Robin et al. demonstrate that chromosome looping brings the telomere close to genes up to 10 Mb away from the telomere when telomeres are long, while the loci become separated when telomeres are short. Many loci, including noncoding RNAs, may be regulated by telomere length. This suggests a potential mechanism for how telomere shortening could contribute to aging and disease initiation/progression in human cells long before the induction of a critical DNA damage response.
Keywords: chromatin, replicative aging, senescence, cancer, age-dependent gene expression, telomerase, chromosome looping
Abstract
While global chromatin conformation studies are emerging, very little is known about the chromatin conformation of human telomeres. Most studies have focused on the role of telomeres as a tumor suppressor mechanism. Here we describe how telomere length regulates gene expression long before telomeres become short enough to produce a DNA damage response (senescence). We directly mapped the interactions adjacent to specific telomere ends using a Hi-C (chromosome capture followed by high-throughput sequencing) technique modified to enrich for specific genomic regions. We demonstrate that chromosome looping brings the telomere close to genes up to 10 Mb away from the telomere when telomeres are long and that the same loci become separated when telomeres are short. Furthermore, expression array analysis reveals that many loci, including noncoding RNAs, may be regulated by telomere length. We report three genes (ISG15 [interferon-stimulated gene 15 kd], DSP [Desmoplakin], and C1S [complement component 1s subcomplement]) located at three different subtelomeric ends (1p, 6p, and 12p) whose expressions are altered with telomere length. Additionally, we confirmed by in situ analysis (3D-FISH [three-dimensional fluorescence in situ hybridization]) that chromosomal looping occurs between the loci of those genes and their respective telomere ends. We term this process TPE-OLD for “telomere position effect over long distances.” Our results suggest a potential novel mechanism for how telomere shortening could contribute to aging and disease initiation/progression in human cells long before the induction of a critical DNA damage response.
Understanding genome organization and how chromatin folds regulate gene transcription has been an emerging area of investigation (The ENCODE Project Consortium et al. 2012). Major technical improvements have allowed an initial description of the global folding of the human genome (de Wit and de Laat 2012). Using a combination of high-end computer algorithms and newly developed chromatin techniques, recent studies have revealed a strikingly well-organized network of three-dimensional (3D) structures (Dixon et al. 2012; Hou et al. 2012; Nora et al. 2012; Giorgetti et al. 2014). Chromatin territories (or fractal globules) separate DNA into multiple topologically associated domains (TADs) (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Giorgetti et al. 2014). Chromatin loops are restricted to one TAD, and current work focuses on determining organizational mechanisms defining these territories. Long-range intrachromosomal chromatin structures of hundreds of kilobases or interchromosomal interactions involving p53 have been reported in Drosophila (Link et al. 2013). In humans, with the exception of the well-known HS40 enhancer and α-globin gene (Zhou et al. 2006; Vernimmen et al. 2007; Bau et al. 2011) forming a 500-kb interaction domain on chromosome 16, precise DNA loops within TADs have been only rarely reported (Dixon et al. 2012; Berlivet et al. 2013). There is growing appreciation of the importance of chromatin organization (Giorgetti et al. 2014) for biological functions (i.e., gene expression). However, investigations on chromatin organization of the genome adjacent to chromosome ends (telomeres) are lacking.
Telomeres [5′-(TTAGGG)n-3′] and associated proteins form a unique DNA–protein structure located at the ends of linear chromosomes (Blackburn 2001; de Lange 2005) and are required for capping/protecting the chromosome ends from being recognized as DNA double-strand breaks. In the absence of compensatory mechanisms, cell division leads to progressive telomere shortening due to incomplete DNA replication (the end replication problem) (Harley et al. 1990), which eventually causes telomeres to generate a DNA damage signal that causes the growth arrest called replicative senescence. In certain cells, telomerase (a ribonucleoprotein enzyme complex) adds telomeric repeats to the ends of chromosomes to compensate for the end replication problem. Telomerase is expressed in developing embryos, in reproductive cells (proliferating germline), in activated immune cells, and transiently in adult stem cells but is turned off in most adult human tissues, initiating the process of telomere shortening. Telomeres shorten in stem cells as a function of age, and it is believed that stem cells express only sufficient telomerase to slow but not prevent telomere shortening (Shay and Wright 2010). Thus, it is generally accepted that this constitutes an important tumor protection mechanism by limiting the number of divisions during which oncogenic mutations can accumulate. However, premalignant cells that are blocked from proliferating by short telomeres can initially bypass this block, usually by first developing defects in the cell cycle DNA damage checkpoint machinery (e.g., p53). This extension of life span is limited, and the premalignant cells go through crisis and die unless they are able to up-regulate telomerase activity so that telomere length can be maintained.
The vast majority of research on telomere shortening has focused on this tumor suppressor function. However, telomere shortening can also alter gene expression (telomere position effect [TPE]) (Gottschling et al. 1990), and this consequence of telomere shortening has remained largely unexplored. Classic TPE involves the spreading of telomeric heterochromatin to silence nearby genes as a function of length. In budding yeast, this can extend a few kilobases toward the subtelomeres (Gottschling et al. 1990; de Bruin et al. 2001). In some cases, yeast telomeres can loop over longer distances (Bystricky et al. 2005; Miele et al. 2009) and repress genes up to 20 kb from the end (Stavenhagen and Zakian 1998; Tham and Zakian 2002). However, the function of TPE in yeast remains largely unexplored (Van de Vosse et al. 2013). Yeast maintain telomere length through the constitutive expression of telomerase, and the biological context for telomere length-dependent modulation of gene expression is lacking except in genetically altered cells.
In contrast to yeast, human telomeres shorten with age (Harley et al. 1990; Daniali et al. 2013). We explored the concept that telomere shortening could be used as a timing mechanism to adjust physiological changes in very long-lived organisms to optimize fitness in an age-appropriate fashion. Initially, we demonstrated that TPE existed on an artificial chromosome end that included a reporter in a chromosome-healing vector in human cells (Baur et al. 2001). However, until recently, we were able to identify only a single gene, ISG15 (interferon-stimulated gene 15 kd; located 1 Mb from the end of chromosome 1p), whose expression changed as a function of telomere length in human fibroblasts (Lou et al. 2009). Many genes between ISG15 and the telomere did not exhibit any telomere length-dependent regulation. Although ISG15 expression increased when telomeres became short, it was unclear whether this was a direct effect or reflected trans-acting changes occurring elsewhere as a function of telomere shortening.
Additional studies of artificial sequences inserted near telomeres showed that expression could be detected in embryonic stem (ES) cells. However, TPE repressed expression upon the induction of ES cell differentiation (Pedram et al. 2006). In a more recent analysis of a complex age-associated disease, FSHD (facioscapulohumeral muscular dystrophy), we showed that TPE extended at least 100 kb into the subtelomeric region of chromosome 4q (the locus linked to the disease), much further than anticipated from model organism studies (Stadler et al. 2013). In turn, this mechanism directly regulates the expression of DUX4, encoding a toxic double-homeobox protein that is a current candidate for the molecular pathogenesis of this complex disease.
In the present studies, we investigated whether long-range chromosome looping involving telomeres could influence gene expression over much larger distances. We report here that TPE-OLD (TPEs over long distances) is a general phenomenon occurring on different human chromosome ends and that it produces widespread changes in gene regulation long before telomeres become short enough to produce DNA damage signals.
