Abstract
The main genetic determinant of soluble IL-6R levels is the missense variant rs2228145, which maps to the cleavage site of IL-6R. For each Ala allele, sIL-6R serum levels increase by ~20 ng/ml and asthma risk by 1.09-fold. However, this variant does not explain the total heritability for sIL-6R levels. Additional independent variants in IL6R may therefore contribute to variation in sIL-6R levels and influence asthma risk. We imputed 471 variants in IL6R and tested these for association with sIL-6R serum levels in 360 individuals. An intronic variant (rs12083537) was associated with sIL-6R levels independently of rs4129267 (P = 0.0005), a proxy SNP for rs2228145. A significant and consistent association for rs12083537 was observed in a replication panel of 354 individuals (P = 0.033). Each rs12083537:A allele increased sIL-6R serum levels by 2.4 ng/ml Analysis of mRNA levels in two cohorts did not identify significant associations between rs12083537 and IL6R transcription levels. On the other hand, results from 16 705 asthmatics and 30 809 controls showed that the rs12083537:A allele increased asthma risk by 1.04-fold (P = 0.0419). Genetic risk scores based on IL6R regulatory variants may prove useful in explaining variation in clinical response to tocilizumab, an anti-IL-6R monoclonal antibody.
Keywords: allergy, eQTL, expression, disease
Introduction
Asthma is a chronic lung disease characterized by airway obstruction, inflammation and hyperresponsiveness.1 It is caused by a combination of both genetic and environmental factors but their interaction is complex and still not fully understood.
IL-6 is a cytokine known to be involved in allergic asthma2–5 and it binds to its target cells through a complex of at least two distinct membrane-bound proteins, an 80 kD ligand binding glycoprotein (IL-6R) and a 130 kD signal transducing glycoprotein (gp130).6, 7 Although gp130 is expressed on most cells, expression of the membrane-bound form of IL-6R (mIL-6R) is limited to hepatocytes and some immune cells, such as neutrophils, monocytes, CD4+ T cells and B cells;8, 9 as such, only these cells are directly stimulated by IL-6. However, IL-6R also exists in a soluble form (sIL-6R) that is either produced by proteolytic cleavage of mIL-6R or by alternative splicing.10–13 sIL-6R can associate with free IL-6 and this complex is then recognized by the membrane-bound gp130, hence providing an alternative IL-6 signalling pathway available to cells that do not express mIL-6R, a process known as trans-signalling.7
The main genetic determinant of sIL-6R levels is thought to be the non-synonymous variant rs2228145 in exon 9,14 which maps to amino acid position 358 on the main cleavage site of IL-6R.15 Specifically, individuals that carry the Ala358 allele have increased sIL-6R levels14 – which in turn are associated with increased levels of allergic inflammation in the lung16 – and also increased risk of asthma.17 However, given that this variant explains ~19% of the variation in sIL-6R levels18 but the total heritability of this trait has been estimated at ~70%,19 we hypothesised that additional variants in or near the IL6R gene contribute to sIL-6R variation independently of rs2228145 and may also influence asthma risk.
To test this hypothesis, in this study we (1) used data from the 1000 Genomes Project20 to comprehensively impute common variants in or near the IL6R gene; (2) tested these variants for association with sIL-6R levels measured in 360 individuals; (3) followed-up the most associated variants in an independent sample of 354 individuals; and (4) tested the replicated variants for association with IL6R transcript levels in two independent cohorts (N = 851 and 5 191) and asthma risk (N = 47 514).
Results
Common variants affecting sIL-6R serum levels independently of rs4129267
To find new common variants associated with sIL-6R levels and possibly with asthma risk, we first measured sIL-6R serum levels in 360 asthmatics (Supplementary Table 1) who had been previously genotyped with Illumina 610K SNP arrays.17 sIL-6R measurements were performed in duplicate, with good agreement between assays (r = 0.95). Assay and plate effects explained 0.7% (P = 0.013) and 13.6% (P = 3.3×10−20), respectively, of the sample variation in sIL-6R levels and so these effects were removed before association analyses. Duplicate measurements were then averaged and the overall distribution normalised using a rank-based inverse-normal transformation. There were no significant effects of age (P = 0.256) or sex (P = 0.932) on sIL-6R levels.
The main genetic determinant of sIL-6R levels is thought to be the non-synonymous variant rs2228145,14 which is in high linkage disequilibrium (r2 = 0.99) with rs4129267 (Figure 1), a SNP that was directly genotyped in all individuals. We confirmed that the rs4129267 SNP has a significant impact on sIL-6R availability, increasing serum levels by 19.7 ng/ml for each copy of the T allele and explaining 30% of the interindividual variation (P = 1.8×10−29, Supplementary Figure 1). However, given that the heritability of sIL-6R levels has been estimated at ~70%,19 we hypothesised that additional variants in or near the IL6R gene contribute to variation in sIL-6R serum levels independently of rs4129267.
