Abstract
Despite the success of combined antiretroviral therapy in controlling viral replication in human immunodeficiency virus (HIV)-infected individuals, HIV-associated neurocognitive disorders, commonly referred to as neuroAIDS, remain a frequent and poorly understood complication. Infection of CD8+ lymphocyte-depleted rhesus macaques with the SIVmac251 viral swarm is a well-established rapid disease model of neuroAIDS that has provided critical insight into HIV-1-associated neurocognitive disorder onset and progression. However, no studies so far have characterized in depth the relationship between intra-host viral evolution and pathogenesis in this model. Simian immunodeficiency virus (SIV) env gp120 sequences were obtained from six infected animals. Sequences were sampled longitudinally from several lymphoid and non-lymphoid tissues, including individual lobes within the brain at necropsy, for four macaques; two animals were sacrificed at 21 days post-infection (p.i.) to evaluate early viral seeding of the brain. Bayesian phylodynamic and phylogeographic analyses of the sequence data were used to ascertain viral population dynamics and gene flow between peripheral and brain tissues, respectively. A steady increase in viral effective population size, with a peak occurring at ~50–80 days p.i., was observed across all longitudinally monitored macaques. Phylogeographic analysis indicated continual viral seeding of the brain from several peripheral tissues throughout infection, with the last migration event before terminal illness occurring in all macaques from cells within the bone marrow. The results strongly supported the role of infected bone marrow cells in HIV/SIV neuropathogenesis. In addition, our work demonstrated the applicability of Bayesian phylogeography to intra-host studies in order to assess the interplay between viral evolution and pathogenesis.
Introduction
Human immunodeficiency virus type 1 (HIV-1)-associated neurocognitive disorders (HANDs), ranging from mild neurological symptoms to the most severe form, HIV-associated dementia (HAD), are serious complications of HIV-1 infection. Since the development of combination antiretroviral therapy (cART), the incidence of the milder forms of HANDs has increased to ~25 % (Woods et al., 2009). In addition, HANDs remain highly prevalent, with an estimated 69 % of the infected population exhibiting some degree of neurological impairment (Simioni et al., 2010), whilst treatment using neuroprotective or anti-inflammatory agents has been unsuccessful (Alfahad & Nath, 2013). Therefore, understanding the underlying pathogenic mechanism of this disease and its progression is of critical importance to the development of new diagnostics and treatment approaches.
Previous studies of HIV-1 infection in the brain have shown that HIV-1 and its non-human primate counterpart, simian immunodeficiency virus (SIV), enter the brain by way of infected monocytes, which act to replenish perivascular macrophages located at the blood–brain barrier, as elegantly reviewed by Koppensteiner et al. (2012). Infection of these cells often allows for the productive infection of nearby microglia and, to a lesser extent, astrocytes, resulting in uncontrolled inflammation and a self-propagating immune reaction in response to the build-up of neurotoxic macrophage byproducts. Whilst HIV-1 does not replicate in either neurons or oligodendrocytes, these cells are directly affected by neuroinflammation and neurotoxic viral proteins produced by infected cells, resulting in the neurodegeneration that is characteristic of the cognitive, motor and behavioural impairment observed for neuroAIDS patients (Alter et al., 2007). Studies show that most cARTs do not entirely clear the central nervous system (CNS) of HIV-1, likely due to poor drug penetration (Zhao et al., 2009). Thus, progressive neuronal loss/dysfunction and CNS inflammation is identified even in well-suppressed patients on long-term cART (Harezlak et al., 2011).
The tissue source of infected cells in the periphery responsible for HIV-1 seeding of the brain is still unclear. Moreover, it is not known whether viral replication observed in the CNS during the later stages of infection and clinical onset of symptomatic HANDs is the result of an initial event occurring during primary infection or the continuous influx of infected monocytes/macrophages over the course of the disease. Many studies have shown compartmentalization of distinct HIV-1 subpopulations within brain and cerebrospinal fluid of infected patients (Lamers et al., 2011a, b, 2012; Salemi et al., 2009a, b; Smith et al., 2009; Wong et al., 1997) as well as of SIV in the rhesus macaque (Macaca mulatta) model (Chen et al., 2006; Kodama et al., 1993; Ryzhova et al., 2002a, b), which may reflect, at least in part, differences in viral subpopulations infecting T-cells (less frequent in the CNS) versus macrophages. However, it has been difficult to establish if compartmentalization is due to the separate evolution of virus in the CNS or the evolution of neurotropic viral variants in peripheral tissues that are increasingly neuroinvasive. Early viral infection of the brain has been described (Kim et al., 2003; Williams & Hickey, 2002), whereas other research has shown that late neuroinvasion of HIV-1 is also possible (Fischer-Smith et al., 2008). Additional studies suggested that neuropathology begins outside of the CNS compartment independent of virus that might already be present in the brain (Burdo et al., 2010). Such hypotheses are difficult to test in human subjects, as the initial infection date is usually unknown, and, for ethical reasons, it is impossible to collect longitudinal samples of lymphoid and non-lymphoid tissues. Alternatively, experimental infection of CD8+ lymphocyte-depleted rhesus macaques with the genetic heterogeneous SIVmac251 viral swarm constitutes a well-established rapid model of neuroAIDS (Burudi & Fox, 2001; Westmoreland et al., 1998; Williams & Burdo, 2012). In this model, ~77 % of the animals develop SIV-associated encephalitis (SIVE) and/or meningitis within 90–120 days post-infection (p.i.) (Williams & Burdo, 2012). The histopathology and neuropathology <1?twb=.3w?>of the macaques closely resembles the development of HAD in humans as both diseases are characterized by the accumulation of perivascular macrophages, microglia and macrophage nodules, multinucleated giant cells, and gliosis (Burudi & Fox, 2001; Lackner, 1994; Mankowski et al., 2002; Zink et al., 1998, 2006). In addition, the SIV/macaque model produces immunological patterns similar to those produced by HIV-1 infection in humans, including the expansion of monocyte subpopulations (Bissel et al., 2006; Kim et al., 2010; Pulliam et al., 1997; Strauss-Ayali et al., 2007). However, little is known about how viral evolution and population dynamics relate to brain infection and the onset and progression of encephalomeningitis in this model of neuroAIDS.
Bayesian phylogenetics and phylogeography are powerful tools used to study viral population dynamics and gene flow (migration) because they can integrate spatial, temporal and demographic inferences by simultaneously accounting for both phylogenetic and mapping uncertainty (Holmes, 2008; Lemey et al., 2009, 2010). These methods have been used primarily to investigate epidemic spread (Bello et al., 2012; Ciccozzi et al., 2012; de Carvalho et al., 2013; Lemey et al., 2009; Salemi et al., 2005, 2007), whereas application at the intra-host level is limited by the quality of zthe datasets, i.e. the lack of longitudinal data and/or sequences derived from several biological locations (Faria et al., 2011). In the present study, we successfully obtained sequences sampled at different time points, from early infection to terminal illness, from several lymphoid and non-lymphoid tissues, thereby enabling the use of Bayesian methods to track the evolution and spread of the virus within the macaque host similar to the tracking of a virus in an epidemic outbreak within a population. The results provided for the first time an in-depth characterization of the intra-host viral evolutionary patterns in this macaque model, and demonstrated the applicability of Bayesian phylogeography to intra-host studies investigating the interplay between viral evolution and pathogenesis.
