Abstract
Background
Induced resistance (IR) can be part of a sustainable plant protection strategy against important plant diseases. β-aminobutyric acid (BABA) can induce resistance in a wide range of plants against several types of pathogens, including potato infected with Phytophthora infestans. However, the molecular mechanisms behind this are unclear and seem to be dependent on the system studied. To elucidate the defence responses activated by BABA in potato, a genome-wide transcript microarray analysis in combination with label-free quantitative proteomics analysis of the apoplast secretome were performed two days after treatment of the leaf canopy with BABA at two concentrations, 1 and 10 mM.
Results
Over 5000 transcripts were differentially expressed and over 90 secretome proteins changed in abundance indicating a massive activation of defence mechanisms with 10 mM BABA, the concentration effective against late blight disease. To aid analysis, we present a more comprehensive functional annotation of the microarray probes and gene models by retrieving information from orthologous gene families across 26 sequenced plant genomes. The new annotation provided GO terms to 8616 previously un-annotated probes.
Conclusions
BABA at 10 mM affected several processes related to plant hormones and amino acid metabolism. A major accumulation of PR proteins was also evident, and in the mevalonate pathway, genes involved in sterol biosynthesis were down-regulated, whereas several enzymes involved in the sesquiterpene phytoalexin biosynthesis were up-regulated. Interestingly, abscisic acid (ABA) responsive genes were not as clearly regulated by BABA in potato as previously reported in Arabidopsis. Together these findings provide candidates and markers for improved resistance in potato, one of the most important crops in the world.
Keywords: Functional annotation, Mevalonate pathway, Phytophthora infestans, Secretome, Solanum tuberosum, Sterol biosynthesis
Background
Potato is today the third largest food crop in the world. One of the largest threats to potato production is late blight disease, caused by Phytophthora infestans, which is currently controlled by frequent use of fungicides. Extensive efforts to find more efficient weapons against late blight are ongoing worldwide. One alternative could be integration of induced resistance (IR) in current management strategies. IR, i.e. exposing plants to abiotic or biotic stress leading to improved resistance to subsequent pathogen attack both locally and systemically [1], is thought to function by activating or enhancing defence signalling [2]. This signalling is suggested to depend either on salicylic acid (SA) or jasmonic acid/ethylene (JA/ET) for the two classical forms of IR: systemic acquired resistance (SAR) and induced systemic resistance (ISR), respectively [3]. Resistance can be induced by treatment of plants with the xenobiotic amino-acid derivative dL-β-aminobutyric acid (BABA) [4,5]. BABA can enhance resistance in a wide range of plants against several types of pathogens, e.g. treatment increases the resistance of potato against P. infestans[4].
We have earlier shown by microscopy that a direct activation of defence responses associated to the formation of HR-like lesions takes place at efficient BABA-IR in the potato Phytophthora system [6]. This is in contrast to a “priming effect” proposed in other plant-pathogen systems [7]. The molecular mechanisms behind BABA-IR remains unclear and a better understanding on the molecular level could lead to the identification of biomarkers which in the future can be implemented in plant protection or breeding strategies.
Two microarray studies investigating BABA-IR have been conducted in Arabidopsis. Tsai et al., 2011 [8], reported that SA-responsive genes were both directly up-regulated by BABA treatment and up-regulated after priming by BABA and subsequent Pseudomonas syringae pv. tomato DC3000 infection. They also showed that BABA inhibits the Arabidopsis response to the bacterial phytotoxin coronatine, thus suppressing the coronatine-associated JA responses. The second microarray study of BABA-IR in Arabidopsis conducted by Zimmerli et al., 2008 [9] showed that BABA priming for increased thermotolerance involved ABA-associated transcription factors. They also showed that neither ET nor SA played a part in the acquired thermotolerance. These two microarray studies in Arabidopsis demonstrate the complexity of BABA-IR mechanisms. Furthermore, BABA has been shown to reduce vegetative growth and to cause major alterations in plant amino acid balance in Arabidopsis, effects that were reversed by L-glutamine treatment [10,11]. Based on these results, it was suggested that BABA provokes a stress-induced morphogenic response (SIMR) in Arabidopsis by activation of a general amino acid inhibition response. Another report indicates that the lectin-receptor kinase VI.2 is required for BABA priming of the PTI response and for BABA-IR against Pseudomonas syringae DC3000 in Arabidopsis [12].
In potato, a very important host for many pathogens such as P. infestans, the only previous RNA study relating to BABA was a time-series study using cDNA-AFLP [13]. Around 60 potato transcripts were found to be differentially expressed after BABA treatment and a clear overlap of genes affected by BABA and Phytophthora inoculation was found. Studies in transgenic NahG expressing potato plants, with an impaired SA metabolism, indicate that BABA-IR against P. infestans is dependent on SA [14]. BABA has also been shown to cause a direct activation of basal defence responses such as reactive oxygen species, hypersensitive response-like spots, phenolics and PR-1 expression in potato [6]. To elucidate molecular defence responses activated by BABA in potato, a genome-wide microarray analysis based on the recently released Solanum tuberosum group phureja genome [15] was performed in combination with label-free quantitative proteomic analysis of the apoplast secretome. Earlier reports showed that mRNA regulation in mammalian cells only explains about 40% of the changes in protein abundance [16] and the apoplast is a major interaction arena between plants and pathogens.
Genome-wide analysis is often hampered by incomplete and outdated functional annotations of genes and derived microarray probes. To this end, we present a network-based approach to retrieve information from orthologous gene families across 26 sequenced plant genomes, which increased the number of annotated probes by close to 9000.
Methods
Plant material and growing conditions
Potato plants (cv. Desiree) were grown under controlled conditions with 16 h/8 h day and night regime with a fluorescent lamp light intensity of 200 μmol m-2 s-1. The temperature was set to 20°C and the relative humidity was 65%. Pots were circulated every week to avoid positional effects. All sampling was done before flowering and only fully expanded leaves from the middle part of the plants were used. Plants were treated with BABA or water 2 days before harvest. For the treatments plants were randomly chosen, labeled and then placed back in the growth chamber evenly dispersed between treatments. For the detached leaf assays we used 29-day-old plants and for the molecular analysis we used 35-day-old plants.
BABA treatment
Plants were treated with BABA (DL-3-aminobutyric acid, Sigma) dissolved in water at a concentration of 1.0 or 10 mM. BABA was sprayed on the plants until saturation (ca 40 mL) and control plants were sprayed with water as earlier described [6].
Phytophthora inoculation and measurement
Four detached leaflets from each of three Desiree plants were infected with P. infestans two days after treatment with either 10 mM BABA or water (control). P. infestans strain SE03058 isolated in mid-Sweden (mating type A1, virulence 1, 2, 3, 4, 7, 10, 11) was used with a sporangial concentration of 36,000 sporangia ml-1. The inoculated leaflets were kept in the dark at 15°C during the first day. On the second day the temperature was changed to 20°C with 16 h light/8 h dark. The size of lesion diameter was measured 7 days post inoculation (dpi). Data from two separate experiments were combined for a total of 18 biological samples. The difference in lesion sizes between the treatments were analysed by Student’s t-test with Minitab v.16.
Gene ontology annotation and enrichment analysis
Genome annotation is commonly done by orthologous inference, the transfer of annotation across genomes based on orthologous relationships between genes in different genomes. There are a number of different algorithms available to determine orthologous relationships [17]. OrthoMCL is an orthology detection method which clusters genes into orthologous gene families based on sequence similarity [18]. However, the OrthoMCL software can currently not be applied to large datasets. Thus a parallel version of OrthoMCL (Parallel-OrthoMCL) was developed in house in order to allow the OrthoMCL algorithm to be applied to large datasets. Orthologous gene families across 26 plant species were constructed using Parallel-OrthoMCL. 25 plant species available from PLAZA [19] were included in the analysis and the protein translations of the longest representative transcript from the Solanum tuberosum group phureja genome was downloaded from the Solanaceae Genomics Resource [ftp://ftp.plantbiology.msu.edu/pub/data/SGR/GO_annotations/]. The OrthoMCL percent match parameter was set to 50% and the MCL inflation value was set to 2.0. These are values commonly used by other genome annotation pipelines such as PLAZA [19]. Once gene families had been constructed, potato genes were annotated using a network-based annotation as follows: (i) all available GO annotations in PLAZA were mapped onto the appropriate genes in each of the 25 species contained in PLAZA; (ii) each potato gene was then assigned the GO terms of all the other genes from the 25 other species that were present in its specific gene family: (iii) the probes from the potato microarray (Agilent JHI Solanum tuberosum 60 k v1) were then annotated by assigning to each probe the GO terms of the genes corresponding to that probe as determined by BLASTing the probe sequences against the nucleotide transcript sequences from the potato genome. The probes of the potato microarray were also annotated using the potato gene GO annotation file downloaded from the Solanaceae Genomics Resource (SGR annotation). As described above, probes were annotated by assigning each probe the GO terms of the genes corresponding to that probe. The two microarray probe annotations were compared to identify the number of probes annotated in each annotation and also to identify the distribution of the number of GO terms assigned to each probe.