Results
Isogenic clonal analysis
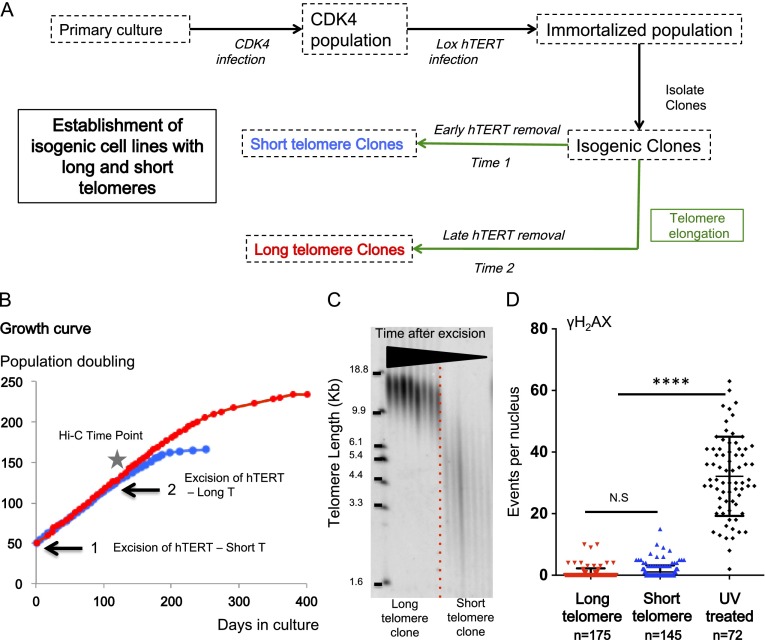
Telomere length in primary cultures is highly variable. Within one cell, the ends of each human chromosome can differ greatly in size. Consequently, there is variation in telomere length between individuals (Barbieri et al. 2009). Primary cultures are formed from tissues containing up to many millions of cells, each with a different proliferative history prior to culture. Clonal succession is a well-established phenomenon in primary cultures, in which individual clones dominate the population only to decline and be replaced in turn by a different clone as the culture is passaged (Martin et al. 1974; Martin 1993). These problems together with multiple other confounding factors make simple comparisons between young and old cells extremely difficult to interpret as a consequence of telomere length. In order to avoid these difficulties and establish a system in which telomere length could be unambiguously controlled, we reversibly immortalized a series of cells using a TERT (telomerase reverse transcriptase) cDNA flanked by LoxP sites (Stadler et al. 2013). The cells could then be cloned, and TERT could be excised at different times to produce isogenic clones in which the only significant difference was the length of the telomeres at the time of TERT excision. These cells could then be cultured for varying periods of time in order to produce cells with intermediate telomere lengths. Figure 1A illustrates the strategy applied to myoblast and fibroblast cell lines.
Figure 1.
Establishment of isogenic clones with long and short telomeres. Isogenic clones with long and short telomeres were produced as described (Stadler et al. 2013). (A) CDK4 is required for myoblasts to prevent growth arrest due to inadequate culture conditions. Telomerase expression preferentially elongates the shortest telomeres so that all telomeres within one cell become normalized to have approximately the same length. Thus, the confounding effects of inheritance (whether a long or short telomere was inherited from one parent) are eliminated, and the specific effects of telomere length can be directly analyzed. An early excision of hTERT produced the “short” telomere subclones, and later excisions yielded subclones with long telomeres. The population doublings for both were similar at the time of experiments. (B) One example of growth curves of isogenic clones. No differences in growth rates were detectable at the time points chosen for the analysis. (C) Telomere length was measured by TRF. The TRF shown has a series of relatively early time points for the long telomere clone and includes several samples from a short telomere clone when cells are entering senescence (cells were used for in situ or expression analysis at least 30 population doublings prior to senescence, when the growth rate had not begun to slow). (D) γH2AX staining. We report the number of events (DNA damage) per nucleus in cells with long and short telomeres along with cells subjected to UV radiation (positive control). No increase in DNA damage was seen in the short telomere cells.
An additional advantage of this strategy is that it normalizes telomere length between different chromosome ends (Londono-Vallejo et al. 2001; Arnoult et al. 2010), thereby reducing the effects of different telomere lengths on different alleles (Bodnar et al. 1998). Telomerase is preferentially recruited to elongate the shortest telomeres (Fakhoury et al. 2010; Cusanelli et al. 2013) so that initially particularly short ends are elongated, while longer telomeres are largely ignored until a uniform elongation of all telomeres takes over. Following the homogenization of telomere length by telomerase and TERT excision, telomere shortening occurs (Fig. 1C). Shortening rates of telomeres may then vary slightly from one chromosome end to another. However, this potential disparity was not detectable in our Southern blot analysis (Fig. 1C). All experiments were performed before replicative senescence (Fig. 1B) and before any DNA damage was detected (Fig. 1D). This strategy thus permits the specific examination of telomere length on gene expression.
Higher-order chromatin organization
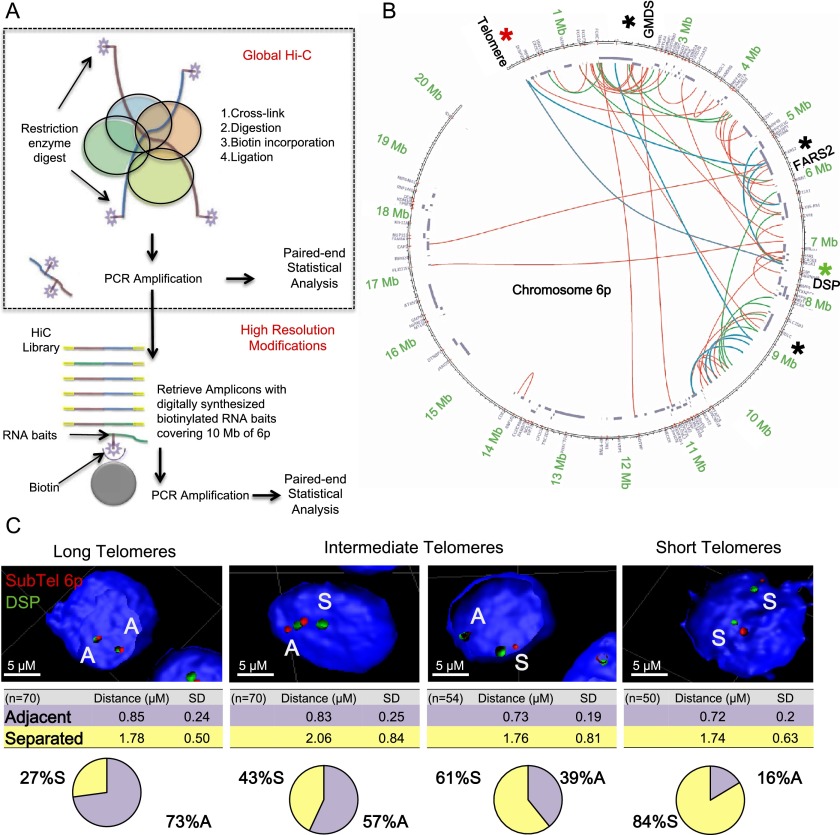
We developed a high-resolution technique to identify specific genomic locations at which interactions involving subtelomeric domains occur. Hi-C (chromosome capture followed by high-throughput sequencing) (van Berkum et al. 2010) is an unbiased chromosome conformation capture (3C) technique that can identify chromosome looping interactions at a low resolution (tens of kilobases scale). We modified this approach to produce a high-resolution map of the interactions occurring at human chromosome 6p (Fig. 2). 6p was chosen because it usually harbors one of the shortest telomeres within a cell (Zou et al. 2004; Meier et al. 2007). We used 80,000 digitally synthesized 120-mers covering the distal 10 Mb of 6p (MYcroarray). The 120-mers were cleaved from the slide and amplified using a primer containing a T4 promoter. Biotinylated RNA probes transcribed from these templates were used to specifically retrieve human chromosome 6p sequences from a global Hi-C preparation, yielding 6p-specific sequences that were then prepared for paired-end high-throughput sequencing (Illumina) (Fig. 2A). Primary myoblasts and fibroblasts (BJs) were transduced with a floxed hTERT, yielding isogenic subclones with experimentally manipulated telomere length (long 15 kb and short 6 kb for myoblasts; long 11 kb and short 5 kb for BJs) (Fig. 1; Stadler et al. 2013). Myoblasts with long telomeres were then analyzed by high-resolution Hi-C. Most of the sequences recovered contained at least one end from 6p (Supplemental Fig. S1). A circular plot (Circos) (Fig. 2B) revealed many long-range looping interactions at the 6p locus in cells with long telomeres. The map in Figure 2B provides only qualitative information about what interactions can occur but does not provide quantitative information about the differences between cells under different conditions. Using this qualitative circular plot, we identified specific interactions that were then quantitated using 3D fluorescence in situ hybridization (3D-FISH) (Fig. 2C) and 3C (Supplemental Fig. S2). 3C is an established technique (de Laat and Dekker 2012) for testing whether a hypothesized chromatin interaction is present above background levels. 3C validations were performed to test a three-point interaction revealed by Hi-C (subtelomeres–FARS2 [phenylalanyl-TRNA synthetase 2]–DSP [Desmoplakin]) (Fig. 2B; Supplemental Fig. S2) for an overall loop distance of 7.5 Mb. The loop was tested by assessing separately an interaction between the subtelomere and the FARS2 gene (5.5 Mb) (Fig. 2B, blue line) and a second interaction between the subtelomere and the DSP gene (7.5 Mb) (Fig. 2B, blue line). Subsequent 3D-FISH experiments confirmed this loop and its telomere length dependence. As telomeres shorten, the particular loop is lost (73% adjacent probes in isogenic clones with long telomeres; 16% adjacent probes in isogenic clones with short telomeres) (Fig. 2C).