Results
SIV infection and persistent CD8+ lymphocyte depletion
Six rhesus macaques were inoculated with the SIVmac251 viral swarm at ~95 months of age (Table 1) and subsequently inoculated with an anti-CD8 antibody (Strickland et al., 2012). Two macaques, referred to as D01 and D02, were sacrificed at 21 days p.i., whilst macaques D03–D06 were allowed to progress until the onset of AIDS-related symptoms. Repeated administration of anti-CD8 antibody resulted in a persistent (>28 days) depletion of CD8+ T-lymphocytes and CD8+ NK cells, as well as sustained elevated viral load (Fig. 1), leading to rapid disease progression and increased incidence of SIVE and/or meningitis as compared with naturally progressing macaques, as expected (Schmitz et al., 1999). CD8+ T-lymphocyte and NK-cell counts over time were monitored by flow cytometry prior to and after antibody administration (Fig. 1, top and middle panels). Both cell types were depleted by 8 days p.i. Whilst considered a persistent depletion, a rebound of CD8+ T-lymphocytes was observed at 42 days p.i. for D06 and 56 days p.i. for D03–D05, whereas a rebound of NK cells was first detected at 21 days p.i., although absolute counts of both cell types did not reach levels observed prior to antibody administration. Viral load patterns over time were very similar amongst macaques and remained consistently elevated after 8 days p.i., although some fluctuation could still be observed, with a peak at ~50 days p.i. detected in all longitudinally sampled animals (Fig. 1 bottom panel).
Table 1. Bayesian coalescent estimates of the SIV molecular clock and demographic parameters for each longitudinally sampled primate.
| Primate ID | Age of infection (years) | Length of infection (days) | TMRCA (days)* | Baseline Ne† | End point Ne† | Growth rate (×10−2 day−1)‡ | Evolutionary rate (×10−4 nt substitutions site−1 day−1)§ |
| D03 | 9.9 | 75 | −4 (4, −13) | 87 (19, 327) | 1877 (909, 4452) | 3.64 (2.65–4.73) | 3.46 (2.87–4.03) |
| D04 | 10.9 | 91 | −14 (−5, −25) | 262 (75, 786) | 2714 (807, 6740) | 3.13 (2.37–3.89) | 2.03 (1.67–2.42) |
| D05 | 5.6 | 95 | −28 (−15, −44) | 859 (304, 2415) | 676 (307, 4463) | 1.39 (0.89–1.87) | 2.53 (2.03–3.01) |
| D06 | 5.7 | 118 | −36 (−21, −53) | 887 (364, 2090) | 1250 (415, 3657) | 1.43 (1.10–1.85) | 2.35 (1.95–2.75) |
Median estimates of TMRCA of SIV in each infected primate. Negative values indicate that the TMRCA does not correspond to the time of inoculation, but rather to the coalescence time of the most recent common ancestor of the SIVmac251 infecting swarm (pre-transmission interval).
Baseline and end point Ne refer to the median estimates of the effective population size at viral load set point (~8 days p.i.) and time of euthanization, respectively.
Ne growth rate parameters inferred by enforcing an exponential population growth demographic model.
SIV gp120 intra-host evolutionary rate estimates.
Fig. 1.
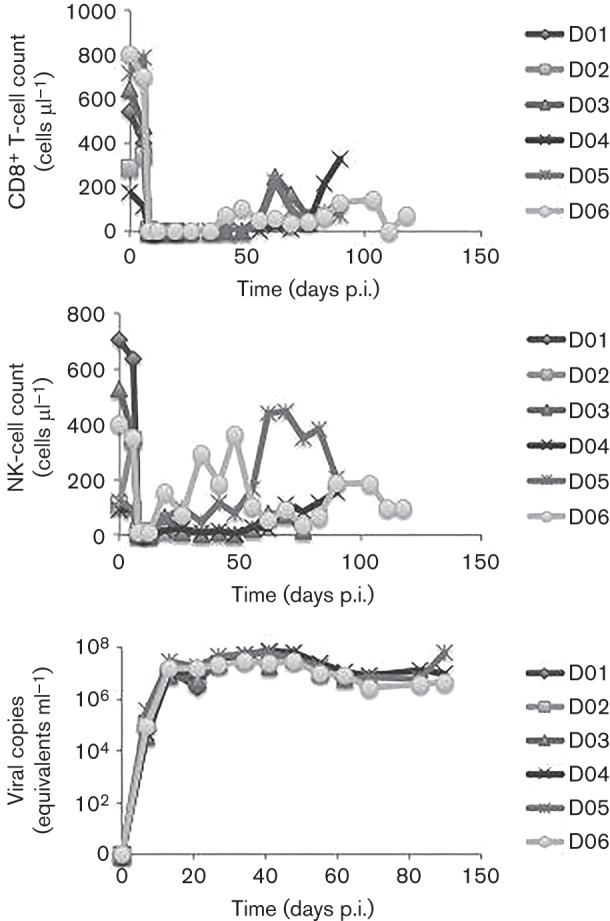
Absolute CD8+ T-lymphocyte and NK-cell counts as well as viral load over time in infected macaques D01–D06. Absolute CD8+ T-lymphocyte (top panel) and NK-cell (middle panel) counts (y-axis) and viral load (bottom panel) were monitored periodically using flow cytometry.
CD8+ lymphocyte depletion was sufficient to produce rapid progression (75–118 days p.i.) to simian AIDS (SAIDS) in all four animals in contrast with naturally progressing macaques, which live on average 1–3 years: D03 was sacrificed at 75 days p.i., D04 at 91 days p.i., D05 at 95 days p.i. and D06 at 118 days p.i. As shown previously, D03–D05 developed SIVE, whereas D06 exhibited perivascular cuffing and meningitis (Strickland et al., 2012).
Bayesian coalescence analysis of SIVmac251 in infected rhesus macaques
The temporal dynamics of SIV intra-host evolution in the infected animals were investigated using the Bayesian coalescence framework implemented in beast (Drummond & Rambaut, 2007; Drummond et al., 2005). In the four macaques sampled longitudinally, Bayes factors (BFs) strongly supported a relaxed molecular clock (lnBF>50) (Kass & Raftery, 1995), which assumes log-normal distributed evolutionary rates along the viral genealogies. The result was also confirmed with the evaluation of the 95 % high-posterior-density (HPD) intervals of the coefficient of variation, which is the evolutionary rate variance scaled by the associated mean. These intervals did not include zero in any of the six SIV datasets (Table S1, available in the online Supplementary Material), indicating significant deviation of the intra-host SIV evolutionary rates from a strict molecular clock. Several demographic models were compared in order to investigate the relative changes of the viral effective population size (Ne) over time. In each infected animal, lnBF strongly supported the Bayesian Skyride model, indicating that a non-parametric change of Ne over time most accurately described SIV intra-host demographic history (Table S1).