RNA sample preparation and microarray analysis
In total one leaflet from each of three plants per treatment was used for RNA analysis. Five leaf discs of 10 mm diameter were punched out of each leaflet, pooled, and immediately frozen in liquid nitrogen. The frozen samples were homogenized for 30 s at 30 rpm in a Retsch Mixer Mill MM 200 (Retsch GmbH, Haan, Germany) followed by extraction of RNA, DNase treatment and purification using RNeasy Mini kit (Qiagen GmbH, Hilden, Germany). RNA concentration and purity (260/280 nm >1.8) was checked by a ND-1000 NanoDrop (Wilmington, USA) and integrity of the samples were analyzed with an Experion™ Automated Electrophoresis System (Bio-Rad Laboratories, Hercules, USA). Each sample was diluted in RNase-free water to a concentration of 200 ng/μl and mRNA expression analysis was done with a custom-made Agilent expression array based on the predicted transcripts of the potato genome (version 3.4). The complete microarray design is available in ArrayExpress (E-MTAB-1655). The arrays were run according to the instructions and recommendations of the supplier (Agilent). The probe intensities were background corrected and normalised using the quantile method in the Limma R-package [20]. Fold changes and standard errors were obtained by fitting a linear model to each gene expression values and standard errors were smoothed based on the empirical Bayes method. Genes with p-values below 0.05 after adjustment (Benjamini-Hochberg) were regarded as significant. The microarray data was deposited in ArrayExpress, accession number: E-MTAB-1545. Significantly differentially expressed genes were analyzed for Enrichment of Gene Ontology Terms in GOEast using default settings on the basis of a custom made GO enrichment list for the microarray. To give an overview of all significantly enriched GO terms ReviGO was applied to cluster terms based on semantics [21]. Based on the potato genome (version 3.4) and manual curation, genes and proteins were functionally characterized by the MapMan mapping file.
Secretome extraction and visualisation
Two days after BABA treatment, three biological replicates in the form of separate plants were subjected to analysis. Four leaflets per plant were harvested on ice. Secretome proteins were extracted by vacuum infiltration with a phosphate buffer directly after harvest followed by centrifugation as described by Ali et al., 2012 [22] and Alexandersson et al., 2013 [23]. For visualisation of the apoplastic proteins, 30 μl of the samples were separated on 14% SDS polyacrylamide gels. The samples were mixed with 6 × SDS sample buffer containing DTT and denatured at 93°C for 3 min prior to electrophoresis. The gels were run for 20 min at 80 V followed by 1.5 h at 100 V and staining overnight in Coomassie Brilliant Blue (10% (v/v) acetic acid, 45% (v/v) methanol, and 0.25% (w/v) Coomassie R-250). Finally, the gels were briefly washed in dH2O before 3 h in destaining solution (10% (v/v) acetic acid and 40% (v/v) methanol).
Identification and quantification of proteins by LC-MS/MS
For identification of the apoplastic proteins, 30 μl of the secretome samples were separated with SDS-PAGE as described for the visualisation of proteins, but the gel was only run 10 mm. The gels were stained with Coomassie Brilliant Blue and the stained portions cut into 1–2 mm2 pieces. Tryptic digestion of the gel pieces and peptide elution was carried out as described by Ali et al., 2012 [22]. The eluted peptides were concentrated to 50 μl by rotary evaporation and stored at -80°C. Six μl of sample was subjected to HPLC-MSMS analysis using an Eksigent nanoLC2D HPLC system coupled to an Orbitrap XL. The peptides were loaded onto a pre-column (Agilent Zorbax 300SB-C18, 0.3 mm ID, 5 mm, 5 μm particle size) connected to an analytical column (Agilent Zorbax 300SB C18, 75 μm ID, 150 mm, 3.5 μm particle size). The analytical column was pre-equilibrated for 10 min using buffer consisting of 0.1% formic acid (FA), 5% acetonitrile (ACN) at a flow rate of 10 μl/min, and the peptides were separated in an 0.1 FA buffer using a 55 min linear gradient from 5% to 40% ACN followed by a 5 min linear gradient from 40% to 80% ACN at a flow rate of 350 nl/min. The eluted peptides were analysed using an LTQ Orbitrap, operated in data-dependent mode to automatically perform Orbitrap-MS and LTQ-MSMS analysis. Survey scan spectra (400–2000 Da) were acquired using the Orbitrap mass analyzer with the resolution R = 60000. Automatic gain control was enabled. The seven most intense ions were selected for fragmentation in the LTQ, using a mass window of 2 Da for precursor ion selection. The precursor ions were fragmented with normalised collision energy of 35 (with activation Q set to 0.25 and an activation time of 30 ms). Dynamic exclusion was enabled with a repeat count of 2, a repeat duration of 20 seconds, exclusion duration of 120 seconds, an exclusion list size of 499 and a 10 ppm exclusion mass width relative to both low and high. This raw data was then converted to mgf files using ProteoWizard [24]. The Proteios software environment [25] was used to search the files with Mascot against a database consisting of the protein sequences from the potato genome and all Solanaceae sequences from UniProt extended with an equal size random part, with conserved protein length and amino acid distribution for the random part. Search tolerances were set to 5 ppm for MS and 0.5 Da for MS/MS. One missed cleavage was allowed. Carbamidomethylation of cysteine residues was selected as a fixed modification and oxidation of methionine residues was selected as a variable modification. Proteios was used to identify hits at an estimated peptide spectrum match false discovery rate (FDR) of 0.01 using the random database mentioned above. For quantitative analysis, a label-free approach based on precursor intensities was used [26], with msInspect feature detection [27], and Proteios alignment as described previously [28]. Quantitative data from different conditions was normalised using a linear transformation procedure where the intensity of each feature was divided by the sum of all features in the same run and then multiplied by the average of the summed intensities of all runs for that replicate. Missing values were set to 1. This data was then log-transformed and analysed using an ANOVA based approach in the Qlucore software (http://www.qlucore.se). Peptides that were differentially abundant were identified by performing a two-group comparison Qlucore with a FDR < 0.1 according to the Benjamini-Hochberg procedure for determining adjusted p-value, q. Only peptides uniquely identifying one protein in the potato genome (version 3.4) were used in order to avoid ambiguous peptides identifying several similar proteins.
Results
P. infestans growth after BABA treatment
To find out if BABA at a concentration of 10 mM could restrict P. infestans growth in potato cv. Desiree, a detached leaf assay was performed. BABA led to significantly reduced lesion sizes of P. infestans in Desiree at 7 dpi (p = 0.012; Figure 1). Small necrotic lesions were visible on the leaves after treatment with BABA at 10 mM but not 1 mM (not shown). These were not observed in the controls, where leaves were treated with water. BABA at 1 mM was not tested since we earlier demonstrated a threshold level of 2.5 mM BABA for successful restriction of P. infestans growth in potato [29].
Figure 1.
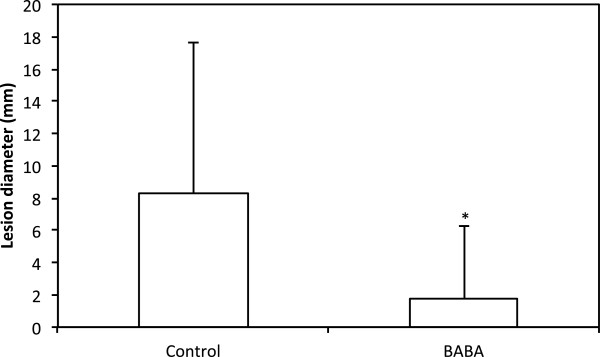
Detached leaf assay. Mean lesion sizes of P. infestans infection in potato leaves, 7 dpi. Leaflets (N = 18) were infected two days after treatment with BABA (10 mM) and water (control). Bars represent Standard Error of the mean. Treatment with a significant effect (p <0.05) is marked with *.
New network-based annotation of potato genes
Network-based annotation of the potato microarray probes and PGSC gene models applying parallelised OrthoMCL across 26 sequenced plant genomes was compared to the annotation of the potato microarray probes provided by Solanaceae Genomics Resource (SGR). Of the available ortholog detection algorithms, OrthoMCL has previously been shown to have the best performance (and the lowest false positive rate) in ortholog detection across multiple genomes [17]. As can be seen from Figure 2, 8616 additional probes out of a total of 45126 receive an annotation based on the 26 genomes with the SGR annotation. The number of ontology terms assigned to a probe is one of the indicators of the amount of information linked to the probe. In comparison to the earlier annotation, the number of GO terms assigned per probe was much larger with a median of 34 GO terms assigned per probe for the new annotation versus 6 for the SGR annotation (Additional file 1: Figure S1). Thus, the new annotation using gene families constructed from 26 plant genomes provides a more complete annotation of the potato microarray than was possible with the annotation file provided by the SGR.
Figure 2.
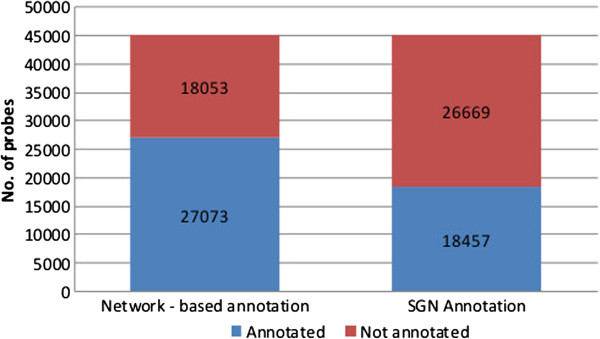
Annotation Coverage. A comparison of the number of annotated and un-annotated probes of the Agilent JHI Solanum tuberosum 60 k v1 microarray, using network based annotation (new annotation) and Solanaceae Genomics Resource (SGR) annotation.
Overall transcript and protein changes
In the microarray analysis, 5378 transcripts were significantly (adj. p-value <0.05) differentially expressed in potato leaf 48 h after treatment with 10 mM BABA (Table 1). In the corresponding apoplast secretome samples, 91 proteins (50 up- and 41 down-regulated) were found to change in abundance (Table 1). BABA applied at the lower concentration (1 mM) had almost no influence on gene expression. Only six differentially expressed transcripts were identified (1 up- and 5 down-regulated), in contrast to 24 apoplastic proteins (11 up- and 13 down-regulated) with a significantly changed abundance (Tables 1 and 2). The only transcript up-regulated by 1 mM BABA, a MutT domain protein (G400017400), was also up-regulated by 10 mM BABA.
Table 1.