Figure 1. Configuration of the interleukin-6 receptor gene (IL6R).
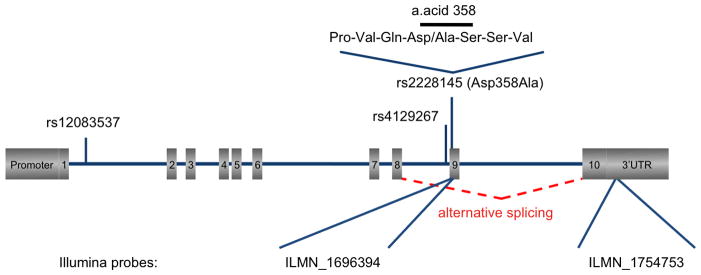
The promoter, exons (1 to 10) and 3′ UTR region are shown in grey boxes, while introns are shown as solid horizontal lines. Also highlighted are the locations of the main genetic regulator of sIL-6R levels (rs2228145), a commonly tested variant in LD with it (rs4129267), the novel regulatory variant identified in this study (rs12083537) and the two Illumina probes used to measure IL6R gene expression levels.
To test this hypothesis, we imputed 452 common variants in IL6R (± 50 kb) in the 360 asthmatics using data from the 1000 Genomes Project as the reference panel. These variants, as well as 19 additional IL6R SNPs that were present in the Illumina 610K array, were then tested for association with sIL-6R levels, after adjustment for the effect of rs4129267. Association results for all 471 SNPs are included in Supplementary Table 2.
The variant with strongest association with rs4129267-adjusted sIL-6R levels was rs12083537 (uncorrected P = 0.0005, Supplementary Table 3 and Supplementary Figure 2); the association remained significant after correcting for the number of SNPs tested through 100 000 permutations (P = 0.0496). The rs12083537 variant is in low linkage disequilibrium with rs4129267 (r2 = 0.03) and has a minor allele (G) frequency of 0.23. An additional three independent (r2 < 0.1) SNPs had a significant (uncorrected P < 0.05) association with sIL-6R levels after adjusting for both rs4129267 and rs12083537 (Supplementary Table 3). However, these associations did not remain significant after correction for multiple SNP testing (P > 0.05).
Replication of the new association between rs12083537 and sIL-6R serum levels
To confirm that the observed association between rs12083537 and sIL-6R levels was real and not a technical or statistical artefact, we performed a replication study using the same experimental procedure in an additional set of 354 unrelated asthmatics (Supplementary Table 4), genotyped for both rs4129267 and rs12083537.
After adjusting for the effects of rs4129267, rs12083537 was significantly associated with sIL-6R levels (P = 0.033), with the same direction of effect observed in the discovery analysis (Supplementary Figure 3). These results thus confirm that rs12083537 is a new quantitative trait locus for sIL-6R serum protein levels.
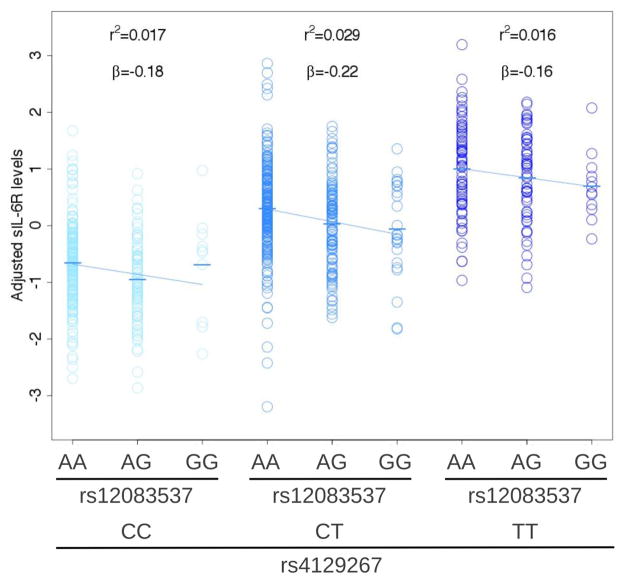
After combining data from the discovery and replication cohorts (combined N = 714), rs12083537 explained 2.2% of the variation in sIL-6R levels after adjustment for the effects of rs4129267 (P = 6.2×10−5). Before adjustment for rs4129267, rs12083537 explained 0.15% of the variation in sIL-6R levels. The rs12083537:G minor allele decreased sIL-6R levels similarly in the three rs4129267 genotype classes (Figure 2).
Figure 2. sIL-6R serum levels as a function of rs4129267 and rs12083537 genotype (N = 714).
The association between sIL-6R levels (after adjusting for technical covariates) and rs12083537 genotype was tested using linear regression and is shown separately for each rs4129267 genotype class, including the proportion of variation in sIL-6R levels explained by rs12083537 (r2), the estimated regression coefficient (β) and the corresponding regression line.