Despite the complexity of the viral demographic history, as indicated by the non-parametric change in Ne over time, similarities for various coalescence parameter estimates were observed amongst the longitudinally monitored macaques. Analyses of these parameter estimates included comparisons of time to the most recent common ancestor (TMRCA), net Ne change and Ne growth rate (Table 1). In each of the longitudinally sampled animals, TMRCA dated back ~4–36 days prior to infection with overlapping 95 % HPD intervals. This was expected because the time at the root of the viral genealogy should not correspond to the time of inoculation, but rather to the pre-transmission interval, i.e. the coalescence time of the most recent common ancestor of the SIVmac251 infecting swarm (Leitner & Albert, 1999). In primates D03 and D04, baseline Ne values calculated at 8 days p.i., the time of set point viral load (Strickland et al., 2012), corresponded to ~175 unique viral variants. This finding indicated the presence of a relatively homogeneous SIV population, the overall diversity of which increased by a mean of ~15 % at the end point (Table 1), prior to euthanization at the onset of AIDS. Differences in baseline and end point Ne were less distinguished in D05 and D06 (mean 0.1 %). A slower viral population growth rate in these macaques was observed in comparison with D03 and D04 (Table 1). Nevertheless, Ne 95 % HPD intervals were similar for all datasets, suggesting a generalized SIV coalescence pattern represented within each infected macaque (Table 1, Fig. 2). In addition, viral evolutionary rates were also similar (Table 1); median nucleotide substitution rates averaged ~2.6×10−4 nucleotide substitutions site–1 day–1 (i.e. 0.10 nt substitutions site–1 year–1). This value is at least 1 log higher than the SIV evolutionary rate estimated in natural hosts (Wertheim & Worobey, 2009), but is likely the result of the impaired innate immune response due to CD8+ T-lymphocyte and NK-cell depletion. It is important to note that despite the relatively short time of infection (75–118 days p.i.), viral sequences sampled at different time points (at 21 days p.i., 60 days p.i. and necropsy) displayed a statistically significant number of nucleotide differences, classifying SIV in the context of this model as a measurably evolving population (Drummond et al., 2003), and allowing for the study of SIV intra-host demographic history and its relationship to brain infection.
Fig. 2.
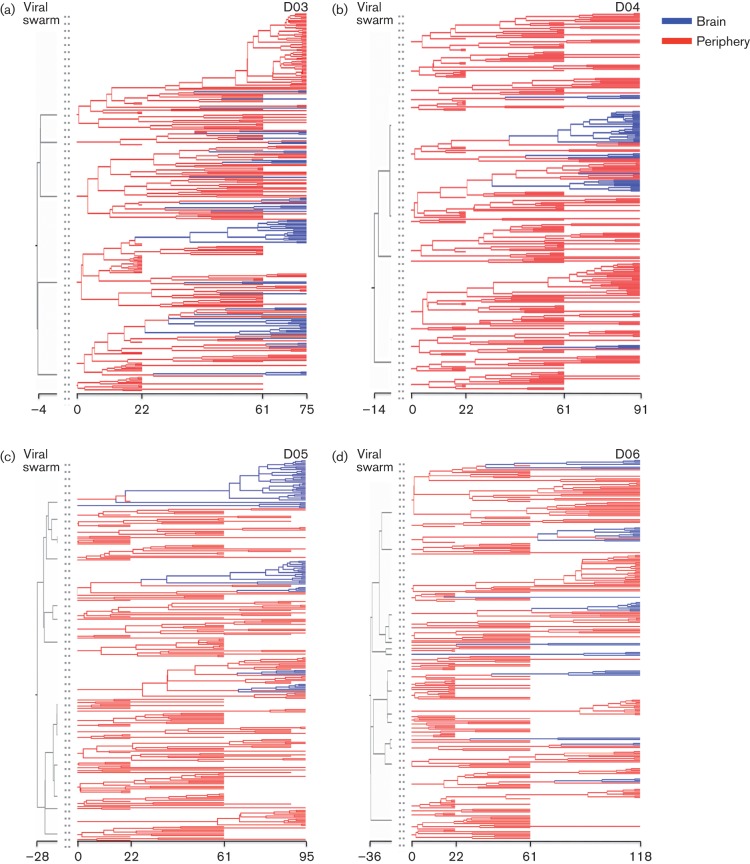
SIV gp120 Bayesian maximum clade credibility trees. The maximum clade credibility tree for each of the primates followed longitudinally (D03–D06) was estimated from the posterior distribution of trees obtained with a Bayesian coalescent framework enforcing a relaxed molecular clock. Branch lengths are scaled in time (days), and are coloured to represent peripheral (red) and brain (blue) tissues. Grey branches represent coalescent events of the viral swarm (pre-transmission interval) prior to inoculation. Viral migration events between the peripheral and brain tissues are represented as colour changes at individual nodes.
Bayesian phylogeography of SIVmac251 brain infection
The spatial dynamics of SIV intra-host evolution in the infected animals was investigated using sequence compartmentalization analyses and the Bayesian phylogeographic framework. SIV migration patterns based on tree topology appeared similar amongst all macaques, with brain-derived sequences tending to cluster in multiple but independent monophyletic clades (Fig. 2). Significant compartmentalization of viral subpopulations infecting the brain for all macaques was confirmed using the Slatkin–Maddison (Fig. S1; Slatkin, 1989) and tree correlation coefficient test (Table S2; Slatkin, 1989). The genealogies consistently indicated multiple gene flow events seeding the brain of each animal, the majority of which were traced back to a common ancestor after 21 days p.i., although a few brain sequences shared common ancestry with peripheral sequences even prior to viral swarm inoculation (Figs 2 and 3a). The latter finding indicated the presence of a small number of early brain entry events (≤21 days p.i.), although an accurate migration time was difficult to infer due to increasing HPD intervals near the root of the tree. Nevertheless, early viral entry into the brain was consistent with previous studies (Kim et al., 2003; Williams & Hickey, 2002), and was also confirmed by the analysis of viral sequences isolated from several brain tissue samples of D01 and D02 sacrificed at 21 days p.i.
Fig. 3.
