Abstract
Purpose
IDH1/2 mutant gliomas harbor a distinct CpG island methylation profile (G-CIMP) that may promote the initiation and progression of secondary pathway gliomas by silencing tumor suppressive genes. The potential role of tumor suppressive miRNAs in this process is not understood.
Experimental Design
To identify potential tumor suppressive microRNAs hypermethylated in glioma, the methylation profiles of IDH1/2WT gliomas (n=11) and IDH1MUT glioma (n=20) were compared by using massively parallel reduced representation bisulfite sequencing (RRBS). The methylation status of selected miRNA was validated by using targeted bisulfite sequencing (BiSEQ) in a large cohort of glioma tissue samples including 219 IDH1WT and 72 IDH1/2MUT samples. The expression of selected miRNAs was determined by using TaqMan qPCR. Functional analyses of miRNA-148a were conducted and target genes were identified.
Results
We identify miR-148a as a novel, G-CIMP associated miRNA whose methylation is tightly correlated with IDH1 mutation and associated with improved survival in malignant glioma patients. We confirm that down-regulation of miR-148a can occur via DNA methylation. We demonstrate that IDH1 mutation provides a mechanism of miR-148a methylation and downregulation, and that restoration of miR-148a reduced tumorigenic properties of glioma cells, possibly by targeting DNMT1.
Conclusions
We identify miR-148a as a novel G-CIMP associated miRNA, and provide results suggesting that miR-148a restoration may have therapeutic implications.
Keywords: IDH1, glioma, tumor suppressor, microRNA, miRNA-148a, methylation, RRBS, whole methylome, CpG island
Introduction
Diffuse gliomas represent the most common type of primary brain cancer in adults and remain incurable (1). Diffuse gliomas are diagnosed histologically according to the WHO classification system; however, a detailed molecular framework can now be overlaid on this existing classification system (2). In particular, mutation of either the isocitrate dehydrogenase 1 or 2 (IDH1/2MUT) gene has now been accepted as an initiating step and molecular marker for gliomas arising along the secondary glioblastoma/oligodendroglioma pathway (2–4).
Several DNA methylation studies have described a coordinated CpG island hypermethylation signature (Glioma-CpG Island Methylator Phenotype, G-CIMP) in gliomas harboring the IDH1/2MUT (5, 6); and accumulating evidence suggests that aberrant IDH1/2MUT protein function contributes to the DNA hypermethylation pattern observed in G-CIMP (7, 8). Through methylation and silencing of potential tumor suppressive genes, the composition and scope of CpG islands/genes within G-CIMP are thought to provide important clues to understanding the initiation and progression of tumors along the secondary glioblastoma/oligodendroglioma pathway (5, 6). The clinical relevance is that re-expression of these silenced genes may be an effective strategy to control these tumors. However, major barriers impeding progress in this direction include both lack of consensus on the exact composition of G-CIMP CpG islands/genes, and lack of detailed functional studies in glioma demonstrating tumor suppressive function of these genes (9).
To address the lack of consensus on G-CIMP composition, we have recently published the first single base resolution methylation profiling characterization of G-CIMP using RRBS (Reduced Representation Bisulfite Sequencing) (6). RRBS is a cost-effective technique for high-resolution methylome sequencing which uses restriction enzyme-based cleavage of genomic DNA in to fragments enriched for CpG islands, that are then sequenced using massively-parallel techniques (10). One of the major limitations of our previous study was the lack of consideration of CpG islands associated with microRNAs (miRNAs). miRNAs are a large family (>1600) of post-transcriptional regulators of gene expression that are 20~22 nucleotides in length and involved in many developmental and cellular processes in eukaryotes (11). Each miRNA is predicted to target and down-regulate many transcripts directly or indirectly (12). miRNA deregulation has been implicated in cancers (13–15) such as glioma (16–18); and similar to coding genes, CpG island methylation-associated silencing of miRNAs with tumor suppressor features have been discovered in human cancer (19–22).
In the present study we sought to determine whether G-CIMP contains CpG islands associated with miRNAs and whether any identified miRNAs have tumor suppressive properties.
Materials and Methods
Cell culture and treatments
Glioma cell lines and HEK293T cell line were generous gift from Dr. Paul Mischel (UCSD) and originally obtained from ATCC. HT1080 cell line was purchased from ATCC (CCL-121). Human neuron stem cells (hNSCs) were obtained from Dr. Harley Kornblum (UCLA). Glioma cell lines were maintained in DMEM/F12 cell culture medium with 10% FBS and penicillin/streptomycin (Invitrogen). HEK293T cell line and HT1080 cell line were maintained in DMEM cell culture medium with 10% FBS and penicillin/streptomycin. hNSCs were maintained in neurobasal medium supplied with EGF (50ng/ml), bFGF (20ng/ml), B27 and penicillin/streptomycin (Invitrogen). All cells were cultured at 37°C and 5% CO2 in tissue culture incubator.
Patient cohorts and tumor specimens
A total of 324 frozen and FFPE tissue specimens were obtained from the UCLA Brain Tumor Translational Resource. Remnant human brain tumor samples were collected from patients undergoing surgical resection and who provided written informed consent. The collection of human brain tumor samples was approved by the UCLA Institutional Review Board. IDH1 was sequenced on all samples and IDH2 was sequenced on selected IDH1WT samples (including all samples used for RRBS analysis and those with miR-148a methylation).
Constructs
Retrovirus constructs for IDH1WT and IDH1R132HMUT expression (pLPCX, pLPCX-IDH1-WT and pLPCX-IDH1-R132H) were generated as previous report (23). Lentivirus constructs for miR-148a expression (pMIF-GFP-zeo-miR-148a and pMIF-GFP-zeo) were generous gift from Dr. Aprelikova. Promoter reporter vector pIS0-DNMT1-3′UTR-WT and pIS0-DNMT1-3′UTR-MUT were generated by cloning 3′UTR sequence of DNMT1 gene using the following primers: forward, 5′ GTAATTCTAGGAGCTCTTCTGCCCTCCCGTCACCCC 3′; reverse, 5′CCGCCCCGACTCTAGATGGTTTATAGGAGAGATTTATTTG 3′. For 3 sites specific mutagenesis of the DNMT1 reporter vector, the following primers were used: forward, 5′CTGGCACCAGGAATCCCCAACAcGaAaTGATGTTGTGTTTTTAAC 3′; reverse, 5′ TCAtTtCgTGTTGGGGATTCCTGGTGCCAGAAACAGGGGTGACG 3′ (mutated sites are indicated with lower case).
Massively Parallel Reduced Representation Bisulfite Sequencing (RRBS) and IDH1 and IDH2 sequencing
Reduced representation bisulfite sequencing and IDH1 and IDH2 sequencing were done using the protocol published previously (6).
Analysis of Methylation and Gene Expression in the Cancer Genome Atlas Dataset
miRNA expression data, measured using the Agilent Sureprint 8×15k Human miRNA microarray (Agilent, Santa Clara, CA), was obtained for 145 TCGA samples. Level 3 expression data, including normalized expression signal per miRNA per sample, was downloaded directly from the TCGA data portal (http://tcga-data.nci.nih.gov/tcga/).
Bisulfite conversion of genomic DNA, target bisulfite sequencing (BiSEQ) and methylation specific PCR (MSP)
Bisulfite conversion of genomic DNA and standard bisulfite sequencing (BiSEQ) utilizing a nested PCR protocol as previously descripted (6). The methylation status of the miR-148a promoter CpG island (region 1: chr7:25991097-25991208; region 2: chr7: 25990178-25990308) were assessed using primer sets shown in Supplementary Table S10. The sequence of each sample was reviewed using Chromas Lite 2.33 (Technelysium Pty Ltd) and CpG sites exhibiting a substantial signal for C (as compared to T) were considered methylated. The samples which had less than 3 methylated CpG sites were considered unmethylated (Unmethyl.), otherwise considered methylated (Methyl.). Methylation specific PCR (MSP) was performed utilizing a nested PCR protocol as previously descripted (24). The methylation specific primer set was shown in Supplementary Table S11.
RNA isolation and quantitative reverse transcription PCR
Total RNA was isolated using Trizol regent according to the manufacturer’s guideline (Invitrogen). The integrity of total RNA was determined by 1% agarose gel electrophoresis. For RT-PCR, reverse transcription of total RNA was performed by Reverse Transcription System (Promega) or TaqMan Reverse Transcription system (Applied Biosystems, UK) according to the manufacturer’s protocol. The qPCR was carried out using LightCycler® 480 System (Roche, Mannheim, Germany) with the universal SYBR Green PCR master mix (Roche) by using β-actin as internal control. All results from 3 independent experiments are presented as mean ± s.e.m. (n=3).
