AT-rich DNA is concentrated in the nucleosome-free regions (NFRs) associated with transcription start sites of most genes. Lorch et al. find that the AT-rich sequences present in many NFRs have little effect on the stability of nucleosomes. These sequences instead facilitate the removal of nucleosomes by the RSC chromatin remodeling complex. RSC activity is stimulated by AT-rich sequences in nucleosomes and inhibited by competition with AT-rich DNA.
Keywords: RSC, poly(dA:dT) tracts, yeast
Abstract
AT-rich DNA is concentrated in the nucleosome-free regions (NFRs) associated with transcription start sites of most genes. We tested the hypothesis that AT-rich DNA engenders NFR formation by virtue of its rigidity and consequent exclusion of nucleosomes. We found that the AT-rich sequences present in many NFRs have little effect on the stability of nucleosomes. Rather, these sequences facilitate the removal of nucleosomes by the RSC chromatin remodeling complex. RSC activity is stimulated by AT-rich sequences in nucleosomes and inhibited by competition with AT-rich DNA. RSC may remove NFR nucleosomes without effect on adjacent ORF nucleosomes. Our findings suggest that many NFRs are formed and maintained by an active mechanism involving the ATP-dependent removal of nucleosomes rather than a passive mechanism due to the intrinsic instability of nucleosomes on AT-rich DNA sequences.
The assembly of promoters in nucleosomes prevents the initiation of transcription in vitro (Lorch et al. 1987), and depletion of nucleosomes leads to gene activation in yeast in vivo (Han and Grunstein 1988). Nucleosomes may be regarded at a fundamental level as general gene repressors. Relief from repression is achieved by the removal of nucleosomes by either the occurrence of nucleosome-free regions (NFRs) at the transcription start sites (TSSs) of TATA-less promoters (∼80% of promoters in yeast) (Yuan et al. 2005; Zhang et al. 2011) or chromatin remodeling of genes with TATA-containing promoters (which may have distinctive nucleosomal configurations, but not NFRs, as defined by size and proximity to TSSs) (Svaren and Horz 1997; Boeger et al. 2003).
The uniform coverage of eukaryote genomes by nucleosomes is punctuated at NFRs by apparently naked DNA regions and by well-defined locations of the nucleosomes nearby. The formation of NFRs is generally believed to be an intrinsic property of the DNA sequence; it is attributed to their high content of AT base pairs and in particular of poly(dA:dT) tracts (Yuan et al. 2005) with consequent destabilization of nucleosomes. Poly(dA:dT) is comparatively rigid and resists bending around the histone core of the nucleosome. The evidence for this passive mechanism of NFR formation comes from nucleosome positioning analysis, chromatin reconstitution, and effects on transcription in vivo. Poly(dA:dT) tracts greater than or equal to seven residues in length are confined to the first two turns of the double helix at the ends of the DNA in chicken nucleosomes (Satchwell et al. 1986). Pure poly(dA:dT) DNA is refractory to nucleosome formation (Kunkel and Martinson 1981; Prunell 1982). Poly(dA:dT) tracts of 17–42 residues stimulate transcription from the HIS3 promoter in yeast (Iyer and Struhl 1995). Poly(dG:dC) tracts have the same effect, leading to the conclusion that a structural property of the sequence element rather than interaction with a protein factor is involved.
The alternative possibility of an active mechanism of NFR formation is suggested by studies of RSC, the most abundant member of the SWI/SNF family of remodelers, which has been directly linked to the removal of nucleosomes in vitro (Lorch et al. 2006) and in vivo (Hartley and Madhani 2009). A partially purified yeast protein fraction reported to reconstitute the formation of NFRs was enriched in RSC (Wippo et al. 2011). It has been suggested that RSC interacts with Reb1, a DNA-binding transcription factor (Gavin et al. 2002). Reb1-binding sites are the most conserved sequence elements in yeast promoters (Elemento and Tavazoie 2005) and are commonly found in NFRs (Fedor et al. 1988; Chasman et al. 1990; Hartley and Madhani 2009). The elimination of either RSC or Reb1 abolishes the formation of numerous NFRs (Hartley and Madhani 2009). Conversely, the introduction of a DNA element, including a Reb1-binding site, brings about the formation of an ectopic NFR (Raisner et al. 2005). On this basis, it has been proposed that Reb1 may in some cases recruit RSC to form an NFR. Reb1-binding sites, however, are found only in a small minority of NFRs. Alternatives for the recruitment of RSC include additional DNA-binding proteins such as ABF1, whose recognition sites also occur in NFRs, although typically at the same low frequency as those for Reb1. Here we reconcile the two opposing views of NFR formation, ruling out the rigorous exclusion of nucleosomes by AT-rich DNA and showing instead that such DNA sequences represent recognition sites for the removal of nucleosomes by RSC.
