Abstract
The phytohormone ethylene is essential to plant growth and development. It plays crucial roles in responses to biotic and abiotic stress. The mulberry tree is an important crop plant in countries in which people rear silkworms for silk production. The availability of the mulberry genome has made it possible to identify mulberry genes involved in ethylene biosynthesis and signal pathways. A total of 145 mulberry genes were identified by both homology-based and hidden Markov model (HMM) search, including 29 genes associated with ethylene biosynthesis and 116 genes in the AP2/ERF family. Studies on gene structure have provided a genetic basis for understanding the functions of these genes. The differences in gene expression were also observed in different tissues. The expression of two mulberry genes in the AP2/ERF family, MaERF-B2-1 and MaERF-B2-2, was found to be associated with the response to flooding stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s10142-014-0403-2) contains supplementary material, which is available to authorized users.
Keywords: Ethylene, Plant hormone, Mulberry, Gene expression
Introduction
Ethylene plays important roles in various aspects of plant growth and development including fruit ripening, germination, flowering, sex determination, leaf senescence, root nodulation, and responses to biotic and abiotic stress (Johnson and Ecker 1998; Lin et al. 2009). As early as 1910, ethylene was used to accelerate the ripening of bananas (Abeles et al. 1992). In Arabidopsis, ethylene signaling has been found to be important to flooding and salt response (Zhang et al. 2011; Voesenek and Blom 1989). Recent work has indicated that ethylene promotes hypocotyl elongation in light and suppresses it in darkness (Zhong et al. 2012).
Ethylene biosynthesis and signaling networks are well understood in plants. Biosynthesis of ethylene occurs in three steps. First, the amino acid methionine is converted to S-adenosylmethionine (AdoMet) by S-adenosylmethionine synthetase (Yang and Hoffman 1984). AdoMet is then converted to 1-aminocyclopropane-1-carboxylic acid (ACC) and 5′-deoxy-5′methylthioadenosine (MTA) by 1-aminocyclopropane-1-carboxylase synthase (ACS) (Adams and Yang 1979). The formation of the ACC is the rate-limiting step in ethylene biosynthesis, and it requires pyrodoxal-5′-phosphate (PLP) as a cofactor. When a large amount of ethylene is produced, MTA is recycled to methionine through the Yang cycle to supplement the methionine pool (Miyazaki and Yang 1987). In the third step, ACC is transformed into ethylene, CO2, and cyanide by 1-aminocyclopropane-1-carboxylase oxidase (ACO). The cyanide is detoxified into β-cyanoalanine by β-cyanoalanine synthase (Blumenthal et al. 1968). Ethylene can be detected by a family of membrane-localized receptors. A copper cofactor is necessary for the transduction of this signal (Rodrıguez et al. 1999). When ethylene is present, the conformation of the compound of ethylene receptor and constitutive triple response 1 (CTR1) changes and the suppression ends (Gao et al. 2003). Ethylene-insensitive 2 (EIN2) is then activated, resulting in the activation of EIN3 transcription factors (Alonso et al. 1999). Proteins EIN3 and EIN3-like 1 (EIL1) are specifically bound to the promoter of ethylene response factor 1 (ERF1) in AP2/ERF family, and ERF1 plays important roles in the transcriptional regulation of a variety of biological processes associated with growth and development and with biotic and abiotic stress responses. ERF1 binds to the promoters that contain the GCC box. These promoters are present in many inducible ethylene genes (Fujimoto et al. 2000). The related apetala 2.2 gene from Arabidopsis (RAP2.2, At3g14230) has an ERF-bearing AP2 domain (Hinz et al. 2010). Overexpression of RAP2.2 has been shown to improve plant survival under hypoxia stress conditions. The ERF-associated amphiphilic repression (EAR) motif is a repression domain in the C-terminal region of repressor-type ERF proteins. These genes are implicated in developmental, hormonal and stress signaling pathways and can be negatively regulated through the EAR motif (Ohta et al. 2001; Kagale et al. 2010).
Mulberry trees are deciduous and woody. They are widely used to produce crops for the sericulture industry. The domesticated silkworm, Bombyx mori, feeds only on mulberry leaves. In this way, the silk industry depends on the quality and quantity of available mulberry leaves. Mulberry fruits are also used to make jam, juice, and wine. Ethylene has been shown to affect seasonal leaf abscission and fruit ripening (Adams-Phillips et al. 2004). The genes involved in ethylene signaling and biosynthesis are important targets for the improvement of leaf and fruit yield. In China’s Three Gorges Reservoir Region, the most serious type of environmental stress limiting mulberry crop productivity is water-logging. ERFs have been reported to play important roles in the regulation of biotic and abiotic stress (Dietz et al. 2010). One ERF subfamily, the VIIERFs, responds to hypoxia stress and promotes the expression of hypoxia-related genes (Hinz et al. 2010). In the present study, we found the expression patterns of two mulberry genes in the AP2/ERF family, MaERF-B2-1 and MaERF-B2-2, changed in association with flooding treatment.