Figure 2.
Telomere shortening causes a change of chromatin organization involving a 7-Mb loop in myoblasts. (A) Schematic of high-resolution Hi-C: A chromatin looping interaction brings two distant parts of a chromosome (brown and blue lines, respectively) close together (colored circles represent proteins). After cross-linking, restriction digest, and fill-in incorporation of biotin, ligation under dilute conditions will connect the two distant DNA fragments (brown and blue lines). The incorporation of biotin during the ligation step allows these interactions to be purified. Because short-range (enhancer/repressor) interactions are very abundant, in global Hi-C, they dominate the paired-end sequencing results, and long-range interactions cannot be identified. In the modified approach, PCR amplification removes the biotin used in global Hi-C. This then allows biotinylated probes specific to the 6p terminus to enrich for interactions involving 6p. (B) Organization of the 6p locus in myoblasts with long telomeres, shown as a Circos representation of the 6p-specific Hi-C experiment. The first 20 Mb of chromosome 6p is shown as a circle, and the locations of known genes are shown as segments in a purple inner circle. Interactions are color-coded ([red] P < 0.05; [green] P < 0.01; [blue] P < 0.001); areas involved in interactions confirmed through further experiments are marked with an asterisk. (C) In situ validation. Confocal images of 3D-FISH ([green] fosmid targeting the gene DSP 7.5 Mb from the telomere of 6p; [red] BAC targeting subtelomeric 6p 200 kb from the telomere of 6p) were processed with IMARIS. n = ∼50 nuclei. Adjacent (A) and separated (S) events are labeled. Isogenic clones with progressively shorter telomeres, corresponding to the four lanes of the 12UB myoblast in Figure 4, were tested. Myoblasts with short telomeres show significant looping differences (independent test between all conditions for binary choice, P < 10 × −7; t-test of the mean distance between centers, P = 0.01).
Global analysis of TPE-OLD
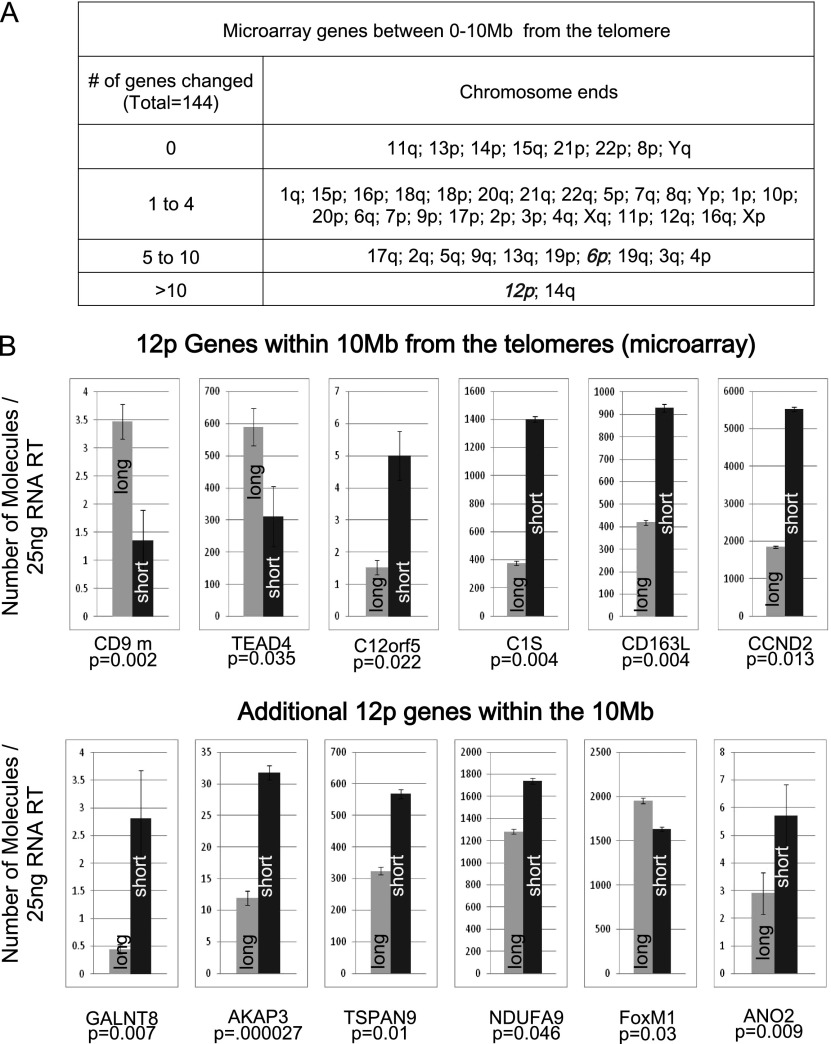
After showing that telomere shortening induces 3D restructuring of the chromatin, we tested whether this results in transcriptional changes and was a general phenomenon involving multiple chromosome ends affecting many different genes. We took an unbiased global approach, analyzing gene expression of myoblasts with long (15-kb) and short (6-kb) telomeres by microarray (in triplicate). The number of significant changes in the level of transcripts (mRNA and lincRNA [long noncoding RNA]) within 10 Mb of the telomeres varied from a minimum of 144 transcripts using eBayes’ moderated t-statistic (Smyth 2005) to 1423 using a standard t-statistic with Benjamin and Hotchberg adjustment (Supplemental Fig. S3; Pollard et al. 2005; http://www.bioconductor.org/packages/release/bioc/html/multtest.html). Figure 3A recapitulates the changes that were observed. The number of transcripts affected by changes in telomere length is likely to be underestimated. Microarrays are informative but poorly sensitive, and subtelomeric transcripts are underrepresented (average of 50% coverage). Consequently, genes previously identified as sensitive to telomere length (such as ISG15) (Supplemental Fig. S4; Lou et al. 2009; Tennen et al. 2011) were not detected as significantly modified in this series of experiments. Irrespectively, some chromosome ends showed no changes (eight of 47), and many showed very few changes (27 of 47), while others exhibited large numbers of transcripts with altered expression (12 of 47). Among the latter, the 6p telomere region showed a total of six modulated transcripts with a 45% array coverage of the 10-Mb region. These microarray results support the concept that telomere length-dependent chromosome conformation may influence the transcription of subtelomeric genes. We called this phenomenon TPE-OLD. TPE-OLD describes the alteration of gene transcription that correlates with telomere length and a change in the chromatin conformation over long distances. We pursued the validation of the array by first using chromosome 12p because it showed >10 changes between short and long telomere mRNA.
Figure 3.
Telomere shortening causes a change in transcription of genes located up to 10 Mb from a telomere in myoblasts. (A) Microarray results comparing transcription in myoblasts with long versus short telomeres. Changes in expression of genes 0–10 Mb from each telomere are grouped by chromosome ends. A microarray was performed using a biological triplicate. (B) ddPCR validation of six of the microarray hits at chromosome 12p and six genes not on the array within the same 10-Mb region. Expression in myoblasts with long telomeres (gray) is compared with an isogenic clone with short telomeres (black). Results are expressed as the number of molecules detected in a 25-ng RT input. P-values indicate significance of the difference for assays done in duplicate using biological triplicates (total of six measurements). All six array genes were validated as significant, and all additional genes tested showed significant differences (P < 0.05, and two-thirds of these showed a >50% change in expression). Many subtelomeric genes were not on the microarray; thus, the table in A provides a significant underestimate of the number of genes whose expression is influenced by telomere length.