Results
As a test of the hypothesis that NFRs exclude nucleosomes by virtue of their DNA structure, we measured affinities of NFR DNAs for nucleosomes. Measurement of affinity was made by exchange of histones between bulk chromatin and 32P-labeled NFR DNA (Thastrom et al. 1999). Exchange was performed in 1 M NaCl followed by stepwise dilution to 0.1 M NaCl. The formation of nucleosomes was revealed by gel electrophoresis and autoradiography and confirmed by gradient centrifugation. The free energy of nucleosome formation was derived from the relative amounts of nucleosomal and naked NFR DNA in the gel. Remarkably, eight of 10 NFRs chosen at random from the Saccharomyces cerevisiae genome (Supplemental Table S1) differed by less than twofold in affinity from the 5S rDNA nucleosome positioning sequence (Table 1, KNFR/K5S), one of the highest-affinity nucleosome-binding sequences known (Thastrom et al. 1999).
Table 1.
Stabilities of nucleosomes formed from NFR DNA
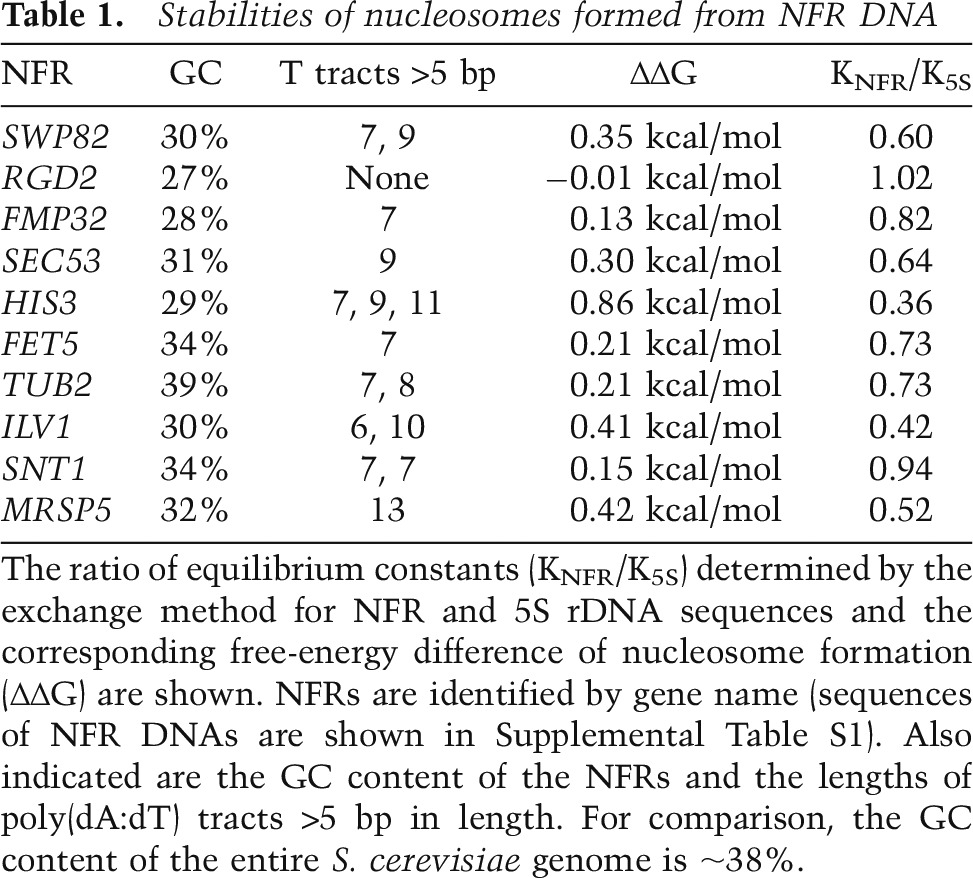
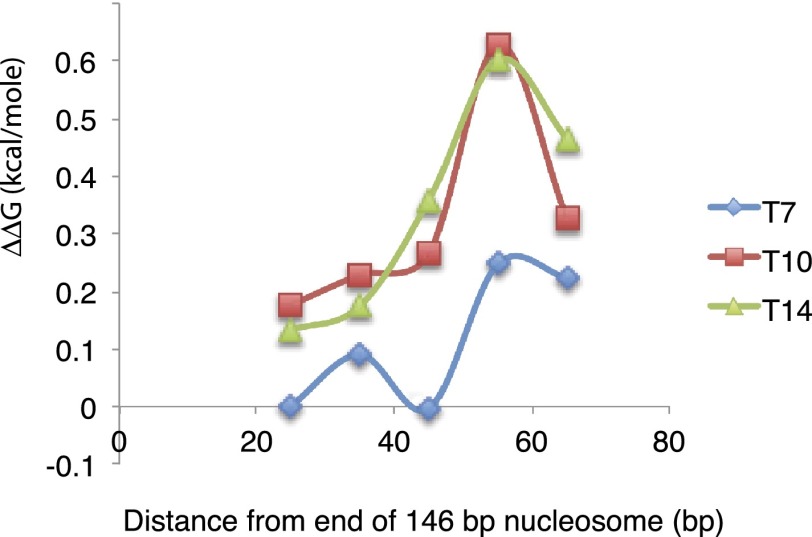
The NFR DNAs studied here exhibited high affinities for nucleosomes despite the occurrence in nine of the 10 NFR DNA sequences of poly(dA:dT) tracts >5 base pairs (bp) in length. To test directly the effect of poly(dA:dT) tracts on affinity for nucleosomes, poly(dA:dT) tracts of seven to 14 residues were inserted at a series of positions across the 5S rDNA nucleosome (Supplemental Table S2). Although poly(dA:dT) tracts did diminish the affinity for nucleosomes, with a peak of instability at about two turns of the DNA double helix from the dyad of the nucleosome (Fig. 1), at no point did a poly(dA:dT) tract even as long as 14 residues reduce the occupancy by a nucleosome by >2.5-fold (ΔΔG of 0.6 kcal/mol).
Figure 1.