Overview of BABA-induced changes
| BABA concentration |
Gene expression |
Secreted proteins |
||
|---|---|---|---|---|
| Up | Down | Up | Down | |
| 1 mM |
1 |
5 |
11 |
15 |
| 10 mM | 3272 | 2106 | 50 | 42 |
Total number differentially expressed transcripts (adj. p-value <0.05) and proteins (FDR < 0.1) with altered abundance in leaves (cv. Desiree) treated with BABA (1 and 10 mM). Samples were analysed two days after treatment of the leaf canopy with BABA.
Table 2.
Differentially expressed transcripts in potato leaves treated with 1 mM BABA
| Protein name | Gene id | Protein id | log2 Fold change |
|---|---|---|---|
|
Protein phosphorylation |
|
|
|
| WAK-like kinase |
G400010032 |
P400017740 |
-1.4 |
|
Metabolic processes |
|
|
|
| MutT/nudix domain protein |
G400017400 |
P400030392 |
3.5 |
| Glucan/water dikinase |
G400016613 |
P400029040 |
-1.0 |
|
Unknown function |
|
|
|
| Hypothetical gene of unknown function |
G400044248 |
P400066352 |
-1.6 |
| Hypothetical gene of unknown function |
G400031825 |
P400055091 |
-1.4 |
| Uncharacterized mitochondrial protein AtMg00030 | G400019855 | P400034471 | -1.3 |
Samples were analysed two days after treatment of the leaf canopy with BABA. Transcripts with significantly changed expression (adj. p-value <0.05) are included in the table.
The overlap of regulation found between the transcript and protein level was low. Of the 91 proteins differentially regulated by 10 mM BABA, only 13 overlapped at the transcript level. Ten out of these were up-regulated both at the transcript and protein levels, i.e. four endochitinases (G400033882, G400008796, G400008673, G400001528), three glucan endo-1,3-beta-glucosidases (G400029830, G402000722, G400010490), two peroxidases (G400012589, G400027614) and one DUF26 domain-containing protein 1 (G400041220). Interestingly, three genes were down-regulated at the protein level but up-regulated at the transcript level, namely pectin acetylesterase (PAE) (G014024140), serine-threonine protein kinase (G400016908) and xylem serine proteinase 1 (G400019147).
Gene ontology analysis and gene expression after 10 mM BABA treatment
With more than 5000 transcripts affected by 10 mM BABA, it is evident that many functional mechanisms were altered. In order to identify processes where gene expressional changes occur, a gene ontology (GO) enrichment analysis was done based on the extended functional annotation described above. The 15 most enriched GO terms are described in Table 3. In all, a large number of GO terms (>500) were significantly enriched (p <0.1; Additional file 2: Table S1). To give an overview of all GO terms we used ReviGO, which clusters GO terms based on semantic similarities [21]. This analysis shows two major clusters of terms associated to stress response and metabolism as well as two minor clusters related to development and metabolism (Figure 3).
Table 3.
Gene ontology enrichment
| GOID | Ontology | Term | p-value |
|---|---|---|---|
| GO:0016023 |
Cellular_component |
Cytoplasmic membrane-bounded vesicle |
1.4E-48 |
| GO:0014070 |
Biological_process |
Response to organic cyclic compound |
9.0E-32 |
| GO:0004674 |
Molecular_function |
Protein serine/threonine kinase activity |
4.6E-25 |
| GO:0006796 |
Biological_process |
Phosphate-containing compound metabolic process |
1.2E-24 |
| GO:0006464 |
Biological_process |
Cellular protein modification process |
1.4E-24 |
| GO:0036211 |
Biological_process |
Protein modification process |
1.4E-24 |
| GO:0004672 |
Molecular_function |
Protein kinase activity |
2.9E-24 |
| GO:0016773 |
Molecular_function |
Phosphotransferase activity, alcohol group as acceptor |
5.5E-24 |
| GO:0016301 |
Molecular_function |
Kinase activity |
3.8E-23 |
| GO:0009751 |
Biological_process |
Response to salicylic acid stimulus |
2.8E-22 |
| GO:0044267 |
Biological_process |
Cellular protein metabolic process |
6.9E-22 |
| GO:0006468 |
Biological_process |
Protein phosphorylation |
4.2E-18 |
| GO:0016310 |
Biological_process |
Phosphorylation |
6.3E-18 |
| GO:0010583 |
Biological_process |
Response to cyclopentenone |
1.2E-17 |
| GO:0004713 | Molecular_function | Protein tyrosine kinase activity | 9.8E-17 |
Gene ontology results of microarray experiment run on leaves (cv. Desiree) two days after treatment of the leaf canopy with BABA (10 mM). The 15 most enriched GO terms are listed.
Figure 3.
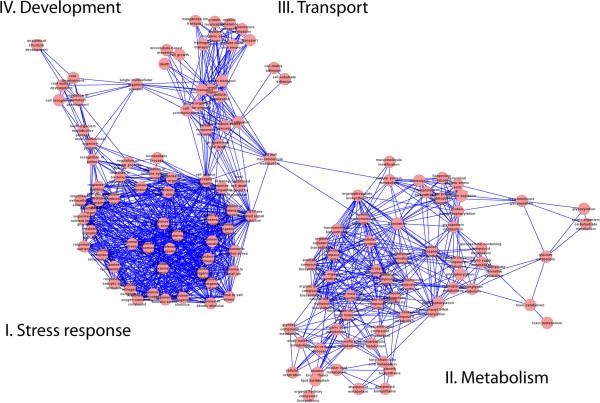
ReviGO analysis of enriched GO terms. Significantly enriched GO terms were clustered by semantic similarities and four overall clusters pointed out.
In Table 4, probes corresponding to the 50 most up- and down-regulated microarray transcripts were divided into functional categories. A full list of significant differentially expressed genes after 10 mM BABA treatment is included in Additional file 3: Table S2. Enriched GO terms after BABA treatment included processes associated with induction of plant defence such as plant-type hypersensitive response, necrotic spotted lesions, cell surface receptor signalling pathways, innate-immune response, defence response to fungus, and systemic acquired resistance (SAR). Examples of genes linked to these processes were two NDR1 homologs (G400032148, G400032231), two EDS1 splice variants (G400033029), two phytoalexin-deficient 4–2 protein splice variants (G400019873), several ethylene receptor splice variants (e.g. G400031819) and PR1 homologs (e.g. G400005112). When categorising genes to cellular compartments one of the most enriched GO terms is cytoplasmic membrane-bound vesicle (Table 3). This category included KiTH-2 (G400008097- G400008100) and Wound-induced protein WIN1 (G400019435) similar to AtPR4, which were among the most up-regulated transcripts after BABA treatment (Table 4). Among these genes was also the potato PEN1 (G400021331) homolog. WIN1 together with an Nb cell death marker (G400010131), osmotin (PR-5; G400003057), and Nectarin 5 (G400031325) were among the four most up-regulated transcripts involved in stress response. One Kunitz proteinase inhibitor and one serine protease inhibitor are among the most down-regulated transcripts related to stress response (G400009512, G400010128). There were no differentially expressed genes among the 17 genes annotated as R-genes on the array. However, the expression of some other LRR transcripts changed significantly. A TIR/NBS/LRR protein (G400001756) is up-regulated 8 times and three NBS-LRR type proteins (G400015872, G400027797, G400043650) are up- or down-regulated about two times. Only subtle expression differences were seen among other members in this large gene family.
Table 4.