Analysis of miRNA expression using TaqMan Reverse Transcription PCR
Expression of mature miR-148a was analyzed using the TaqMan® MicroRNA Assays (Applied Biosystems). Expression of RNU6B (Applied Biosystems) was used as an endogenous control. miR-148a expression was measured relative to RNU6b (internal control) and quantified by the relative Ct method (2ΔΔCt). All the results are from 3 independent experiments done in duplicate. 2 commercially avoidable normal brain cDNA libraries were used as normal control (Invitrogen, Grand Island, NY; Biochain, Hayward, CA). The TaqMan qPCR was carried out using LightCycler® 480 System (Roche) with the TaqMan universal PCR master mix (Applied Biosystems). All results from 3 independent experiments were performed in duplicate are presented as mean ± s.e.m. (n=3).
Western blot analysis
Western blot analysis was carried out using standard methods. The following primary antibodies were used: anti-DNMT1 (Abcam) at 1:800 dilution; anti-α-tubulin (Sigma) at 1:2000 dilution; anti-FLAG (Sigma) at 1:2000 dilution; anti-IDH1 (Santa Cruz) at 1:750 dilution; anti-IDH1R132H (Dianova, German) at 1:750. The following secondary antibodies were used:horse radish peroxidase (HRP)-conjugated goat anti-rabbit (1:4000) IgG (Santa Cruz Biotech, Santa Cruz, CA), horse radish peroxidase (HRP)-conjugated donkey anti-goat (1:8000) IgG (Santa Cruz Biotech, Santa Cruz, CA), horse radish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:10 000, Jackson Immuno Research, West Grove, PA). An enhanced chemiluminescence detection kit (Pierce, Rockford, IL) was used for the detection of HRP. Densitometry was performed with Gel-Pro Analyzer 4.0 software (Media Cybernetics, Bethesda, MD).
Luciferase reporter assay
After closely examination of miR-148a precursor gene and analysis of miR-148a promoter activity by using bioinformatics method, we have identified that miR-148a associated CpG island located at −407~−1716 upstream of miR-148a precursor region, and it is overlapped with the predicted TSS (−1112) as shown in Fig. 3D. 1.623 kb DNA fragment localized at the upstream of predicted miR-148a TSSs (−1038~−2661) was cloned into pGL4.17 vector (pGL4.17-miR-148a-P). Wild type or mutant 3′ UTR of DNMT1 were cloned into firefly luciferase reporter pIS0 vector (Addgene plasmid, Cat 12178). Renilla luciferase vector pRL-SV40 (Promega) was co-trasnfected in each experiment as internal control. Transfection was carried out by using X-Treme GENE HP Transfection Reagent (Roche, German) according to the manufactory protocol. Firefly and Renilla luciferase activity was measured by dual-luciferase assay kit (Promega) on a Wallac Victor2 plate reader (Perklin, Finland) and data were normalized to Renilla activity 24 hours after the transfection. All results from 3 independent experiments were performed in 6 repeats and are presented as mean ± s.e.m. (n=3).
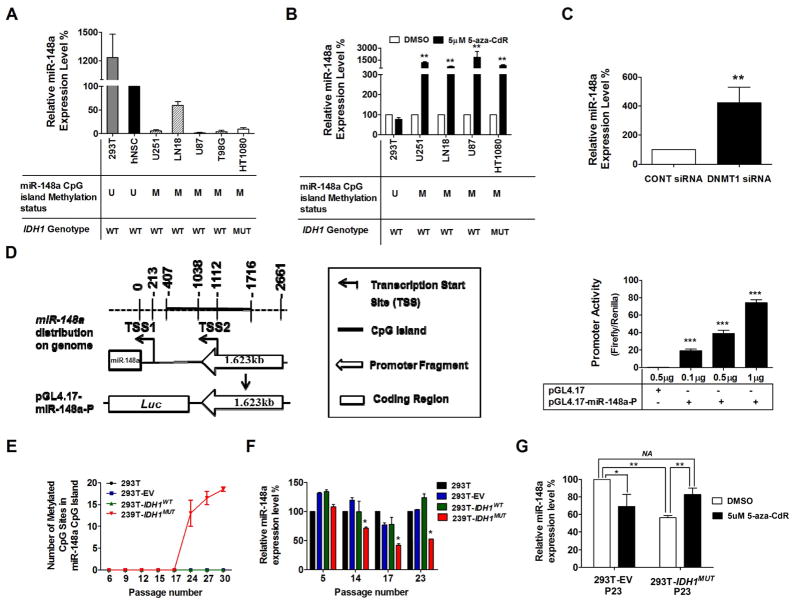
Figure 3. miR-148a is methylated and downregulated in glioma cell lines and upregulated following treatment with DNA demethylation agent or DNMT1-siRNA.
A and B, TaqMan RT-PCR analysis of miR-148a expression in HEK293T cells, human neural stem cells (hNSC), glioma cell lines (U251, LN18, U87 and T98G) and fibrosarcoma cell line (HT1080) without (A) or with (B) 5-aza-CdR treatment for 72 hours. miR-148a expression was determined by TaqMan qPCR. Data were standardized to the mean value of hNSC (A) or DMSO (B), which were set as 100%. C, miR-148a expression level in U251 cells transfected with DNMT1-siRNA was determined by TaqMan qPCR. D, left, 1.623 kb DNA fragment localized at the upstream of predicted miR-148a TSSs (−1038~−2661) was cloned into pGL4.17 vector as shown in the scheme; right, promoter activity of miR-148a CpG island was measured as the mean value of Firefly/Renilla luciferase activity ± SD (n=4). E, total number of methylated CpG sites determined by targeted BiSEQ of region 1 and 2 of miR-148a CpG island in parent HEK293T cells (parental 293T), empty vector control HEK293T cells (293T-EV), IDH1WT expressing HEK293T cells (293T-IDH1WT) or IDH1MUT expressing 293T cells (293T-IDH1MUT). Data shown represents mean value from 2 independent clones for each construct. F and G, miR-148a expression levels were measured in parent 293T cells, 293T-EV cells, 293T-IDH1WT and 293T-IDH1MUT. Results were standardized to the mean value for parent 293T cells in each passage (F) or the mean value for 293T-EV treated with DMSO (G), which was set as 100%. Each bar compared to HEK293T-EV treated with DMSO.*, P<0.05; **, P<0.01.
Stably overexpression of IDH1 mutant protein in HEK293T cells
Isogenic HEK293T cell lines stably overexpressing mutant IDH1 (R132H), wild-type IDH1 and empty vector (EV) were generated by using retrovirus constructs for IDH1WT or IDH1R132HMUT as previous description (23).
Transient and stable transfection of miR-148a
miR-148a mimics and negative control mimics were purchased from Ambion (Austin, TX) and transient transfection (30uM microRNA mimics) was performed using siPORT™ NeoFX™ transfection region according to the manufactory’s instruction. Stable overexpression of miR-148a was performed using Lentivirus constructs for miR-148a. The packaging of lentivirus was conducted by using Lentivector Expression System (SBI, Mountain View, CA) according to the manufactory’s instruction.
DNMT1 knockdown
Silencer select® DNMT1 siRNA (10nM) and negative control siRNA (10nM) were purchased from Ambion (Carlsbad, CA) and transfected by using Lipofectamine® RNAiMAX (Invitrogen) according to the manufactory’s instruction at day 1, day 3 and day 5 after seeding into 24-well plates. Total RNA was isolated 48 hours after the last transfection.
MTT assay
Cells were seeded into 24-well plates. Cell viability was determined by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma) assay as previously described (23). Absorbance at 535 nm was measured on a Wallac Victor2 plate reader (Perklin, Finland) with a background reference filter at 660 nm. All results from 3 independent experiments were performed in 6 repeats and are presented as mean ± s.e.m. (n=3).
Soft agar assay
Growth in soft agar was measured by colony assay as previously described (23). Briefly, 2 ml under-layers of 0.6% agar medium was prepared in 6-well plates by combining equal volumes of 1.2% Noble agar (Fisher, Fair Lawn, NJ, USA) and DMEM with 20% fetal bovine serum. Cells (2 × 103) were plated in 0.3% agar medium and cultured for 2–3 weeks. Colonies were then photographed, and colonies larger than 50 μm in diameter were counted in 5 random microscopic fields and tabulated. All results from 3 independent experiments were performed in 3 repeats and are presented as mean ± s.e.m. (n=3).