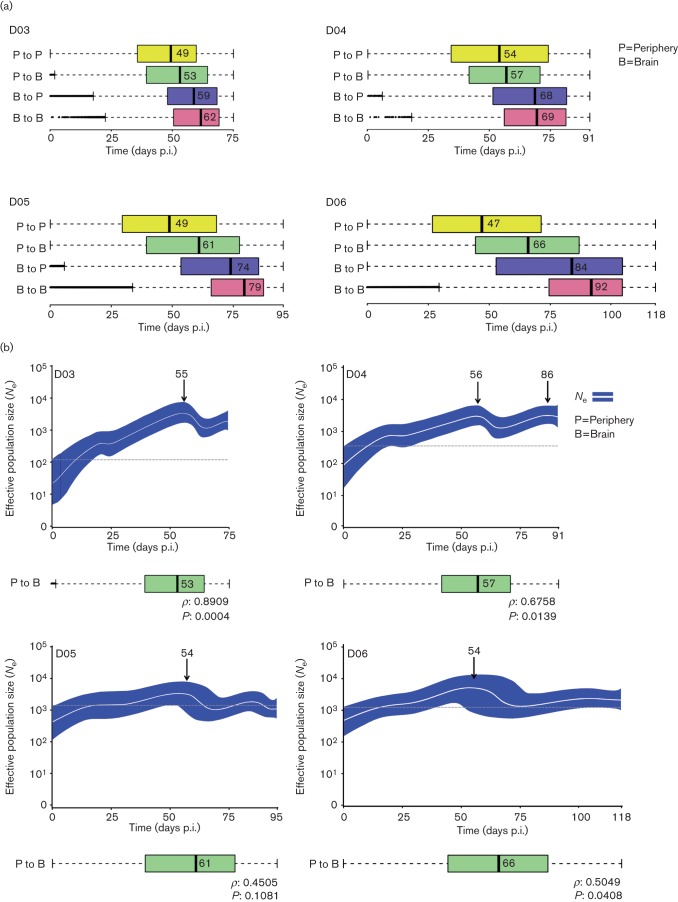
SIV phylogeographic and demographic patterns within primates followed longitudinally. (a) Viral gene flow (migration) events amongst different tissues within infected primates. Box plots represent the 95 % confidence intervals of the distribution of migration times, in days p.i., estimated by phylogeographic analysis. Broken lines represent outliers. Migrations within peripheral tissues (P to P) are displayed in yellow, from peripheral tissues to brain (P to B) in green, from brain to peripheral tissues (B to P) in blue and within brain tissues (B to B) in pink. Time intervals were inferred from a posterior distribution of trees obtained using a Bayesian coalescent framework enforcing a log-normal relaxed molecular clock with a Bayesian Skyride demographic prior and discrete phylogeographic parameter. The tissues are ordered according to the median migration time given by the number adjacent to the black vertical bar inside each box. (b) Bayesian Skyride plots with SIV migration times from peripheral tissues to the brain below. Bayesian Skyride plots show the change in Ne over time (days p.i.). The white line represents the median Ne with the surrounding blue area representing 95 % HPD intervals. Arrows indicate the time of the highest peak in Ne during the course of the infection. The dashed line highlights the fluctuations of Ne over time. Correlation between periphery to brain viral migration times and the Ne peak in the Skyride plots was analysed using Spearman’s rank order (correlation coefficient ρ and P value are given at the bottom of each plot).
In order to study in detail the timing of viral gene flow events with respect to initial infection, we compared the 95 % confidence intervals of the distributions of migration times between different tissues and cell types inferred from the posterior distribution of trees. SIV intra-host phylogeographic patterns were similar amongst macaques and revealed an intriguing temporal order: the initial migration amongst peripheral tissues occurred within 47–54 days p.i., followed by migration from peripheral tissues to brain 53–66 days p.i., from brain back to peripheral tissues within 59–84 days p.i., and ended with migrations confined to subcompartments within the brain ~62–92 days p.i. (Figs 3a and S2). The median time of viral migration from the periphery to the brain correlated significantly with the time at which peak viral Ne was observed for three of the four longitudinally monitored macaques (Fig. 3b).
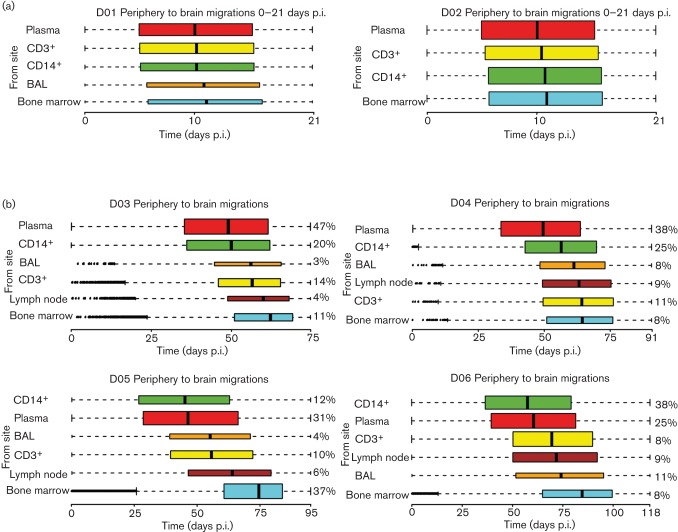
The temporal pattern of viral migrations from individual peripheral tissues to the brain was also assessed (Fig. 4). SIV strains infecting the brain originated from different tissues and/or cell types even prior to 3 weeks p.i. Early viral entry into the CNS from plasma, peripheral blood CD3+ and CD14+ cells, cells within the bronchoalveolar lavage fluid (BAL), and bone marrow was already evident in the two animals sacrificed at 21 days p.i., with a median migration time from periphery to brain occurring ~12 days p.i. (Fig. 4a). Interestingly, SIV phylogeographic analysis in macaques sampled longitudinally revealed that the majority of migration events from the periphery to the brain occurred later in infection (>12 days p.i.), and followed a specific and consistent temporal order (Fig. 4b). Initial viral migrations to the brain originated from plasma and CD14+ cells, trailed by contribution from cells within the BAL, CD3+ cells and lymph nodes, and followed, in all cases, by a final migration of viral strains from the bone marrow within 62–85 days p.i. for all longitudinally monitored macaques. In contrast, for the two macaques euthanized at 21 days p.i., brain infection from peripheral tissues did not exhibit a particular temporal order (Fig. 4a).
Fig. 4.
SIV gene flow events from peripheral tissues to the brain. Box plots show the 95 % confidence intervals of the distribution of migration times (days p.i.), estimated by phylogeographic analysis, from peripheral tissues to brain. Broken lines represent outliers. Time intervals were inferred from a posterior distribution of trees obtained with a Bayesian coalescent framework enforcing a relaxed molecular clock and a discrete phylogeographic parameter. The colour of the bar indicates a specific tissue or cell type: plasma (red), CD3+ T cells from peripheral blood (yellow), CD14+ monocytes from peripheral blood (green), BAL (orange), lymph nodes (brown) and bone marrow (cyan). The tissues are ordered according to the median time of brain entry (black vertical bar), with the earliest at the top and the latest at the bottom. The thickness of the bar is proportional to the percentage of the observed migrations (shown to the right of each bar for primates D03–D06). (a) Primates D01 and D02 euthanized at 21 days p.i. (b) Primates D04–D06 followed longitudinally until development of SIVE.
Discussion
Infection of CD8+ lymphocyte-depleted rhesus macaques with SIV is a well-established rapid model of neuroAIDS. In the present study, longitudinal samples derived from a multitude of animal peripheral as well as post-mortem brain tissues were used to evaluate the contribution of viral intra-host evolution and gene flow (migration) to the establishment and maintenance of SIV infection in the brain. Results of the Bayesian phylodynamic and phylogeographic analyses for macaques with varying forms of neuropathology provided significant insight into the interplay between viral evolution and disease progression.