Differentially expressed transcripts in potato leaves treated with 10 mM BABA
| Protein name | Gene id | Protein id | log2 Fold change |
|---|---|---|---|
|
Transport |
|
|
|
| Glucose-6-phosphate/phosphate translocator 2 |
G400025495 |
P400044203 |
5.2 |
| P-rich protein EIG-I30 |
G400004797 |
P400008501 |
6.2 |
| Tetrapyrrole synthesis |
|
|
|
| Gamma aminobutyrate transaminase isoform 2 |
G400024281 |
P400042007 |
-3.0 |
|
Stress |
|
|
|
| Nb cell death marker |
G400010131 |
P400017936 |
5.5 |
| Nectarin 5 |
G400031325 |
P400054574 |
5.2 |
| Osmotin |
G400003057 |
P400005490 |
5.5 |
| Wound-induced protein WIN1 |
G400019435 |
P400033767 |
7.5 |
| Chitin-binding lectin 1 |
G400019517 |
P400033919 |
-4.7 |
| Kunitz-type proteinase inhibitor |
G400009512 |
P400016823 |
-4.5 |
| Major latex |
G400028725 |
P400050037 |
-3.0 |
| Major latex |
G400028724 |
P400050036 |
-3.2 |
| Serine protease inhibitor 7 |
G400010128 |
P400017933 |
-5.6 |
|
Secondary metabolism |
|
|
|
| Tryptophan decarboxylase |
G400018358 |
P400031995 |
5.1 |
| Tryptophan decarboxylase |
G400018359 |
P400031996 |
5.5 |
| Linalool synthase |
G401011252 |
P400019916 |
-3.0 |
| Phenylcoumaran benzylic ether reductase |
G400003691 |
P400006583 |
-3.9 |
|
RNA |
|
|
|
| AT-HSFB3 (Arabidopsis thaliana heat shock transcription factor B3) |
G401008167 |
P400014364 |
5.7 |
| CHP-rich zinc finger protein |
G400028967 |
P400050478 |
5.1 |
| MYB8 |
G400020071 |
P400034798 |
-3.9 |
| MYC1 |
G401010822 |
P400019120 |
-3.3 |
| Zinc finger protein |
G401014997 |
P400026368 |
-3.0 |
|
Protein |
|
|
|
| E3 ubiquitin-protein ligase RMA1H1 |
G400021683 |
P400037599 |
-4.1 |
| Phytophthora-inhibited protease 1 |
G400004380 |
P400007779 |
-3.5 |
| Nucleotide metabolism |
|
|
|
| MutT domain protein |
G400017400 |
P400030392 |
7.8 |
|
Lipid metabolism |
|
|
|
| Non-specific lipid-transfer protein |
G400032250 |
P400055449 |
5.8 |
| C-4 sterol methyl oxidase 2 |
G400002156 |
P400003856 |
-3.7 |
| C-4 sterol methyl oxidase |
G400012763 |
P400022612 |
-4.8 |
| Non-specific lipid-transfer protein 2 |
G400011953 |
P400021168 |
-3.2 |
| 3-ketoacyl-CoA synthase |
G400014549 |
P400025626 |
-3.4 |
|
Hormone metabolism |
|
|
|
| Conserved gene of unknown function |
G400001001 |
P400001896 |
5.7 |
| Copalyl diphosphate synthase |
G400001948 |
P400003472 |
5.5 |
| Lipoxygenase |
G400010859 |
P400019187 |
6.5 |
| DWARF1/DIMINUTO |
G400021142 |
P400036651 |
-4.8 |
| ERF1 |
G400025989 |
P400045082 |
-4.9 |
| Sterol delta-7 reductase DWF5 |
G400005931 |
P400010516 |
-3.4 |
|
Development |
|
|
|
| KiTH-2 |
G400008097 |
P400014246 |
8.6 |
| KiTH-2 |
G400008098 |
P400014247 |
6.4 |
| KiTH-2 |
G400008099 |
P400014248 |
9.2 |
| KiTH-2 |
G400008100 |
P400014249 |
6.3 |
| Nodulin |
G400030381 |
P400052889 |
-3.3 |
| Patatin-2-Kuras 4 |
G400014104 |
P400024808 |
-5.5 |
| Patatin-04/09 |
G402017090 |
P400029876 |
-5.3 |
| Patatin group O |
G400029247 |
P400050915 |
-5.0 |
|
Cell wall |
|
|
|
| Glucan endo-1,3-beta-glucosidase, basic isoform 1 |
G400040260 |
P400062364 |
5.1 |
| Major pollen allergen Ory s 1 |
G400026220 |
P400045511 |
5.7 |
| Cellulose synthase |
G400011752 |
P400020836 |
-4.9 |
| Fasciclin-like arabinogalactan protein 19 |
G400017376 |
P400030347 |
-3.1 |
|
Biodegradation of Xenobiotics |
|
|
|
| 2-Hydroxyisoflavanone dehydratase |
G400031849 |
P400055127 |
6.2 |
|
Oxidation-reduction process |
|
|
|
| Cationic peroxidase |
G400012589 |
P400022299 |
6.6 |
| DC1 domain-containing protein |
G401008889 |
P400015622 |
5.4 |
| Polyphenol oxidase |
G400018916 |
P400032951 |
6.2 |
| Oxidoreductase, 2OG-Fe(II) oxygenase family protein |
G402003479 |
P400006181 |
-5.1 |
| Oxidoreductase, 2OG-Fe(II) oxygenase family protein |
G400021383 |
P400037071 |
-4.3 |
| Oxidoreductase, 2OG-Fe(II) oxygenase family protein |
G400032208 |
P400055421 |
-5.2 |
|
Not assigned/Misc |
|
|
|
| Ankyrin repeat-containing protein |
G400013763 |
P400024321 |
6.1 |
| Arachidonic acid-induced DEA1 |
G400039214 |
P400061318 |
5.5 |
| ATP-binding component of a transport system |
G400019445 |
P400033804 |
6.5 |
| Citrate binding protein |
G400003993 |
P400007118 |
7.3 |
| Conserved gene of unknown function |
G400024991 |
P400043372 |
5.3 |
| Conserved gene of unknown function |
G400031326 |
P400054575 |
5.6 |
| Conserved gene of unknown function |
G400002292 |
P400004083 |
6.5 |
| Cytochrome P450 |
G400043512 |
P400065616 |
6.5 |
| Gene of unknown function |
G400025446 |
P400044135 |
5.6 |
| Hypothetical repeat protein |
G400028412 |
P400049430 |
5.2 |
| Lichenase |
G400020017 |
P400034720 |
5.2 |
| P-rich protein EIG-I30 |
G400004737 |
P400008397 |
7.9 |
| Primary amine oxidase |
G400030082 |
P400052390 |
5.7 |
| Abhydrolase domain containing |
G400014029 |
P400024703 |
-4.0 |
| Blue (Type 1) copper domain |
G400013271 |
P400023469 |
-3.4 |
| Conserved gene of unknown function |
G400015196 |
P400026658 |
-3.0 |
| Conserved gene of unknown function |
G400015948 |
P400027933 |
-3.6 |
| Cytochrome P-450 |
G400011750 |
P400020830 |
-3.7 |
| Family 1 glycosyltransferase |
G400027200 |
P400047278 |
-4.2 |
| Gene of unknown function |
G400040677 |
P400062781 |
-4.0 |
| Glucosyltransferase |
G400005960 |
P400010581 |
-3.2 |
| Glucosyltransferase |
G402027210 |
P400047294 |
-5.3 |
| SGA | G400011740 | P400020813 | -5.7 |
The 50 up- or down-regulated transcripts with the highest- fold change (including splice variants, not shown in table) in potato leaves following treatment with 10 mM BABA. Samples were analysed two days after treatment of the leaf canopy with BABA. Transcripts with significantly changed expression (adj. p-value <0.05) are included in the table.
Significant enrichments were found for genes associated with the hormones ethylene, ABA, SA, JA and cyclopentenone, but of the processes related to hormone metabolism, only gibberellin biosynthesis was enriched. In this pathway, several splice variants of copalyl diphosphate synthases, which act early in biosynthesis of gibberellins, were sharply up-regulated (Additional file 2: Table S1 and Additional file 3: Table S2). Furthermore, several gibberellin 20-oxidases (G400036251, G400039968, G400040776, G400036003), which catalyze the conversion of different gibberellin forms, as well as gibberellin 2-oxidase 1, which is able to de-activate gibberellins [30], were down-regulated by BABA, except for one transcript (G400015796) that was slightly up-regulated. In addition, several gibberellic acid insensitive dwarf (GID) proteins (G400022493, G400022492, G401008665, G400018153, G400015446) which are GA-receptors regulating the stability of DELLA proteins, were also affected by BABA treatment, and two splice variants of DELLA proteins (GAI) (G400019212) were up-regulated.
In JA-regulation, two Jaz proteins were oppositely regulated, with transcripts representing Jaz1 (G400002930) up-regulated and Jaz3 (G400032119) down-regulated. Several splice variants of MYC1 (G401010822) and MYC2 (G404010822 G400005525, G402010822) transcription factors were down-regulated, whereas two splice variants of a JA-induced WRKY (G400019824) were up-regulated. JAZs act as negative regulators of the JA signalling pathway by binding to MYC transcription factors [31]. In JA biosynthesis (although not significant in the GO analysis), allene oxide cyclase (AOC) (G401012679), was down-regulated, whereas splice variants of lipoxygenase (LOX) (G400010859, G400007793) and Acyl-coenzyme A oxidase (G400010548, G400020620) were up-regulated.
Ethylene response factor 1 (ERF1) (G400025989), a positive regulator of JA and ET signalling [32], was down-regulated. However, two splice variants of 1-aminocyclopropane-1-carboxylate oxidase 2 (ACC) (G400013894), involved in the last step of ET biosynthesis, were up-regulated by 10 mM BABA.
In the brassinosteroid (BR) pathway, several brassinosteroid insensitive 1-associated receptor kinase 1 s (BAKs) were up-regulated. Among the 50 most down-regulated are transcripts encoding two forms of the DWARF1/DIMINUTO protein (G400021142) which are involved in early brassinosteroid biosynthesis. The Arabidopsis DWARF1 knock-out mutant has stunted growth and an altered secondary cell wall [33].
In the auxin pathway, TIR1 (G400020729) along with several auxin-response factors (ARFs) were down-regulated by BABA, but one ARF 5 (G400003771) was up-regulated. The transcript of auxin-induced SAUR (G400013321) was also up-regulated.
Treatment with 10 mM BABA also affected the regulation of several transcripts involved with amino acid metabolism e.g. related to proline, arginine and glutamine biosynthesis in the glutamate pathway. The most profound up-regulation was found in transcripts for biosynthesis of aromatic amino acids, e.g. chorismate mutase 1 (G400001438), arogenate dehydratase (G400007122) and arogenate dehydrogenase 1 (G400030683, G400020334) involved in phenylalanine and tyrosine biosynthesis [34]. Several transcripts of tryptophan decarboxylase (TDC) (G400018359, G400018358, G400018357, G400005444, G400035067) and one transcript of strictosidine synthase (G402017288) were highly up-regulated.
Two C-4 sterol methyl oxidase (SMO; G400002156, G400012763) transcripts were found among the 50 most down-regulated by 10 mM BABA (Table 4). Other enzyme-coding transcripts involved in sterol biosynthesis and glucosylation and in part of the mevalonate pathway, for example sterol delta-7 reductase DWF5 (G400014722, G400005931), DWARF1/DIMINUTO (G400021142), delta-7-sterol-C5(6)-desaturase (G400026401), squalene epoxidase (G400003324), delta-14-sterol reductase (G400002x720), and a UDP-glucose:sterol glucosyltransferase (G400015817), were also down-regulated. In contrast, transcripts encoding several sesquiterpene synthase 2 (G400010587, G400010585, G400010593, 400029200, G400029201, G400009854, G400018066, G400019782, G400009832) and vetispiradiene synthase 1 (G400018245), enzymes involved in sesquiterpene phytoalexin biosynthesis and belonging to another branch of the mevalonate pathway, were strongly up-regulated (Figure 4).
Figure 4.