Colony formation assay
One thousand cells were seeded onto 60-mm dishes and incubated in the tissue culture incubator for 14 days. Cells were fixed with methanol and stained with 0.25% crystal violet. Colonies containing >50 cells were counted under a dissecting microscope. The results are reported as a percentage of the colonies in untreated cultures of each corresponding clone. All results from 3 independent experiments were performed in 6 repeats and are presented as mean ± s.e.m. (n=3).
Wound-healing assay
Wound-healing assay was performed previously described (23), cells were plated in 6-well plates coated with 0.1% gelatin to create a confluent monolayer. The scratch wound was observed using contrast microscopy (Olympus, IX41), and images were taken 0 h, 6 h, 9 h, 12 h, 24 h and 36h after the initial scratch. Scratch wound distance was quantitated using Adobe Photoshop software. All results from 3 independent experiments were performed in 6 repeats and are presented as mean ± s.e.m. (n=3).
FACS analysis
Cells were dissociated with Trypsin and washed twice in ice cold PBS before fixed with 70% ethanol/PBS at 4°C overnight. Cells were washed twice in ice cold PBS and resuspended in staining solution (0.1% TritonX-100, 0.01% Propidium iodide 0.002% Ribonuclease A in PBS) for at least 30 min on ice. Cells were gone through a filter and placed in a tube before measuring with FACS can analyzer (Becton Dickinson) in Flow Cytometry Core Facility at UCLA. All results from 3 independent experiments were performed in 6 repeats and are presented as mean ± s.e.m. (n=3).
Xenograft model
All animal experiments were approved by the University of California at Los Angeles Institutional Animal Care and Use Committee. Two groups of male NOD-scid mice (6–8 weeks old) were used for either subcutaneous injection or intracranial injection as follows: (i) U251-EV cells (empty vector control) (n = 6); and (ii) U251-miR-148a cells (stably transfected with miR-148a) (n = 6). For the subcutaneous injection, mice were anesthetized and injected with 106 glioma cells in 100 ul 10% matrix gel/PBS into the right flank. Tumor volume was monitored over time as determined from calculation (0.4 × width2 × length) using two perpendicular measurements with a caliper. Mice were killed 30 days after injection, and tumors from both groups were excised and weighed. The mean volume or tumor mass SEM was calculated. For the stereotactic intracranial injection, the surgical site was shaved and prepared with 70 % ethyl alcohol. A midline incision was made and a 1 mm diameter right parietal burr hole, centered 2 mm posterior to the coronal suture and 2 mm lateral to the sagittal suture, was drilled. Mice were placed in a stereotactic frame and 2.5 × 105 glioma cells in 2 μL 10% matrix gel/PBS were intracranially-injected with a 26-gauge needle at a depth of 3 mm. The needle was removed and the skin incision was closed by holding the edges with forceps and applying one clip. Mice were monitored daily and killed 60 days after injection. Survival was estimated using the Kaplan-Meier method, and comparisons between groups were performed using the log–rank test. The presence of macroscopic brain metastases was analyzed by H&E tissue staining.
Statistical analysis
Differentially methylated CpG islands located in miRNA promoter regions were identified by performing a Student’s T-test between IDH1/2MUT and IDH1/2WT samples. To reduce the number of false positive results a minimum threshold of 0.2 was set for the difference of means between IDH1/2MUT and IDH1/2WT samples. In addition, to control for multiple testing, we set a significance threshold of Q≤.05, evaluated per Storey et al (25). All other quantitative comparisons were done using the Student’s t-test, the Wilcoxon test (non-parametric paired analysis), and the Mann-Whitney U test (non-paired analysis). Qualitative variables were analyzed using the Chi-Square test and Fisher’s exact test. All tests were two-sided, and a P-value of less than 0.05 was considered statistically significant. The other in vitro and in vivo data were analyzed using the GraphPad Prism 5.0 statistical software (CA, US). Quantitative variables were analyzed using Student’s test, Wilcoxon test (nonparametric paired analysis), and Mann-Whitney U test (nonpaired analysis). Qualitative variables were analyzed using χ2 test or Fisher’s exact test. A two-sided P-value of <0.05 was regarded as significant.
Results
Identification of hypermethylated CpG islands associated with miRNAs in gliomas harboring the IDH1 mutation
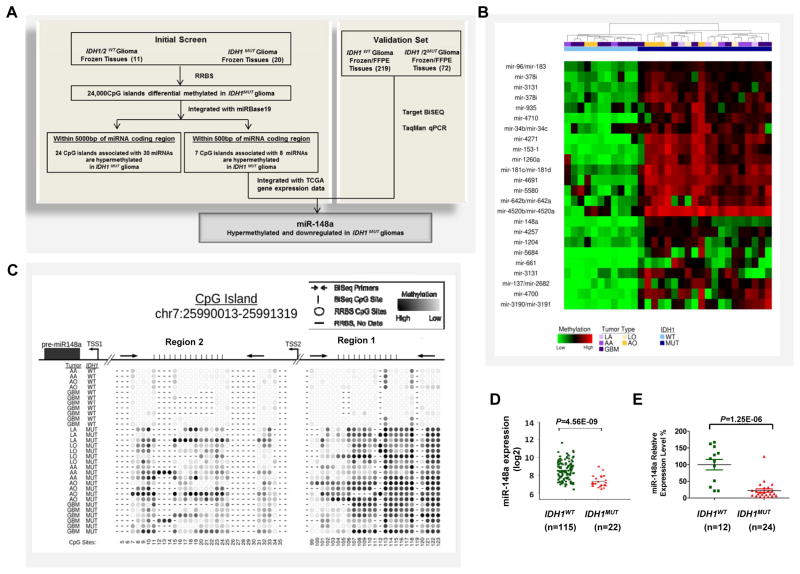
To identify hypermethylated CpG islands proximally associated with miRNAs in IDH1MUT gliomas, we performed RRBS as previously described (6) to profile methylomes of 11IDH1/2WT and 20 IDH1MUT patient glioma tissues (Fig. 1A). This included 5 IDH1/2WT and 5 IDH1MUT samples previously reported (6). The clinical characteristics of these 31 patients are listed in Supplementary Table S1, and the coverage details of the RRBS methylation data are shown in Supplementary Table S2. Overall, we generated methylation data on approximately 2.1 million CpG sites per sample covering 22,646 CpG islands, with an average of 56.9% CpG sites covered per island and at least 5x oversampling. Unsupervised comparison of RRBS clearly demonstrates G-CIMP within IDH1MUT gliomas (data not shown). By mapping the most current list of ~1600 miRNAs (miRBase 19) (26) to known CpG islands (27), we found that 605 CpG islands were located within 5000 base pairs upstream or downstream of 491 pre-miRNA regions, and of these, 216 CpG islands were located within 500 base pairs upstream or downstream of 231 pre-miRNA regions. RRBS covered 510 CpG islands within 5000 base pairs of 430 miRNAs and 182 CpG islands within 500 base pairs of 198 miRNA (Supplementary Table S3). For each sample, a methylation score for each CpG island was calculated as previously described (6). Using an absolute difference in CpG island methylation greater than 20%, a maximum mean methylation of 25% among hypomethylated samples, and a significance value of Q≤0.05 (25), we found that 24 CpG islands within 5000 base pairs of pre-miRNA regions, associated with 30 miRNAs, were significantly hypermethylated in IDH1MUT tumors; 7 CpG islands within 500 base pairs of pre-miRNA regions, associated with 8 miRNAs (miR-148a, miR-935, miR-137, miR-2682, miR-4520a, miR-4520b, miR-3131, and miR-4710), were significantly hypermethylated in IDH1MUT tumors (Fig. 1B, Table 1 and Supplementary Table S4). Among them, the miRNA-148a associated CpG island, spans a 1.3 kb region between −407 ~−1716 upstream of the pre-miR-148a region.
Figure 1. Discovery of hypermethylated miRNAs in glioma CpG Island Methylator Phenotype (G-CIMP).
