression Z scores ranging from –0.04 to –0.26 (P = 0.0068–0.63). This one sample, 2674, may have had an undetected or complex area of AI not detected by our microsatellite analysis.
Table 1. Summary table of microsatellite analysis, average 3p21–23 expression Z scores, 22q expression Z scores, and associated P values for seven primary HNSC using χ2 analysis for trend to determine the expression in these regions compared to Z scores for the remaining genome.
| Tumor identification
|
|||||||
|---|---|---|---|---|---|---|---|
| .2304 | 2674 | 2700 | 2291 | 2313 | 2397 | 2717 | |
| 3p microsatellite marker | |||||||
| D3S1100 | □ | NI | □ | NI | NI | ▪ | NI |
| D3S1211 | □ | □ | □ | ▪ | NI | ▪ | ▪ |
| D3S1277 | NI | NI | NI | NI | NI | NI | NI |
| D3S1537 | NI | NI | NI | NI | ▪ | NI | NI |
| D3S1619 | □ | □ | NI | ▪ | ▪ | NI | ▪ |
| D3S2405 | □ | □ | □ | ▪ | ▪ | ▪ | ▪ |
| Avg 3p21-23 | -0.22 | -0.26 | -0.04 | -0.59 | -0.68 | -0.52 | -0.47 |
| Z score | |||||||
| P value | 0.07 | 0.007 | 0.63 | <0.0001 | <0.0001 | <0.0001 | 0.0003 |
| 22q microsatellite marker | |||||||
| D22S264 | □ | □ | NI | □ | NI | NI | □ |
| D22S343 | NI | NI | NI | NI | □ | □ | NI |
| D22S450 | ▪ | □ | □ | ▪ | □ | □ | □ |
| D22S526 | □ | □ | NI | NI | ▪ | □ | ▪ |
| Avg22q Z score | 0.53 | 0.36 | 0.35 | 0.42 | 0.58 | 0.20 | 0.44 |
| P value | 0.001 | 0.10 | 0.17 | 0.05 | <0.0001 | 0.19 | 0.02 |
Tumor samples underwent microsatellite analysis at 3p21-23 and 22q using markers identified in the left column. Those markers demonstrating retention (open squares), noninformativity (NI), and allelic imbalance indicative of chromosomal loss or gain (filled squares) at a particular chromosomal locus are indicated. Tumors with documented allelic imbalance demonstrate significant up- or down-regulation appropriate to a site of previously reported chromosomal copy number alteration.
Four of the seven tumors (samples 2291, 2304, 2313, and 2717) demonstrated AI on 22q11 in at least one of the four microsatellite markers chosen (Table 1). The remaining three samples (samples 2397, 2674, and 2700) failed to demonstrate evidence of AI on 22q11 (Table 1). To establish the relationship between expression and AI on 22q, the average expression scores of all genes in this region were calculated for individual tumors and compared to the results of the microsatellite analysis. Again, statistical significance of expression scores was determined by using χ2 tests for trend. Table 1 summarizes our results and demonstrates a clear average expression Z score demarcation between those samples with and those without AI. Samples 2291, 2304, 2313, and 2717, all with documented AI, demonstrated significant overexpression with average expression Z scores that ranged from 0.42 to 0.58 (P = <0.0001–0.022). The three samples without AI on this arm did not demonstrate significant overexpression with average Z scores that ranged from 0.2 to 0.36 (P = 0.1–0.19).
Statistically significant underexpression, as determined by χ2 analysis, was found in four individual tumor samples within a subregion on chromosomal arm 3p (3p21–23). Furthermore, this pattern of global underexpression was associated with AI in the same subregion in these four tumors. In addition, significant global overexpression was found in four individual tumors on 22q11 by using χ2 analysis. This alteration in expression was also found to be associated with AI on the same chromosomal arm in these four samples.
Discussion
Gene expression microarray technology has allowed for the rapid screening of several thousand gene products and the prospect of identifying those genes responsible for tumor progression or formation. However, such a large quantity of data has proven to be difficult to manipulate and analyze. The database referencing of array genes online (dragon) is a useful annotation tool that allows for quick and efficient mapping of large numbers of genes used in array-based analyses. By combining microarray technology and this Web-accessible annotation device, global patterns of gene expression for chromosomal regions can be determined for any given tissue type and even individual tumors. We were able to analyze global expression for chromosomal arms and found concordance for multiple chromosomal arms among expression changes in a small set of primary HNSC and alterations in DNA copy number found on CGH analyses reported previously. A smaller number of chromosomal arms showed discordant results when CGH data were compared to expression in this analysis, possibly due to the small size of our analyzed sample. It is more likely that CGH tends to average copy number signal over large stretches of DNA and results in oversimplification of AI that may result in discrepancies with expression data.