Effect of poly(dA:dT) tracts on the stability of nucleosomes. Poly(dA:dT) tracts of 7, 10, and 14 bp were substituted for the sequence of the 5S rDNA nucleosome positioning sequence at the locations indicated (sequences of the DNAs assembled in nucleosomes are shown in Supplemental Table S2). Stabilities of nucleosomes were determined by the exchange method (average of six determinations).
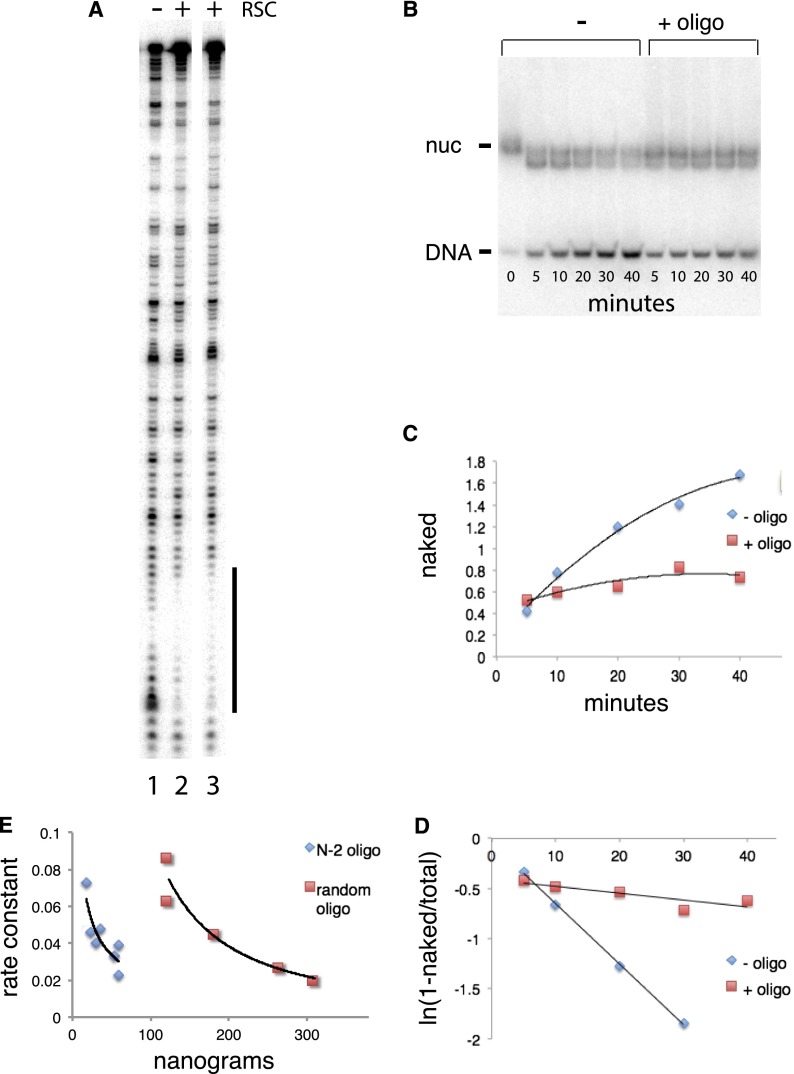
An alternative mechanism through which AT-rich sequences might engender NFR formation was suggested by an observation on the PHO5 gene of S. cerevisiae. Previous studies showed that transcriptional activation of PHO5 leads to the depletion of promoter nucleosomes (Svaren and Horz 1997) and that this depletion is due to the complete disruption of nucleosomes (Boeger et al. 2003). The effect is most pronounced for the second nucleosome (N-2) upstream of the TSS, whose occupancy in the activated state is only 18% of that in the transcriptionally repressed state (Boeger et al. 2003). We investigated the interaction of RSC with the N-2 nucleosome by DNase I digestion and observed a footprint of the protein on the nucleosomal DNA (Fig. 2A, indicated by a vertical line, experiment performed in the absence of ATP). The footprint encompassed the sequence T4ACT7, located 4–19 bp from the end of the 160-bp nucleosomal DNA. A footprint was also obtained with the same sequence in naked rather than nucleosomal DNA (data not shown).
Figure 2.
DNase I footprint of RSC and effect of oligonucleotides on RSC activity. (A) The N-2 nucleosome of the PHO5 promoter, with a footprint over a T-rich element indicated by a vertical line. (Lane 1) Without RSC. (Lanes 2,3) With RSC. Lane 3 also contained 1 mM β,γ-methylene ATP. (B) Inhibition by oligonucleotides of the disassembly of