Association between rs12083537 with IL6R mRNA levels
The main genetic determinant of sIL-6R levels, rs2228145, is located on the main cleavage site of IL-6R and so it is thought to affect protein levels through a post-transcriptional mechanism.15 To provide some insight into the molecular mechanisms underlying the new rs12083537 association with sIL-6R protein levels, we investigated if this variant influenced IL6R mRNA levels measured in whole blood in a population-based study of adolescent twins and their relatives (N = 851). There was no significant association between rs12083537 and IL6R mRNA levels in this cohort, either measured by a probe targeting the 3′UTR (beta = −0.050 for the G allele, SE = 0.062, P = 0.4234) or exon 9 (beta = −0.062 for the G allele, SE = 0.063, P = 0.3209). On the other hand, we observed a significant association between rs4129267 (a proxy for rs2228145) and IL6R gene transcription (exon 9 probe, beta = −0.120 for T allele, SE = 0.051, P = 0.0187). Counterintuitively, the rs4129267:T allele that increases sIL-6R protein levels was associated with decreased mRNA expression.
To confirm that the lack of association between rs12083537 and mRNA levels was not due to of low power or incomplete coverage of IL6R expression patterns, we performed a similar analysis in a larger independent study (N = 5 191) with IL6R gene expression levels in whole blood measured by 32 individual Affymetrix probes, targeting exons 5, 7, 8 and the 3′UTR. In this cohort, the rs12083537 variant was also not associated with IL6R transcription levels after a Bonferroni correction for the number of probes tested (P < 0.0016, Supplementary Table 5). The strongest association was with a probe mapping to the 3′UTR region (P = 0.0031), with the rs12083537:A allele that increases sIL-6R protein levels being associated with decreased mRNA expression, as noted above for rs4129267.
Association between rs12083537 and asthma risk
The rs2228145:C (or rs4129267:T) allele that increases sIL-6R levels14 also increases asthma risk.17 We therefore tested the hypothesis that rs12083537 is also a genetic risk factor for asthma. Specifically, we hypothesised that, as for rs2228145, the rs12083537:A allele that is associated with higher sIL-6R levels would be predisposing for asthma. Association results were available for 47 514 individuals from three independent studies, including 16 705 doctor-diagnosed asthmatics and 30 809 controls. A consistent predisposing effect was observed for the rs12083537:A allele in all three studies (Table 1). Overall, the association between rs12083537 and asthma risk was weak but statistically significant (OR = 1.039, 95% C.I. = 1.002 – 1.078, P = 0.0419). Results remained largely unchanged when the association was tested while adjusting for rs4129267 (OR = 1.050, 95% C.I. = 1.012 – 1.089, P = 0.0088).
Table 1.
Association between rs12083537 and asthma risk in three independent studies.
| Study | N cases | N Controls | Before conditioning on rs4129267 | After conditioning on rs4129267b | ||||
|---|---|---|---|---|---|---|---|---|
| ORa | SE | P-value | ORa | SE | P-value | |||
| AAGC | 2110 | 3857 | 1.095 | 0.0528 | 0.0846 | 1.109 | 0.0527 | 0.0507 |
| 23andMe | 4230 | 10842 | 1.036 | 0.0333 | 0.2888 | 1.044 | 0.0334 | 0.2012 |
| GABRIELc | 10365 | 16110 | 1.028 | 0.0249 | 0.2726 | 1.041 | 0.0247 | 0.1052 |
| All samples | 16705 | 30809 | 1.039 | 0.0186 | 0.0419 | 1.050 | 0.0186 | 0.0088 |
Odds ratio corresponds to the major allele, A.
For the AAGC and 23andMe studies, rs4129267 genotype (coded as allelic dosage: 0, 1 or 2) was included as a covariate in a logistic regression analysis modelling the association between asthma status and rs12083537 allelic dosage. Only GWAS summary statistics were available for the GABRIEL; as such, for this study, the conditional analysis was performed using the Yang et al approach implemented in GCTA. 21
For the GABRIEL study, results are shown for rs1386821 (T allele), a proxy SNP (r2 = 0.94) for rs12083537. The rs1386821:T allele is in phase with the rs12083537:A allele.
Discussion
We recently identified a variant (rs4129267) in the IL6R gene with a modest (OR = 1.09) but genome-wide significant association with asthma risk.17 This variant is in near complete LD with the exon 9 missense variant rs2228145, and is associated with a 1.4-fold increase in sIL-6R levels for each copy of the T allele.22 The goal of this study was to test the hypotheses that (1) other variants in IL6R regulate gene expression and (2) that these also associate with asthma risk.