However, correlations among expression values, CGH profiles, and AI for specific chromosomal regions were made and found to be highly statistically significant for individual tumors. This indicates that, although this expression analysis reflects the effects of chromosomal status on expression for individual tumors, a larger sample size may improve the ability of chromosomal locus annotated expression analysis to consistently define areas of chromosomal copy alteration for a set of primary tumors. We anticipate that further refinement of chromosomal locus-specific expression analysis would occur with increased primary tumor sample size, more complete expression array coverage, and improved mapping of chromosomal copy number alterations by technologies such as single-nucleotide polymorphism arrays and bacterial artificial chromosome arrays.
Several studies have looked at gains or losses of DNA copy numbers at different chromosomal loci; however, to the best of our knowledge, there have not been any attempts to draw correlations between DNA copy number and expression at specific chromosomal arms or regions on specific chromosomal arms. Our study found that significant losses or gains of entire chromosomal arms can result in altered expression of large numbers of genes. Because loss of chromosomal arm 3p has been established as a genetic alteration in head and neck cancer, this arm was of particular interest (15). Chromosomal arm 3p remained consistently underexpressed when looking at all tumors and significantly so in selected tumors with documented 3p AI. In fact, the expression profile for 3p remained remarkably consistent even when significant differentially expressed genes (as determined by SAM) were removed from analysis, suggesting chromosomal aberrations result in alterations of expression of both significant and nonsignificant genes over large chromosomal regions.
Analysis of all annotated genes on chromosomal arm 22q demonstrated significant overexpression. Statistically significant overexpression was found when looking at all seven tumors as a whole and again in selected tumors, especially in the 22q11.1–11.23 region. This pattern of expression persisted even after omitting genes on 22q, demonstrating significant expression differences from the analysis. Further correlations were found between expression patterns and AI on loci on chromosomal arm 22q for individual tumors. This locus has recently been identified as an area of amplification in HNSC with prognostic significance (28).
The implications of these data are that global dysregulation of multiple genes at a physical chromosomal location occurs as a result of alterations in chromosomal number in individual tumors. This raises the question of the significance of expression alterations in genes adjacent to tumor suppressor genes or oncogenes targeted by alterations in chromosomal copy number by deletion or amplification. From our analysis, it is likely that expression alterations may occur in bystander genes that are physically located close to target genetic or chromosomal alterations, and this effect is more widespread than previously recognized. The effect of altered expression of bystander genes adjacent to genes critical to malignant progression may result in an erroneous assumption that these bystander gene expression alterations are involved in tumor progression.
Alternatively, loss of a significant portion of a chromosomal arm may provide a selective growth advantage by expression alterations in multiple genes that individually have little to no effect on selective growth, but in aggregate provide significant growth advantage by additive effects in cellular pathways.
Finally, expression mapping may be used as a supportive technique in addition to other means by which significant genetic or expression alterations are physically located on the genome. This technique may be used in combination with other techniques, including single-nucleotide polymorphism or other analyses, to characterize targets of genomic loss and amplification in individual tumors or groups of tumors. Clearly, this technique is exploratory in nature when used in this fashion and must be validated by more conventional genomic-, expression-, and protein-based analyses to confirm gene targeting in the process of carcinogenesis.
Supplementary Material
Acknowledgments
J.C. is a Damon Runyon–Lilly Clinical Investigator supported by the Damon Runyon Cancer Research Foundation (CI-#9) and by a Clinical Innovator Award from the Flight Attendant Medical Research Institute. This work was supported by Specialized Programs of Research Excellence Grant P50 CA96784 from the National Cancer Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HNSC, head and neck squamous cell carcinoma; AI, allelic imbalance; SAM, significance analysis of microarrays; CGH, comparative genomic hybridization.