Materials and methods
Database searches and sequence analysis
The mulberry genome database was downloaded from MorusDB (http://morus.swu.edu.cn/morusdb/). The Arabidopsis genome database was obtained from TAIR (http://www.arabidopsis.org/). The majority of amino acid sequences of the Arabidopsis thaliana genes involved in the ethylene biosynthesis and signaling pathways were retrieved from Uniprot (http://www.uniprot.org/). The A. thaliana AP2/ERF genes were downloaded from the DATF website (http://datf.cbi.pku.edu.cn/index.php). All of the A. thaliana sequences were used as templates in a search of the mulberry genome using BLASTP. Hidden Markov model (HMM) was used to identify motifs of mulberry proteins. The presence of AP2 domains in the mulberry sequences identified in this way was determined by (http://smart.embl-heidelberg.de/) (SMART) analyses (Letunic et al. 2012).
Sequence alignments and construction of the phylogenetic tree
Multiple alignment analysis was performed with ClustalX and subsequently adjusted manually by Genedoc (Larkin et al. 2007). A phylogenetic tree of AP2/ERF was constructed using maximum-likelihood method in PhyML 3.0 using a Perl script. Web tools at the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/help.php) were used to draw the structures of the genes (Guo et al. 2007).
Analysis of the expression of the genes involved in ethylene biosynthesis and signal transduction
The reads per kilobase of exon model per million mapped reads (RPKM) was used to analyze the expression levels of all predicted mulberry genes using RNA sequencing data (Mortazavi et al. 2008). The clustering analyses were carried out using Gene Cluster 3.0. The data were adjusted by log transformation and the mean center method. Hierarchical clustering (HCL) with average linkage was set to calculate K-medians with five clusters. Heat maps illustrating the gene expression data were generated using Java TreeView.
RNA extractions and quantitative RT-PCR
Total RNA was isolated from mulberry roots, barks, leaves, male flowers, female flowers, and fruits using RNAiso Plus (Takara, Japan) according to the manufacturer’s instructions. The quality and concentration of RNA samples were measured using a ND-1000 UV spectrophometer (Nanodrop Technologies, USA). Reverse transcription was performed following the manufacturer’s instructions (Takara, Japan). Primers were designed using Primer Premier 5 software (http://www.primer-e.com/). Polymerase chain reactions were performed in a 96-well plate with a StepOne Plus System apparatus, amplified with SYBR® Green II (Takara, Japan) according to the manufacturer’s instructions. Cycling conditions were as follows: 90 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Amplification specificity was verified by a dissociation curve. To compare data from different PCR runs, cycle threshold (C T) values for samples were normalized to that of the mulberry ribosomal protein gene (Morus024083). The cDNA quantity was calculated using ; here, ΔC T is the difference in the number of cycles between tests and controls. The change in gene expression after flooding treatment was measured using , where ΔΔCT = (ΔCT sample − ΔCT control). Gene-specific primers used for the real-time RT-PCR are listed in Supplementary Table 2. All results are representative of three independent experiments.
Prediction of cis-acting elements
To identify cis-regulatory elements, upstream regions within about 2 kb of the transcription start sites of three genes, MnERF-B1, MnERF-B2, and MnERF-B3, were analyzed. All promoter sequences were placed in the PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html), which has 435 different plant transcription binding sites. The search results are divided into four categories including stress response, hormone response, light response, and others (Lescot et al. 2002).
Plant growth and flooding treatment
Mulberry species, Husang-32 (Morus alba L. var. multicaulis), maintained at the Mulberry Germplasm Nursery of Southwest University (China), was used in the present study. Husang-32 seedlings were grown in plastic pots filled with soil, vermiculite, and perlite mixture (6:1:1) in incubators (25 °C; 16 h day, 8 h night; 75 % relative humidity). After being incubated for 2 months, the seedlings with 15-cm length were subjected to flooding treatment. Plants in the flooding-treated group were submerged 2–3 cm under the surface of the water. The roots and leaves from both treated and control groups of Husang-32 were collected after the treatments lasting 1 h, 3 h, 5 h, 7 h, 1 day, 3 days, 5 days, and 7 days. Plant materials were frozen in liquid N2, and stored at −80 °C until further use for total RNA isolation.
Results
Identification of genes involved in the mulberry ethylene biosynthesis and signaling pathways
In total, 145 genes were predicted to belong to the ethylene biosynthesis and signaling pathways. Of these, 29 genes were found to be involved in ethylene biosynthesis and signal transduction (Table 1). The AP2/ERF family contains 116 genes (Supplemental Table 1). The MnACS family contains 7 genes, of which MnACS10 and 12 are transaminases. Four of the remaining five MnACS genes each had four exons. MnACS5 only had three exons (Fig. 1). Sequence alignment indicated that each MnACS protein contained seven conserved boxes also found in ACS proteins from other plant species. The conserved glutamate (E) residue in box 1, which determines substrate specificity, is present in all the members of the ACS gene family. Except for MnACS5, the MnACS proteins all shared a Ser residue in the C-terminal, which produced a shorter C-terminus than other ACS isozymes (Supplemental Figure 1). ACO is a member of the Fe II-dependent family of oxidases. They require ascorbate as a substrate and Fe (II) as a cofactor to exert enzymatic activity. Four MnACOs were identified in the mulberry genome. MnACO1 and MnACO4 have three exons, and MnACO2/3 has four exons (Fig. 1). The motifs for binding the cofactor (H-X-D-X-H) and the cosubstrate (R-X-S) are also conserved in MnACOs. In the C-terminus, E-R-E is essential to enzyme activity, particularly R, which may be involved in the mechanism of CO2 activation (Supplementary Figure 2).