A single variant (rs12083537) located in intron 1 of IL6R, 2.9 kb away from exon 1, was found to associate with sIL-6R levels independently of rs4129267 and after correction for multiple SNP testing. A significant and consistent association for this variant was then observed in our replication panel. Furthermore, at the time of submission, we also noted that R. Ferreira et al.23 independently reported two new genetic associations for sIL-6R levels, one of which (rs1386821) is correlated with rs12083537 (r2 = 0.94). The alleles that increase sIL-6R levels for these two SNPs (rs12083537:A and rs1386821:T) are in phase and so our results are consistent with those of R. Ferreira et al. Together, these results establish rs12083537, or a variant in LD with it, as a new regulatory variant for sIL-6R serum levels.
The rs12083537 variant is located in a region with high H3K4Me1 lysine methylation in four cell lines, including lung fibroblasts and keratinocytes.24 This histone modification is associated with enhancers and promoters and so it is plausible that rs12083537 may influence gene transcription. However, analysis of mRNA levels extracted from whole blood in two independent cohorts did not identify any significant associations between rs12083537 and IL6R transcription levels, after correcting for multiple testing. Thus, the molecular mechanism underlying the association between rs12083537 and sIL-6R levels remains to be elucidated. A potential caveat of our analysis was that we identified the association between rs12083537 and sIL-6R protein levels in asthma patients but then analysed mRNA levels in two studies that included mostly non-asthmatics. However, this difference is unlikely to explain the lack of association between rs12083537 and mRNA levels because IL6R regulatory variants have been shown to influence protein levels similarly in population-based samples 22, healthy individuals 14, 25 and patients with different diseases 23, 25, 26. Gene expression studies in individual immune cell types or upon allergen stimulation may help characterise the function of this variant in greater detail.
Interestingly, we observed that the rs4129267:T allele that is associated with increased sIL-6R protein levels22 was associated with decreased IL6R mRNA levels, which may appear contradicting. The Illumina probe for which we observed an association with rs4129267 maps to exon 9 (containing the IL-6R trans-membrane domain), which is skipped in the main differentially spliced IL6R isoform that directly produces sIL-6R. Therefore, this probe measures the levels of the full length isoform, which necessarily decrease if splicing of exon 9 increases. Given that the rs4129267:T allele, or an allele associated with it (e.g. rs2228145:C), strongly increases the splicing of exon 9,23 it is in fact not surprising that consequently this allele is associated with decreased mRNA levels of the full length isoform. Unlike our results, R. Ferreira23 did not observe a significant effect of rs2228145 on the levels of the full length isoform. However, as the authors noted, splicing of exon 9 is a rare event. Therefore, the lack of association in that study likely reflected the low power provided by a small sample size (N = 88) to detect small differences in the levels of the full length isoform between genotype classes.
Lastly, we found rs12083537 to be significantly associated with asthma risk. Based on our analysis, each rs12083537:A allele increased sIL-6R serum levels by 2.4 ng/ml and increased asthma risk by 1.04-fold. For comparison, each rs4129267:T allele increased sIL-6R serum levels by 19.7 ng/ml and increases asthma risk by 1.09-fold.17 The association between rs12083537 and asthma risk was consistent across the three cohorts analysed but only marginally statistically significant in the overall analysis. Replication of our results by well powered studies is therefore warranted; over 23 000 cases and 23 000 controls (or, for example, 18 000 cases and 36 000 controls) are required to achieve 80% power to detect (α = 0.05) the effect we observed.
In summary, our study demonstrates that at least two independent genetic variants influence sIL-6R serum levels. Consequently, pharmacogenetic studies of clinical response to tocilizumab, an anti-IL-6R monoclonal antibody approved to treat rheumatoid arthritis34 and suggested to treat asthma,16, 17 should consider the effects of not only the major genetic determinant of sIL-6R levels (rs2228145) but also of other confirmed regulatory variants such as rs12083537. For this purpose, multi-SNP scores that summarise an individual’s combined genetic risk of having high sIL-6R levels may prove useful.
Methods
Study participants
Genetic variants in or near the IL6R gene were tested for association with sIL-6R protein levels, mRNA levels and asthma risk in the cohorts described below. Informed consent was obtained from all participants and the study protocols were approved by the appropriate ethics committees.
Association with IL-6R protein levels – discovery cohort
To find new variants in the IL6R gene that affect sIL-6R levels, we first studied 360 unrelated asthmatic subjects from the 1995–1998 Asthma and Allergy study, which is described in detail elsewhere.27 Briefly, 3 073 subjects were recruited from 802 families that were registered on the Australian Twin Registry and had at least one twin who previously reported a history of wheezing in studies conducted at QIMR and by collaborators elsewhere in Australia. Participants completed a questionnaire that was designed to validate the diagnosis of asthma and to obtain data on respiratory symptoms, environmental exposures and family history of asthma. In addition, participants underwent clinical testing, including lung function and skin prick tests. For the present study, we selected 360 unrelated individuals with available DNA and a serum sample, and who answered “Yes, told to me by a doctor” to the question “Have you ever had asthma?”. Demographics and clinical characteristics for this discovery sample are summarised in Supplementary Table 1. These individuals were included in a recent GWAS of asthma.17
Association with IL-6R protein levels – replication cohort
To confirm associations between SNPs and IL-6R protein levels observed in the discovery sample, we studied an additional sample of 354 unrelated asthmatics recruited through the QIMR Asthma Genetics Study between 2011 and 2013. In this study, individuals with a self-reported doctor-diagnosis of asthma were recruited from the general community through media appeals (newspapers, radio, television and social media) to participate in a study of genetic risk factors for asthma. Participation included completing a brief online questionnaire about asthma symptoms, medication and severity, and the provision of a blood sample for genetic and immune function testing. Blood was collected at a local pathology laboratory and shipped to QIMR by overnight courier. Demographics and clinical characteristics for this replication sample are summarised in Supplementary Table 4.