Droplet digital PCR (ddPCR) is a nucleic-acid-counting technique that gives an absolute number of molecules per input (in this case, per 25 ng of RNA used in the RT reaction) and does not need to be normalized to an arbitrary set of housekeeping genes that can vary significantly between cell types and experimental conditions (Supplemental Fig. S5; Loven et al. 2012). We selected six genes on 12p that were differentially expressed according to the microarray data and confirmed these differences with ddPCR (Fig. 3B; Supplemental Table S1) and six other 12p genes that were not on the array but in the same 10-Mb region. All of these additional genes also showed differential expression patterns with ddPCR (Fig. 3B). As expected from the behavior of chromosome looping involving enhancers and repressors elsewhere in the genome, we hypothesized that TPE-OLD could alter expression by either increasing or decreasing the level of transcripts when telomeres became short. Transcript levels of all candidates were modified (P-values < 0.05), with changes ranging from a threefold increase (complement component 1s subcomplement [C1S], P = 0.004; Cyclin D2 -[CCND2], P = 0.013) to a 1.3-fold decrease (Forkhead box M1 [FoxM1], P = 0.03).
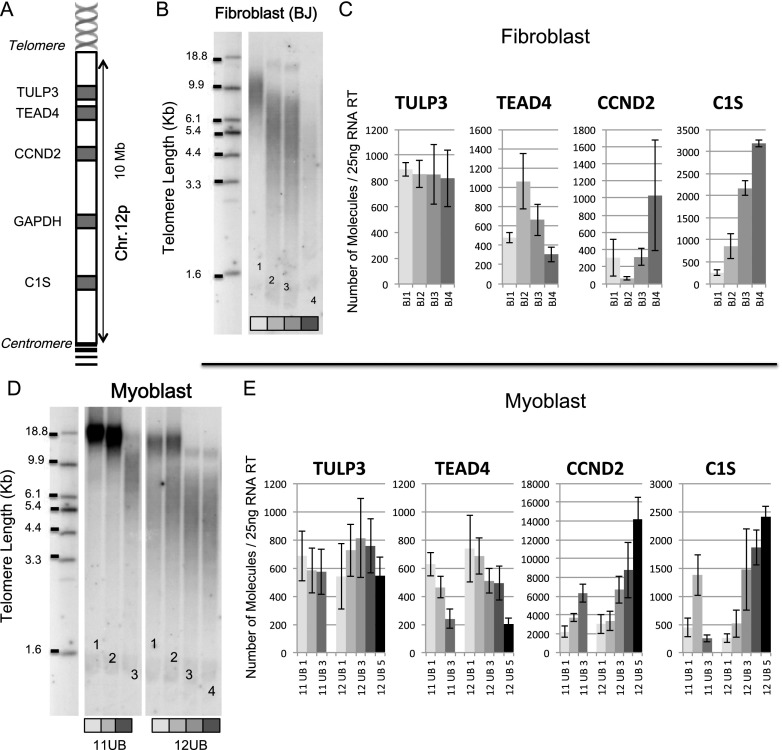
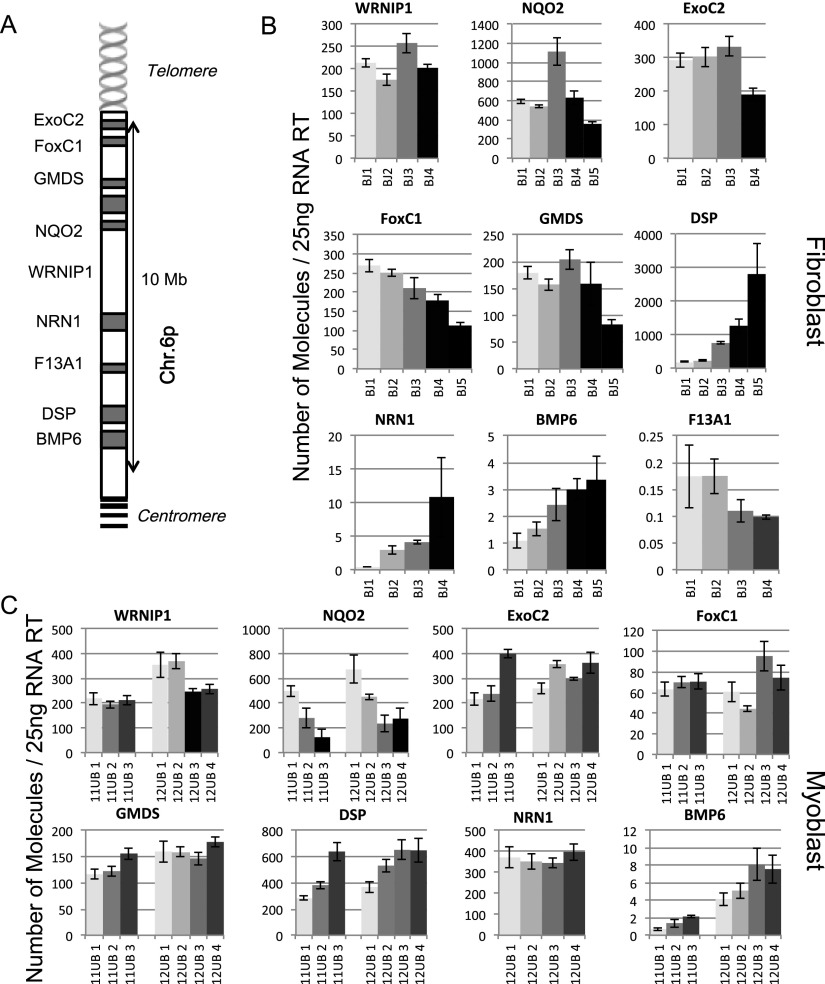
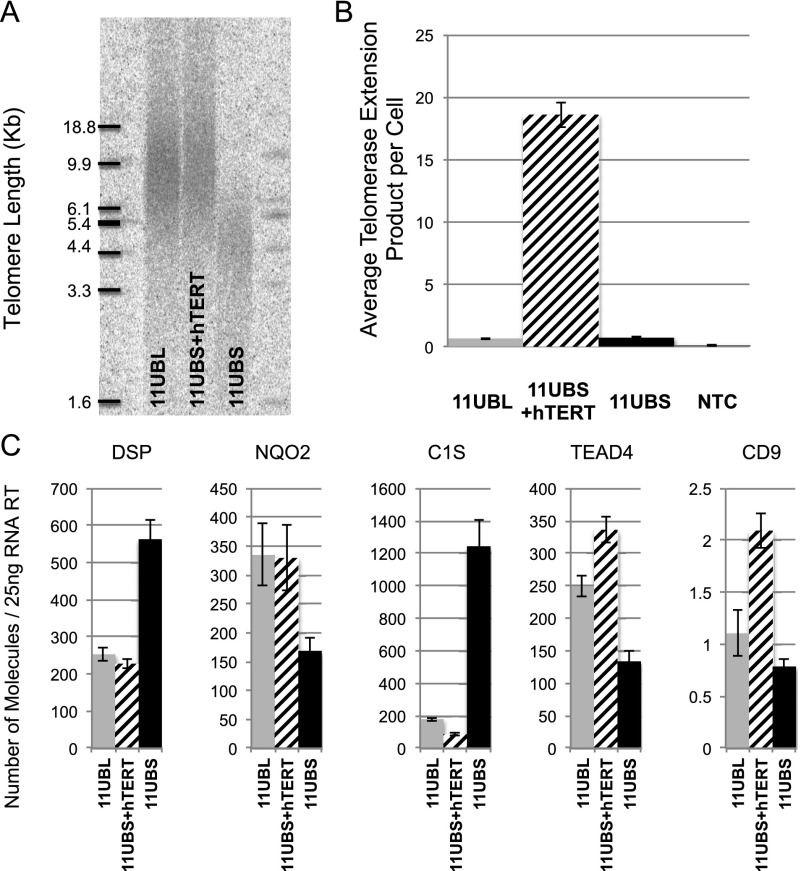
We next analyzed multiple genes from 12p, 6p, and 1p in a series of additional isogenic subclones. As described previously, hTERT was excised at different time points in order to obtain clones with a panel of different telomere lengths and identical time in culture. For myoblasts, two series were produced arising from two different immortalized clones: 11UB1, 11UB2, and 11UB3 (18 kb, 15 kb, and 9 kb, respectively) and 12UB1, 12UB2, 12UB3, 12UB4, and 12UB5 (15 kb, 12 kb, 6 kb, 5 kb, and 4 kb, respectively). Another series from fibroblasts was reported as BJ1, BJ2, BJ3, BJ4, and BJ5 (11 kb, 6.2 kb, 5.4 kb, 4.3 kb, and 2.7 kb, respectively) (Fig. 4). The two sets of myoblast clones and the series of fibroblast clones were used to test 14 genes (Figs. 4, 5; Supplemental Fig S4––S6). At the 12p ends, we observed a clear increase in C1S expression (12-fold between BJ cells with long and short telomeres and ninefold for the myoblast), unlike the Tubby-like protein 3 (TULP3), where no significant changes between any conditions were seen (Fig. 4). At the 6p telomere, the Werner helicase-interacting protein 1 (WRNIP1), the Exocyst complex component 2 (ExoC2), and the GDP-mannose 4,6-dehydratase (GMDS) gene expressions do not appear to be regulated by TPE-OLD (Fig. 5). The FoxC1 and the Neutrin 1 (NRN1) gene expression show changes in fibroblasts but not in myoblasts. The bone morphogenetic protein 6 (BMP6) and DSP gene exhibit changes in their transcription in both fibroblasts and myoblasts (e.g., DSP 3000-fold increase in BJs and threefold increase in myoblasts). TPE-OLD regulation is thus variable depending on the specific cell type, suggesting that additional complex mechanisms are in play in a cell-specific manner. Finally, we analyzed two genes at 1p, including ISG15 (Supplemental Figs. S4, S6). As reported in previous studies, ISG15 transcription was found to be up-regulated with telomere shortening (900-fold between BJ cells with long and short telomeres and sevenfold for the myoblasts). Interestingly, the sterile α motif domain-containing 11 (SAMD11) gene, located in between ISG15 and the 1p telomere, was not regulated by telomere shortening. To determine the nature of the relationship between telomere length and TPE-OLD gene expression, we reintroduced hTERT (Fig. 6). Telomerase activity reversed the expression of TPE-OLD genes, a finding that corroborates observations made in a recent study of senescent-related gene expression (Lackner et al. 2014).