Table 1.
Genes involved in ethylene biosynthesis and signaling pathways in the M. notabilis genome
| Genes | Accession no. | Protein size | CDS length | Gene length | Exons | Scaffold. no. (strand) | Start codon | Stop codon |
|---|---|---|---|---|---|---|---|---|
| MnSAM1 | Morus003267 | 393 | 1182 | 1182 | 1 | 1843(+) | 66,189 | 67,370 |
| MnSAM2 | Morus013867 | 393 | 1182 | 1182 | 1 | 498(+) | 76,865 | 78,046 |
| MnSAM3 | Morus025140 | 393 | 1182 | 1182 | 1 | 172(+) | 571,546 | 572,727 |
| MnSAM4 | Morus022555 | 390 | 1173 | 1173 | 1 | 124(+) | 13,770 | 14,942 |
| MnACS1 | Morus012919 | 496 | 1491 | 2732 | 4 | 298(+) | 185,423 | 188,154 |
| MnACS2 | Morus024218 | 486 | 1461 | 2196 | 4 | 297(+) | 4647 | 6842 |
| MnACS3 | Morus007775 | 471 | 1416 | 1782 | 4 | 300(−) | 229,933 | 231,714 |
| MnACS4 | Morus007092 | 467 | 1404 | 2044 | 4 | 991(+) | 117,590 | 119,633 |
| MnACS5 | Morus027243 | 446 | 1341 | 1660 | 3 | 329(−) | 186,543 | 188,202 |
| MnACS10a | Morus023174 | 557 | 1674 | 3428 | 5 | 87(+) | 408,302 | 411,729 |
| MnACS12a | Morus012967 | 504 | 1515 | 1791 | 4 | 399(+) | 330,934 | 332,724 |
| MnACO1 | Morus027261 | 306 | 921 | 1127 | 3 | 329(+) | 408,750 | 409,876 |
| MnACO2 | Morus014137 | 322 | 969 | 1526 | 4 | 1292(−) | 16,575 | 18,100 |
| MnACO3 | Morus004820 | 319 | 960 | 1682 | 4 | 528(−) | 23,279 | 24,960 |
| MnACO4 | Morus013401 | 310 | 933 | 1272 | 3 | 1379(+) | 391,862 | 393,133 |
| MnETR1 | Morus018344 | 738 | 2217 | 5082 | 6 | 521(−) | 266,232 | 271,313 |
| MnETR2 | Morus008145 | 793 | 2382 | 2949 | 2 | 606(−) | 247,113 | 250,061 |
| MnERS1 | Morus007485 | 617 | 1854 | 2872 | 5 | 1526(+) | 209,537 | 212,408 |
| MnEIN4 | Morus024538 | 764 | 2295 | 2558 | 2 | 205(−) | 594,177 | 596,734 |
| MnCTR1 | Morus003569 | 861 | 2586 | 6563 | 17 | 548(+) | 106,465 | 113,027 |
| MnCTR2 | Morus016797 | 666 | 2001 | 6117 | 12 | 112(−) | 213,548 | 219,664 |
| MnEIN2 | Morus024376 | 1306 | 3921 | 5614 | 7 | 342(−) | 402,131 | 407,744 |
| MnEBF1 | Morus000805 | 697 | 2094 | 2590 | 2 | 1037(+) | 4387 | 6976 |
| MnEBF2 | Morus002248 | 642 | 1929 | 2397 | 2 | 1513(−) | 59,843 | 62,239 |
| MnEIN3 | Morus002490 | 617 | 1854 | 1854 | 1 | 3287(−)b | 31,544 | 33,397 |
| MnEIL1 | Morus002491 | 607 | 1824 | 1824 | 1 | 3287(−)b | 50,362 | 52,185 |
| MnEIL3 | Morus007978 | 611 | 1836 | 1836 | 1 | 18(−) | 151,180 | 153,015 |
| MnEIL4 | Morus016592 | 478 | 1437 | 1910 | 2 | 841(+)b | 82,684 | 84,593 |
| MnEIL5 | Morus016593 | 543 | 1632 | 1632 | 1 | 841(+)b | 86,287 | 87,918 |
SAM S-adenosylmethionine synthase, ACS 1-aminocyclopropane-1-carboxylate synthase, ACO 1-aminocyclopropane-1-carboxylate oxidase, ETR ethylene receptor, CTR1 constitutive triple response-1, EIN2 ethylene-insensitive 2, EIN3 ethylene-insensitive 3, EBF EIN3-binding F-box protein, ERF ethylene-responsive element binding factor.
aThe transaminases in ACS family
bThe genes located at the same scaffold
Fig. 1.