A, schematic strategy used to identify target miRNAs in G-CIMP. B, unsupervised hierarchical clustering of differentially methylated CpG islands (P<0.05, unpaired t-test) that were identified by comparing IDH1MUT (MUT) and IDH1WT (WT) glioma patient samples using Reduced Representative Bisulfite Sequencing (RRBS). All CpG islands within 5000 bp of pre-miRNA regions are shown. C, methylation profile of the miR-148a associated CpG island via RRBS. Top panel, map of the miR-148a CpG island (chr7:25990013-25991319), position of pre-miR-148a region (black box, chr7:25989539-25989606), predicted putative transcription start sites (TSSs) and PCR products used for bisulfite sequencing in region 1 and 2 (Black arrow); bottom panel, representative CpG site methylation pattern of IDH1WT gliomas or IDH1MUT gliomas determined by RRBS. D, differential expression data for miR-148a in IDH1WT and IDH1MUT GBM from The Cancer Genome Atlas (TCGA) dataset. E, TaqMan qPCR analysis of relative miR-148a expression in IDH1WT and IDH1MUT glioma tissue samples from validation cohort. Data are standardized to the mean value for IDH1WT samples, which was set as 100%.
Table 1.
Hypermethylated CpG islands at least partially within 500 bp of miRNAs in IDH1MUT versus IDH1/2WT gliomas (G-CIMP).
| miRNA ID | Associated CpG Island Location | RRBS Methylation | TCGA Gene Expression | TaqMan qPCR | ||||
|---|---|---|---|---|---|---|---|---|
| chr | Start-end | Diff. of Mean (WT-MUT) | P-value | Fold change (WT/MUT) | P-value | Fold change (WT/MUT) | P-value | |
| miR-148a | 7 | 25990012_25991320 | −0.24 | 1.30E-10 | 2.44 | 2.40E-09 | 4.50 | 1.26E-06 |
| miR-935 | 19 | 54485303_54486322 | −0.29 | 1.32E-05 | NA | NA | 1.90 | 0.481 |
| miR-137 | 1 | 98510966_98511710 | −0.38 | 6.52E-08 | 1.09 | 0.44 | 2.66 | 0.22 |
| miR-2682 | 1 | 98510966_98511710 | −0.38 | 6.52E-08 | NA | NA | NA | NA |
| miR-4520b | 17 | 6558221_6558441 | −0.61 | 1.23E-07 | NA | NA | NA | NA |
| miR-4520a | 17 | 6558221_6558441 | −0.61 | 1.23E-07 | NA | NA | NA | NA |
| miR-3131 | 2 | 219922154_219923130 | −0.32 | 1.79E-09 | NA | NA | Undetectable in all samples | -- |
| 219923669_219923911 | −0.30 | 3.18E-05 | ||||||
| miR-4710 | 14 | 105144247_105144503 | −0.42 | 4.60E-10 | NA | NA | 0.03 | 0.262 |
RRBS: Reduced Representative Bisulfite Sequencing; TCGA, The Cancer Genome Alta
Integrated analysis of expression and methylation identifies miR-148a as a hypermethylated and silenced miRNA in IDH1MUT gliomas
Based on the notion that functionally important hypermethylated miRNAs should display decreased expression via epigenetic silencing, we integrated miRNA expression data from The Cancer Genome Atlas (TCGA) (http://tcga-data.nci.nih.gov/tcga/) with our RRBS methylation data to determine methylated miRNAs with down-regulated expression. To do so, we calculated mean miRNA expression levels in IDH1MUT (n=22) versus IDH1WT (n=115) tumors (n.b. IDH2 status not available). The TCGA miRNA microarray analysis provided data for 2 of the 8 miRNA candidates with hypermethylated CpG islands within 500 base pairs of their precursor region (Table 1); therefore, we randomly selected 3 of the remaining 6 possible miRNA candidates (miR-935, miR-3131 and miR-4710) and performed expression analysis using TaqMan qPCR on a set of glioma tissue samples. Among these five candidate miRNAs with expression data, only miR-148a showed significantly reduced expression in IDH1MUT tumors compared with IDH1WT tumors (Table 1, Fig. 1D and E).
To confirm the DNA methylation status of the miR-148a associated CpG island in a larger validation set, we performed direct targeted bisulfite sequencing (BiSEQ) in 17 autopsy normal brain tissue samples, 219 IDH1WT and 72 IDH1/2MUT glioma tissue samples (including 2 IDH2MUT samples) (Supplementary Table S5). The miR-148a associated CpG island is located on chromosome 7 (chr7: 25990012-25991320) (28), 407 base pairs upstream of pre-miR-148a region (−407~−1716), and contains 123 CpG sites and 2 predicted transcription start sites (TSS) (TSS1: −213; TSS2: −1038) (http://www.cbs.dtu.dk/services/Promoter/) (28). ~90 of 123 CpG sites were covered by RRBS (Supplementary Fig. S1). The representative CpG island methylation pattern of miR-148a characterized by RRBS, along with the localization of predicted TSSs and PCR products for BiSEQ, is shown in Fig. 1C. Overall, we found that miR-148a was hypermethylated in Region 1 in 0% (0 out of 17) normal brain tissues samples, 5.4 % (12 out 219) IDH1WT glioma tissue samples, and 97.2%(70 out of 72) IDH1/2MUT glioma tissue samples (Supplementary Table S5). To validate that differential methylation was also present in Region 2 of the miR-148a CpG island, we performed bisulfite sequencing in Region 2 on a subset of patient tissue samples and obtained similar results as in Region 1 (Supplementary Table S6). These results show that miR148a promoter hypermethylation is associated with IDH1/2 mutation within all grades and histology.
To confirm that methylation was associated with decreased expression in our samples as observed in the TCGA dataset, we analyzed the expression of miR-148a in the available frozen glioma tissues within the BiSEQ validation set by using TaqMan qPCR. We found that miR-148a expression level was downregulated in IDH1MUT gliomas in tight correlation with its CpG island methylation status (Fig. 1E).
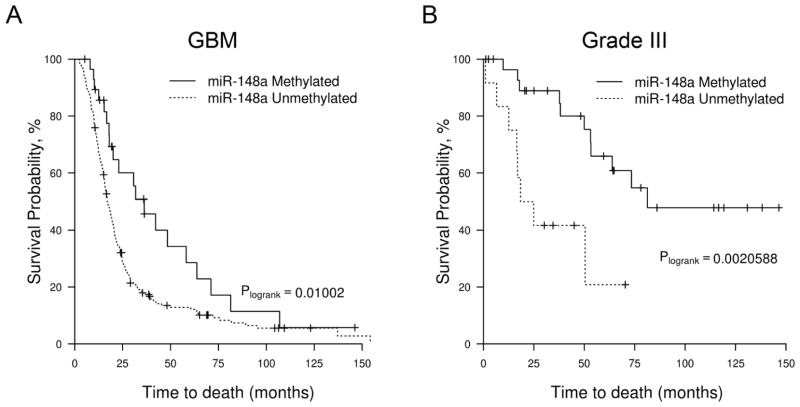
miR-148a methylation is prognostic of increased survival in malignant gliomas
To determine if miR-148a methylation can be used as a prognostic biomarker, we correlated miR-148a methylation with overall survival in the 224 primary GBM patients and 42 grade III glioma patients in our cohort who had treatment-naïve tumor samples (Supplementary Table S7). In the GBM patients, miR-148a-Unmethylated patients (n = 195) had decreased median OS of 17.0 months vs 36.3 months (log-rank, P= 0.01) (Fig. 2A). By univariate Cox analysis, the miR-148a-Unmethylated GBM patients had a hazard ratio =1.82 (confidence interval=1.14 to 2.90, PCox=0.011) (Supplementary Table S8). In the grade III glioma patients, miR-148a-Unmethylated patients (n=12) had a decreased median OS of 21.6 months vs 81.3 months (log-rank, P=0.002) (Fig. 2B); for the miR-148a-Unmethylated patients, the hazard ratio =4.28 (confidence interval=1.58 to 11.59, PCox=0.004) (Supplementary Table S8). Using multivariate analysis with age, sex, performance status, and extent of resection, miR-148a-Unmethylation was confirmed as an independent prognostic marker in grade III glioma (HR=5.83, confidence interval=1.47 to 23.08, PCox =0.012) and trended towards being an independent prognostic marker in GBM (HR=1.59, confidence interval= 0.98 to 2.6, PCox=0.059) (Supplementary Table S8). Similar to our previous finding, IDH1/2 mutational status was an independent prognostic marker in an overlapping dataset (Supplementary Fig. S2) (6). However, when miR-148a-Methylated patients were stratified by IDH1/2 status, the methylated IDH1/2WT patients (GBM: n=10; grade III: n=2) had similar poor survival to miR-148a-Unmethylated IDH1WT patients (data not shown). This suggests that miR-148a methylation is unlikely to add to IDH1/2 mutational status in prognostication.
Figure 2. The methylation status of miR-148a is prognostic in GBM and Grade III gliomas.