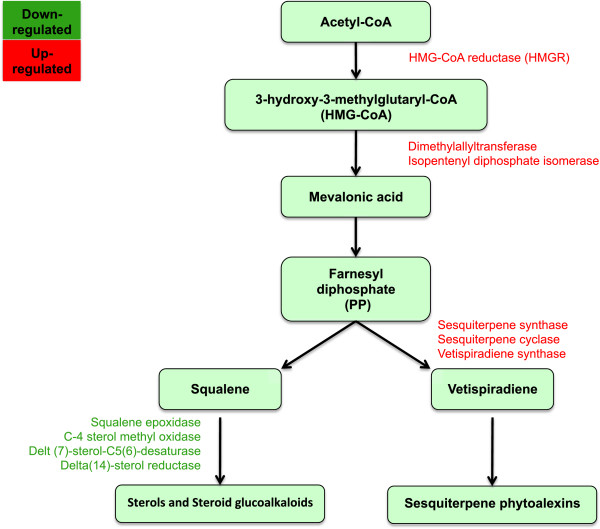
BABA-influenced changes within the mevalonate pathway. The effect of 10 mM BABA on transcripts encoding genes within the mevalonate pathway in leaves of potato. BABA caused a down-regulation of several transcripts involved with sterol biosynthesis, whereas transcripts involved with sesquiterpene phytoalexin biosynthesis were up-regulated. Transcripts with significantly changed expression (adj. p-value <0.05) are denoted in red (up-regulation) and green (down-regulation).
BABA also seemed to influence genes involved in cell wall structure, e.g., several cellulose synthase genes were slightly up-regulated whereas one was found among the 50 most down-regulated transcripts (Table 4).
Secretome SDS-PAGE and LC-MS/MS analysis
The quality of the apoplast samples were checked by separation on SDS-polyacrylamide gels prior to protein identification by mass spectrometry (Figure 5). A thick band previously identified by us as PR-1 [35] was found after application of 10 mM BABA. A faint PR-1 band was seen after 1 mM BABA whereas no PR-1 band was detected in the water control.
Figure 5.
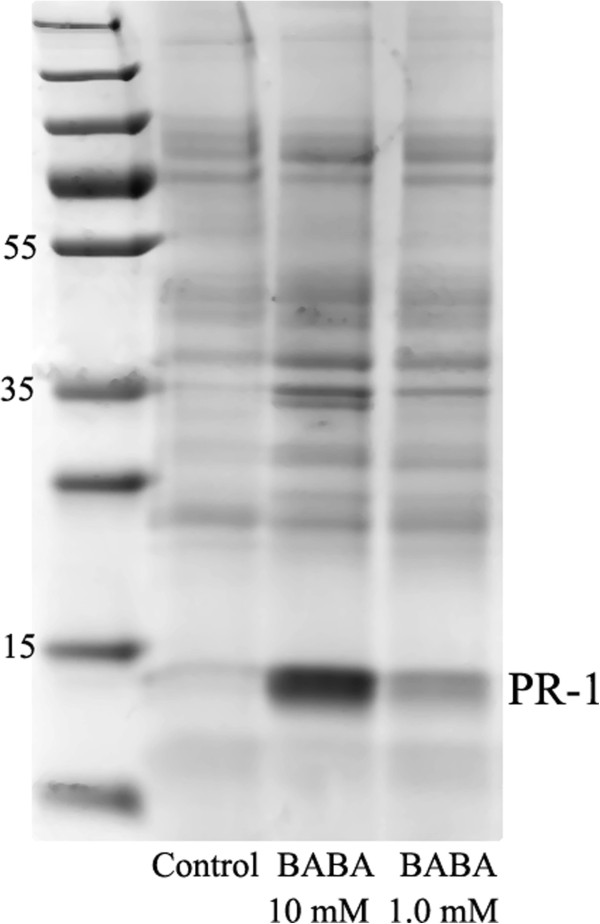
SDS-PAGE of apoplast from Desiree leaves treated with BABA. Separation on a SDS-PAGE gel, showing apoplast proteins extracted two days after treatment with BABA in cv. Desiree. From left: control plant treated with water, plant treated with 10 mM BABA and plant treated with 1 mM BABA. The ladder used was Page Ruler™ Plus Prestained Protein Ladder (SM1811, Fermentas).
In the apoplast secretome, we identified 24 proteins with altered abundance in potato after treatment with 1 mM BABA (Table 1; Table 5), compared to 91 proteins with 10 mM (Table 1; Table 6). Almost half of the 24 proteins affected by 1 mM BABA were affected in the same way by 10 mM BABA. Among these, six were up-regulated: two glucan endo-1, 3-beta-D-glucosidases (Q70BW9, G400012702), two endochitinases (G400008796, G400001528), pathogen-and wound-inducible antifungal protein CBP20 (PR-4; G400019437) and one annotated as a resistance gene-like (Q93YA6); and five down-regulated: 24 K germin (G400013010), kunitz trypsin inhibitor (G400027052), PAE (G401024140), beta-galactosidase (G402009228) and subtilisin-type protease (G400009947). Under both BABA conditions, there was a dose dependent response where 10 mM led to 1.4 to 11 fold higher protein abundance. The four most abundant secreted proteins affected by 10 mM BABA, were an Nb cell death marker (G400010131) with low sequence similarity to a Kunitz-type protease in Arabidopsis (At1G17860), a Sn-1 protein (G400011716), an acidic class II 1,3-beta-glucanase (PR-2) (Q8GUR4) and a pathogenesis-related leaf protein 6 (P04284) annotated as a basic PR-1 protein (PR1b1) in tobacco. Overall, proteins like thaumatin, glucanases, glucosidases, P69 proteins, peroxidases, PRp27 protein, pathogen-and wound-inducible antifungal protein CBP20 and chitinases, all belonging to pathogenesis-related (PR) protein families, were the most abundant among the proteins up-regulated by BABA.
Table 5.
Proteins with differential abundance in the apoplast of potato leaves treated with 1 mM BABA
| Protein name | External IDs | log2 Fold change (peptide median) 2 |
|---|---|---|
| 1,3-beta-glucan glucanohydrolase |
Q70BW9 |
20 |
| Conserved gene of unknown function |
P400042337 |
19 |
| Acidic class II 1,3-beta-glucanase (Fragment) |
Q8GUR3 |
17 |
| Glucan endo-1,3-beta-D-glucosidase |
P400022509 |
17 |
| Resistance gene-like |
Q93YA6 |
17 |
| Basic 30 kDa endochitinase |
P400015454 |
14 |
| Leucine aminopeptidase 1, chloroplastic (DR57) |
Q10712 |
13 |
| Pathogen-and wound-inducible antifungal protein CBP20 |
P400033771 |
1.3 |
| Glucan endo-1,3-beta-D-glucosidase |
P400018563 |
1.2 |
| Strictosidine synthase |
P400030201 |
1.1 |
| Endochitinase |
P400002757 |
0.9 |
| Miraculin |
P400018006 |
-1.1 |
| Kunitz trypsin inhibitor |
P400046980 |
-16 |
| Peptidyl-prolyl cis-trans isomerase |
F4K2G0 |
-17 |
| Heparanase-2 |
P400042675 |
-18 |
| Thioredoxin |
P400021964 |
-18 |
| PAE |
P400041742 |
-18 |
| Peroxidase |
P400026173 |
-18 |
| Subtilisin-type protease |
P400017566 |
-18 |
| Beta-galactosidase |
P400016282 |
-18 |
| P69B protein |
P400007007 |
-19 |
| Leucine-rich repeat family protein |
P400011041 |
-19 |
| 24 K germin |
P400023053 |
-20 |
| Major latex | P400046294 | -21 |
Samples were analysed two days after treatment of the leaf canopy with BABA. Proteins with significantly changed abundance (FDR <0.1) are included in the table.
Table 6.