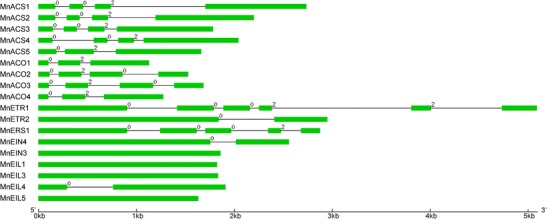
Exon/intron structures of ACS, ACO, ETR, and EIN3 genes in mulberry plants. Gene structures were plotted using the GSDS server. The intron phases indicate the position of the intron within a codon. The scale bar indicates 1 kb
Ethylene signal transduction is important to ethylene synthesis. Many genes take part in this process. Ethylene receptors (ETRs) form a family of membrane-localized receptors. There are four MnETRs in the mulberry genome. The N-terminal of MnETRs includes three highly conserved transmembrane regions. This region has been demonstrated to be essential to the ethylene binding activity of MnETRs. The C-terminal region of all MnETRs except MnERS1, which has a truncated C-terminal, contains a receiver domain. In addition, the N-terminal regions of MnETR2 and MnEIN4 are much longer than those of other receptors. SMART prediction revealed that they contain four transmembrane regions (Supplementary Figure 3). EIN3 and EIN3-like proteins are essential to the ethylene signal pathway. They are positive regulators, and they bind to the promoter regions of the downstream genes to regulate their expression. Five EIN3s were found here in the mulberry genome. They all have a conserved amino-terminal acidic domain (AD), pro-rich region (PR), and five small basic domains (BDI-V). The Gln-rich and Asn-rich regions, found in mung bean plants, are only conserved in MnEIN3 and MnEIL1 (Supplementary Figure 4). MnEIN3, MnEIL1, MnEIL4, and MnEIL5 were all located on the same scaffold in the mulberry genome (Table 1).
Expression of mulberry ethylene biosynthesis and signal pathway genes
Genes whose expression was detected by qPCR are listed in Fig. 2. Results showed that the genes in these four families have diverse expression profiles across different tissues. MnACS2, MnACO2, MnACO3, MnETR2, MnEIN3, and MnEIL1 have higher levels of expression. On the contrary, the expression of MnACS3, MnACS4, MnACO4, MnERS1, MnEIN4, MnEIL3, and MnEIL4 was relatively repressed. In addition, MnACS5 exhibited tissue-specific expression in female flowers. Among ACO genes, MnACO1 and MnACO2 exhibited tissue-biased expression in fruit, although the expression levels of MnACO1 were only about one tenth of those of MnACO2. MnEIN3 and MnEIL1 showed more expression than the rest of the EIN3 family in roots and fruits.
Fig. 2.
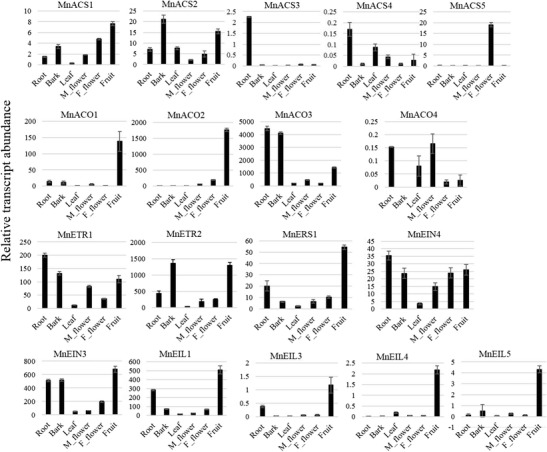
Expression patterns of 18 genes involved in the ethylene biosynthesis and signal transduction pathways. Six tissues were used. Relative levels of gene expression by qRT-PCR were normalized against the mulberry ribosomal protein gene Morus024083. Data are represented as mean ± standard error of three replicates
Phylogenetic and structural analysis of the AP2/ERF transcription factor family
In the present study, 116 AP2/ERF transcription factors were identified in the mulberry genome. According to the number of AP2 domains and their structural features, these 116 proteins can be divided into five subfamilies. There are 58 genes in the ERF subfamily, 33 genes in the DREB subfamily, 21 genes in the AP2 subfamily, 3 genes in the RAV subfamily, and 1 gene in the Soloist subfamily. A phylogenetic tree was constructed using the amino acid sequences of AP2/ERF transcription factors of Morus and Arabidopsis. This tree was separated into 15 groups. Groups I–IV represent the DREB subfamily, and V–X represent the ERF subfamily (Supplementary Figure 5). All groups except for the V group have lower numbers of genes in mulberry tree than those in Arabidopsis. There are 11 genes in the V group of mulberry and 5 in the V group of Arabidopsis.
The pattern of exon/intron splicing usually provides information useful to understanding of the emergence and evolution of a gene family. Structural analyses of genes indicated that all genes in the AP2 subfamily had different numbers of introns ranging from zero to twelve. However, 28 of the 33 genes in the DREB subfamily, 44 of the 58 genes in the ERF subfamily, and 2 of the 3 genes in the RAV subfamily had no introns. Results showed that most genes in the AP2 family shared similar patterns of exon/intron splicing (Supplementary Figure 6).
Genes in the AP2/ERF family containing the EAR motif
The ERF-associated amphiphilic repression (EAR) motif (DLNxxP or LXLXL) has been reported to be a repression domain in repressor-type ERF proteins. These proteins negatively regulate genes involved in developmental, hormonal, and stress signaling pathways. There are three types of EAR motifs in mulberry plants, DLNXXP, LXLXL, and LDLNLXPP. These three motifs have been found in ERF, DREB, AP2, and RAV subfamily (Fig. 3). All of MnERF proteins bearing EAR motif belong to MnERF-B1 group except MnERF-B4-3 and MnERF-B6-8. Only four genes in MnDREB subfamily contain EAR motif, and the DLNXXP motif has been identified in MnDREB-A5-4 and MnDREB-A5-5. Sequence analyses revealed the 19 genes that bear LXLXL motif found in mulberry proteins.