A, Kaplan–Meier overall survival analysis of GBM patients in the validation cohort (n=224). Survival among miR-148a-Methylated (n=29, solid line) and miR-148a-Unmethylated (n=195, dotted line) patients is shown. B, Kaplan–Meier overall survival analysis of Grade III glioma patients in the validation cohort (n=42, solid line). Survival among miR-148a-Methylated (n=30) and miR-148a-Unmethylated (n=12, dotted line) patients is shown.
Downregulated miR-148a associated with promoter CpG island hypermethylation can be re-expressed by either pharmacologic or genetic demethylation
To provide additional evidence that CpG island promoter methylation of miR-148a is responsible for silenced expression, we evaluated methylation and expression of miR-148a in a variety of cell lines including glioma cell lines (U251, LN18, T98G and U87), human neural stem cells (hNSC), human HT1080 fibrosarcoma cells (IDH1MUT) and human HEK293T epithelia kidney cells. We found that miR-148a was hypermethylated in all of the glioma and HT1080 cells, and hypomethylated in hNSC and HEK293T cells. These results indicate that miR-148a hypermethylation developed in the IDH1/2WT glioma cell lines by mechanisms independent of IDH1/2 mutant protein expression. Next, we analyzed the expression of miR-148a in the cell lines by using TaqMan RT-PCR, and found, as expected, that there was silencing of expression in the methylated cell lines (Fig. 3A). To determine whether pharmacologic demethylation could restore miR-148a expression in hypermethylated cell lines, we treated the cells with 5-aza-CdR and observed upregulation of miR-148a in glioma cell lines with hypermethylated miR-148a, but not in HEK293T or hNSC cells with hypomethylated miR-148a (Fig. 3B). To confirm that 5-aza-CdR treatment resulted in demethylation, we used the same conditions as in Fig. 3B (5 μM 5-aza-CdR treatment for 4 days) and found that there was qualitatively a slight decrease in methylation using Methylation specific PCR (MSP). We followed up this experiment using prolonged low dose 5-aza-CdR treatment based on published reports (29) and found that 0.625 μM 5-aza-CdR (9 days treatment) resulted in dramatically decreased demethylation using both BiSEQ (Fig. S3. A) and MSP (Fig. S3.B). Significant increase of miR-148a expression in the long term 5-aza-CdR treated U251 cells was also detected by TaqMan qPCR (Fig. S3. C). These results provide evidence that 5-aza-CdR increases expression of miR-148a via demethylation but do not rule out the possibility of methylation-independent effects of 5-aza-CdR. Consistent with these results, we found that knockdown of DNA (cytosine-5-)-methyltransferase 1 (DNMT1) using siRNA also resulted in upregulated expression of miR-148a in U251 glioma cells (Fig. 3C, Supplementary Fig. S4). Lastly, using a luciferase reporter assay conducted in HEK293T, we confirmed that a 1623bp long fragment upstream of pre-miR-148a gene (−1038~−2661) overlapping the CpG island (−407~ −1716) possesses promoter activity (Fig. 3D). Taken together, these results strongly suggest that the expression of miR-148a can be silenced through promoter CpG island methylation in glioma.
Overexpression of IDH1 mutant protein in HEK293T cells results in the hypermethylation and silencing of miR-148a
As shown in Fig. 3A, all glioma derived cell lines demonstrate miR-148a hypermethylation, and therefore cannot be used to demonstrate causality between IDH1 mutation and miR-148a hypermethylation. Therefore, to provide evidence linking hypermethylation of miR-148a to IDH1 mutant protein expression, we generated isogenic HEK293T cell lines stably overexpressing mutant IDH1 (R132H), wild-type IDH1 and empty vector (EV) (Fig. S5). By monitoring methylation longitudinally using targeted bisulfite sequencing, we detected miR-148a promoter CpG island hypermethylation in HEK293T-IDH1R132HMUT after passage 17 but not in the parent HEK293T cells, HEK293T-EV cells or HEK293T-IDH1WT cells assayed in parallel at the same passage (Fig. 3E). TaqMan RT-PCR analysis revealed that the expression of miR-148a was downregulated in HEK293T-IDH1R132HMUT cells as well (Fig. 3F), and that the expression level of miR-148a in later passage (p23) HEK293T-IDH1R132HMUT cells could be restored by demethylation treatment with 5-aza-CdR (Fig. 3G). Interestingly, we observed downregulation of miR-148a in HEK293T cells treated with 5-aza-CdR in both Fig. 3B (parental HEK293T cells) and Fig. 3G (stable HEK293T clone expressing vector only or IDH1R132H), indicating that this is a common observance with HEK293T when treated with 5 μM 5-aza-CdR. Although the cause of this effect is unknown, we speculate that it may reflect direct toxicity.
In order to determine whether miR-148a overexpression could prevent the establishment of the hypermethylator phenotype induced by mutant IDH1 (R132H), we stably overexpressed miR-148a in an early passage of HEK293T stable cell lines (passage 6) which were transfected with a vector control, IDH1WT or IDH1R132H respectively. The following stable cell lines were generated and cultured for an additional 20 passages: HEK293T-Control, HEK293T-IDH1WT, HEK293T-IDH1R132H, HEK293T-Control+miR148a, HEK293T-IDH1WT+miR148a, HEK293T-IDH1R132H+miR148a. The overexpression of miR-148a was monitored by GFP fluorescence and confirmed by TaqMan real-time PCR (Fig. S6A). By using targeted bisulfite sequencing (BiSEQ), we observed that G-CIMP genes, including RBP1 and miR-148a, remained unmethylated in passage 6 of HEK293T-Control, HEK293T-IDH1WT, and HEK293T-IDH1R132H stable cell lines, which confirmed our previous finding. At passage 26, RBP1 and miR-148a remained unmethylated in the HEK293T-Control and HEK293T-IDH1WT cell lines regardless of the overexpression of miR-148a, however, hypermethylation occurred in the HEK293T-IDH1R132H cell line in the context of overexpression of miR-148a (Fig. S6B and C). Interestingly, by using real time PCR, we were able to detect the increased expression of RBP1 in HKE293T-IDH1R132H cells when miR-148a was overexpressed (p26), this suggests that partial reduction in methylation of the RBP1 CpG island may be achieved although not detectable using BiSEQ method (data not shown).
miR-148a shows tumor suppressive features in glioma cell lines in vitro and in vivo
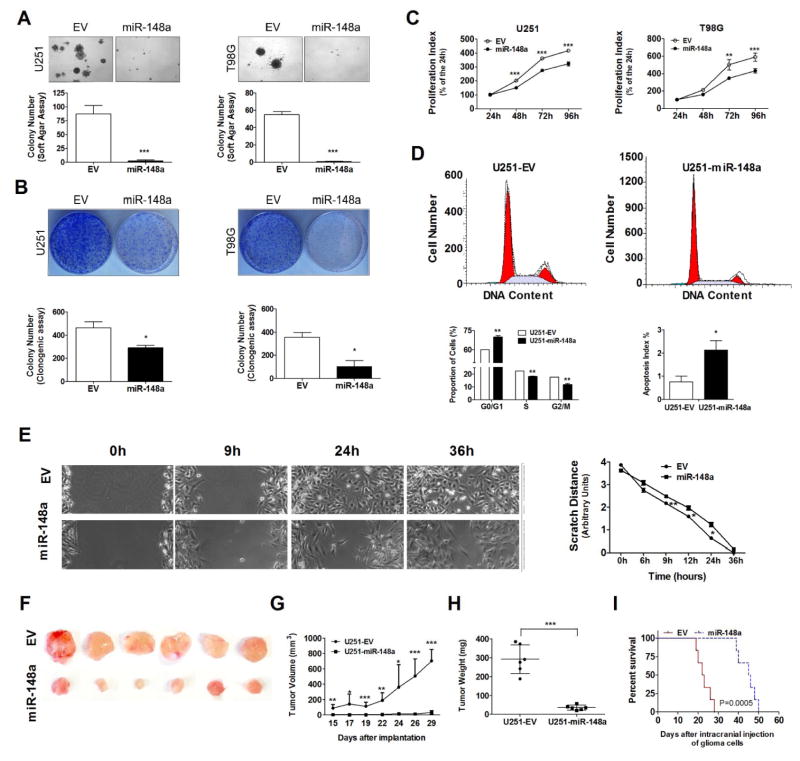
To examine the biologic effects of miR-148a in glioma cell lines, we overexpressed miR-148a in 2 glioma cell lines (U251 and T98G, previously determined to have basally methylated miR-148a and low miR-148a expression (Fig. 3A) and compared cell growth, migration, cell cycle and apoptosis. The efficiency of the transfection was determined by measuring the mature miRNA levels by TaqMan qPCR (Supplementary Fig. S7) and monitored by GFP fluorescence (data not shown). The miR-148a expression level relative to that of hNSC was 8~9-fold, raising the possibility that supra-physiologic miR-148a levels were achieved. We found that overexpression of miR-148a inhibited the anchorage-independent growth in soft agar (Fig. 4A), colony formation ability (Fig. 4B) and cell proliferation (Fig. 4C) of U251 and T98G glioma cells as compared to control cells. Using FACS analysis, we found that overexpression of miR-148a resulted in significant G0/G1 cell cycle arrest and a distinct decrease of S-phase cells in U251 and T98G cells as compared to control cells (Fig. 4D, Supplementary Fig. S8), and we also observed 2.8 fold (P<0.05) increase in apoptosis in miR-148a overexpressed U251 cells (Fig. 4D). Using a wound healing assay, we measured the migration capability of the miR-148a transfected U251 cells compared with control cells and found that overexpression of miR-148a inhibited cell migration (Fig. 4E).