Similar demographic patterns were observed for all of the longitudinally sampled macaques, consisting of a steady increase in Ne, i.e. number of genomes effectively contributing to the next generation, that culminated at ~55 days p.i. This similarity might reflect the shared characteristic of impaired innate and adaptive immunity in the CD8+ lymphocyte-depleted model. However, the population growth rate was elevated for primates D03 and D04 compared with D05 and D06, possibly related to the difference in age between macaques at the time of virus inoculation, in agreement with previous findings indicating that increased rate of disease progression correlates with increase in age at seroconversion (Langford et al., 2007). However, patterns in viral load over time were consistent amongst all of the longitudinally sampled macaques, with a peak occurring at ~55 days p.i., as with the peak in Ne. This is not unexpected, as changes over time in Ne are not linearly related to viral load measurements (Pennings, 2012). Individual phylogenetic tree topologies and compartmentalization statistics for each of the macaques also appeared similar, with significantly compartmentalized virus within the brain, particularly for sequences from the temporal and parietal lobes, comprising monophyletic clades within the tree seeded by peripheral tissues. This contribution from all of the peripheral tissues to virus in the brain was also observed for the gene flow analysis and occurred throughout the course of infection for all four longitudinally sampled macaques. The majority of these migrations occurred during 53–66 days p.i., which correlated significantly with the temporal peak in Ne for three of the four longitudinally monitored macaques. This correlation suggests a relationship between viral migration to the brain and an increase in viral diversity. However, the median time of viral migration from each of the individual tissues for the longitudinally sampled macaques appeared to be ordered temporally, with migration from the bone marrow consistently occurring last. This pattern was not observed in macaques euthanized early, which may reflect a lower resolution of the phylogenetic reconstruction inferred from sequences sampled at a single time point. However, the rapid expansion of the viral population that occurs during the primary stage of infection differs remarkably from the population dynamics during chronic infection. Therefore, our data likely indicate that albeit viral seeding of the brain from several peripheral tissues, including the bone marrow, occurs in early infection, subsequent waves of migration events to the brain take place during progression to SAIDS, following a specific temporal order that can only be discerned by longitudinal sampling. Overall, the combined results of the phylodynamic and phylogeographic analyses indicate two important events during rapid neuroAIDS progression in these animals: (1) a peak in viral diversity related to the migration of virus from peripheral tissues to the brain, consistently followed by (2) viral migration from the bone marrow to the brain.
The observation of both temporal and spatial patterns with respect to the establishment and maintenance of SIV in the brain is informative considering the previous historical roadblocks in assessing early HIV evolution in humans, in which the timing of infection and viral source is often unknown, and the collection of longitudinal biopsies for experimental research would be unethical. Furthermore, due to the growing body of research incorporating SIV infection in the CD8+ lymphocyte-depleted rhesus macaque model, studies concerning how the virus evolves in this model are increasingly critical to our understanding of infection and immune dynamics (Annamalai et al., 2010a, b; Burdo et al., 2012; Kim et al., 2010; Ratai et al., 2011; Soulas et al., 2011; von Gegerfelt et al., 2010; Williams & Burdo, 2012). In our investigation, we were able to show that, even within the relatively short lifespan of these macaques, meaningful estimates of evolutionary rates and population dynamics over time could be obtained. Whilst increased viral evolutionary rate, massive infection and poor control of viral replication were expected given the impaired innate immune response (Burdo et al., 2010; Kim et al., 2003, 2010; Ratai et al., 2011), the consistency of demographic patterns amongst different animals strengthens confidence in the results.
The rapid disease model also provides a unique platform for assessing the application of an adapted Bayesian phylogeographic framework to the study of intra-host viral evolution. The relatedness of intra-host sequences often hinders the ability to determine accurate phylogeographic inferences (Faria et al., 2011); however, the use of an extensive sequence dataset comprised of longitudinally collected sequences from several different tissues enabled us to infer viral transition times and pathways amongst different tissues and/or cell types within the macaque hosts, as well as the contribution of these patterns to varying forms of AIDS-related neuropathology.
NeuroAIDS has long been considered a macrophage-mediated disease, characterized by HIV/SIV infection of brain-derived macrophages, resulting in activation and the production of inflammatory cytokines and other soluble proteins that impact the overall integrity of CNS cells (Lamers et al., 2012). Studies have shown that there is an increased turnover of CNS-derived activated perivascular macrophages, as well as an increase in the rate and magnitude of monocyte trafficking from the bone marrow in animals that develop SIVE (Burdo et al., 2010; Williams & Burdo, 2012). Data shown here provide further support for these findings and a critical insight into the temporality of this occurrence. The late migration time of virus from the bone marrow to the brain observed in this study is consistent with the late clinical onset of symptomatic HANDs, commonly associated with the onset of AIDS (McCombe et al., 2013). Although early viral entry and continual seeding of the brain from peripheral tissues may contribute to neuropathology, our findings suggest a more central role for SIV-infected cells within the bone marrow in the progression or even onset of neuroAIDS during the later stage of the infection. In addition, once the virus had entered the brain, our data not only revealed compartmentalization of virus within this tissue, but also viral gene flow back to peripheral tissues later in infection – an observation highlighting the significance of the CNS as a potential viral reservoir capable of reseeding the body during cART.
In conclusion, the present work provides a better understanding of neuroAIDS pathogenesis in SIV-infected CD8+ lymphocyte-depleted macaques and reveals that SIV-infected cells within the bone marrow may contribute significantly to neuropathogenesis. Targeting infection and migration of bone marrow cells should be a focus in future studies related to treatment and prevention of HIV/SIV-associated neurological impairment. Furthermore, characterization of the specific infected cell types and microenvironment of the bone marrow could provide insight into the environmental factors and mechanisms driving the emergence of neurovirulent variants within this tissue prior to migration to the CNS.
Methods
Study population.
These studies were performed with the approval of the Tulane University’s Institutional Animal Care and Use Committee (IACUC). Macaques were housed at the Tulane National Primate Research Center in accordance with standards of the American Association for Accreditation of Laboratory Animal Care and Tulane IACUC protocol 3497. All possible measures were taken to minimize discomfort of the animals and the guidelines for humane euthanasia of rhesus macaques were followed. Ketamine (10–20 mg kg−1, intramuscularly) was used for anaesthetization prior to euthanization by intravenous injection of pentobarbital overdose and exsanguination.
The study design timeline is outlined in Fig. S3 and has previously been described in detail (Strickland et al., 2012). A total of six rhesus macaques of Indian origin were intravenously inoculated with the SIVmac251 (1 ng SIV p27) viral swarm at ~95 months of age. In order to achieve rapid disease progression and increased incidence of SIVE, animals were depleted of CD8+ lymphocytes via administration of mouse–rhesus chimeric anti-CD8a antibody, cM-T807 (10 mg kg−1), at 6, 8 and 12 days p.i. Antibody cMT807 was provided by the National Institutes of Health Non-human Primate Reagent Resource (RR016001, A1040101). CD8+ lymphocyte depletion was monitored by flow cytometry prior to antibody treatment and weekly thereafter, as described previously (Autissier et al., 2010; Burdo et al., 2010). Primates D01 and D02 were euthanized at 21 days p.i. to evaluate early evolutionary events. Primates D03, D04 and D05 were euthanized at the onset of SAIDS at 75, 91 and 95 days p.i., respectively, with confirmed SIVE at autopsy. D06 was euthanized at 118 days p.i. due to SAIDS, and showed perivascular cuffing and inflammation of meninges at autopsy (Strickland et al., 2012).
Sample collection and sequence generation.