Fig. 3.
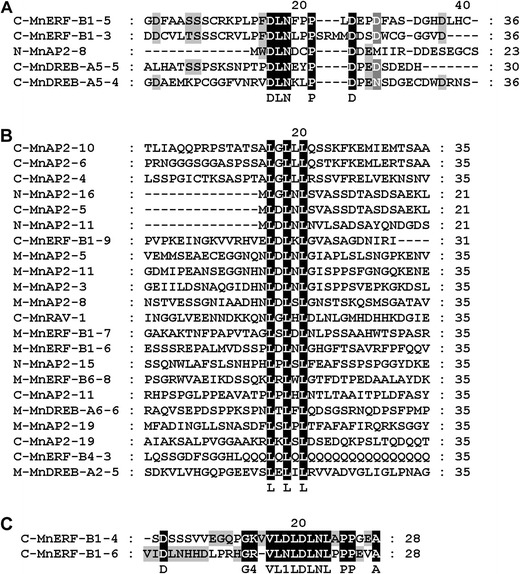
EAR motif-like sequences of AP2/ERF in mulberry plants. a The genes contain DLNXXP motif. b The genes contain LXLXL motif. c The genes contain LDLNLXPP motif. Multiple sequence alignments were performed using ClustalX. The letters C, M, and N prior to protein names indicate location in the C-terminal, middle of the sequence, and N-terminal
Expression of MnERF genes
To investigate the levels of expression of AP2/ERF genes in different tissues, RPKM data of these genes were analyzed in roots, barks, leaves, flowers, and buds. Heat maps were constructed based on the RPKM data. Sixteen MnERF genes, including five genes in MnERF-B1, four in MnERF-B3, three in MnERF-B2, two in MnERF-B4, and one each in MnERF-B5 and MnERF-B6, had relatively high levels of expression in five tissues (Fig. 4a). This was also true of 8 of the 33 genes in the DREB subfamily, 6 of the 21 genes in the AP2 subfamily, and 1 of the 3 genes in the RAV subfamily. In addition, several genes were also found to be expressed in a tissue-biased manner. For example, MnERF-B3-21 was expressed solely in male flowers (Fig. 4a). MnDREB-A4-7 was more abundantly expressed in leaves than in other tissues (Fig. 4b). MnAP2-5 had high levels of expression in male flower (Fig. 4c). However, the expression of nine genes (MnERF-B6-1/8, MnDREB-A3-1, MnDREB-A4-4, MnDREB-A6-1/2, MnAP2-13/18/21) was not detectable in any tissue.
Fig. 4.
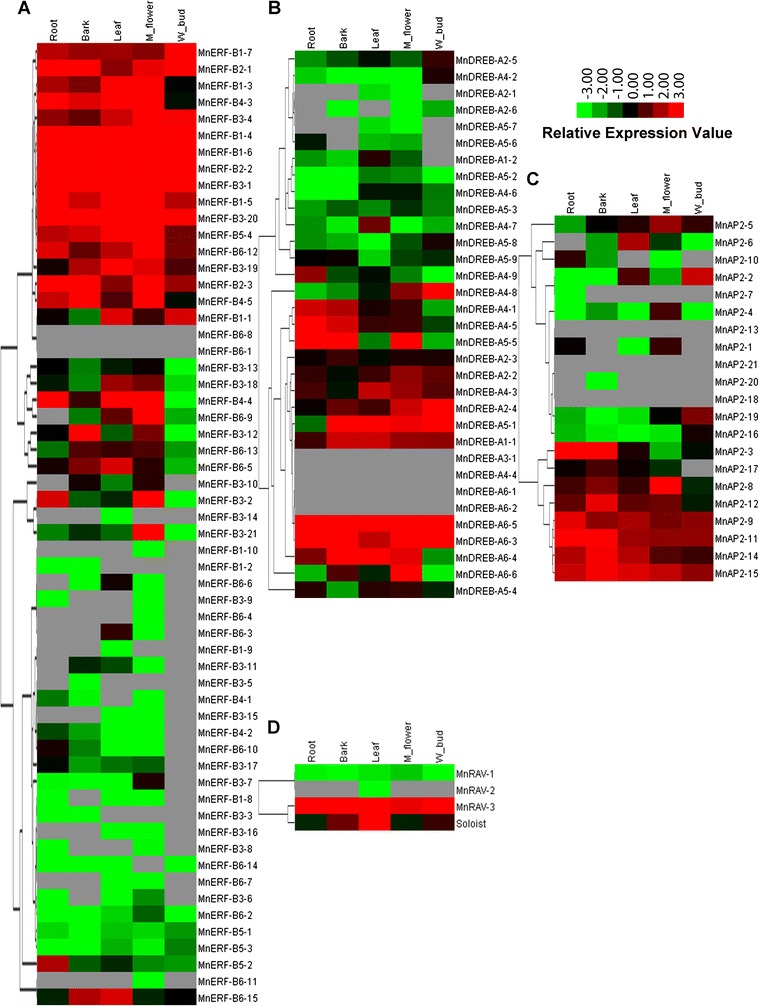
Heat maps of hierarchical clustering of mulberry genes in the AP2/ERF family. a ERF subfamily; b DREB subfamily; c AP2 subfamily; and d RAV and Soloist subfamilies. The clustering analyses were carried out using Gene Cluster 3.0. The data were adjusted by log transformation and the mean center method. Hierarchical clustering (HCL) with average linkage was set to calculate K-medians with five clusters. Heat maps illustrating the gene expression data were generated using Java TreeView
Response of MaERF-B2-1 and MaERF-B2-2 to flooding stress