Association with IL6R mRNA levels
To provide some insight into the molecular mechanisms underlying SNP-protein associations, SNPs confirmed to associate with sIL-6R protein levels were then tested for association with mRNA levels measured in 851 individuals from 262 families who participated in a genome-wide gene-expression study reported recently.28 Briefly, adolescent MZ and DZ twins, their siblings, and their parents have been recruited over a 21-year period into an ongoing study of the genetic and environmental factors influencing pigmented nevi and the associated risk of developing skin cancer and cognition (Twin Moles study)29. Results obtained in this cohort were confirmed by analysing expression levels from an additional 5 191 individuals who participated in two large-scale longitudinal studies: the Netherlands Twin Register (N = 3 170) and the Netherlands Study of Depression and Anxiety (N = 2 021); henceforth we refer to this as the NTR-NESDA eQTL study. The individual NTR and NESDA cohorts are described in detail elsewhere.30, 31
Association with asthma risk
SNPs associated with sIL-6R protein levels were also tested for association with asthma risk in 21 039 unrelated individuals of European ancestry who participated in two independent studies. First, 5 967 individuals who participated in the Australian asthma GWAS,17 including 2 110 doctor-diagnosed asthmatics and 3 857 controls. Sample ascertainment and patient characteristics for this study are described in detail in Ferreira.17 Second, 15 072 customers of 23andMe, Inc., a personal genetics company, who had been genotyped as part of the 23andMe Personal Genome Service®. We selected 4 230 cases and 10 842 controls of European ancestry who had taken a survey about asthma. Cases (mean age 48, 54% female, 55% with onset of asthma at or before age 16) gave positive responses to the question “Have you ever been diagnosed by a doctor with asthma or bronchial asthma?”, and also checked a box indicating that they had ever had “allergic rhinitis (stuffed or dripping nose caused by allergies)”. Controls (mean age 49, 39% females) gave negative responses to both questions. Association results from these 6 340 cases and 14 699 controls were meta-analysed with publicly available results from 10 365 asthmatics and 16 110 controls included in the GABRIEL GWAS,32 for a combined total sample size of 47 514 individuals, including 16 705 cases and 30 809 controls.
sIL-6R serum protein measurements
A 40 mL venous blood sample was collected from all consenting subjects. After spinning 10 mL of blood in a serum tube at 805g for 10 min, the serum layer was extracted and stored at −80°C until analysis. ELISA kits (R&D Systems, Minneapolis, MN, USA) were used to measure sIL-6R levels according to manufacturer’s procedures. The optical density was determined using a PowerWave XS2 microplate spectrophotometer (BioTek Instruments, Inc.) at both 450 and 540 nm wavelengths. Results and standard curves were acquired with Gen5 2.0 Data Analysis Software. sIL-6R levels were measured in duplicate for each sample.
IL6R mRNA measurements
Whole blood for expression profiling was collected from 851 individuals from the Twin Moles study and processed as described in detail by Powell et al.28 Briefly, total RNA was extracted from PAXgene™ tubes using the QIAGEN whole blood gene RNA purification kit. RNA quality and concentration was estimated with Agilent Bioanalyzer and subsequently converted to cDNA, amplified and purified using the Ambion Illumina TotalPrep RNA Amplification Kit (Ambion). Expression profiles were generated by hybridising 750 ng of cRNA to Illumina HumanHT-12 v4.0 Beadchip according to Illumina whole-genome gene expression direct hybridization assay guide (Illumina Inc, San Diego, USA). Beadchips were then washed and stained and subsequently scanned to obtain fluorescence intensities. Samples were scanned using an Illumina Bead Array Reader. Samples were randomised across chips and chip positions, with check for balance across families, sex and generation. For this study, we restricted our analysis to two probes targeting the IL6R gene, specifically ILMN_1754753 on the 3′-end and ILMN_1696394 on exon 9 (Supplementary Table 6).