a nucleosome by RSC, Nap1, and ATP. PAGE of reaction containing 5S rDNA nucleosome without (−) or with (+) T4ACT7 oligonucleotide (132 ng in the reaction) (Supplemental Table S3). Times of reaction were, from left, 0, 5, 10, 20, 30, 40, 5, 10, 20, 30, and 40 min. Bands due to nucleosomes (nuc) and naked DNA are indicated. The band below that for nucleosomes is due to the removal of H2A–H2B dimers. A PhosphorImager scan of the gel is shown. (C) Appearance of naked DNA during the reaction of 5S rDNA nucleosome with RSC, Nap1, and ATP. Intensities of bands due to naked DNA at the times indicated are plotted for reactions without (blue diamonds) and with (red squares) T4ACT7 oligonucleotide (Supplemental Table S3). (D) Disappearance of nucleosomes during the reaction in B. (E) First-order rate constant (in units of per minute) for the disappearance of nucleosomes as a function of the amount of T4ACT7 oligonucleotide (blue diamonds) or random sequence oligonucleotide (red squares) in the reaction.
We investigated a possible influence of the T4ACT7 element upon chromatin remodeling by RSC. In the presence of the Nap1 histone chaperone and ATP, histones are removed from nucleosomes by RSC (Lorch et al. 2006). The progress of such a reaction, performed with the 5S rDNA nucleosome, was monitored by gel electrophoresis, which revealed the appearance of naked DNA (Fig. 2B,C). The reaction followed first-order kinetics (Fig. 2D), and the rate constant was diminished by competition with a 30-bp oligonucleotide containing the T4ACT7 element (Fig. 2D,E). Other AT-rich oligonucleotides (Supplemental Table S3), such as T3GT6 from the GAL2 promoter, were even more inhibitory (Table 2), whereas a “random” oligonucleotide lacking an AT-rich element had little effect (Fig. 2C–E).
Table 2.