Genes in MnERF-B subfamily were expressed at relatively higher levels in different tissues. All members of this subfamily have a conserved N-terminal motif MCGGAV/II, which is considered to be involved in the regulation of low-oxygen response in plants (Fig. 5). Analyses of the promoter regions of MnERF-B2-1, MnERF-B2-2, and MnERF-B2-3 showed that they contain consensus sequences of transcription factor binding sites. As shown in Table 2, four catalogues of putative regulatory elements (stress response, hormone response, light response, and others) were detected in the flanking regions of three genes. GARE, CE3, ABRE, TCA, and TGACG-motifs are involved in the signaling of mulberry hormones. Typical heat shock element (HSE) and low temperature stress response element (LTR) were also observed. It is noteworthy that ARE and GC motifs, which are known to be responsive to the hypoxemia and anaerobic conditions, were detected in the promoters of MnERF-B2-1 and MnERF-B2-3. In this context, two of these three genes from Husang-32, MaERF-B2-1 and MaERF-B2-2, were cloned, sequenced, and their expression patterns were further investigated. Mulberry cultivar Husang-32 is widely planted in the Three Gorges Reservoir area. As shown in Fig. 6a, MaERF-B2-2 has higher expression than MaERF-B2-1 or MaERF-B2-3. To understand the changes in gene expression that takes place after flooding, the expression of the MaERF-B2-1 and MaERF-B2-2 was detected after treatment lasting 1 h, 3 h, 5 h, 7 h, 1 day, 3 days, 5 days, and 7 days (Fig. 6c). Results showed that MaERF-B2-1 was up-regulated in roots and leaves after 1 day of flooding. The change took place faster in leaves than that in roots. MaERFB2-2 showed fast up-regulation in roots after 1 h of flooding. MaERF-B2-1 showed a greater fold change than MaERF-B2-2. These results indicated that MaERF-B2-1 and MaERFB2-2 may play important roles in Husang-32’s response to flooding.
Fig. 5.
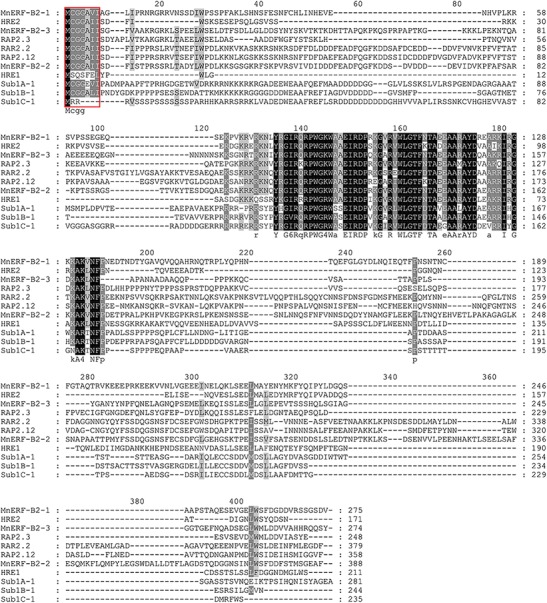
Alignment of amino acid sequences of ERF VII subfamily genes from mulberry, Arabidopsis and rice using the ClustalX program. HRE1/2, RAP2.2, RAP2.3, and RAP2.12 belong to VII group of Arabidopsis. Sub1A-1, Sub1B-1, and Sub1C-1 belong to Oryza sativa
Table 2.
Cis-elements in MnERF-B2-1, MnERF-B2-2, and MnERF-B2-3 promoters
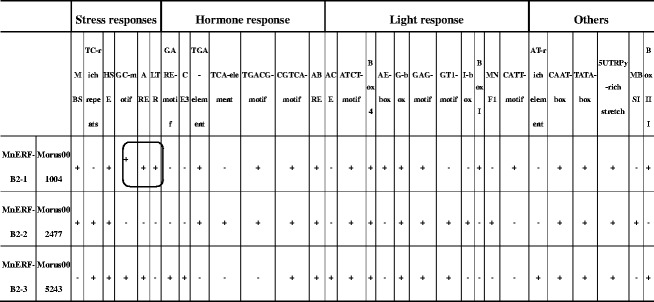
Fig. 6.