As described previously (Strickland et al., 2012), plasma, CD3+ lymphocytes, CD14+ monocytes, unelicited BAL, lymph nodes and bone marrow were collected at three time points (21, 61 and 75–118 days), whilst meninges and brain tissues from parietal, frontal and temporal lobes were collected at necropsy. SIV gp120 sequences (SMM239 coordinates 6706–8049) were amplified, cloned and sequenced from each sampled tissue as described previously (Strickland et al., 2012). Although potential limitations of clonal analysis have been described, such as in vitro recombination, Taq polymerase-induced errors and resampling bias (Liu et al., 1996; Palmer et al., 2005; Shriner et al., 2004), it has been shown recently that similar levels of genetic diversity for HIV-1 sequences are inferred from sequences obtained by PCR/cloning and single-genome sequencing (Jordan et al., 2010). Moreover, as described previously (Strickland et al., 2012), our protocol uses a high-fidelity Taq polymerase, nested PCRs were optimized to reduce the impact of PCR-induced recombination by reducing the number of templates present in each reaction and potential recombinant sequences were removed from the dataset to avoid bias in the phylogeny inference (see below).
Sequences were aligned using the clustal (Thompson et al., 1997) algorithm implemented in BioEdit (Hall, 1999); the alignment was further modified by a manual optimization protocol taking into account conserved glycosylation motifs (Lamers et al., 1996). The highly variable region of the V1 domain was removed so as not to confound the genetic analysis (Salemi et al., 2009b). All alignments were gap-stripped for further analysis. Intra-host recombinants were determined using SplitsTree (Huson, 1998) as described previously (Salemi et al., 2009a) and were omitted from the dataset. Resulting sequences (SMM239 coordinates 6706–8049) are summarized in Table S3 (GenBank accession numbers JF764947–JF766081 and JQ608488–JQ609071).
Bayesian phylodynamic analysis.
SIV intra-host population dynamics, depicted as relative changes of the viral effective population size (Ne) over time, were investigated using the beast software package (Drummond & Rambaut, 2007; Drummond et al., 2005). Model testing was performed for both parametric and non-parametric demographic coalescent models, enforcing either a strict or relaxed molecular clock (Baele et al., 2012). A detailed description of the analysis is given in the Supplementary Methods.
Compartmentalization and intra-host Bayesian phylogeographic analysis.
Compartmentalization of SIV subpopulation(s) in brain tissues was evaluated by a modified version of the Slatkin–Maddison test for intra-host viral gene flow (Salemi et al., 2005; Slatkin, 1989), implemented in MacClade v4 (http://macclade.org/macclade.html), by using the posterior distribution of trees obtained from the beast analysis. This analysis was followed by a tree correlation coefficient test (Critchlow et al., 2000) implemented in HyPhy (Pond et al., 2005) for the maximum clade credibility trees for each animal. Statistical significance for the tree correlation coefficient test was determined based on a null distribution of coefficients generated using 1000 permutations.
Temporal and spatial gene flow patterns of SIV amongst different tissues were inferred using the discrete phylogeography coalescent framework also implemented in beast. Two non-parametric demographic models (BSP and Bayesian Skyride; see Supplementary Methods) were used, enforcing a relaxed molecular clock (selected as the best-fitting model) and a discrete phylogeographic parameter representing the sampled tissue and/or cell type. An additional computational tool was implemented in beast r5017 to allow a special history log file to be generated, reporting within a specified period (migration period) the inferred transition along the branches at different time points, as drawn from their posterior distribution. The code to generate the special history report is shown in Supplementary Document DS1 and complete .xml files used for the study are available upon request.
Statistical analysis.
The Wilcoxon rank-sum test was used to assess the significance of the correlation of the timing of viral gene flow (migrations) from/to different tissues and the peak in Ne. Statistical significance was assumed with P<0.05. All tests were performed in using the SAS statistical analysis software and r v2.15.2 (http://www.R-project.org/; r Core Team, 2012).
Acknowledgements
This work was supported by National Institutes of Health (R01 NS063897-01A2). We are grateful to Tyler Strickland for assistance in writing computational algorithms to facilitate this study and to Rebecca Rose for assistance with analytical pipeline development.
Footnotes
Supplementary material is available with the online version of this paper.
References
- Alfahad T. B., Nath A. (2013). Update on HIV-associated neurocognitive disorders. Curr Neurol Neurosci Rep 13, 387. 10.1007/s11910-013-0387-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G., Suscovich T. J., Teigen N., Meier A., Streeck H., Brander C., Altfeld M. (2007). Single-stranded RNA derived from HIV-1 serves as a potent activator of NK cells. J Immunol 178, 7658–7666. 10.4049/jimmunol.178.12.7658 [DOI] [PubMed] [Google Scholar]
- Annamalai L., Bhaskar V., Pauley D. R., Knight H., Williams K., Lentz M., Ratai E., Westmoreland S. V., González R. G., O’Neil S. P. (2010a). Impact of short-term combined antiretroviral therapy on brain virus burden in simian immunodeficiency virus-infected and CD8+ lymphocyte-depleted rhesus macaques. Am J Pathol 177, 777–791. 10.2353/ajpath.2010.091248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai L., Westmoreland S. V., Domingues H. G., Walsh D. G., Gonzalez R. G., O’Neil S. P. (2010b). Myocarditis in CD8-depleted SIV-infected rhesus macaques after short-term dual therapy with nucleoside and nucleotide reverse transcriptase inhibitors. PLoS ONE 5, e14429. 10.1371/journal.pone.0014429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autissier P., Soulas C., Burdo T. H., Williams K. C. (2010). Immunophenotyping of lymphocyte, monocyte and dendritic cell subsets in normal rhesus macaques by 12-color flow cytometry: clarification on DC heterogeneity. J Immunol Methods 360, 119–128. 10.1016/j.jim.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G., Lemey P., Bedford T., Rambaut A., Suchard M. A., Alekseyenko A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29, 2157–2167. 10.1093/molbev/mss084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello G., Zanotto P. M., Iamarino A., Gräf T., Pinto A. R., Couto-Fernandez J. C., Morgado M. G. (2012). Phylogeographic analysis of HIV-1 subtype C dissemination in Southern Brazil. PLoS ONE 7, e35649. 10.1371/journal.pone.0035649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissel S. J., Wang G., Trichel A. M., Murphey-Corb M., Wiley C. A. (2006). Longitudinal analysis of activation markers on monocyte subsets during the development of simian immunodeficiency virus encephalitis. J Neuroimmunol 177, 85–98. 10.1016/j.jneuroim.2006.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo T. H., Soulas C., Orzechowski K., Button J., Krishnan A., Sugimoto C., Alvarez X., Kuroda M. J., Williams K. C. (2010). Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog 6, e1000842. 10.1371/journal.ppat.1000842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo T. H., Orzechowski K., Knight H. L., Miller A. D., Williams K. (2012). Dorsal root ganglia damage in SIV-infected rhesus macaques: an animal model of HIV-induced sensory neuropathy. Am J Pathol 180, 1362–1369. 10.1016/j.ajpath.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burudi E. M., Fox H. S. (2001). Simian immunodeficiency virus model of HIV-induced central nervous system dysfunction. Adv Virus Res 56, 435–468. 10.1016/S0065-3527(01)56035-2 [DOI] [PubMed] [Google Scholar]