Proteins with differential abundance in the apoplast of potato leaves treated with 10 mM BABA
| Protein name | External IDs | log2 Fold change (peptide median) |
|---|---|---|
| Pathogenesis-related leaf protein 6 (P6) (PR protein) |
P04284 |
23 |
| Acidic class II 1,3-beta-glucanase (Fragment) |
Q8GUR4 |
23 |
| Nb cell death marker |
P400017936 |
22 |
| Sn-1 protein |
P400020769 |
21 |
| Cell wall peroxidase |
P400048009 |
21 |
| 1,3-beta-glucan glucanohydrolase |
Q70BW9 |
20 |
| Resistance gene-like |
Q93YA6 |
20 |
| Citrate binding protein |
Q8H9C1 |
20 |
| Cytoplasmic aconitate hydratase |
Q56WE8 |
20 |
| Glucan endo-1,3-beta-D-glucosidase |
P400022509 |
19 |
| Acetylornithine deacetylase |
P400030587 |
18 |
| Transketolase, chloroplastic |
P400038185 |
18 |
| Beta-galactosidase |
P400000661 |
17 |
| Peptidyl-prolyl cis-trans isomerase |
P400002952 |
14 |
| Cationic peroxidase |
P400022299 |
4.3 |
| ADP-ribosylation factor 3 |
P400047760 |
3.8 |
| Gene of unknown function |
P400068931 |
3.8 |
| Class III peroxidase |
P400001015 |
3.8 |
| Beta-1,3-glucanase, acidic |
P400022914 |
3.8 |
| Glycerophosphodiesterase |
P400038623 |
3.4 |
| Chlorophyll a-b binding protein 6A, chloroplastic |
P400040595 |
3.3 |
| Peroxidase |
P400055404 |
3.0 |
| Thaumatin |
P400007581 |
2.9 |
| Retinoid-inducible serine carboxypeptidase |
P400000831 |
2.8 |
| Glucan endo-1,3-beta-D-glucosidase |
P400051976 |
2.7 |
| AT5G66190 protein (Fragment) |
B9DI26 |
2.7 |
| Basic 30 kDa endochitinase |
P400015454 |
2.6 |
| Pathogen-and wound-inducible antifungal protein CBP20 |
P400033771 |
2.5 |
| NtPRp27-like protein |
Q84XQ4 |
2.5 |
| Glucan endo-1,3-beta-D-glucosidase |
Q42890 |
2.4 |
| Glucan endo-1,3-beta-D-glucosidase |
O82063 |
2.3 |
| Putative glucan endo-1,3-beta-D-glucosidase |
Q2HPL1 |
2.2 |
| NtPRp27 |
P400011122 |
2.2 |
| Glutathione reductase, cytosolic (GR) (GRase) |
P48641 |
2.1 |
| Endochitinase (Chitinase) |
P400015232 |
2.0 |
| Peroxidase |
Q43774 |
2.0 |
| P69E protein |
P400007009 |
1.9 |
| DUF26 domain-containing protein 1 |
P400063324 |
1.9 |
| Cystatin |
P400014844 |
1.8 |
| Patatin 3 |
P400017707 |
1.7 |
| Endochitinase (Chitinase) |
P400002757 |
1.5 |
| Peroxidase |
P400043335 |
1.5 |
| Acidic class II 1,3-beta-glucanase |
P400018562 |
1.4 |
| 1,3-beta-D-glucan glucanohydrolase (Glucan endo-1,3-beta-glucosidase a) |
Q70C53 |
1.4 |
| STS14 protein |
P400038079 |
1.4 |
| Acidic endochitinase |
P400056271 |
1.3 |
| Lignin-forming anionic peroxidase (TOPA) |
P11965 |
1.3 |
| Glucan endo-1,3-beta-glucosidase |
P400001406 |
1.1 |
| Uncharacterized protein |
K4ASJ5 |
1.1 |
| Reticuline oxidase |
P400031346 |
0.9 |
| Aspartic proteinase nepenthesin-1 |
P400009908 |
-0.6 |
| Kunitz-type proteinase inhibitor group B1 (Fragment) |
Q58I49 |
-0.7 |
| Conserved gene of unknown function |
P400010730 |
-0.8 |
| Catechol oxidase B, chloroplastic |
P400051502 |
-0.8 |
| 24 K germin |
P400023053 |
-0.8 |
| Beta-hexosaminidase 1 |
P400054228 |
-0.9 |
| Pectinesterase-2 |
P400049953 |
-1.1 |
| Reticuline oxidase |
P400042020 |
-1.1 |
| Alpha-L-arabinofuranosidase |
Q76LU4 |
-1.1 |
| Acidic 27 kDa endochitinase |
P400002758 |
-1.4 |
| Phosphoesterase family protein |
P400039390 |
-1.6 |
| Kunitz trypsin inhibitor |
P400046981 |
-1.9 |
| Hydrolase, hydrolyzing O-glycosyl compounds |
P400016965 |
-2.0 |
| Peroxidase N |
P400041612 |
-2.1 |
| Zinc finger protein |
P400004464 |
-16 |
| H+transporting two-sector ATPase, alpha/beta subunit, central region |
P400001921 |
-16 |
| Periplasmic beta-glucosidase |
P400016780 |
-16 |
| Subtilisin-like protease |
P400026165 |
-16 |
| Kunitz trypsin inhibitor |
P400046980 |
-16 |
| Erg-1 |
P400022271 |
-16 |
| Ferredoxin-dependent glutamate synthase 1 |
P400017124 |
-16 |
| Glycolate oxidase |
P400048099 |
-17 |
| Cucumisin |
P400010997 |
-17 |
| Serine-threonine protein kinase, plant-type |
P400029544 |
-17 |
| Auxin-induced beta-glucosidase |
P400039774 |
-18 |
| ATP synthase subunit alpha, chloroplastic |
P00823 |
-18 |
| PAE |
P400041742 |
-18 |
| Cysteine protease inhibitor 1 |
P400017942 |
-18 |
| Sn-1 protein |
P400013688 |
-18 |
| Salicylic acid-binding protein 2 |
P400001464 |
-18 |
| Beta-galactosidase |
P400016282 |
-18 |
| Subtilisin-type protease |
P400017566 |
-19 |
| Hydrolase |
P400031772 |
-19 |
| Cellulase containing protein |
P400015021 |
-20 |
| Alpha-galactosidase/alpha-n-acetylgalactosaminidase |
P400043893 |
-20 |
| Beta-galactosidase |
E3UVW7 |
-20 |
| GDSL-like Lipase/Acylhydrolase family protein |
P400011471 |
-20 |
| Uncharacterized protein |
K4CNZ8 |
-20 |
| Receptor protein kinase CLAVATA1 |
P400020982 |
-20 |
| Xyloglucan endotransglycosylase LeXET2 |
Q9FZ05 |
-21 |
| Xylem serine proteinase 1 | P400033261 | -22 |
Samples were analysed two days after treatment of the leaf canopy with BABA. Proteins with significantly changed abundance (FDR < 0.1) are included in the table.
Discussion
Novel functional annotation and overall changes
In this paper we provide the potato research community with a more comprehensive functional annotation of the potato PGSC gene models and potato microarray probes (Agilent JHI Solanum tuberosum 60 k v1; Additional files 4 and 5). This new functional annotation was used for the interpretation of the BABA-IR response.
BABA at a concentration of 10 mM alters the expression of over 5000 transcripts and secretion of over 90 proteins to the apoplast. This is in contrast to the finding that only six transcripts (1 up- and 5 down-regulated) and 24 proteins (11 up- and 13 down-regulated) were affected by 1 mM BABA. This may explain the results from an earlier study by Liljeroth et al., 2010 [29], where 5 mM BABA was required to affect P. infestans infection rate, whereas BABA applied at concentrations below 2.5 mM failed to restrict P. infestans growth in the three tested potato cultivars. In agreement with this, we found that treatment of potato cv. Desiree with 10 mM BABA has a clear inhibitory effect of P. infestans infection. Whether the differences seen after treatment with 1 mM BABA, especially in the apoplast, lead to an increased resistance against other pathogens still has to be tested.
Furthermore, we can confirm the up-regulation of family members of proteinase inhibitor type-2:s, ubiquitin-conjugating proteins, glutathione S-transferases, lipoxygenases, histone H3.2 and a sesquiterpene synthase 2 previously identified by Li et al., 2009 [13] who reported 65 up-regulated transcripts based on cDNA-AFLPs after treatment with 4 mM BABA in potato.
The only up-regulated transcript by 1 mM BABA, a MutT/nudix domain protein, was also up-regulated by 10 mM BABA. Several MutT/nudix were also found to be up-regulated after BABA treatment in Arabidopsis [9]. This family is not well characterised functionally, but some members use their hydrolase activity to maintain cell homeostasis and have been implied to play a role in plant defence. For example, over-expression of AtNUDX2 resulted in increased pyrophosphatase activity toward ADP-ribose and increased tolerance to oxidative stress [36] and AtNUDX7 has been identified as an SA-independent factor in Enhanced Disease Susceptibility1 (EDS1)-controlled plant defence and programmed cell death [37]. In line with these observations, we found an ADP-ribose polymerase 2 and several other ADP-ribosylation factors as well as EDS1 to be transcriptionally up-regulated by 10 mM BABA in potato.
When comparing the transcripts and apoplastic protein abundance after 10 mM BABA treatment, regulation of 13 of the 70 proteins with peptides matching the predicted potato genome models overlapped. The samples are taken two days after treatment with BABA in both the transcript and protein analysis, but the overlap might have been greater if the RNA samples were taken earlier since there is an expected time lag between transcript and protein synthesis. There are also differences in transcript and protein stability. Another reason for these differences might be that protein abundance in the apoplast is at least partially regulated by protein transport and vesicle trafficking, and thus not directly dependent on transcript levels. Cytoplasmic membrane-bound vesicle was the most enriched GO term and several transcript encoding proteins in vesicle trafficking, such as clathrin assembly protein, Epsin-2 and PEN1 were all found to be between 2.5- and 11-fold up-regulated. The low overlap between transcript regulation and apoplast protein abundance confirms the value of combining the two methods.
Immune responses and sterols
Several typical stress-response genes up-regulated by BABA in potato (e.g. PR-1, PR-2, PAD4, thaumatin, aspartyl protease family protein) have previously been found to be up-regulated in an Arabidopsis microarray study, where 46 genes in total were up-regulated about 3 days after treatment with 0.25 mM BABA [8]. In the present study, no known annotated R-genes and only a few other NBS-LRR proteins were up-regulated by BABA, which might indicate that BABA-IR is not mediated by increased expression of these genes through R-gene recognition and effector-triggered immunity (ETI). Instead most of the up-regulated genes and proteins affected by BABA were PR-proteins and components involved in hormone signalling and defence signalling such as PAMP triggered immunity, PTI. The up-regulated potato PEN1 homolog, which encodes membrane associated syntaxin, is involved in PTI, where it plays a part in the vesicle-based secretion system [38]. EDS1, PAD4 and SAG101, genes implicated to play a part in non-host resistance [39], were also up-regulated by 10 mM BABA. However, none of them have been shown to be required for the defence against P. infestans in Arabidopsis [40-42]. Another gene among the four most up-regulated transcripts involved in stress response encodes osmotin (PR-5). This protein might play a role in BABA-IR to drought stress, as its abundance was increased in the leaf proteome of crabapple seedlings that were treated with BABA prior to drought stress exposure [43].