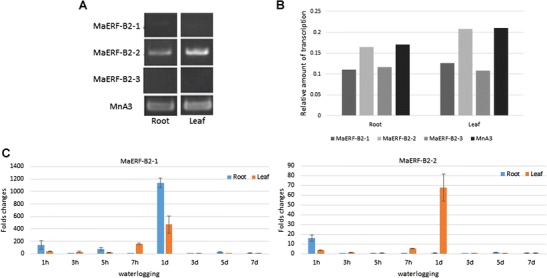
Expression analyses of MaERF-B2-1/2/3 genes. Expression of MaERF-B2-1, MaERF-B2-2, and MaERF-B2-3 in roots and leaves (a, b). The qPCR data of MaERF-B2-1 and MaERF-B2-2 in roots and leaves after flooding treatment (c). Fold inductions are calculated by dividing the value of treatment with that of control in corresponding time point. Data are represented as mean ± standard error of three replicates
Discussion
Genes such as ACS, ACO, and EIN3 are essential targets in the biosynthesis of ethylene and the transduction of this signal. In Arabidopsis, ACS is considered a rate-limiting synthase in ethylene biosynthesis (Jakubowicz and Sadowski 2002). The phosphorylation sites of ACS are important to rapid turnover during the response to various kinds of stimulation (Joo et al. 2008). Previous research indicated CmACS-7 affects the development of the stamina in female flowers (Boualem et al. 2008). The expression of MnACS5 was restricted to female flowers in mulberry plants, suggesting that MnACS5, rather than an ACC synthase, may play a different role. ACO genes are responsible for ethylene production. In the present study, the expression profiles of four putative MnACOs were analyzed and variation was observed in their levels of expression. Mulberry ACO genes, such as MnACO1 and MnACO2, whose expression takes place mainly in fruit, might be involved in the regulation of ethylene during fruit ripening. In ethylene signal transduction, EIL and EIN3 proteins are crucial transcriptional factors. They are bound to the promoters of downstream genes. In this way, they regulate a wide range of stress responses (Chao et al. 1997). MnEIN3, MnEIL1, MnEIL3, and MnEIL5 have no introns. MnEIN3, MnEIL1, MnEIL4, and MnEIL5 form tandem arrays in the mulberry genome, and the genes in these two pairs are not only similar in sequence but in pattern of expression, suggesting the existence of complicated regulation affecting those processes.
The AP2/ERF family includes plant-specific transcription factors called ethylene response element binding proteins (EREBPs). EREBPs were originally identified through their ability to bind to the GCC box, a DNA motif associated with ethylene and pathogen-induced gene expression (Fujimoto et al. 2000). In the present study, 116 of AP2/ERF genes were identified in the mulberry genome. A phylogenetic analysis of AP2/ERF genes in mulberry plants and Arabidopsis showed mulberry plants to have more AP2/ERF genes in V group. Two research groups have shown that the overexpression of WIN1/SHN1 (At1g15360), an Arabidopsis AP2/ERF gene in V group, resulted in the enhanced accumulation of epidermal wax (Aharoni et al. 2004; Broun et al. 2004). The expansion of mulberry AP2/ERF genes in V group is of particular interest.
Flooding is a serious matter in China’s Three Gorges Reservoir Region. It has a profound effect on mulberry growth and development. Previous studies on rice have shown that the genes at the Submergence1 (Sub1) locus improve tolerance to submergence. This locus includes three ERF transcriptional factors (Sub1A, Sub1B, and Sub1C) (Jung et al. 2010). In Arabidopsis, the RAP2.12 and RAP2.2 genes became active in response to water-logging. In flooding, the expression of RAP2.12 is up-regulated and the product accumulates in the nucleus to activate the expression of hypoxia genes (Licausi et al. 2011). The overexpression of RAR2.2 improves plant survival under hypoxia stress conditions (Hinz et al. 2010). Three mulberry genes, MaERF-B2-1, MaERF-B2-2, and MaERF-B2-3, appear to be the counterparts of RAP2.12, RAP2.2, and RAP2.3 based on the sequences. All of them contain conserved N-terminal sequences, which are important in response to hypoxia. Moreover, the presences of ARE and GC cis-elements in the promoters of MnERF-B2-1 and MnERF-B2-3 could lead to a proposal that these two genes might be involved in dealing with hypoxia. Our data further indicated that the patterns of expression of MaERF-B2-1 and MaERF-B2-2 changed in response to flooding treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 23 kb)
(DOCX 14 kb)
Alignment of amino acid sequences of five MnACS genes using the ClustalX program. Conserved residues are shaded in black. The 11 black circles designate the residues that represent the conserved amino acids in aminotransferases. The conserved glutamate residue (E) is marked with an grey circle. Three black arrows indicate the Val and Ser residues which are the sites for phosphorylation. The Ser residues that are targets of the MPK6 kinase are marked with asterisks. (JPEG 5318 kb)
Alignment of amino acid sequences of MnACO genes. Fe (II)-binding motif and two cosubstrate-binding motifs are marked with red dash box, and red boxes, respectively. The triangles designate the sites of the cysteines. The other important amino acid are marked with the asterisks. (JPEG 4846 kb)
Alignment of amino acid sequences of MnETR genes. Regions I, II, and III represent three transmembrane fragments. (JPEG 6306 kb)
Alignment of amino acid sequences of MnEIN3 genes. The amino-terminal acidic domain (AD), pro-rich region (PR), and five small basic domain (BDI-V) are labeled. The poly-Gln and Poly-Asn repeats at the C-terminal portion are marked with triangles. The arrow indicates the Lys residue essential for the function of EIN3. (JPEG 4085 kb)
Phylogenetic tree of the predicted mulberry AP2/ERF proteins. A phylogenetic tree of AP2/ERF was constructed using maximum-likelihood method in PhyML 3.0 with a Perl script. 15 groups are marked. (JPEG 3240 kb)
Exon/intron structures of mulberry AP2/ERF genes. Different exon/intron structures of genes were plotted using the GSDS server. The intron phase indicates the position of the intron within a codon. If the codon is not located within a codon or is located between two codons, the phase is 0. Phase 2 designates introns between the first and second bases of a codon and phase 2 designates introns between the second and third bases of a codon. (JPEG 1571 kb)
Acknowledgments
This project was funded by the research grants from the National Hi-Tech Research and Development Program of China (No. 2013AA100605-3), the Fundamental Research Funds for the Central Universities (No. 2362014xk05), the “111” Project (B12006), and the Science Fund for Distinguished Young Scholars of Chongqing (Grant No. cstc2011jjjq0010).