Figure 4. miR-148a inhibits the proliferation and migration of glioma cells.
In vitro cell growth of miR-148a overexpressing glioma cells (U151 and T98G) are determined by soft agar assay (A), colony formation assay (B) and MTT assay (C); cell cycle fraction and apoptosis index are determined by FACS assay (D); cell migration ability is determined by monolayer cell migration scratch assay in a non-FBS culture condition as shown in representative phase micrographs (E); in vivo tumorigenesis of miR-148a overexpressing glioma cells (U151) is determined in NOD-scid mouse subcutaneous xenograft model (F) by measuring of tumor size (G) and weight (H), or in intracranial xenograft model by using Kaplan-Meier overall survival (OS) analysis (I).*, P<0.05; **, P<0.01; ***, P<0.001.
To confirm these tumor suppressive properties of miR-148a in vivo, we tested whether miR-148a could reduce tumorigenicity in both subcutaneous and intracranial glioma xenograft models. When U251 cells stably overexpressing the control empty vector (U251-EV control cells) or pMIF-GFP-zeo-miR-148a vector (U251-miR-148a cells) were injected s.c. in to the flank of NOD-scid mice, we found that the miR-148a overexpressing cell line showed much less tumor growth than that seen with control cells (Fig. 4F–G). When animals were sacrificed at 30 days after injection we found that the tumor derived from the U251-miR-148a cells were significantly smaller than those derived from U251-EV control cells (Fig. 4H). To confirm these results in an intracranial xenograft model with overall survival (OS) as the primary endpoint, we injected U251-EV control cells or U251-miR-148a cells in to the brains of NOD-scid mice. Using Kaplan-Meier analysis, we found that the median overall survival of mice injected with U251-miR-148a cells (45.5 days) was greater than those of mice injected with U251-EV control cells (22.5 days) (log rank, P=0.0005) (Fig. 4I).
miR-148a directly suppresses DNMT1 expression in glioma resulting in re-expression of silenced G-CIMP genes
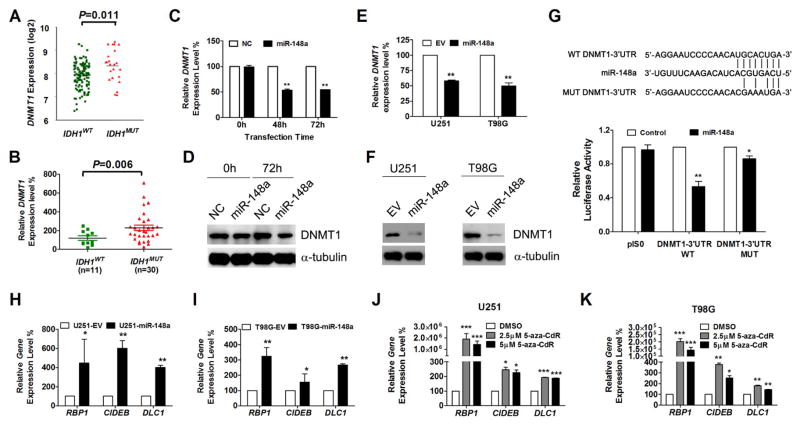
To investigate miR-148a target genes that could mediate observed tumor suppression, we identified candidate target genes by combining 4 well recognized miRNA databases including Target Scan (www.targetscan.org) (30), microT-CDS (DianaTools/index.php?r=microtv4/index.), microRNA (www.microrna.org) and miRDB (www.mirdb.org), and refined the candidate gene list by annotating these genes with TCGA gene expression data (http://tcga-data.nci.nih.gov/tcga/) analyzed for mean expression difference between IDH1MUT and IDH1WT tumors. We found 16 genes present on all four lists that were downregulated in IDH1MUT gliomas based on analysis of the TCGA gene expression database; from these genes, we selected DNMT1 for additional study (Supplementary Table S9). Consistent with this finding, it has been shown that miR-148a was involved in the regulation of DNA methylation by targeting DNMT1 in breast cancer (31), gastric cancer (32) and lupus CD4+T cells (33). Therefore, to confirm the relationship between DNMT1 expression and miR-148a level in glioma, we examined the expression level of DNMT1 in glioma patient tissue samples. We confirmed that the expression of DNMT1 was slightly upregulated in IDH1MUT tissue samples (Fig. 5A and B), suggesting that there are miR-148a independent regulators of DNMT1 expression. No apparent differences in DNMT1 level was observed amongst IDH1MUT samples separated by grade (data not shown). To determine whether miR-148a downregulates the expression of DNMT1 in glioma cells, we transiently transfected U251 glioma cells with miR-148a mimics or negative control mimics, and measured the mRNA and protein levels of DNMT1 by quantitative real-time PCR and Western blot, respectively. Our quantitative RT-PCR results showed that in U251 cells, transfection of miR-148a mimics resulted in 47% (P<0.01) reduction of endogenous DNMT1 mRNA level after 48 hours (Fig. 5C). Westernblot analysis confirmed that transfection of miR-148a mimics resulted in a reduction of DNMT1 protein expression after 72 hours (Fig. 5D). Additionally, downregulation of DNMT1 expression by miR-148a was also observed in the glioma cells (U251 and T98G) stably overexpressing miR-148a (Fig. 5E and F). By searching the Target Scan database (http://www.targetscan.org), we found that the 3′-UTR of DNMT1 contained one conserved site for miR-148a (34). To determine whether this region was required for miR-148a regulation, we cloned the DNMT1 3′ UTR (WT and MUT) in to the pIS0 reporter vector and performed dual luciferase assays in U251 cells, and observed a reduction of the DNMT1 3′ UTR-WT but not the MUT reporter gene expression when treated with miR-148a mimics (50nM) (Fig. 5G). This result was consistent with a previous study conducted in Hepa cells (35).
Figure 5. DNMT1 is the direct target of miR-148a in glioma.
A and B, the expression level of DNMT1 in IDH1WT and IDH1MUT GBM patients from the TCGA data set (A) and our validation patient cohort (B). C–F, DNMT1 expression was determined by qPCR (C and E) and Western Blot (D and F) in U251 cells transiently (C and D) and stably (E and F) overexpressing miR-148a. G, human DNMT1 3′-UTR region contain miR-148a binding sites and dural luciferase assays were conducted using DNMT1 wild type (WT) and mutant (MUT) 3′-UTR reporter constructs. H and I, gene expression of hypermethylated RBP1, CIDEB and DLC1 in glioma cell lines U251 (H) and T98G (I) overexpressing miR-148a was determined by qPCR. Data were standardized to EV which was set as 100%. J and K, qPCR analysis of gene expression level in U251 (J) or T98G (K) cells treated with DMSO or 5-aza-CdR for 72 hours. Data were standardized to vehicle control (DMSO) which was set as 100%. *, P<0.05; **, P<0.01; ***, P<0.001.
We next investigated the functional consequence of miR-148a regulated DNMT1 expression in terms of maintenance of hypermethylation and downregulation of other potentially important G-CIMP component genes. To do so, we selected and confirmed that representative G-CIMP genes (RBP1, DLC1 and CIDEB) with potential involvement in tumorigenesis were downregulated in IDH1MUT gliomas (Ref. 6 Fig. 3B and Supplementary Fig. S9) (6). Upon overexpression of miR-148a in U251 and T98G cells, we found that the mRNA levels of RBP1, DLC1 and CIDEB were upregulated compared to control cells (Fig. 5H and I). In addition, the expression of RBP1, CIDEB and DLC1 in U251 and T98G cells could be also restored by demethylation treatment with 5-aza-CdR (Fig. 5J and K). These results suggest that miR-148a can diminish G-CIMP hypermethylation via DNMT1 down-regulation.