Inhibition by oligonucleotides of chromatin remodeling by RSC
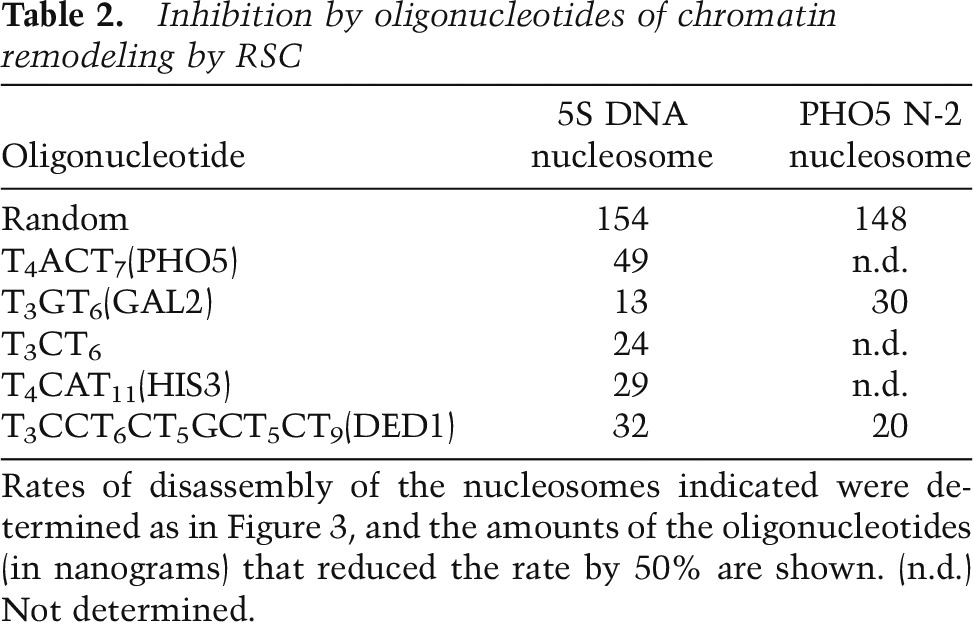
The inhibition of chromatin remodeling by competing oligonucleotides containing AT-rich elements raised the possibility of a stimulatory effect on remodeling by such elements in a nucleosome. To this end, we incorporated AT-rich elements in the 5S rDNA nucleosome 19–29 residues from the end of the nucleosome (Supplemental Table S4). The formation of nucleosomes was confirmed by gradient centrifugation, and the locations of nucleosomes were confirmed by exonuclease III mapping (data not shown). Incorporation of an AT-rich element resulted in a footprint of RSC closely similar to that observed for the PHO5 N-2 nucleosome (data not shown). The incorporation of T3GT6 stimulated remodeling (Fig. 3A). A seven-residue poly(dA:dT) tract was almost as effective, as were nine- and 11-residue poly(dA:dT) tracts, while a five-residue poly(dA:dT) tract was stimulatory but less so (data not shown).
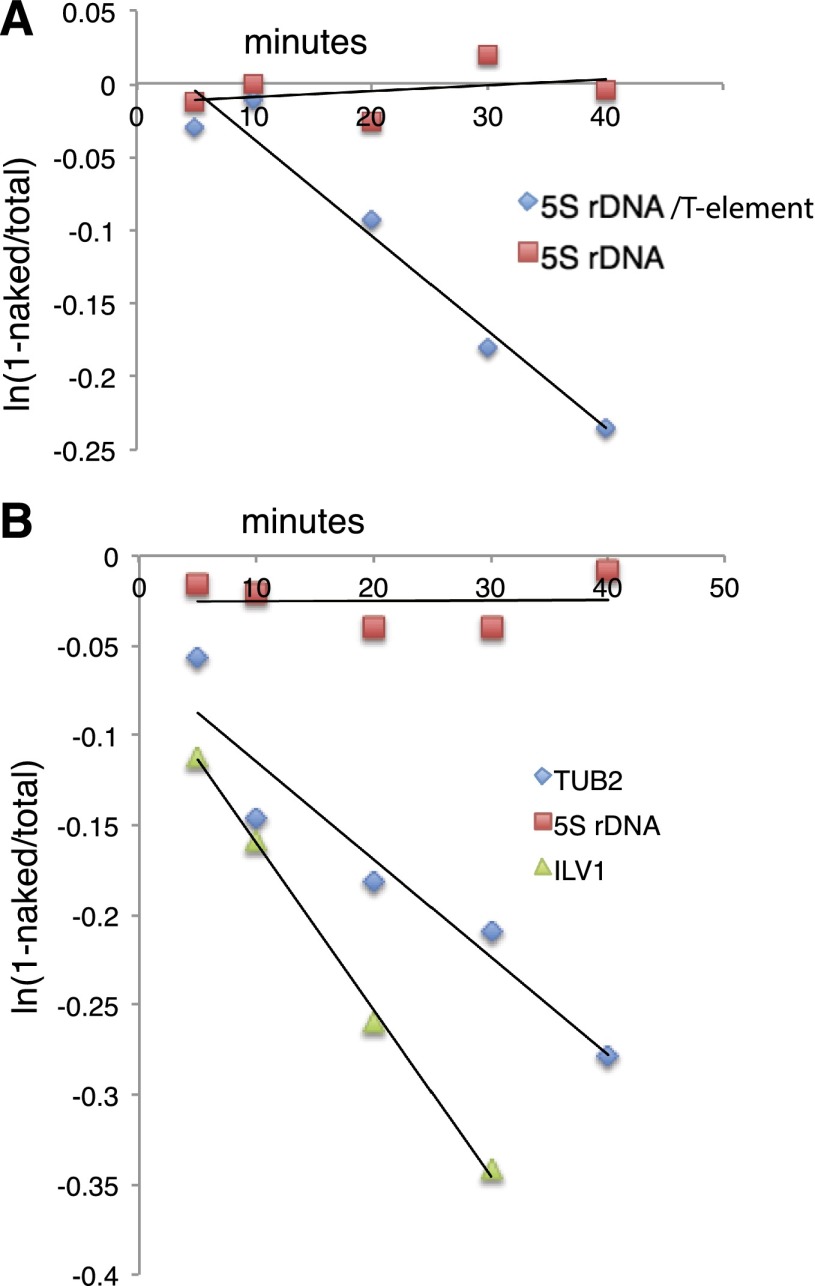
Figure 3.
Disassembly by RSC, Nap1, and ATP of nucleosomes containing a T-rich element or an NFR. (A) Disappearance of 5S rDNA nucleosomes containing a T3GT6 element (blue diamonds) (sequence in Supplemental Table S4) or not (red squares) in reactions with RSC and ATP and 250 ng of T3GT6 oligonucleotide as competitor. (B) The disappearance of nucleosomes containing TUB2 or ILV1 NFR DNA (blue diamonds and green triangles) (sequences in Supplemental Table S1) and the disappearance of 5S rDNA nucleosomes (red squares) in reactions with RSC, Nap1, ATP, and 370 ng of H1-depleted rat liver chromatin as competitor.