Figure 4.
TPE-OLD at chromosome 12p. The same procedure described for myoblasts in Figure 1 was used for four isogenic fibroblast clones (BJ1, BJ2, BJ3, and BJ4, each indicated by a different shade of gray) and two series of isogenic myoblast clones (11UB and 12UB). All clones were analyzed during logarithmic growth at least 30 population doublings prior to senescence. The location of each gene is shown in A in a schematic representation of the short arm (p) of chromosome 12 and different genes present in the 10-Mb subtelomeric region. (B) TRF analysis of fibroblast isogenic clones. (C) Expression in fibroblasts with long telomeres (light gray) is compared with those with shorter telomeres (grayscale). ddPCR analysis of four genes on chromosome 12p. Results are expressed as the number of molecules detected in a 25-ng RT input. (D) TRF analysis of myoblast isogenic clones. (E) The same genes in the myoblasts are changing as were seen in fibroblast (C1S and TEAD4). This region contains three genes with observed changes (TEAD4, CCND2, and C1S), a housekeeping gene (GAPDH) (Supplemental Fig. S6), and a gene with no changes (TULP3). Each assay was performed in biological triplicate and technical duplicate.
Figure 5.
TPE-OLD at chromosome 6p. Gene expression of four isogenic fibroblast clones (BJ1, BJ2, BJ3, and BJ4, each indicated by a different shade of gray) and two series of isogenic myoblast clones (11UB and 12UB). All clones were analyzed during logarithmic growth at least 30 population doublings prior to senescence. The location of each gene is shown in A in a schematic representation of the short arm (p) of chromosome 6 and different genes present in the 10-Mb subtelomeric region. Nine genes located within the first 10 Mb of chromosome 6 were examined in fibroblasts (B) and myoblasts (C). Expression with long telomeres is compared with a series of isogenic clones with shorter telomeres. Results are expressed as the number of molecules detected in a 25-ng RT input. Each assay was performed in biological triplicate and technical duplicate.
Figure 6.
Reverted expression of TPE-OLD genes by telomerase. (A) TRF analysis of isogenic myoblast clones with long (11UBL), re-elongated (11UBS+hTERT), and short (11UBS) telomeres. (B) hTERT overexpression was assessed by measuring telomerase activity, as reported by the ddTRAP assay. Results are expressed as average telomerase extension products per cell from biological duplicates and technical triplicates. (NTC) Nontemplate control. (C) Gene expression analysis of TPE-OLD genes in myoblasts from A. All clones were analyzed during logarithmic growth at least 30 population doublings prior to senescence. Results are expressed as the number of molecules detected in a 25-ng RT input. Each assay was performed in biological duplicate and technical duplicate.
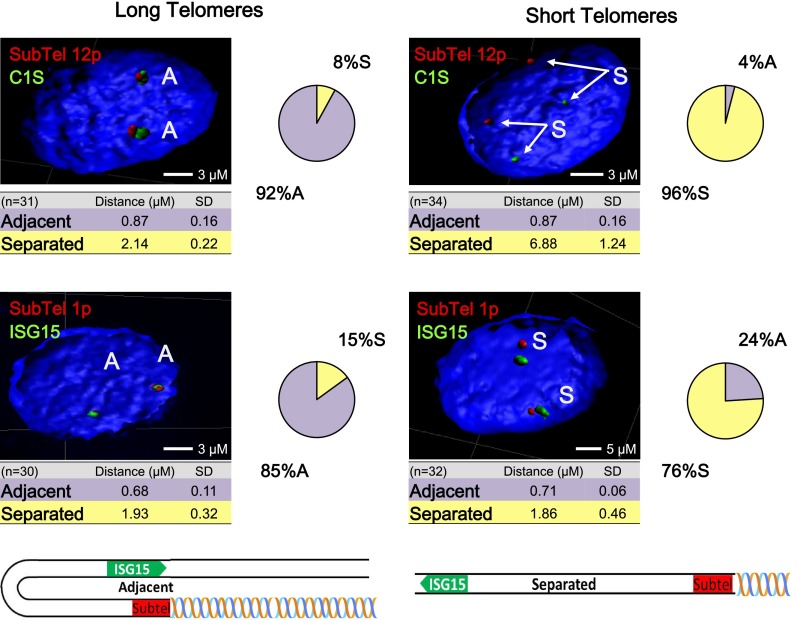
We examined chromosome reorganization in TPE-OLD on 12p and 1p by 3D-FISH. For 12p, we used C1S (Fig. 7, 7.5 Mb from the 12p end, green) and a subtelomeric 12p probe (Fig. 7, 200 kb from the end of 12p, red). For 1p, we used an ISG15 probe (Fig. 7, 1 Mb from the 1p36 end, green) along with a subtelomeric 1p probe (Fig. 7, 20 kb from the 1p36 end, red). Figure 7 demonstrates a dramatic change in the distribution of the two loci in cells with long versus short telomere lengths. In both cases, the two probes are adjacent in cells with long telomeres (12p, 92% of the signals are adjacent; 1p, 85% of the signals are adjacent). The situation shifts in cells with short telomeres where probes are separated (12p, 96% separated; 1p, 76% separated). Strikingly, the two 12p probes are separated by a distance of up to 7 μm in cells with short telomeres, suggesting that shortening of the 12p telomere induces a major reorganization of the distal chromosomal region. A simplified model of how this may be occurring is illustrated at the bottom of Figure 7.
Figure 7.
Telomere shortening causes a change of chromatin organization. Confocal images of 3D-FISH after IMARIS processing. (Top) Telomere shortening causes a change in chromatin organization at chromosome 12p. A Bac containing the C1S probe (7.5 Mb from the telomere; green) and a fosmid containing the most unique subtelomeric region of 12p (∼200 kb from the telomeres; red) showed a dramatic change from >90% within 2 μm of each other to >95% separated by almost 7 μm. The loop identified by 3D-FISH is 7.5 Mb long. (Bottom) Chromatin organization at chromosome 1p using a fosmid ISG15 probe (1 Mb from the telomere; green) and another (∼20 kb from the telomere; red) containing the most unique subtelomeric region of chromosome 1p. An average of 30 nuclear z-stacks per condition were processed. Adjacent (A) and separated (S) are labeled. A test of independence confirms the binary observation based on the distance associated (P < 0.00001). No significant differences were seen in the amount of separation with telomere length, only in the fraction of adjacent or separated locations. A schematic representation of the loop is attached.