Discussion
In this study, we expanded our previous RRBS characterization of G-CIMP (6) to examine CpG islands associated with miRNAs. We identified 7 hypermethylated CpG islands at least partially within 500 base pairs of 8 pre-miRNAs regions. This study represents the first recognition of miRNAs as part of G-CIMP and the first functional validation of tumor suppressive effects of a G-CIMP gene, miR-148a, in glioma.
We used the next generation sequencing technique, RRBS, to perform whole methylome profiling of IDH1MUT and IDH1/2WT patient glioma tissues. By annotating our CpG island sequence data with the latest microRNA database, we were able to identify hypermethylated microRNAs in IDH1MUT gliomas showing that G-CIMP is comprised of miRNAs in addition to coding genes. This observation was made possible by the adaptability of the RRBS dataset to annotation with previously unrecognized features. Another advantage of RRBS is conferred by the evaluation of contiguous CpG site coverage within CpG islands instead of sampling single CpG sites. However, because RRBS coverage relies on the location of naturally-occurring restriction sites, we were able to cover ~85% of all CpG islands associated with miRNAs; additionally, our annotation does not account for the possibility that remotely-located CpG islands can regulate miRNAs. Our list of hypermethylated miRNAs contains miRNAs previously found to be hypermethylated in glioma or other tumors. For example, miR-137 has been shown to be downregulated in high grade glioma possibly through DNA hypermethylation (36); miR-34b has been shown to be silenced by promoter hypermethylation in multiple myeloma (37); and hypermethylation and/or downregulation of miR-148a has been observed in multiple cancer types including gastric, pancreatic, lung, breast and colorectal cancer but until now, has not been reported in gliomas (31, 32, 38–40).
To screen for functionally important hypermethylated/downregulated miRNAs, we integrated gene expression data either from the TCGA GBM dataset or from our own targeted expression analysis in our tissue samples. We required that functionally important miRNAs show reduced mean expression in IDH1MUT versus IDH1WT GBMs. In doing so, we found that aside from miR-148a, none of the other four hypermethylated miRNAs with expression data demonstrated correlation with decreased expression. We did not perform expression analysis on the remaining three miRNAs. That miR-148a is the only 1 of 5 hypermethylated miRNAs demonstrating reduced expression in our screen led us to explore its functional significance. Other studies have shown that hypermethylated miRNAs are often not associated with reduced expression (41, 42). Aside from the obvious explanation that a hypermethylated CpG island is not involved in transcriptional regulation of the associated miRNA, another explanation is that the expression of miRNAs is too low to be detected or correlated with methylation data as seen with miR-3131 in this study. Alternatively, there may be compensatory restoration of miRNAs after hypermethylation. Lastly, correlation of methylation and expression pairwise in each sample, which was not possible in our study, may enable increased sensitivity of the expression filter, particularly in candidates that may be hypermethylated in only a subset of IDH1MUT samples. Therefore, the use of our ‘expression’ filter may eliminate consideration of hypermethylated miRNA that may have functional importance. Perhaps more importantly, each hypermethylated CpG island could retain prognostic or predictive biomarker value, independent of changes in expression.
Our results show that miR-148a possesses tumor suppressive properties and is epigenetically regulated in glioma. The CpG island hypermethylation of miR-148a and reduced expression has been reported in other types of cancers including lymph node metastatic cancer (40), gastric cancer (32), colon cancer (39), breast cancer (31). This convergence of various cancer types demonstrating epigenetic miR-148a silencing indicates that the likely importance of miR-148a dysregulation as a commonly employed tumor mechanism. To address the lack of detailed understanding of the transcriptional regulation of miR-148a, we demonstrated the promoter activity of a portion of the miR-148a CpG island and confirmed the strong inverse correlation between promoter CpG island hypermethylation and gene expression in glioma. However, determination of the DNA hydroxymethylation levels, the accessibility of miR-148a promoter to transcription factors, and the methylation/acetylation level of miR-148a promoter associated histones will be required to fully understand the mechanism of miR-148a inactivation in IDH1MUT gliomas.
We show that miR-148a methylation is prognostic of better outcome in newly diagnosed GBMs receiving standard chemo-radiation. In contrast, it has been suggested in advanced colorectal cancer that miR-148a methylation status correlated inversely with its expression, and was associated with poor overall survival (OS) in stage IV colorectal cancer (39). From our results, it does not appear that miR-148a methylation provides independent prognostication above IDH1/2 testing. In particular, there appears to be a ‘false’ positive rate in which hypermethylation is detected in ~5% IDH1/2WT glioma patients. Although small in number, these patients do not share improved survival seen in IDH1/2MUT patients. Further study is necessary to determine the correlation of methylation with expression in this subset, and it remains to be seen whether miR-148a expression level, itself, is prognostic either in IDH1/2WT or IDH1/2MUT gliomas.
Our experiments in glioma cells (U251 and T98G) clearly demonstrate the tumor suppressive features of overexpressed miR-148a in vitro and in vivo. These results are consistent with previous reports documenting tumor suppressor features of miR-148a in other cancer types (40). We utilized U251 and T98G cells after determining that all established GBM cell lines tested demonstrated miR-148a hypermethylation and downregulated expression despite absence of IDH1/2 mutation. These results, along with the finding of a small percentage of IDH1/2WT patient tumor samples that are miR-148a-methylated, indicates that miR-148a methylation can occur in the absence of IDH1/2 mutation and may be important for the growth advantages necessary for these tumors or cell lines. Nonetheless, the use of non-IDH1/2MUT glioma cell lines may limit our interpretation of miR-148a’s tumor suppressive function.
In order to identify miR-148a target genes involved in tumor suppressive effects, we took advantage of publicly available computational-based miRNA target prediction tools and the TCGA gene expression data, looking for target candidates whose expression was significantly upregulated in IDH1MUT glioma. We identified several miR-148a targets including E2F3 and ROCK1, with oncogenic features in other cancer types (40, 43). In this study, we selected DNA (cytosine-5)-methyltransferase 1 (DNMT1) for further investigation. DNMT1 is an enzyme considered to be the key maintenance CpG methyltransferase in mammals (44, 45). In other cancer types, miR-148a was silenced by hypermethylation and was found to interact with DNMT1 in gastric cancer and breast cancer (31, 32). In our study, we found the expression of DNMT1 was increased in IDH1MUT glioma and inversely correlated with miR-148a expression level. We found that overexpression of miR-148a directly downregulated DNMT1 expression by targeting its 3′-UTR and restored the expression of hypermethylated G-CIMP genes in glioma cells. Further experiments involving tandem overexpression of miR-148a and DNMT1 will be important to confirm the importance of this relationship. Our findings suggest that miR-148a is not only regulated by DNA methylation, but itself might be involved in the maintenance of G-CIMP by targeting DNMT1. Similarly, miR-29b was found to be involved in the regulation of DNA methylation by targeting DNA methylation machinery in multiple diseases including AML, lung cancer and lupus (46).
In order to determine whether miR-148a overexpression could prevent the establishment of hypermethylator phenotype induced by mutant IDH1 (R132H), we stably overexpressed miR-148a in early passage of HEK293T stable cell lines which had been transfected with vector control, IDH1WT or IDH1R132H respectively and monitored methylation after 20 passages. Based on this one timepoint, it appears that the hypermethylation induced by IDH1 mutant protein is not blocked by overexpression of miR-148a. It is also possible that the kinetics of hypermethylation may be altered which could be detected by monitoring development of hypermethylation over several timepoints. Nevertheless, these results are consistent with studies in which blockade of 2-HG generation by the mutant IDH1 enzyme could not reverse the G-CIMP (47) Overall, these results suggest that methylation of miR-148a is part of G-CIMP and not the cause, although its role in maintenance of G-CIMP remains unclear.
Further study is required to determine effects of miR-148a overexpression on global methylation profiles and whether G-CIMP genes preferentially undergo hypomethylation. In addition, the possibility remains whether miR-148a expression may have some impact on TET demethylase activity, whose inhibition by the IDH1/2 mutant product, 2-HG, is widely hypothesized to contribute to G-CIMP hypermethylation by (48, 49). However, by the searching microRNA target gene databases, none of the TET family (TET1, TET2 and TET3) genes were found to be targets of miR-148a or any of the miRNA candidates on Table 1 or Table S4. We also examined the expression level of TET family genes in miR-148a transient transfected U251 and T98G cells, and found that the expression level of TET1, TET2, and TET3 were nearly undetectable at baseline and transient transfection of miR-148a mimic did not alter TET family gene expression level (data not shown). This suggests that miR-148a might not be a direct factor that regulates TET family.