Gene expression association results were confirmed by analysing a further 5 191 individuals from the NTR-NESDA eQTL study. In this study, venous blood samples were drawn in the morning after an overnight fast; heparinised whole blood was transferred within 20 minutes of sampling into PAXgene™ tubes and stored at −20°C. RNA extraction and analysis was performed at the Rutgers University Cell and DNA Repository (USA). RNA was extracted using Qiagen Universal liquid handling system (PAXgene extraction kits following the manufacturer’s protocol). Total RNA was measured by spectroscopy (Trinean DropSense) to determine purity and concentration, while RNA fidelity was measured by the Agilent Bioanalyzer analysis. RNA samples were hybridized to Affymetrix U219 array plates (GeneTitan), which contains 530 467 probes, each 25 bases in length. Array hybridization, washing, staining, and scanning were carried out in an Affymetrix GeneTitan System following the manufacturer’s protocol. Non-uniquely mapping probes (hg19) and probes containing a polymorphic SNP based on snp137 (UCSC) were removed. Expression values were obtained using RMA normalization implemented in Affymetrix Power Tools (APT, v 1.12.0). Thirty-two probes targeting IL6R were selected for analysis (Supplementary Table 5).
IL6R SNP genotyping
For association with sIL-6R protein levels – discovery cohort
Genome-wide SNP data for the 360 individuals were obtained using Illumina 610-Quad Beadchip as part of the Australian Asthma Genetics Consortium GWAS, which is described in detail elsewhere.17 For this study, we selected 254 post-QC (minor allele frequency [MAF] > 1%, call rate > 95%, Hardy-Weinberg equilibrium P-value > 10−6) SNPs in or within 1 000 kb of IL6R, including rs4129267. To expand the genetic coverage of IL6R, these 254 directly genotyped SNPs were then used to impute 530 common variants (MAF >= 1%) from the 1000 Genomes Project (March 2012 release, 1,092 samples of all ancestries) using Impute2.33 After QC (MAF > 1%, imputation information score > 0.3), there were 471 SNPs located in or within 50 kb of IL6R available for analysis, of which 19 had been directly genotyped.
For association with sIL-6R protein levels – replication cohort
Genotypes for two SNPs (rs4129267 and rs12083537) were obtained for the 354 individuals with Sequenom MassARRAY iPLEX platform; 12.5 ng of DNA isolated from buffy coats with a salt precipitation method were used per assay. SNP sequences were downloaded from the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and cross checked. Design of the PCR and iPLEX Extension assays was done using the Sequenom Design Suite (https://mysequenom.com/). The assays were then manually adjusted to increase the level of multiplexing. SNPs were typed using iPLEXTM Gold chemistry and analysed using a Sequenom MassARRAY Compact Mass Spectrometer (Sequenom Inc, San Diego, CA, USA). The PCR, SAP and iPLEX reactions were performed using half the volume of reagents recommended in the manufacturer’s specifications. The post-PCR products were spotted on a Sequenom SpectroChip 2, and the data was processed and analysed using Sequenom MassARRAY TYPER 4.0 software. The primers used to amplify both SNPs are included in Supplementary Table 7.
For association with IL6R mRNA levels
DNA samples from the Twin Moles study (N = 851) were genotyped by the Scientific Services Division at deCODE Genetics, Iceland, using the Illumina 610-Quad Beadchip. Genotypes were called with the Illumina BeadStudio software. Full details of genotyping procedures are given in Medland et al.34 DNA extraction for the NTR-NESDA eQTL study (N = 5 191) has been described before.30 Genotyping was performed on multiple SNP array platforms, including Perlegen 5.0, Illumina 370, Illumina 660, Illumina Omni Express 1M and Affymetrix 6.0. Standard pre-imputation quality control filters were applied first within and then between chip platforms; subsequently, data were merged into a single dataset. Genome-wide SNP imputation was done with Impute2 33 using the 1000 Genomes phase I Interim Jun 2011 release. This included the rs12083537 variant analysed in this study, which was imputed with high confidence (information score > 0.9).
For association with asthma risk
We accessed individual-level genotype data for a single SNP (rs12083537) from the Australian GWAS (N = 5 967) and the 23andMe cohort (N = 15 072). This SNP was imputed in the Australian GWAS with high confidence (information score 0.97), using Impute2 and genotype data from the combined 1000 Genomes (60 individuals with northern and western European ancestry from the Centre d’Etude du Polymorphisme Humain collection [CEU], March, 2010 release) and HapMap 3 (955 individuals from 11 populations, February, 2009 release) projects as the reference panel. In the 23andMe cohort, rs12083537 was imputed with high confidence (r2 = 0.95) using the August 2010 release of the 1000 Genomes reference haplotypes.
Statistical analyses
QC of sIL-6R ELISA data
Each sample was measured in duplicate (two assays), with up to 80 serum samples included per ELISA plate. No outlier observations (located six standard deviations from the mean) were observed for either assay. The same procedure was used for the discovery and replication cohorts to estimate and remove the effects of both technical and biological effects on sIL-6R levels. Briefly, we (1) used linear regression to adjust for assay and plate effects; (2) normalized the distribution using a rank-based inverse-normal transformation; and (3) tested and, if significant (P < 0.05), adjusted for the effects of age and sex. Subsequent association analyses were carried out using the residuals obtained from (3).