In a recent study, Kopischke et al. [44] reported that sterol balance plays a role in Arabidopsis non-host resistance to P. infestans. In their study, the erp1 mutant lacking sterol ester accumulation experienced increased cell death. Oomycetes lack the ability to synthesize sterols, which are essential for their reproduction, and therefore depend on their hosts for sterol compounds [45]. The sterol biosynthesis within the mevalonate pathway was clearly down-regulated in the present study, whereas the biosynthesis of another branch of the pathway, namely sesquiterpene phytoalexin, was up-regulated (Figure 3). These results are in line with earlier findings in potato, where wound-induced sterol and steroid glycoalkaloid synthesis are suppressed in favour of sesquiterpenoid phytoalexin biosynthesis when potato tubers are exposed to elicitors or pathogens that induce HR [46,47]. This redirection of the isoprenoid biosynthetic pathway has been linked to changes in expression of 3-hydroxy-3 methylglutaryl-coenzyme A reductase (HMGR) genes, where hmg2 and hmg3 are up-regulated whereas hmg1 is suppressed [48,49]. In the present study two HMGRs were up-regulated by 10 mM BABA. In a study conducted by Ros et al. [50], sesquiterpene synthase and vetispiradiene synthase were up-regulated after infection with P. infestans in two field-resistant cultivars, Bettina and Indira. It had earlier been shown that arachidonic acid, a fatty acid present in the lipids of oomycetes, can suppress steroid-glycoalkaloids [51] and act as an elicitor of sesquiterpene phytoalexins [52] with the involvement of LOX [53,54] in potato tubers. LOX is also among the 50 most up-regulated transcripts by 10 mM BABA in this study. However, BABA-IR in potato has been shown to require SA but not 9-LOX-derived oxylipins [14].
When comparing the present secretome data with that obtained from three genotypes, SW93-1015, Sarpo Mira (P. infestans incompatible) and Desiree (P. infestans compatible), 24 and 72 hpi with P. infestans (Ashfaq Ali, personal communication), patatin 3 (P400017707) increased in abundance after BABA treatment as well as infected SW93-1015 and Sarpo Mira, but not in susceptible Desiree. This is in line with previous findings that tobacco NTPAT1-3 is involved in cell death during HR [55], since HR are formed in SW93-1015 and Sarpo Mira but not Desiree. A role for patatin-like proteins in cotton HR has also been proposed [56].
Amino acid metabolism
Earlier studies in Arabidopsis have revealed that BABA treatment alters amino acid composition and can be linked to an amino acid associated alternation of growth [10,11]. The BABA-mediated growth reduction and IR to Pseudomonas syringae pv. tomato DC3000 was compromised by treatment with L-glutamine [11]. The amino acid L-serine and the stress-responsive amino acids proline and asparagine were reported to be induced by BABA in Arabidopsis, whereas the aspartic acid content decreased (Singh et al., 2010). Transcripts involved in the proline biosynthesis were also observed after treatment with 10 mM BABA in the present study. Recently, a study in Arabidopsis revealed decreased abundance of several amino acids, including serine and glutamic acid, after treatment with potassium phosphite [57], and of aspartic acid after BABA treatment. However, whether a BABA-induced altered amino acid balance leads to changed leaf morphology in potato similar to those seen in Arabidopsis, remains to be investigated.
Another up-regulated transcript was arginine decarboxylase, which is involved in the biosynthesis of the polyamine putrescine by catalyzing the synthesis of agmatine from arginine. Polyamines are involved in a wide range of stress responses, partly as reactive oxygen scavengers [58]. Arginine decarboxylase has previously been reported to increase systemically in tobacco leaves after BABA treatment [59], which suggests a role for polyamines in BABA-IR.
Hormones
SA has previously been shown to be important for basal defence against P. infestans and required for BABA-IR in the same potato pathosystem [14,60]. In the SA signalling pathway, transcripts of a phenlyalanine ammonia lyase (PAL) homolog were two fold up-regulated, whereas isochorismate synthase (ICS) did not change significantly. ICS, in contrast to PAL, is directly involved in the SA biosynthesis and has been shown to be up-regulated after BABA-treatment of Arabidopsis [8,9]. In contrast to SA, genes involved in JA, ET, GA and ABA biosynthesis were affected by BABA, and GO term analysis pointed to an enrichment of genes in GA biosynthesis.
In the present study, two splice variants of DELLA proteins, involved in GA signalling, were up-regulated. DELLA proteins play a role in resistance to necrotrophs and susceptibility to biotrophs by modulating the SA/JA signalling [61]. This modulation is accomplished by competitive binding of DELLAs to JAZ proteins, which prevents interaction between JAZ and MYC2, thus promoting MYC2-induced responses such as LOX [61]. Navarro et al. (2008) further suggested that GA might change this balance by degradation of DELLAs, thus promoting resistance to biotrophs and susceptibility to necrotrophs. Thus, it would be interesting to further investigate the role of DELLAs in BABA-IR in potato.
9-cis-epoxy-carotenoid dioxygenase 2 (NCED2), which is involved in the biosynthesis of, for example, ABA, was up-regulated. In a previous study in Arabidopsis, BABA treatment was shown to lead to the accumulation of several ABA responsive elements and it was suggested that crosstalk occurs between BABA and ABA signalling [9]. The ABA responsive transcription factors, identified as BABA-induced in Arabidopsis e.g. ABI1, 2 and 3, were not differentially expressed in BABA-treated potato. Instead, the only ABA-related transcription factor we could identify as differentially regulated, a bZIP, was repressed after BABA treatment.
Several genes related to BRs and auxins, which can affect plant development and cell wall formation, changed expression after treatment with BABA e.g. TIR1 and several auxin-response factors (ARFs), but not ARF5, were down-regulated. Repression of auxin-related processes has repeatedly been observed in defence signalling. In Arabidopsis, Wang et al. [62] found that a number of auxin responsive genes were down-regulated after treatment with the SA analog benzothiadiazole S-methyl ester (BTH). With some exceptions, e.g. auxin-induced SAUR and auxin responsive factor 5 (ARF5), this is also true in our data. Furthermore, the over-expression of a rice IAA-amino synthetase, GH3, led to an enhanced disease resistance to the rice pathogen Xanthomonas oryzae, possibly due to down-regulation of expansins, leading to decreased loosening of the cell walls [62,63]. However, we see a clear up-regulation of several GH3 isoforms, but no significant change in expression of expansin transcripts in potato.
Secreted proteins
Data obtained from the analysis of the apoplastic proteins revealed a high number of proteins related to defence and metabolism in the secretome of BABA-treated leaves. Several pathogenesis-related (PR) proteins were found to be highly abundant following BABA treatment. These proteins are used as markers for pathogen response and are induced by different biotic and abiotic stresses [64]. Among these were an acidic class II 1, 3-beta glucanase (PR-2) and thaumatin. PR-2, but not thaumatin, also showed increased abundance after BABA treatment of drought-stressed crabapple seedlings [43]. Another highly induced protein of less known biological function was the alginate-lyase motif containing citrate binding protein not present in the Arabidopsis genome [65]. The transcript encoding the citrate binding protein was also among the most up-regulated and has been described to be induced in potato after inoculation with P. infestans [50].
A cell wall peroxidase was highly induced after BABA treatment. These are known to be involved in cell wall strengthening following stress challenge and function by combatting degradation enzymes produced by pathogens [66]. Furthermore, peroxidases are involved in a number of defence-related responses such as HR, cross-linking of phenolics, and phytoalexin production. Peroxidases can be induced by different stresses and some plant defence activators including BABA [67]. A Snakin (Sn-1) protein was also highly up-regulated. Snakin proteins are known for their antimicrobial role in plant defence [68,69]. In addition, Sn-1 was recently found to play a role in leaf primary metabolism and cell wall composition in potato, suggesting a possible role in plant growth and development [70].
Conclusions
Overall, at a concentration of BABA that gives reduced Phytophthora growth on potato, we find a global effect on the transcriptome and a large number of secreted proteins with increased abundance. Several processes related to classical plant hormones and amino acid metabolism seem to be affected and many known defence proteins are up-regulated. However, unlike the situation for Arabidopsis, ABA responsive genes seem not to be clearly regulated by BABA in potato. It is tempting to speculate that the strong down-regulation of the biosynthesis of sterols and steroid glucoalkaloids, in contrast to the up-regulation of important enzymes involved in sesquiterpene phytoalexin biosynthesis, could explain some of the BABA-IR against P. infestans.
Taken together, the clear direct effects of BABA treatment in potato might suggest a yield penalty. However, in our earlier study [29], the tuber yield was unaffected by weekly applications of BABA to field grown potatoes. The same result was reported for the tuber yield after repeated BABA treatments to potato grown in greenhouse conditions [71]. In fact, the tuber yield was improved in two of the cultivars used in the latter experiment. Thus, the individual genes and proteins, as well as general processes identified in this study, are possible candidates or markers for improved resistance without major influence on potato yield.
Availability of supporting data
The microarray data was deposited in ArrayExpress, accession number: E-MTAB-1545.
Abbreviations
ABA: Abscisic acid; ACN: Acetonitrile; AFLP: Amplified fragment length polymorphism; ANOVA: Analysis of variance; ARFs: Auxin-response factors; BABA: ß-aminobutyric acid; BR: Brassinosteroid; BAKs: Brassinosteroid insensitive 1-associated receptor kinase 1 s; cDNA: Complementary DNA; cv.: Cultivar; dpi: Days post infection; dH2O: Destilled water; DTT: Dithiothreitol; ETI: Effector-triggered immunity; ET: Ethylene; FDR: False discovery rate; GA: Gibberellic acid; GID: Gibberellic acid insensitive dwarf; Gene ontology terms: GO terms; HPLC: High-performance liquid chromatography; HR: Hypersensitive response; IR: Induced resistance; ISR: Induced systemic resistance; JA: Jasmonic acid; LRR: Leucine rich repeats; LTQ: Linear trap quadropole; MS: Mass spectrometry; mRNA: Messenger RNA; NBS: Nucleotide binding site; PTI: PAMP triggered immunity; PAMP: Pathogen-associated molecular patterns; PR protein: Pathogenesis-related protein; P. infestans: Phytophthora infestans; PAGE: Polyacrylamide gel electrophoresis; R-gene: Resistance gene; SA: Salicylic acid; SDS: Sodium dodecyl sulfate; SGR: Solanaceae genomics resource; SIMR: Stress-induced morphogenic response; SAR: Systemic acquired resistance; PGSC: The potato genome sequencing consortium; TIR: TOLL/interleukin-1 receptor.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TB, EOA, EL, EA planned the experiments and wrote the paper. LIM assisted in the lab experiment. SR and FL performed the proteomics and helped with the analysis. PH performed the microarray analysis. EOA and EPW helped with the bioinformatics analysis. DW and DJ designed and developed Parallel OrthoMCL and analysed the output data together with DDB. All authors read and approved the final manuscript.