References
- Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in plant biology. New York: Academic; 1992. [Google Scholar]
- Adams D, Yang S. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci U S A. 1979;76(1):170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Phillips L, Barry C, Giovannoni J. Signal transduction systems regulating fruit ripening. Trends Plant Sci. 2004;9(7):331–338. doi: 10.1016/j.tplants.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16(9):2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284(5423):2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Blumenthal S, Hendrickson H, Abrol Y, Conn EE. Cyanide metabolism in higher plants III. The biosynthesis of β-cyanoalanine. J Biol Chem. 1968;243(20):5302–5307. [PubMed] [Google Scholar]
- Boualem A, Fergany M, Fernandez R, Troadec C, Martin A, Morin H, et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonecy in melons. Science. 2008;321:836. doi: 10.1126/science.1159023. [DOI] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101(13):4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89(7):1133–1144. doi: 10.1016/S0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Vogel MO, Viehhauser A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma. 2010;245:3–14. doi: 10.1007/s00709-010-0142-8. [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12(3):393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003;278(36):34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29(8):1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010;153(2):757–772. doi: 10.1104/pp.110.155077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowicz M, Sadowski J. 1-aminocyclopropane-1-carboxylate synthase—genes and expression. Acta Physiol Plant. 2002;24(4):459–478. doi: 10.1007/s11738-002-0043-3. [DOI] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;1:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Joo S, Yd L, Lueth A, Zhang S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 2008;54(1):129–140. doi: 10.1111/j.1365-313X.2008.03404.x. [DOI] [PubMed] [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC. The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol. 2010;152(3):1674–1692. doi: 10.1104/pp.109.152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152(3):1109–1134. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(1):302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, Perata P, van Dongen JT. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature. 2011;479(7373):419–422. doi: 10.1038/nature10536. [DOI] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60(12):3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- Miyazaki JH, Yang SF. The methionine salvage pathway in relation to ethylene and polyamine biosynthesis. Physiol Plant. 1987;69(2):366–370. doi: 10.1111/j.1399-3054.1987.tb04302.x. [DOI] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13(8):1959–1968. doi: 10.1105/tpc.13.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrıguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283(5404):996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Voesenek L, Blom C. Growth responses of Rumex species in relation to submergence and ethylene. Plant Cell Environ. 1989;12(4):245–253. doi: 10.1111/j.1365-3040.1989.tb01959.x. [DOI] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- Zhang L, Li Z, Quan R, Li G, Wang R, Huang R. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 2011;157(2):854–865. doi: 10.1104/pp.111.179028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi YP, Li JG, Quail PH, Deng XW, Guo HW. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol. 2012;22(16):1530–1535. doi: 10.1016/j.cub.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 23 kb)
(DOCX 14 kb)
Alignment of amino acid sequences of five MnACS genes using the ClustalX program. Conserved residues are shaded in black. The 11 black circles designate the residues that represent the conserved amino acids in aminotransferases. The conserved glutamate residue (E) is marked with an grey circle. Three black arrows indicate the Val and Ser residues which are the sites for phosphorylation. The Ser residues that are targets of the MPK6 kinase are marked with asterisks. (JPEG 5318 kb)
Alignment of amino acid sequences of MnACO genes. Fe (II)-binding motif and two cosubstrate-binding motifs are marked with red dash box, and red boxes, respectively. The triangles designate the sites of the cysteines. The other important amino acid are marked with the asterisks. (JPEG 4846 kb)
Alignment of amino acid sequences of MnETR genes. Regions I, II, and III represent three transmembrane fragments. (JPEG 6306 kb)
Alignment of amino acid sequences of MnEIN3 genes. The amino-terminal acidic domain (AD), pro-rich region (PR), and five small basic domain (BDI-V) are labeled. The poly-Gln and Poly-Asn repeats at the C-terminal portion are marked with triangles. The arrow indicates the Lys residue essential for the function of EIN3. (JPEG 4085 kb)
Phylogenetic tree of the predicted mulberry AP2/ERF proteins. A phylogenetic tree of AP2/ERF was constructed using maximum-likelihood method in PhyML 3.0 with a Perl script. 15 groups are marked. (JPEG 3240 kb)
Exon/intron structures of mulberry AP2/ERF genes. Different exon/intron structures of genes were plotted using the GSDS server. The intron phase indicates the position of the intron within a codon. If the codon is not located within a codon or is located between two codons, the phase is 0. Phase 2 designates introns between the first and second bases of a codon and phase 2 designates introns between the second and third bases of a codon. (JPEG 1571 kb)