TPE-OLD and chromatin structure
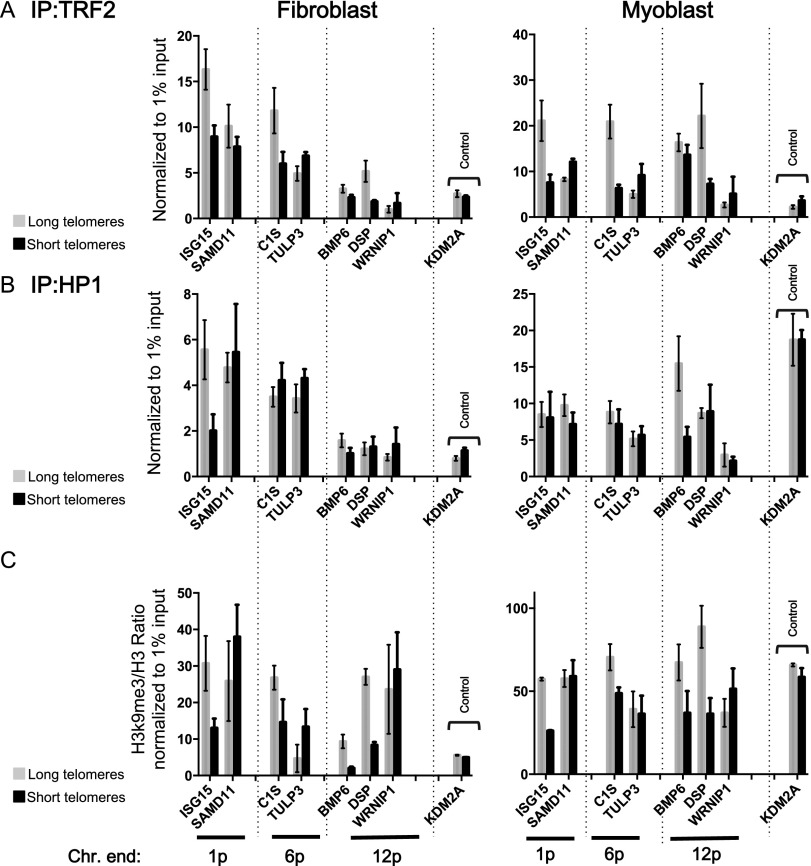
Next, using chromatin immunoprecipitation (ChIP), we investigated whether changes in the higher-order organization of TPE-OLD-sensitive subtelomeres correlate with epigenetics modifications. First, we tested whether TPE-OLD affects whole-chromatin compaction. Using two probes far from the telomeres of chromosome 1, we observed that telomere length did not induce any changes in the global compaction of chromatin (Supplemental Fig. S8). Next, we focused on three different TPE-OLD genes (ISG15, DSP, and C1S) from three different chromosome ends (1p, 6p, and 12p) to query their chromatin states. This set of experiments led to a dual observation. First, after immunoprecipitation using antibodies against the shelterin telomeric repeat-binding factor 2 (TRF2) protein, a major component of telomere chromatin (Smogorzewska et al. 2000), we observed TRF2 enrichment at the promoter sequences of our target genes in cells with long telomeres (Fig. 8). For each subtelomere, we complemented our ChIP analysis by including internal controls by designing primers for different genes (respectively, SAMD11, WRNIP1, and TULP3) located between the telomeres and the TPE-OLD-sensitive genes. Expression of these genes is not modulated by telomere length. With the exception of a moderate 0.7-fold enrichment increase for TULP3 after TRF2 ChIP in myoblasts with short telomeres, no enrichment was detected for control genes. Thus, TRF2 association at specific promoters suggests a gene-specific effect rather than a global spreading of the shelterin marks toward the entire subtelomeric region. In addition, the decreased H3K9me3 (histone H3 Lys9 trimethylation)/H3 ratio in cells with short telomeres suggests that modulation of TPE-OLD gene expression correlates with local chromatin decompaction and a decrease in heterochromatin marks (Fig. 8C).
Figure 8.
Effect of telomere shortening on histone modifications and TRF2. Antibodies against H3, trimethylated H3K9, HP1α, and TRF2 were used in ChIP. Immunoprecipitated DNA was analyzed by ddPCR using primers designed against the promoter region of TPE-OLD genes (ISG15, C1S, BMP6, and DSP) of three chromosome ends (1p, 6p, and 12p). For each chromosome end, a control gene (located in between the telomere and the TPE-OLD gene: SAMD11 for 1p, WRNIP1 for 6p, and TULP3 for 12p) whose expression was not affected by telomere length was assayed. An external gene located far from the telomeres was added as an additional control (KDM2A; 70 Mb away from 11q); no telomere length-dependent variation in the control genes was observed. Each assay was performed in biological duplicate and technical triplicate. (A,B) A decrease of TRF2 enrichment was seen in cells with short telomeres (loss of the loop). HP1 showed little change, suggesting that this marker of constitutive heterochromatin may not be involved in TPE-OLD. (C) Effect of telomere length on the heterochromatic signature of TPE-OLD genes. TPE-OLD genes exhibit a significant decrease of H3K9me3 in cells with short telomeres, underlying a loss of this repressive transcription marker.
Enrichment in the H3K9me3 heterochromatin mark is often associated with the presence of the heterochromatin protein 1 (HP1) proteins at telomeres and centromeres that is believed to contribute to the maintenance of a heterochromatin state (Sharma et al. 2003; Prasanth et al. 2010). Immunoprecipitation analysis using anti-HP1α antibodies suggests that this protein is not involved in TPE-OLD. Indeed, we only observed a significant decrease in HP1α enrichment between cells with long and short telomeres for the ISG15 promoter (chromosome 1p; 2.9-fold decrease) in fibroblasts and the BMP6 promoter (chromosome 12p; threefold decrease) in myoblasts. This led us to conclude that HP1α is not a central player in the TPE-OLD phenomenon.
Discussion
We show here that telomere length influences the formation of chromatin loops at a distance of up to 10 Mb from the chromosome ends. We identified 15 endogenous genes regulated by telomere length in fibroblasts and myoblasts. We named this phenomenon TPE-OLD in reference to the previously described classic TPEs (Stavenhagen and Zakian 1998; Doheny et al. 2008; Ottaviani et al. 2008). Transcriptional changes correlated with chromatin decompaction and modification of the higher-order chromatin organization. Long telomeres bring TPE-OLD genes in direct proximity of the telomere in a loop structure that can extend at least 10 Mb from the chromosome end. However, upon telomere shortening, looping diminishes, separating the TPE-OLD genes from the telomere and its heterochromatic signature. In turn, this phenomenon is associated with increased TPE-OLD gene expression. Loop disruption occurs long before telomere shortening induces DNA damage response.
Previously, telomere shortening and cellular senescence have been studied mainly in diseases or during the aging process. Most of these studies have focused on understanding the link between critically short telomeres and induction of DNA damage signals (d’Adda di Fagagna et al. 2004; Zou et al. 2004; Shay and Wright 2011). Telomere shortening as a tumor suppressor pathway and the role of telomerase and telomere maintenance in cancer are of obvious importance and have justifiably stimulated major efforts to understand the mechanistic details of and develop therapeutics targeting telomerase activity in proliferative tumor cells. However, in a physiological context, telomere shortening occurs in different cell types throughout the human life span and could also be responsible for additional levels of regulation. Our results demonstrate that the expression of a subset of subtelomeric genes is dependent on the length of telomeres and that widespread changes in gene expression are induced by telomere shortening long before telomeres become rate-limiting for division or before short telomeres initiate DNA damage signaling. These changes include up-regulation and down-regulation of gene expression levels.