In this study, we identify miR-148a as a tumor suppressive miRNA component within G-CIMP. The ability of miR-148a to have similar tumor suppressive function across several cancer types indicates that miR-148a silencing may be a broadly employed mechanism. Based on our findings, we propose the following model for tumor promotion of IDH1 mutant gliomas (Fig. S10) in which IDH1 mutant protein promotes a hypermethylation phenotype that includes hypermethylation and silencing of miR-148a. In the poorer prognosis IDH1/2 WT gliomas in which miR-148a expression is not lowered, other potent tumorigenic pathways are likely to be activated. Silenced miR-148a results in upregulated DNMT1 levels, ordinarily suppressed by miR-148a, which contributes to maintenance of hypermethylated tumor suppressive genes within G-CIMP. Thus re-introduction of miR148a, in this DNMT1-dependent manner can lead to re-expression of tumor suppressors. As evident from the miR-148a target list, there could be direct oncogenic targets that have increased expression in the context of silenced miR-148a. Further experiments are necessary to substantiate this potential mechanism and its relative importance compared to downregulation of tumor suppressors via maintenance of G-CIMP. More importantly, further experiments are necessary to determine whether silencing of miR-148a itself can be transforming or tumor promoting. Ideally, better IDH1/2 mutant cellular model systems will be available to confirm our model. Our results provide a pharmacological rationale for the potential use of synthetic miR-148a mimics as an approach for the treatment of IDH1MUT gliomas. In addition, as demonstrated by glioma cell lines and rare IDHWT gliomas, there may be IDH1/2MUT independent mechanisms of miR-148a methylation that may also be benefitted by miR-148a restoration. Together with recent papers demonstrating this potential in IDH1MUT gliomas (46, 50), our results suggests possible therapeutic benefit of pharmacological DNMT1 inhibition by FDA approved agents to achieve miR-148a re-expression in low miR-148a expressing glioma.
Supplementary Material
Statement of Translational Relevance.
IDH1/2mutant gliomas harbor a distinct CpG island methylation phenotype (G-CIMP) hypothesized to play a role in the development and progression of secondary pathway gliomas by silencing tumor suppressive genes. Although various protein-coding genes have been characterized as G-CIMP members, non-coding miRNAs have not yet been recognized within G-CIMP. More importantly, the functional validation of the G-CIMP genes, whether coding or non-coding, contributing to glioma progression and DNA methylation deregulation is still lacking. The identification and characterization of hypermethylation silenced miRNAs within G-CIMP could therefore contribute to a better understanding of the underlying mechanisms of the development and progression of secondary pathway gliomas, and provide a pharmacological rationale for the potential use of demethylating agents and synthetic miR-148a mimics as approaches for the treatment of IDH1 mutant gliomas.
Acknowledgments
The authors thank the support of UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards CA-16042 and AI-28697; Dr. Olga Aprelikova for the pMIF-GFP-zeo-miR-148a and pMIF-GFP-zeo constructs; Dr. Soraya Yekta (Cambridge, MA) for pIS0 plasmid; Dr. Paul S Mischel for glioma cell lines; Drs. Andreas von Deimling and Jörg Balss for helping with enzymatic assay for quantitative analysis of D-2-hydroxyglutarate (D-2-HG); Silicon Med for clinical data aggregation; UCLA Brain Tumor Translational Resource for logistical and technical support. This study is supported by National Institutes of Health (K08CA124479 and R01CA179071 to A.L.), Art of the Brain Foundation, University of California Cancer Research Coordinating Committee, American Brain Tumor Association Basic Research Fellowship (to S.L. and A.C.).
Footnotes
Disclosure of Potential Conflicts of interest: No potential conflicts of interest were disclosed.
References
- 1.Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY. Primary brain tumours in adults. Lancet. 2003;361:323–31. doi: 10.1016/S0140-6736(03)12328-8. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:764–72. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 3.Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4482–90. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou AP, Chowdhury R, Li S, Chen W, Kim AJ, Piccioni DE, et al. Identification of retinol binding protein 1 promoter hypermethylation in isocitrate dehydrogenase 1 and 2 mutant gliomas. Journal of the National Cancer Institute. 2012;104:1458–69. doi: 10.1093/jnci/djs357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan CG, Barwick BG, Jin G, Rago C, Kapoor-Vazirani P, Powell DR, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome research. 2012;22:2339–55. doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirozzi CJ, Reitman ZJ, Yan H. Releasing the block: setting differentiation free with mutant IDH inhibitors. Cancer cell. 2013;23:570–2. doi: 10.1016/j.ccr.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature protocols. 2011;6:468–81. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and cellular biology. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & development. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Molecular neurobiology. 2013;47:131–44. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiota K, Takahashi M, Masaki T, Sudo K. Calcium metabolism, serum thyroid-stimulating hormone, prolactin, and growth hormone in spontaneously hypercholesterolemic rats. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1989;190:229–33. doi: 10.3181/00379727-190-42853. [DOI] [PubMed] [Google Scholar]
- 20.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–34. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 21.Mazar J, DeBlasio D, Govindarajan SS, Zhang S, Perera RJ. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS letters. 2011;585:2467–76. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Molecular cancer. 2010;9:167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Chou AP, Chen W, Chen R, Deng Y, Phillips HS, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro-oncology. 2013;15:57–68. doi: 10.1093/neuonc/nos261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalezari S, Chou AP, Tran A, Solis OE, Khanlou N, Chen W, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro-oncology. 2013;15:370–81. doi: 10.1093/neuonc/nos308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–98. [Google Scholar]
- 26.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic acids research. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardinergarden M, Frommer M. Cpg Islands in Vertebrate Genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 28.Knudsen S. Promoter 2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–61. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- 29.Borodovsky A, Salmasi V, Turcan S, Fabius AW, Baia GS, Eberhart CG, et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget. 2013;4:1737–47. doi: 10.18632/oncotarget.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu A, Xia J, Zuo J, Jin S, Zhou H, Yao L, et al. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Medical oncology. 2012;29:2701–9. doi: 10.1007/s12032-011-0134-3. [DOI] [PubMed] [Google Scholar]
- 33.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. Journal of immunology. 2010;184:6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Kutay H, Klepper C, Wang B, Hsu SH, Datta J, Yu L, et al. Reduced susceptibility of DNA methyltransferase 1 hypomorphic (Dnmt1N/+) mice to hepatic steatosis upon feeding liquid alcohol diet. PloS one. 2012;7:e41949. doi: 10.1371/journal.pone.0041949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC medicine. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong KY, Yim RL, So CC, Jin DY, Liang R, Chim CS. Epigenetic inactivation of the MIR34B/C in multiple myeloma. Blood. 2011;118:5901–4. doi: 10.1182/blood-2011-06-361022. [DOI] [PubMed] [Google Scholar]
- 38.Hanoun N, Delpu Y, Suriawinata AA, Bournet B, Bureau C, Selves J, et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clinical chemistry. 2010;56:1107–18. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, et al. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PloS one. 2012;7:e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gort EH, Suijkerbuijk KP, Roothaan SM, Raman V, Vooijs M, van der Wall E, et al. Methylation of the TWIST1 promoter, TWIST1 mRNA levels, and immunohistochemical expression of TWIST1 in breast cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:3325–30. doi: 10.1158/1055-9965.EPI-08-0472. [DOI] [PubMed] [Google Scholar]
- 42.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7574–83. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 44.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem : a European journal of chemical biology. 2011;12:206–22. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 45.Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A, et al. Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Current medicinal chemistry. 2010;17:4052–71. doi: 10.2174/092986710793205372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudley KJ, Revill K, Whitby P, Clayton RN, Farrell WE. Genome-wide analysis in a murine Dnmt1 knockdown model identifies epigenetically silenced genes in primary human pituitary tumors. Molecular cancer research : MCR. 2008;6:1567–74. doi: 10.1158/1541-7786.MCR-08-0234. [DOI] [PubMed] [Google Scholar]
- 47.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sevin Turcan AWMF, Borodovsky Alexandra, Pedraza Alicia, Cameron Brennan JH, Viale Agnes, Riggins Gregory, Chan Timothy A. Oncotarget. 2013. Efficient Induction of Differentiation and Growth Inhibition in IDH1 Mutant Glioma Cells by the DNMT Inhibitor Decitabine. online. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.