- Chen M. F., Westmoreland S., Ryzhova E. V., Martín-García J., Soldan S. S., Lackner A., González-Scarano F. (2006). Simian immunodeficiency virus envelope compartmentalizes in brain regions independent of neuropathology. J Neurovirol 12, 73–89. 10.1080/13550280600654565 [DOI] [PubMed] [Google Scholar]
- Ciccozzi M., Equestre M., Costantino A., Marascio N., Quirino A., Lo Presti A., Cella E., Bruni R., Liberto M. C. & other authors (2012). Hepatitis C virus genotype 4d in Southern Italy: reconstruction of its origin and spread by a phylodynamic analysis. J Med Virol 84, 1613–1619. 10.1002/jmv.23384 [DOI] [PubMed] [Google Scholar]
- Critchlow D. L., Li S., Nourijelyani K., Pearl D. K. (2000). Some statistical methods for phylogenetic trees with application to HIV disease. Math Comput Model 32, 69–81 10.1016/S0895-7177(00)00120-5 [DOI] [Google Scholar]
- de Carvalho L. M., Santos L. B., Faria N. R., de Castro Silveira W. (2013). Phylogeography of foot-and-mouth disease virus serotype O in Ecuador. Infect Genet Evol 13, 76–88. 10.1016/j.meegid.2012.08.016 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Pybus O. G., Rambaut A., Forsberg R., Rodrigo A. G. (2003). Measurably evolving populations. Trends Ecol Evol 18, 481–488 10.1016/S0169-5347(03)00216-7 [DOI] [Google Scholar]
- Drummond A. J., Rambaut A., Shapiro B., Pybus O. G. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22, 1185–1192. 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- Faria N. R., Suchard M. A., Rambaut A., Lemey P. (2011). Toward a quantitative understanding of viral phylogeography. Curr Opin Virol 1, 423–429. 10.1016/j.coviro.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Smith T., Bell C., Croul S., Lewis M., Rappaport J. (2008). Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies. J Neurovirol 14, 318–326. 10.1080/13550280802132857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41, 95–98. [Google Scholar]
- Harezlak J., Buchthal S., Taylor M., Schifitto G., Zhong J., Daar E., Alger J., Singer E., Campbell T. & other authors (2011). Persistence of HIV-associated cognitive impairment, inflammation, and neuronal injury in era of highly active antiretroviral treatment. AIDS 25, 625–633. 10.1097/QAD.0b013e3283427da7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. (2008). Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol 62, 307–328. 10.1146/annurev.micro.62.081307.162912 [DOI] [PubMed] [Google Scholar]
- Huson D. H. (1998). SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73. 10.1093/bioinformatics/14.1.68 [DOI] [PubMed] [Google Scholar]
- Jordan M. R., Kearney M., Palmer S., Shao W., Maldarelli F., Coakley E. P., Chappey C., Wanke C., Coffin J. M. (2010). Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J Virol Methods 168, 114–120. 10.1016/j.jviromet.2010.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E. (1995). Bayes factors. J Am Stat Assoc 90, 773–795 10.1080/01621459.1995.10476572 [DOI] [Google Scholar]
- Kim W. K., Corey S., Alvarez X., Williams K. (2003). Monocyte/macrophage traffic in HIV and SIV encephalitis. J Leukoc Biol 74, 650–656. 10.1189/jlb.0503207 [DOI] [PubMed] [Google Scholar]
- Kim W. K., Sun Y., Do H., Autissier P., Halpern E. F., Piatak M., Jr, Lifson J. D., Burdo T. H., McGrath M. S., Williams K. (2010). Monocyte heterogeneity underlying phenotypic changes in monocytes according to SIV disease stage. J Leukoc Biol 87, 557–567. 10.1189/jlb.0209082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T., Mori K., Kawahara T., Ringler D. J., Desrosiers R. C. (1993). Analysis of simian immunodeficiency virus sequence variation in tissues of rhesus macaques with simian AIDS. J Virol 67, 6522–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppensteiner H., Brack-Werner R., Schindler M. (2012). Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology 9, 82. 10.1186/1742-4690-9-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner A. A. (1994). Pathology of simian immunodeficiency virus induced disease. Curr Top Microbiol Immunol 188, 35–64. [DOI] [PubMed] [Google Scholar]
- Lamers S. L., Sleasman J. W., Goodenow M. M. (1996). A model for alignment of Env V1 and V2 hypervariable domains from human and simian immunodeficiency viruses. AIDS Res Hum Retroviruses 12, 1169–1178. 10.1089/aid.1996.12.1169 [DOI] [PubMed] [Google Scholar]
- Lamers S. L., Gray R. R., Salemi M., Huysentruyt L. C., McGrath M. S. (2011a). HIV-1 phylogenetic analysis shows HIV-1 transits through the meninges to brain and peripheral tissues. Infect Genet Evol 11, 31–37. 10.1016/j.meegid.2010.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers S. L., Poon A. F., McGrath M. S. (2011b). HIV-1 nef protein structures associated with brain infection and dementia pathogenesis. PLoS ONE 6, e16659. 10.1371/journal.pone.0016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers S. L., Fogel G. B., Singer E. J., Salemi M., Nolan D. J., Huysentruyt L. C., McGrath M. S. (2012). HIV-1 Nef in macrophage-mediated disease pathogenesis. Int Rev Immunol 31, 432–450. 10.3109/08830185.2012.737073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford S. E., Ananworanich J., Cooper D. A. (2007). Predictors of disease progression in HIV infection: a review. AIDS Res Ther 4, 11. 10.1186/1742-6405-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T., Albert J. (1999). The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc Natl Acad Sci U S A 96, 10752–10757. 10.1073/pnas.96.19.10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Drummond A. J., Suchard M. A. (2009). Bayesian phylogeography finds its roots. PLOS Comput Biol 5, e1000520. 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Welch J. J., Suchard M. A. (2010). Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol 27, 1877–1885. 10.1093/molbev/msq067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Rodrigo A. G., Shankarappa R., Learn G. H., Hsu L., Davidov O., Zhao L. P., Mullins J. I. (1996). HIV quasispecies and resampling. Science 273, 415–416. 10.1126/science.273.5274.415 [DOI] [PubMed] [Google Scholar]
- Mankowski J. L., Clements J. E., Zink M. C. (2002). Searching for clues: tracking the pathogenesis of human immunodeficiency virus central nervous system disease by use of an accelerated, consistent simian immunodeficiency virus macaque model. J Infect Dis 186 (Suppl 2), S199–S208. 10.1086/344938 [DOI] [PubMed] [Google Scholar]
- McCombe J. A., Vivithanaporn P., Gill M. J., Power C. (2013). Predictors of symptomatic HIV-associated neurocognitive disorders in universal health care. HIV Med 14, 99–107. 10.1111/j.1468-1293.2012.01043.x [DOI] [PubMed] [Google Scholar]
- Palmer S., Kearney M., Maldarelli F., Halvas E. K., Bixby C. J., Bazmi H., Rock D., Falloon J., Davey R. T., Jr & other authors (2005). Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 43, 406–413. 10.1128/JCM.43.1.406-413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings P. S. (2012). Standing genetic variation and the evolution of drug resistance in HIV. PLOS Comput Biol 8, e1002527. 10.1371/journal.pcbi.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S. L., Frost S. D., Muse S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- Pulliam L., Gascon R., Stubblebine M., McGuire D., McGrath M. S. (1997). Unique monocyte subset in patients with AIDS dementia. Lancet 349, 692–695. 10.1016/S0140-6736(96)10178-1 [DOI] [PubMed] [Google Scholar]