Association analyses
Individual SNPs in IL6R, directly genotyped or imputed, were tested for association with sIL-6R protein levels (discovery and replication cohorts), mRNA levels and asthma risk. In all cases, we used additive allelic tests of association; for imputed SNPs, we used the inferred allelic dosage.
The association between the 471 SNPs in IL6R and protein levels was tested using linear regression in R. Association analyses were performed before and after adjusting sIL-6R levels for the effects of rs4129267, a SNP highly correlated with the missense variant rs2228145 (r2 = 0.99), the main genetic determinant of sIL-6R protein serum levels.22 To adjust for the effects of rs4129267, we (1) performed a linear regression of sIL-6R levels (dependent 410 variable) on rs4129267 allelic dosage (independent variable); (2) extracted the residuals from this analysis, which represent rs4129267-adjusted sIL-6R levels; and (3) performed a linear regression of rs4129267-adjusted sIL-6R levels (dependent variable) on the allelic dosage of individual SNPs (independent variable). To correct the observed asymptotic association P-value (Pobs) of a given SNP for the number of (and correlation between) SNPs tested, we (1) permuted rs4129267-adjusted sIL-6R levels between the 360 asthmatics; (2) tested the permuted sIL-6R levels for association individually with each of the 471 SNPs and recorded the asymptotic P-value for the most significantly associated SNP (Pmin); (3) repeated steps (1) and (2) 100 000 times; and (4) calculated for each SNP the empirical association P-value corrected for the number of SNPs tested as the proportion of 100 000 replicates for which Pmin ≤ Pobs.
Association with mRNA levels in the Twin Moles study was tested using MACH2QTL,35, 36 which takes into account the family structure of the data. In the NTR-NESDA eQTL study, association with mRNA levels was tested using a linear mixed model to correct for family structure (as random effects) as well as sex, age, body mass index, smoking status, technical covariates and ancestry (as fixed effects).
Lastly, association with asthma risk was tested using logistic regression in R, including covariates to adjust for sample origin in the Australian GWAS and, in the 23andMe study, for age, sex and ancestry. Association analysis conditional on rs4129267 were performed in both studies by including this SNP (coded as allelic dosage: 0, 1 or 2) as a covariate in the logistic regression model. Only GWAS summary statistics were available for the GABRIEL study; as such, for this study, the conditional analysis was performed using the Yang et al approach implemented in GCTA.21 Fixed-effects meta-analysis of results from the Australian GWAS, 23andMe and the publicly available GABRIEL GWAS was performed with METAL.37
Supplementary Material
Acknowledgments
We thank all the participants of the Asthma and Twin moles studies; Ann Eldridge, Marlene Grace, Kerrie McAloney (sample collection); Melinda Richter, Lisa Bowdler, Steven Crooks (DNA processing); David Smyth, Harry Beeby, Daniel Park (IT support). Funding was provided by the Australian National Health and Medical Research Council (NHMRC, 613627). MARF is supported by a Career Development Fellowship from the NHMRC.
Footnotes
Conflict of Interest
We declare that there are no conflicts of interest relating to the work described in this paper.
Supplementary information is available 443 at the Gene and Immunity’s website.
References
- 1.Murphy DM, O’Byrne PM. Recent advances in the pathophysiology of asthma. Chest. 2010;137(6):1417–26. doi: 10.1378/chest.09-1895. [DOI] [PubMed] [Google Scholar]
- 2.Doganci A, Sauer K, Karwot R, Finotto S. Pathological role of IL-6 in the experimental allergic bronchial asthma in mice. Clinical reviews in allergy & immunology. 2005;28(3):257–70. doi: 10.1385/CRIAI:28:3:257. [DOI] [PubMed] [Google Scholar]
- 3.Neveu WA, Allard JL, Raymond DM, Bourassa LM, Burns SM, Bunn JY, et al. Elevation of IL-6 in the allergic asthmatic airway is independent of inflammation but associates with loss of central airway function. Respiratory research. 2010;11:28. doi: 10.1186/1465-9921-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rincon M, Irvin CG. Role of IL-6 in asthma and other inflammatory pulmonary diseases. International journal of biological sciences. 2012;8(9):1281–90. doi: 10.7150/ijbs.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Homer RJ, Chen Q, Elias JA. Endogenous and exogenous IL-6 inhibit aeroallergen-induced Th2 inflammation. Journal of immunology (Baltimore, Md: 1950) 2000;165(7):4051–61. doi: 10.4049/jimmunol.165.7.4051. [DOI] [PubMed] [Google Scholar]
- 6.Hibi M, Murakami M, Saito M, Hirano T, Taga T, Kishimoto T. Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell. 1990;63(6):1149–57. doi: 10.1016/0092-8674(90)90411-7. [DOI] [PubMed] [Google Scholar]