Finally, we investigated the remodeling by RSC, Nap1, and ATP of nucleosomes formed on NFR DNAs. NFRs containing one or two poly(dA:dT) tracts of seven or eight residues and with affinities for nucleosomes only 20%–25% less than that of the 5S rDNA were remodeled at rates comparable with the 5S rDNA nucleosome containing a T3GT6 element (Fig. 3B). It is noteworthy that remodeling was performed in the presence of a 400-fold excess of chromatin over NFR nucleosomes and a 10-fold molar excess of nucleosomes over RSC. The vast excess of chromatin simulated conditions in vivo, reducing the effective concentration of RSC. Remodeling was evidently highly specific for NFR nucleosomes.
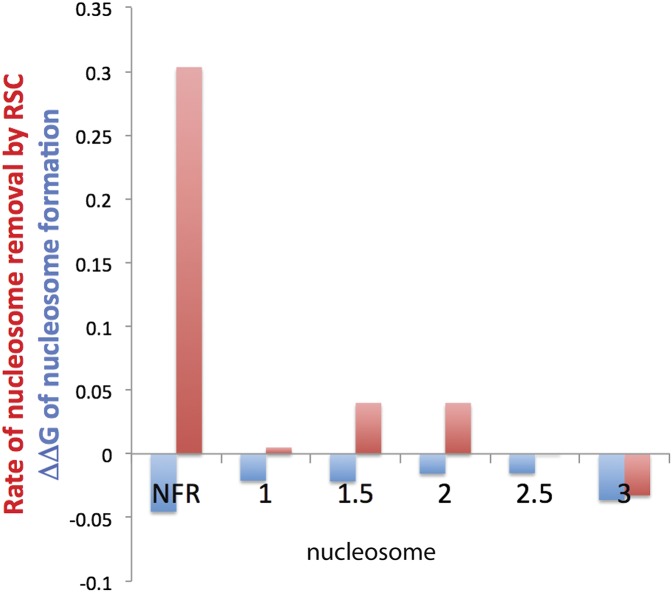
The question of specificity pertains especially to remodeling at the NFR relative to remodeling of the adjacent ORF. Can remodeling by RSC account for the virtual absence of nucleosomes from an NFR despite the full occupancy of nucleosomes on the adjacent ORF? We compared both stabilities and remodeling rates for nucleosomes on the NFR and the ORF of the ILV1 gene (Fig. 4). The stabilities of nucleosomes were very similar, all within 0.2 kcal/mol of that of the reference 5S rDNA nucleosome. In contrast, the rate of removal of a nucleosome was far greater at the NFR than at positions within the ORF. Therefore, RSC may well be responsible for the contrasting occupancies by nucleosomes of the NFR and the ORF.