- r Core Team (2012). r: A Language and Environment for Statistical Computing. Vienna: r Foundation for Statistical Computing. [Google Scholar]
- Ratai E. M., Pilkenton S., He J., Fell R., Bombardier J. P., Joo C. G., Lentz M. R., Kim W. K., Burdo T. H. & other authors (2011). CD8+ lymphocyte depletion without SIV infection does not produce metabolic changes or pathological abnormalities in the rhesus macaque brain. J Med Primatol 40, 300–309. 10.1111/j.1600-0684.2011.00475.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzhova E., Whitbeck J. C., Canziani G., Westmoreland S. V., Cohen G. H., Eisenberg R. J., Lackner A., González-Scarano F. (2002a). Rapid progression to simian AIDS can be accompanied by selection of CD4-independent gp120 variants with impaired ability to bind CD4. J Virol 76, 7903–7909. 10.1128/JVI.76.15.7903-7909.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzhova E. V., Crino P., Shawver L., Westmoreland S. V., Lackner A. A., González-Scarano F. (2002b). Simian immunodeficiency virus encephalitis: analysis of envelope sequences from individual brain multinucleated giant cells and tissue samples. Virology 297, 57–67. 10.1006/viro.2002.1395 [DOI] [PubMed] [Google Scholar]
- Salemi M., Lamers S. L., Yu S., de Oliveira T., Fitch W. M., McGrath M. S. (2005). Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J Virol 79, 11343–11352. 10.1128/JVI.79.17.11343-11352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M., Burkhardt B. R., Gray R. R., Ghaffari G., Sleasman J. W., Goodenow M. M. (2007). Phylodynamics of HIV-1 in lymphoid and non-lymphoid tissues reveals a central role for the thymus in emergence of CXCR4-using quasispecies. PLoS ONE 2, e950. 10.1371/journal.pone.0000950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M., Lamers S. L., Huysentruyt L. C., Galligan D., Gray R. R., Morris A., McGrath M. S. (2009a). Distinct patterns of HIV-1 evolution within metastatic tissues in patients with non-Hodgkins lymphoma. PLoS ONE 4, e8153. 10.1371/journal.pone.0008153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M., Vandamme A.-M., Lemey P. (2009b). The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. Cambridge: Cambridge University Press. [Google Scholar]
- Schmitz J. E., Kuroda M. J., Santra S., Sasseville V. G., Simon M. A., Lifton M. A., Racz P., Tenner-Racz K., Dalesandro M. & other authors (1999). Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283, 857–860. 10.1126/science.283.5403.857 [DOI] [PubMed] [Google Scholar]
- Shriner D., Rodrigo A. G., Nickle D. C., Mullins J. I. (2004). Pervasive genomic recombination of HIV-1 in vivo. Genetics 167, 1573–1583. 10.1534/genetics.103.023382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simioni S., Cavassini M., Annoni J. M., Rimbault Abraham A., Bourquin I., Schiffer V., Calmy A., Chave J. P., Giacobini E. & other authors (2010). Cognitive dysfunction in HIV patients despite long-standing suppression of viremia. AIDS 24, 1243–1250. [DOI] [PubMed] [Google Scholar]
- Slatkin M. (1989). Detecting small amounts of gene flow from phylogenies of alleles. Genetics 121, 609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. M., Zárate S., Shao H., Pillai S. K., Letendre S. L., Wong J. K., Richman D. D., Frost S. D., Ellis R. J., HNRC Group (2009). Pleocytosis is associated with disruption of HIV compartmentalization between blood and cerebral spinal fluid viral populations. Virology 385, 204–208. 10.1016/j.virol.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulas C., Conerly C., Kim W. K., Burdo T. H., Alvarez X., Lackner A. A., Williams K. C. (2011). Recently infiltrating MAC387+ monocytes/macrophages a third macrophage population involved in SIV and HIV encephalitic lesion formation. Am J Pathol 178, 2121–2135. 10.1016/j.ajpath.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss-Ayali D., Conrad S. M., Mosser D. M. (2007). Monocyte subpopulations and their differentiation patterns during infection. J Leukoc Biol 82, 244–252. 10.1189/jlb.0307191 [DOI] [PubMed] [Google Scholar]
- Strickland S. L., Gray R. R., Lamers S. L., Burdo T. H., Huenink E., Nolan D. J., Nowlin B., Alvarez X., Midkiff C. C. & other authors (2012). Efficient transmission and persistence of low-frequency SIVmac251 variants in CD8-depleted rhesus macaques with different neuropathology. J Gen Virol 93, 925–938. 10.1099/vir.0.039586-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gegerfelt A., Valentin A., Alicea C., Van Rompay K. K., Marthas M. L., Montefiori D. C., Pavlakis G. N., Felber B. K. (2010). Emergence of simian immunodeficiency virus-specific cytotoxic CD4+ T cells and increased humoral responses correlate with control of rebounding viremia in CD8-depleted macaques infected with Rev-independent live-attenuated simian immunodeficiency virus. J Immunol 185, 3348–3358. 10.4049/jimmunol.1000572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J. O., Worobey M. (2009). Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLOS Comput Biol 5, e1000377. 10.1371/journal.pcbi.1000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland S. V., Halpern E., Lackner A. A. (1998). Simian immunodeficiency virus encephalitis in rhesus macaques is associated with rapid disease progression. J Neurovirol 4, 260–268. 10.3109/13550289809114527 [DOI] [PubMed] [Google Scholar]
- Williams K., Burdo T. H. (2012). Monocyte mobilization, activation markers, and unique macrophage populations in the brain: observations from SIV infected monkeys are informative with regard to pathogenic mechanisms of HIV infection in humans. J Neuroimmune Pharmacol 7, 363–371. 10.1007/s11481-011-9330-3 [DOI] [PubMed] [Google Scholar]
- Williams K. C., Hickey W. F. (2002). Central nervous system damage, monocytes and macrophages, and neurological disorders in AIDS. Annu Rev Neurosci 25, 537–562. 10.1146/annurev.neuro.25.112701.142822 [DOI] [PubMed] [Google Scholar]
- Wong J. K., Ignacio C. C., Torriani F., Havlir D., Fitch N. J., Richman D. D. (1997). In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol 71, 2059–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods S. P., Moore D. J., Weber E., Grant I. (2009). Cognitive neuropsychology of HIV-associated neurocognitive disorders. Neuropsychol Rev 19, 152–168. 10.1007/s11065-009-9102-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Galligan D. C., Lamers S. L., Yu S., Shagrun L., Salemi M., McGrath M. S. (2009). High level HIV-1 DNA concentrations in brain tissues differentiate patients with post-HAART AIDS dementia complex or cardiovascular disease from those with AIDS. Sci China C Life Sci 52, 651–656. 10.1007/s11427-009-0085-5 [DOI] [PubMed] [Google Scholar]
- Zink M. C., Spelman J. P., Robinson R. B., Clements J. E. (1998). SIV infection of macaques–modeling the progression to AIDS dementia. J Neurovirol 4, 249–259. 10.3109/13550289809114526 [DOI] [PubMed] [Google Scholar]
- Zink M. C., Laast V. A., Helke K. L., Brice A. K., Barber S. A., Clements J. E., Mankowski J. L. (2006). From mice to macaques – animal models of HIV nervous system disease. Curr HIV Res 4, 293–305. 10.2174/157016206777709410 [DOI] [PubMed] [Google Scholar]