TPE-OLD: mechanism
It remains unknown at the present time how telomere–chromatin loops are formed in human cells. However, partial mechanisms of chromosome looping within and between chromosomes have been described in a large number of model systems (Murrell et al. 2004; Spilianakis et al. 2005; Ling et al. 2006; Lomvardas et al. 2006; Zhao et al. 2006; Apostolou and Thanos 2008; Hu et al. 2008). Many protein factors have been identified that influence general chromosomal looping, and many of the same factors are almost certainly involved in the general chromosome movements used by TPE-OLD. For example, nuclear myosin-I (NMI) (Chuang et al. 2006; Hu et al. 2008) and dynein light chain-1 (DLCI) (Hu et al. 2008) have both been implicated in interchromosomal interactions. Oligomerized G-actin is present in many transcriptional complexes (Rizk and Walczak 2005; Hofmann and de Lanerolle 2006; Percipalle and Visa 2006; Hofmann 2009; Percipalle 2013), and the demonstration that NMI can be required for chromosome looping suggests that oligomerized nuclear G-actin may also be involved. Latruculin (Rizk and Walczak 2005) blocks actin polymerization, jasplakinolide inhibits actin depolymerization (Holzinger 2009), and both have been found to inhibit chromosome looping (Hu et al. 2008). The chromatin-modifying factor BAF5331 and a variety of coactivators are involved in estrogen receptor α (ERα)-mediated looping interactions (CBP/p300 and the p160 coactivator SRC1/pCIP) (Hu et al. 2008). Knockdown of any of these factors has been shown to block interchromosomal interactions. Histone lysine demethylase-1 (LSD1) did not inhibit ERα-mediated looping but did inhibit the association of the looped genes with nuclear speckles, which are sites of RNA splicing. Thus, due to the overwhelming possible factors involved in the TPE-OLD mechanism and the level of analysis required to decipher between the causes and consequences of chromatin modifications, we chose to focus our study on known telomeric chromatin markers.
In addition to the general mechanisms regulating chromatin domains, chromatin movement and enhancer/repressor interactions might be involved at the telomere sequences and influence TPE-OLD. To gain further insights into such a mechanism, we performed ChIP assays with telomeric factors such as TRF2 or histone heterochromatic markers such as H3K9me3 or HP1α enriched at the subtelomeres and telomere regions (Garcia-Cao et al. 2004; Mikkelsen et al. 2007). Remarkably, loss of heterochromatin marks at the chromosome ends results in telomere dysfunction, increased telomere length, and chromosomal instability (Schoeftner and Blasco 2009; Palacios et al. 2010). In humans, when telomeres shorten, a decrease in TRF1, TRF2, and, surprisingly, H3 binding was observed with a decrease in heterochromatin marks such as H3K9me3, H3K20me3, and H3K27me3 (O’Sullivan et al. 2010; Ernst et al. 2011). In contrast, we observed no differences between H3 and H3K9me3 binding (Fig. 8; Supplemental Fig. S9). These differing results can be due to cell/tissue-specific effects, length of telomeres (i.e., telomeres so short as to initiate DNA damage signaling), and the subtelomeric region chosen to normalize results.
The phenomenon that we describe here at specific ends might be chromosome-specific and depend on the composition of the subtelomeric sequences. Chromosome ends that exhibit no expression changes with telomere length might be protected by elements (such as CTCF or insulators) preventing the ends from forming contacts with subtelomeric regions. Alternatively, insulators or matrix attachment sites might maintain those contacts regardless of telomere length. By high-resolution Hi-C (Fig. 2), we identified many TPE-OLD sites of contact. These contacts are present in the same cell at the same time and might be maintained in a cooperative manner. Alternatively, some interactions may be weaker and lost, while others are preserved but disappear upon further telomere shortening. It is reasonable to invoke the shelterin complex as an important participant, but this multifunctional complex is present at all telomeres, and it is clear that all telomeres do not behave in the same fashion. TERRA (telomere repeat-containing RNA) (Azzalin et al. 2007; Caslini et al. 2009; Nergadze et al. 2009; Deng et al. 2012) is an RNA transcribed from the telomeric sequence that has been hypothesized to regulate telomere replication and is possibly involved in silencing effects. It is thought that TERRA is primarily produced from very short telomeres, and its cis versus trans effects on other telomeres are complex and not yet understood. The number of possibilities is large, and much more needs to be discovered about TPE-OLD before deciphering the telomere-specific regulatory mechanisms from our initial work.
In summary, TPE-OLD as described in this study establishes a new paradigm for the mechanism by which telomere shortening could influence human biology, including aging and cancer. Length-regulated long-range (megabases) telomere chromatin conformation changes may alter gene expression to optimize fitness in long-lived species such as humans and may profoundly affect human physiology, aging, life span, and disease
Materials and methods
Cells and reversible immortalization
CD56-positive myoblasts from primary muscle biopsies were infected with retroviral constructs containing CDK4 and a floxable human TERT as described (Stadler et al. 2013). Individual clones were then treated with Cre-recombinase at different times and cultivated for approximately the same number of population doublings to generate isogenic subclones with different telomere lengths (Fig. 1).
Chromatin organization
The unbiased Hi-C technique to determine looping interactions was modified by enriching interactions involving chromosome 6p (van Berkum et al. 2010) with synthesized specific biotinylated RNA baits. The results were validated using 3C and 3D-FISH (de Laat and Dekker 2012).
Probes
Probes for FISH were produced following the manufacturer’s instructions (nick translation kit, Abbott Molecular). Good nick translation reactions occurred when the DNA smear ranged ∼300 bp. The purified probe was resuspended in a 50-μL mixture of hybridization solution B (Cytocell). Templates were chosen with the University of California at Santa Cruz Genome Browser and ordered from the CHORI Institute (ISG15, G248P88202G4; subtelomeric 1p, G248P8237G10; DSP, RP11-1078C15; subtelomeric 6p, RP11-116D13; C1S, RP11-434L20; and subtelomeric 12p, RP11-945G3). BACS and fosmids were verified by PCR and/or metaphase spreads.
3D-FISH
Slide preparation
Eight-thousand cells were plated in four-chamber well slides (VWR) previously coated with 0.1% gelatin 1 d earlier. The next day, cells were washed once in 1× PBS, chambers were removed, and the slides were incubated for 5 min in 1× PBS at room temperature and fixed in a 4% freshly made PFA for 10 min at room temperature under a fume hood. After three 5-min washes with 1× PBS, nuclei were permeabilized in 0.5% Triton X-100 in PBS for 20 min at room temperature. A first incubation in 20% glycerol and PBS for 30 min at room temperature was done prior to six cycles of successive incubations between liquid nitrogen and glycerol. Following six cycles, slides were again incubated for 1 min in 20% glycerol at room temperature followed by three 5-min washes with 1× PBS and a 0.1 M HCl incubation for 10 min. Finally, slides were incubated for 5 min in 2× SSC and stored at 4°C in 50% formamide (pH 7) in 2× SSC in a Coplin jar wrapped in aluminum foil.
Slide hybridization
After a minimum of 5 d of storage at 4°C in formamide, slides were used for hybridization. Excess solution was removed from the slide, and a 10-μL mixture of 3 μL of each probe (6 μL total) + 4 μL of hybridization solution B (Cytocell) was used for each well. The slides were covered with a coverglass (FisherBrand) and sealed with rubber cement. The slides were then codenatured (with the probe mix) for 5 min at 80°C in a hybridization oven prior to an overnight incubation at 37°C in a humidification chamber. The next day, the rubber cement was removed, and the slides were incubated for 5 min at 70°C in 0.5× SSC and 0.1% SDS. Slides were then sequentially washed in 0.001% PBS-Tween20 for 5 min at room temperature and then in 1× PBS for 10 min at room temperature. Excess solution was removed, and a 10-μL DAPI (0.125 μg/mL in VectaShield) solution was dropped onto each slide.
Image processing
Images were acquired with a confocal scanning laser system (LSM 780 from Zeiss). A 40× plan-apochromat oil immersion objective was used to capture optical sections at intervals of 0.24 μm. The pinhole was set to 1 airy unit with optical slices in all wavelengths with identical thickness. Images were then generated into .lsm files and processed through Imaris software (Bitplane). After 3D reconstruction, at least 30 nuclei were examined per condition, and the volume of the probes and the distance between their gravity centers (distance between the closest probes of each target) were determined and used for statistical analysis. Cells with more or less than four signals, irregular DAPI staining (e.g., mitotic cells), and abnormal shape were excluded from the analysis.