Figure 4.
Profiles of nucleosome stability and disassembly rate across the NFR and ORF of the ILV1 gene. Stabilities of nucleosomes were determined by the exchange method for 160-bp DNA fragments (Supplemental Table S5) centered on the NFR; the published positions of the +1, +2, and +3 nucleosomes of the ILV1 gene (Yuan et al. 2005); and positions halfway between the +1 and +2 nucleosomes (1.5) and halfway between the +2 and +3 nucleosomes (2.5). Values of ΔΔG (kilocalories per mole) are multiplied by 0.25 in the plot. Rates of disassembly by RSC, Nap1, and ATP were determined for nucleosomes formed on the same DNA fragments as described. First-order rate constants (per minute) are multiplied by −10 in the plot.
Discussion
The notable findings of this study are the large stimulatory effect of short AT-rich DNA sequences upon the removal of nucleosomes by RSC and the lack of effect of such sequences upon the stability of nucleosomes. AT-rich DNA brings about the formation of NFRs by an active rather than passive mechanism by enhancement of chromatin remodeling rather than through an effect of DNA structure. We present two lines of evidence against the hypothesis of NFR formation by DNA structure: First, many NFR DNAs form exceptionally stable nucleosomes in vitro, and second, poly(dA:dT) tracts as long as 14 bp diminish the stability of nucleosomes by at most a factor of two relative to the most stable nucleosome known. It may also be noted that many NFRs contain no AT-rich element. NFRs would therefore be mostly, if not fully, occupied by nucleosomes if the DNA sequence played a dominant role.
We found instead that AT-rich DNA stimulates the removal of nucleosomes by RSC in an ATP-dependent manner. Poly(dA:dT) tracts of 7 bp are effective in this regard, tracts of 9 or 11 bp are not more effective, and tracts of 5 bp are less so. This length dependence may explain the occurrence of a poly(dA:dT) tract of 7 bp adjacent to a binding site for the Reb1 protein that recruits RSC to NFRs (Angermayr et al. 2003; Raisner et al. 2005). The combination of a poly(dA:dT) tract and a Reb1 site proved sufficient to induce the formation of an NFR at an ectopic site in chromatin in vivo (Raisner et al. 2005).
Remodeling is essential for NFR formation and function. Previous mapping of nucleosomes in vivo has shown that occupancies of NFRs by nucleosomes are very low, at least 100-fold less than for neighboring DNA regions. NFRs may be essentially naked to allow interaction with transcription factors. A reduction in nucleosome occupancy by a factor of two or less due to DNA structure alone (Table 1) would be insufficient for this purpose. The removal of nucleosomes by an active mechanism is required. An abundance of AT-rich DNA in NFRs supports this mechanism not primarily by the exclusion of nucleosomes but rather by the recruitment of RSC for the removal of nucleosomes.
A recent bioinformatics analysis of poly(dA:dT) tracts in NFRs revealed several features that are more consistent with the stimulation of chromatin remodeling than with destabilization of nucleosomes (Wu and Li 2010). For example, poly(dA:dT) tracts occur predominantly 20–25 bp from the center of the NFR and in opposite orientations, with poly(dT) upstream of and poly(dA) downstream from the center. Such dyad symmetry is suggestive of protein–nucleic acid interaction, whereas the orientation of poly(dA:dT) tracts is not obviously relevant to the destabilization of nucleosomes.