References
- 1.Landis, S. H., Murray, T., Bolden, S. & Wingo, P. A. (1998) CA Cancer J. Clin. 48, 6–29. [DOI] [PubMed] [Google Scholar]
- 2.Parkin, D. M., Laara, E. & Muir, C. S. (1988) Int. J. Cancer 41, 184–197. [DOI] [PubMed] [Google Scholar]
- 3.Redon, R., Muller, D., Caulee, K., Wanherdrick, K., Abecassis, J. & du Manoir, S. (2001) Cancer Res. 61, 4122–4129. [PubMed] [Google Scholar]
- 4.Bergamo, N. A., Rogatto, S. R., Poli-Frederico, R. C., Reis, P. P., Kowalski, L. P., Zielenska, M. & Squire, J. A. (2000) Cancer Genet. Cytogenet. 119, 48–55. [DOI] [PubMed] [Google Scholar]
- 5.Bockmuhl, U., Wolf, G., Schmidt, S., Schwendel, A., Jahnke, V., Dietel, M. & Petersen, I. (1998) Head Neck 20, 145–151. [DOI] [PubMed] [Google Scholar]
- 6.Huang, Q., Yu, G. P., McCormick, S. A., Mo, J., Datta, B., Mahimkar, M., Lazarus, P., Schaffer, A. A., Desper, R. & Schantz, S. P. (2002) Genes Chromosomes Cancer 34, 224–233. [DOI] [PubMed] [Google Scholar]
- 7.Singh, B., Gogineni, S. K., Sacks, P. G., Shaha, A. R., Shah, J. P., Stoffel, A. & Rao, P. H. (2001) Cancer Res. 61, 4506–4513. [PubMed] [Google Scholar]
- 8.Wolff, E., Girod, S., Liehr, T., Vorderwulbecke, U., Ries, J., Steininger, H. & Gebhart, E. (1998) Oral Oncol. 34, 186–190. [DOI] [PubMed] [Google Scholar]
- 9.Alevizos, I., Mahadevappa, M., Zhang, X., Ohyama, H., Kohno, Y., Posner, M., Gallagher, G. T., Varvares, M., Cohen, D., Kim, D., et al.. (2001) Oncogene 20, 6196–6204. [DOI] [PubMed] [Google Scholar]
- 10.Belbin, T. J., Singh, B., Barber, I., Socci, N., Wenig, B., Smith, R., Prystowsky, M. B. & Childs, G. (2002) Cancer Res. 62, 1184–1190. [PubMed] [Google Scholar]
- 11.Boyle, J. O., Hakim, J., Koch, W., van der Riet, P., Hruban, R. H., Roa, R. A., Correo, R., Eby, Y. J., Ruppert, J. M. & Sidransky, D. (1993) Cancer Res. 53, 4477–4480. [PubMed] [Google Scholar]
- 12.Kamb, A., Gruis, N. A., Weaver-Feldhaus, J., Liu, Q., Harshman, K., Tavtigian, S. V., Stockert, E., Day, R. S., III, Johnson, B. E. & Skolnick, M. H. (1994) Science 264, 436–440. [DOI] [PubMed] [Google Scholar]
- 13.Nobori, T., Miura, K., Wu, D. J., Lois, A., Takabayashi, K. & Carson, D. A. (1994) Nature 368, 753–756. [DOI] [PubMed] [Google Scholar]
- 14.Nawroz, H., van der Riet, P., Hruban, R. H., Koch, W., Ruppert, J. M. & Sidransky, D. (1994) Cancer Res. 54, 1152–1155. [PubMed] [Google Scholar]
- 15.Maestro, R., Gasparotto, D., Vukosavljevic, T., Barzan, L., Sulfaro, S. & Boiocchi, M. (1993) Cancer Res. 53, 5775–5779. [PubMed] [Google Scholar]
- 16.Reed, A. L., Califano, J., Cairns, P., Westra, W. H., Jones, R. M., Koch, W., Ahrendt, S., Eby, Y., Sewell, D., Nawroz, H., et al. (1996) Cancer Res. 56, 3630–3633. [PubMed] [Google Scholar]
- 17.Wu, C. L., Sloan, P., Read, A. P., Harris, R. & Thakker, N. (1994) Cancer Res. 54, 6484–6488. [PubMed] [Google Scholar]
- 18.Califano, J., van der Riet, P., Westra, W., Nawroz, H., Clayman, G., Piantadosi, S., Corio, R., Lee, D., Greenberg, B., Koch, W., et al.. (1996) Cancer Res. 56, 2488–2492. [PubMed] [Google Scholar]
- 19.Speicher, M. R., Howe, C., Crotty, P., du Manoir, S., Costa, J. & Ward, D. C. (1995) Cancer Res. 55, 1010–1013. [PubMed] [Google Scholar]
- 20.Yamaguchi, K., Wu, L., Caballero, O. L., Hibi, K., Trink, B., Resto, V., Cairns, P., Okami, K., Koch, W. M., Sidransky, D. & Jen, J. (2000) Int. J. Cancer 86, 684–689. [DOI] [PubMed] [Google Scholar]
- 21.Colantuoni, C., Henry, G., Zeger, S. & Pevsner, J. (2002) Bioinformatics 18, 1540–1541. [DOI] [PubMed] [Google Scholar]
- 22.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouton, C. M. & Pevsner, J. (2000) Bioinformatics 16, 1038–1039. [DOI] [PubMed] [Google Scholar]
- 24.Mao, R., Zielke, C. L., Ronald Zielke, H. & Pevsner, J. (2003) Genomics 81, 457–467. [DOI] [PubMed] [Google Scholar]
- 25.Ha, P. K., Benoit, N. E., Yochem, R., Sciubba, J., Zahurak, M., Sidransky, D., Pevsner, J., Westra, W. H. & Califano, J. (2003) Clin. Cancer Res. 9, 3058–3064. [PubMed] [Google Scholar]
- 26.Roz, L., Wu, C. L., Porter, S., Scully, C., Speight, P., Read, A., Sloan, P. & Thakker, N. (1996) Cancer Res. 56, 1228–1231. [PubMed] [Google Scholar]
- 27.Squire, J. A., Bayani, J., Luk, C., Unwin, L., Tokunaga, J., MacMillan, C., Irish, J., Brown, D. & Kamel-Reid, S. (2002) Head Neck 24, 874–887. [DOI] [PubMed] [Google Scholar]
- 28.Ashman, J. N., Patmore, H. S., Condon, L. T., Cawkwell, L., Stafford, N. D. & Greenman, J. (2003) Br. J. Cancer 89, 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.