ChIP
ChIP for TRF2 (NB110-57130, Novus), HP1 (109028, AbCam), H3 (17-10254, Milipore), and H3K9me3 (17-10242, Milipore) was performed according to the manufacturer’s instructions (TruChIP, Covaris; and Magna ChIPA/G, Milipore). A series of controls (i.e., IgG, RabSe, no cross-linking condition) was run for each preparation (Supplemental Fig. S8). Primers were designed against the promoter region of each chosen gene.
Supplementary Material
Acknowledgments
We thank John Abrams (University of Texas Southwestern) for valuable discussions, the FSH Society, and Genila Bibat (Kennedy Krieger Institute, Baltimore, MD) for subject recruitment. We also thank Jean-Marie Rouillard Biodiscovery, LLC (Ann Arbor, MI) for valuable reagents. This work was supported by the National Institute of Aging ( AG01228 to W.E.W.), Lung Cancer Specialized Programs of Research Excellence (SPORE) (P50CA70907), the National Institutes of Health (NIH) Post-doctoral Training Fellowship (T32 CA124334 to A.T.L.), the Austrian Science Fund, and the American Federation for Aging Research (G.S.). This work was performed in laboratories constructed with support from NIH grant C06 RR30414. J.D.R helped design and execute the experiments and contributed to the writing of the manuscript, W.E.W. and J.W.S. designed the project and wrote the manuscript, A.T.L. helped develop and perform the ddPCR, K.B. analyzed the microarrays and performed the bioinformatics analysis of Hi-C results, F.M. supervised the in situ colocalization technique, K.R.W. obtained muscle biopsies, G.S. helped generate the reversibly immortalized muscle cultures with different telomere lengths, and all authors edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.251041.114.
References
- Apostolou E, Thanos D. 2008. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell 134: 85–96. [DOI] [PubMed] [Google Scholar]
- Arnoult N, Schluth-Bolard C, Letessier A, Drascovic I, Bouarich-Bourimi R, Campisi J, Kim SH, Boussouar A, Ottaviani A, Magdinier F, et al. 2010. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet 6: e1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801. [DOI] [PubMed] [Google Scholar]
- Barbieri M, Paolisso G, Kimura M, Gardner JP, Boccardi V, Papa M, Hjelmborg JV, Christensen K, Brimacombe M, Nawrot TS, et al. 2009. Higher circulating levels of IGF-1 are associated with longer leukocyte telomere length in healthy subjects. Mech Ageing Dev 130: 771–776. [DOI] [PubMed] [Google Scholar]
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. 2011. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE. 2001. Telomere position effect in human cells. Science 292: 2075–2077. [DOI] [PubMed] [Google Scholar]
- Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. 2013. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet 9: e1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. 2001. Switching and signaling at the telomere. Cell 106: 661–673. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352. [DOI] [PubMed] [Google Scholar]
- Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL. 2009. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol 29: 4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825-831. [DOI] [PubMed] [Google Scholar]
- Cusanelli E, Romero CAP, Chartrand P. 2013. Telomeric noncoding RNA TERRA Is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell 51: 780–791. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Teo SH, Jackson SP. 2004. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev 18: 1781–1799. [DOI] [PubMed] [Google Scholar]
- Daniali L, Benetos A, Susser E, Kark JD, Labat C, Kimura M, Desai K, Granick M, Aviv A. 2013. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun 4: 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D, Zaman Z, Liberatore RA, Ptashne M. 2001. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409: 109–113. [DOI] [PubMed] [Google Scholar]
- de Laat W, Dekker J. 2012. 3C-based technologies to study the shape of the genome. Methods 58: 189–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110. [DOI] [PubMed] [Google Scholar]
- Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, et al. 2012. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J 31: 4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. 2012. A decade of 3C technologies: insights into nuclear organization. Genes Dev 26: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny JG, Mottus R, Grigliatti TA. 2008. Telomeric position effect–a third silencing mechanism in eukaryotes. PLoS ONE 3: e3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhoury J, Marie-Egyptienne DT, Londono-Vallejo JA, Autexier C. 2010. Telomeric function of mammalian telomerases at short telomeres. J Cell Sci 123: 1693–1704. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. 2014. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157: 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460. [DOI] [PubMed] [Google Scholar]
- Hofmann WA. 2009. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol 273: 219-263. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, de Lanerolle P. 2006. Nuclear actin: to polymerize or not to polymerize. J Cell Biol 172: 495–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A. 2009. Jasplakinolide: an actin-specific reagent that promotes actin polymerization. Methods Mol Biol 586: 71–87. [DOI] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 48: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon Y, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. 2008. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci 105: 19199–19204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Hayashi MT, Cesare AJ, Karlseder J. 2014. A genomics approach identifies senescence-specific gene expression regulation. Aging Cell 13: 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272. [DOI] [PubMed] [Google Scholar]
- Link N, Kurtz P, O’Neal M, Garcia-Hughes G, Abrams JM. 2013. A p53 enhancer region regulates target genes through chromatin conformations in cis and in trans. Genes Dev 27: 2433–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, DerSarkissian H, Cases L, Thomas G. 2001. Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res 29: 3164–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Wei J, Riethman H, Baur JA, Voglauer R, Shay JW, Wright WE. 2009. Telomere length regulates ISG15 expression in human cells. Aging 1: 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. 2012. Revisiting global gene expression analysis. Cell 151: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM. 1993. Clonal attenuation: causes and consequences. J Gerontol 48: B171–B172. [DOI] [PubMed] [Google Scholar]
- Martin GM, Sprague CA, Norwood TH, Pendergrass WR. 1974. Clonal selection, attenuation and differentiation in an in vitro model of hyperplasia. Am J Pathol 74: 137–154. [PMC free article] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP. 2007. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J 26: 2707–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Bystricky K, Dekker J. 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet 5: e1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893. [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. 2009. CpG-island promoters drive transcription of human telomeres. RNA 15: 2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17: 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Gilson E, Magdinier F. 2008. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 90: 93–107. [DOI] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. 2010. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol 191: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram M, Sprung CN, Gao Q, Lo AW, Reynolds GE, Murnane JP. 2006. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol 26: 1865–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P. 2013. Co-transcriptional nuclear actin dynamics. Nucleus 4: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Visa N. 2006. Molecular functions of nuclear actin in transcription. J Cell Biol 172: 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Dudoit S, van der Laan MJ. 2005 Multiple testing procedures: the multtest package and applications to genomics. In Bioinformatics and computational biology solutions using R and Bioconductor (ed. Gentleman R, et al.), pp. 249–271. Springer, New York. [Google Scholar]
- Prasanth SG, Shen Z, Prasanth KV, Stillman B. 2010. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci 107: 15093–15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Walczak CE. 2005. Chromosome dynamics: actin’s gone fishing. Curr Biol 15: R841-R842. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. 2009. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, Hwang KK, Pandita RK, Gupta A, Dhar S, Parenteau J, Agarwal M, Worman HJ, Wellinger RJ, Pandita TK. 2003. Human heterochromatin protein 1 isoforms HP1(Hsα) and HP1(Hsβ) interfere with hTERT–telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol 23: 8363–8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. 2010. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett 584: 3819–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. 2011. Role of telomeres and telomerase in cancer. Semin Cancer Biol 21: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. 2000. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and Bioconductor (ed. Gentleman R, et al.), pp. 397–420. Springer, New York. [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645. [DOI] [PubMed] [Google Scholar]
- Stadler G, Rahimov F, King OD, Chen JC, Robin JD, Wagner KR, Shay JW, Emerson CP Jr, Wright WE. 2013. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat Struct Mol Biol 20: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen JB, Zakian VA. 1998. Yeast telomeres exert a position effect on recombination between internal tracts of yeast telomeric DNA. Genes & Dev 12: 3044–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennen RI, Bua DJ, Wright WE, Chua KF. 2011. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun 2: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Zakian VA. 2002. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21: 512–521. [DOI] [PubMed] [Google Scholar]
- van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES. 2010. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152: 969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347. [DOI] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. 2006. Active chromatin hub of the mouse α-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol 26: 5096–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Sfeir A, Gryaznov SM, Shay JW, Wright WE. 2004. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol Biol Cell 15: 3709–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.