Further support for a role of remodeling rather than DNA structure in the formation of NFRs comes from genome-wide studies of chromatin reconstitution in vitro and chromatin formation in vivo. Reconstitution experiments showed at most a twofold reduction in nucleosome occupancy over all NFRs in S. cerevisiae (Kaplan et al. 2009), consistent with our measurements on the nucleosome affinities of selected NFRs (Table 1). When chromatin was formed on Kluyveromyces lactis DNA in S. cerevisiae in vivo, NFRs were observed in the same locations as in K. lactis to a greater extent on sequences containing poly(dA:dT) tracts than those deficient in them. From these and other observations, it was concluded that NFRs are formed either as a result of the rigidity of poly(dA:dT) DNA or due to species-specific chromatin remodeling factors (Hughes et al. 2012; Struhl and Segal 2013). More direct evidence for a role of chromatin remodeling factors in NFR formation has come from genome-wide assembly of chromatin by yeast extract and ATP (Zhang et al. 2011).
Our findings support a primary role for chromatin remodeling in the formation of NFRs, with recognition by RSC of AT-rich sequences rather than their structure as a basis for their involvement. The rigidity of poly(dA:dT) tracts may play a role but only a minor one, as the extent of nucleosome exclusion from NFRs is far beyond what can be achieved by DNA structure alone. The complete exposure of all NFRs, likely essential for function, cannot be achieved except through an enzyme-catalyzed reaction with the consumption of energy in the process.
Materials and methods
Nucleosomes were prepared as described (Lorch et al. 2005) with the use of 160-bp DNA (Supplemental Table S2) labeled with 32P at one 5′ end by PCR. RSC and Nap1 were prepared as described (Lorch and Kornberg 2003; Wittmeyer et al. 2004). DNase I footprint analysis was performed as described (Lorch et al. 2010) except with 0.6 μg rather than 0.3 μg of RSC. Nucleosome disassembly reactions contained nucleosomes (1 ng of DNA), 160 ng of RSC, 250 ng of Nap1, 0.5 mM ATP, 15 mM HEPES (pH 8.0), 3 mM MgCl2, 0.1 mg/mL BSA, and oligonucleotides (Supplemental Table S1) as indicated in a total volume of 15 μL. Following incubation for the times indicated at 30°C, reaction products were separated by electrophoresis in 3.2% polyacrylamide gels (19:1 acrylamide:bisacrylamide) in TE. Measurement of nucleosome stability by the exchange method was performed as described (Thastrom et al. 1999).
Supplementary Material
Acknowledgments
We are most grateful to Weihong Xu and Wenzhong Xiao for computer programs to calculate the frequencies of AT-rich elements. This research was supported by National Institutes of Health grant GM36659.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.250704.114.
References
- Angermayr M, Oechsner U, Bandlow W. 2003. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J Biol Chem 278: 17918–17926. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD. 1990. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev 4: 503–514. [DOI] [PubMed] [Google Scholar]
- Elemento O, Tavazoie S. 2005. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol 6: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ, Lue NF, Kornberg RD. 1988. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol 204: 109–128. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Han M, Grunstein M. 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55: 1137–1145. [DOI] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K. 2012. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell 48: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GR, Martinson HG. 1981. Nucleosomes will not form on double-stranded RNA or over poly(dA)•poly(dT) tracts in recombinant DNA. Nucleic Acids Res 9: 6869–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Kornberg RD. 2003. Isolation and assay of the RSC chromatin-remodeling complex from Saccharomyces cerevisiae. Methods Enzymol 377: 316–322. [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. 1987. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49: 203–210. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Davis B, Kornberg RD. 2005. Chromatin remodeling by DNA bending, not twisting. Proc Natl Acad Sci 102: 1329–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci 103: 3090–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. 2010. Mechanism of chromatin remodeling. Proc Natl Acad Sci 107: 3458–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A. 1982. Nucleosome reconstitution on plasmid-inserted poly(dA). poly(dT). EMBO J 1: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA. 1986. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191: 659–675. [DOI] [PubMed] [Google Scholar]
- Struhl K, Segal E. 2013. Determinants of nucleosome positioning. Nat Struct Mol Biol 20: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Horz W. 1997. Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci 22: 93–97. [DOI] [PubMed] [Google Scholar]
- Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol 288: 213–229. [DOI] [PubMed] [Google Scholar]
- Wippo CJ, Israel L, Watanabe S, Hochheimer A, Peterson CL, Korber P. 2011. The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes. EMBO J 30: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Saha A, Cairns B. 2004. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol 377: 322–343. [DOI] [PubMed] [Google Scholar]
- Wu R, Li H. 2010. Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters. Genome Res 20: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.