- 7.Taga T, Hibi M, Hirata Y, Yamasaki K, Yasukawa K, Matsuda T, et al. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell. 1989;58(3):573–81. doi: 10.1016/0092-8674(89)90438-8. [DOI] [PubMed] [Google Scholar]
- 8.Chalaris A, Rabe B, Paliga K, Lange H, Laskay T, Fielding CA, et al. Apoptosis is a natural stimulus of IL6R shedding and contributes to the proinflammatory trans-signaling function of neutrophils. Blood. 2007;110(6):1748–55. doi: 10.1182/blood-2007-01-067918. [DOI] [PubMed] [Google Scholar]
- 9.Oberg HH, Wesch D, Grussel S, Rose-John S, Kabelitz D. Differential expression of CD126 and CD130 mediates different STAT-3 phosphorylation in CD4+CD25− and CD25high regulatory T cells. International immunology. 2006;18(4):555–63. doi: 10.1093/intimm/dxh396. [DOI] [PubMed] [Google Scholar]
- 10.Horiuchi S, Koyanagi Y, Zhou Y, Miyamoto H, Tanaka Y, Waki M, et al. Soluble interleukin-6 receptors released from T cell or granulocyte/macrophage cell lines and human peripheral blood mononuclear cells are generated through an alternative splicing mechanism. European journal of immunology. 1994;24(8):1945–8. doi: 10.1002/eji.1830240837. [DOI] [PubMed] [Google Scholar]
- 11.Lust JA, Donovan KA, Kline MP, Greipp PR, Kyle RA, Maihle NJ. Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine. 1992;4(2):96–100. doi: 10.1016/1043-4666(92)90043-q. [DOI] [PubMed] [Google Scholar]
- 12.Mullberg J, Schooltink H, Stoyan T, Gunther M, Graeve L, Buse G, et al. The soluble interleukin-6 receptor is generated by shedding. European journal of immunology. 1993;23 (2):473–80. doi: 10.1002/eji.1830230226. [DOI] [PubMed] [Google Scholar]
- 13.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. The Biochemical journal. 1994;300 (Pt 2):281–90. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes and immunity. 2004;5(6):513–6. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- 15.Mullberg J, Oberthur W, Lottspeich F, Mehl E, Dittrich E, Graeve L, et al. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. Journal of immunology (Baltimore, Md: 1950) 1994;152(10):4958–68. [PubMed] [Google Scholar]
- 16.Doganci A, Eigenbrod T, Krug N, De Sanctis GT, Hausding M, Erpenbeck VJ, et al. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. The Journal of clinical investigation. 2005;115(2):313–25. doi: 10.1172/JCI22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rafiq S, Frayling TM, Murray A, Hurst A, Stevens K, Weedon MN, et al. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes and immunity. 2007;8(7):552–9. doi: 10.1038/sj.gene.6364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raggi P, Su S, Karohl C, Veledar E, Rojas-Campos E, Vaccarino V. Heritability of renal function and inflammatory markers in adult male twins. American journal of nephrology. 2010;32(4):317–23. doi: 10.1159/000319449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Ferreira T, Morris AP, Medland SE, et al. Genetic Investigation of ATC, Replication DIG. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature genetics. 2012;44(4):369–75. S1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS genetics. 2008;4(5):e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira RC, Freitag DF, Cutler AJ, Howson JM, Rainbow DB, Smyth DJ, et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS genetics. 2013;9(4):e1003444. doi: 10.1371/journal.pgen.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nature genetics. 2013;45(7):730–8. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hingorani AD, Casas JP. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379(9822):1214–24. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira MA, O’Gorman L, Le Souef P, Burton PR, Toelle BG, Robertson CF, et al. Variance components analyses of multiple asthma traits in a large sample of Australian families ascertained through a twin proband. Allergy. 2006;61(2):245–53. doi: 10.1111/j.1398-9995.2005.00954.x. [DOI] [PubMed] [Google Scholar]
- 28.Powell JE, Henders AK, McRae AF, Caracella A, Smith S, Wright MJ, et al. The Brisbane Systems Genetics Study: genetical genomics meets complex trait genetics. PLoS One. 2012;7 (4):e35430. doi: 10.1371/journal.pone.0035430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu G, Montgomery GW, James MR, Trent JM, Hayward NK, Martin NG, et al. A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur J Hum Genet. 2007;15(1):94–102. doi: 10.1038/sj.ejhg.5201729. [DOI] [PubMed] [Google Scholar]
- 30.Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D, et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16(3):335–42. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- 31.Willemsen G, Vink JM, Abdellaoui A, den Braber A, van Beek JH, Draisma HH, et al. The Adult Netherlands Twin Register: twenty-five years of survey and biological data collection. Twin Res Hum Genet. 2013;16(1):271–81. doi: 10.1017/thg.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85 (5):750–5. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.