Supplementary Material
Distribution of number of Gene Ontology terms. The number GO terms per probe of the Agilent JHI Solanum tuberosum 60 k v1 microarray annotated using either Parallel-OrthoMCL across 26 plant genomes or the Solanaceae Genomics Resource annotation.
Complete list of enriched GO terms. Significant differentially expressed genes after 10 mM BABA treatment were analysed by GOEast and GO terms with p >0.1 were considered significant.
Significant differentially expressed transcripts. A full list of significant differentially expressed transcripts (adj. p-value <0.05) in potato leaves following treatment with 10 mM BABA. Samples were analysed two days after treatment of the leaf canopy with BABA.
Text file 1. Extended gene ontology terms for microarray probes. Extended gene ontology terms for the custom-made Agilent expression array (JHI Solanum tuberosum 60 k v1). The file is formatted for use in GOEast.
Text file 2. Extended gene ontology terms for PGSC gene models v.3.4. Extended gene ontology terms for the PGSC gene models. The file is formatted for use in GOEast.
Contributor Information
Therese Bengtsson, Email: therese.bengtsson@slu.se.
Deborah Weighill, Email: daweighill@sun.ac.za.
Estelle Proux-Wéra, Email: estelle.wera@slu.se.
Fredrik Levander, Email: fredrik.levander@immun.lth.se.
Svante Resjö, Email: svante.resjo@slu.se.
Dharani Dhar Burra, Email: dharani.burra@slu.se.
Laith Ibrahim Moushib, Email: laith.moushib@slu.se.
Pete E Hedley, Email: pete.hedley@hutton.ac.uk.
Erland Liljeroth, Email: erland.liljeroth@slu.se.
Dan Jacobson, Email: jacobson@sun.ac.za.
Erik Alexandersson, Email: erik.alexandersson@slu.se.
Erik Andreasson, Email: erik.andreasson@slu.se.
Acknowledgements
The Swedish foundation for Strategic research, Crafoord foundation, The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning are thanked for financial support. The bioinformatical part of the study was supported by the Swedish Ministry of Foreign Affairs as part of its special allocation on global food security. We thank Mia Mogren for excellent technical support, Karin Hansson for performing the LC-MS analysis, Jenny Morris for microarray experiments, Björn Andersson, SLU Ultuna, and Larsvikens Lantbruks AB (Sweden) for providing us with P. infestans isolate and potato tubers, respectively.
References
- Walters D, Walsh D, Newton A, Lyon G. Induced resistance for plant disease control: maximizing the efficacy of resistance elicitors. Phytopathology. 2005;95:1368–1373. doi: 10.1094/PHYTO-95-1368. [DOI] [PubMed] [Google Scholar]
- Ton J, De Vos M, Robben C, Buchala A, Metraux JP, Van Loon LC, Pieterse CM. Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J. 2002;29:11–21. doi: 10.1046/j.1365-313x.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen YR. beta-aminobutyric acid-induced resistance against plant pathogens. Plant Dis. 2002;86:448–457. doi: 10.1094/PDIS.2002.86.5.448. [DOI] [PubMed] [Google Scholar]
- Justyna P-G, Ewa K. Induction of resistance against pathogens by β-aminobutyric acid. Acta Physiol Plant. 2013;35:1–14. doi: 10.1007/s11738-012-1069-9. [DOI] [Google Scholar]
- Bengtsson T, Holefors A, Witzell J, Andreasson E, Liljeroth E. Activation of defence responses to Phytophthora infestans in potato by BABA. Plant Pathol. 2013;63(1):193–202. [Google Scholar]
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Metraux J-P, Mauch-Mani B. Dissecting the beta-aminobutyric acid-induced priming phenomenon in arabidopsis. Plant Cell. 2005;17:987–999. doi: 10.1105/tpc.104.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CH, Singh P, Chen CW, Thomas J, Weber J, Mauch-Mani B, Zimmerli L. Priming for enhanced defence responses by specific inhibition of the Arabidopsis response to coronatine. Plant J. 2011;65:469–479. doi: 10.1111/j.1365-313X.2010.04436.x. [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Hou BH, Tsai CH, Jakab G, Mauch-Mani B, Somerville S. The xenobiotic beta-aminobutyric acid enhances Arabidopsis thermotolerance. Plant J. 2008;53:144–156. doi: 10.1111/j.1365-313X.2007.03343.x. [DOI] [PubMed] [Google Scholar]
- Singh P, Wu CC, Zimmerli L. beta-aminobutyric acid priming by stress imprinting. Plant Signal Behav. 2010;5:878–880. doi: 10.4161/psb.5.7.11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-C, Singh P, Chen M-C, Zimmerli L. L-Glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. J Exp Bot. 2010;61:995–1002. doi: 10.1093/jxb/erp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Kuo YC, Mishra S, Tsai CH, Chien CC, Chen CW, Desclos-Theveniau M, Chu PW, Schulze B, Chinchilla D, Boller T, Zimmerli L. The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell. 2012;24:1256–1270. doi: 10.1105/tpc.112.095778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tian Z, Liu J, Xie C. Comparative cDNA-AFLP analysis reveals that DL-beta-amino-butyric acid induces resistance through early activation of the host-defense genes in potato. Physiol Plant. 2009;136:19–29. doi: 10.1111/j.1399-3054.2009.01209.x. [DOI] [PubMed] [Google Scholar]
- Eschen-Lippold L, Altmann S, Rosahl S. DL-beta-aminobutyric acid-induced resistance of potato against Phytophthora infestans requires salicylic acid but not oxylipins. Mol Plant Microbe Interact. 2010;23:585–592. doi: 10.1094/MPMI-23-5-0585. [DOI] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J, Orjeda G, Guzman F, Torres M, Lozano R, Ponce O, Martinez D, De la Cruz G, Chakrabarti SK, Patil VU, Skryabin KG, Kuznetsov BB, Ravin NV, Kolganova TV, Beletsky AV, Mardanov AV, Di Genova A, Bolser DM, Martin DM, Li G, Yang Y. et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell Online. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. "Limma: linear models for microarray data." Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Moushib LI, Lenman M, Levander F, Olsson K, Carlson-Nilson U, Zoteyeva N, Liljeroth E, Andreasson E. Paranoid potato phytophthora-resistant genotype shows constitutively activated defense. Plant Signal Behav. 2012;7:400–408. doi: 10.4161/psb.19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersson E, Ali A, Resjö S, Andreasson E. Plant secretome proteomics. Front Plant Sci. 2013;4:9. doi: 10.3389/fpls.2013.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkinen J, Vincic G, Mansson O, Warell K, Levander F. The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J Proteome Res. 2009;8:3037–3043. doi: 10.1021/pr900189c. [DOI] [PubMed] [Google Scholar]
- Sandin M, Krogh M, Hansson K, Levander F. Generic workflow for quality assessment of quantitative label-free LC-MS analysis. Proteomics. 2011;11:1114–1124. doi: 10.1002/pmic.201000493. [DOI] [PubMed] [Google Scholar]
- Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang R, Lin C, Chen J, Goodlett D, Whiteaker J, Paulovich A, McIntosh M. A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics. 2006;22:1902–1909. doi: 10.1093/bioinformatics/btl276. [DOI] [PubMed] [Google Scholar]
- Sandin M, Ali A, Hansson K, Månsson O, Andreasson E, Resjö S, Levander F. An adaptive alignment algorithm for quality-controlled label-free LC-MS. Mol Cell Proteomics. 2013;12:1407–1420. doi: 10.1074/mcp.O112.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeroth E, Bengtsson T, Wiik L, Andreasson E. Induced resistance in potato to Phytphthora infestans-effects of BABA in greenhouse and field tests with different potato varieties. Eur J Plant Pathol. 2010;127:171–183. doi: 10.1007/s10658-010-9582-4. [DOI] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L, Witters E, Puga MI, Paz-Ares J, Goossens A, Reymond P, De Jaeger G, Solano R. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z, McGarvey B, Amyot L, Gruber M, Jung J, Hannoufa A. DIMINUTO 1 affects the lignin profile and secondary cell wall formation in Arabidopsis. Planta. 2012;235:485–498. doi: 10.1007/s00425-011-1519-4. [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant. 2010;3:956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- Moushib LI, Witzell J, Lenman M, Liljeroth E, Andreasson E. Sugar beet extract induces defence against Phytophthora infestans in potato plants. Eur J Plant Pathol. 2013;136(2):261–271. doi: 10.1007/s10658-012-0160-9. [DOI] [Google Scholar]
- Ogawa T, Ishikawa K, Harada K, Fukusaki E, Yoshimura K, Shigeoka S. Overexpression of an ADP-ribose pyrophosphatase, AtNUDX2, confers enhanced tolerance to oxidative stress in Arabidopsis plants. Plant J. 2009;57:289–301. doi: 10.1111/j.1365-313X.2008.03686.x. [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Huckelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 2011;16:117–125. doi: 10.1016/j.tplants.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310(5751):1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou B-H, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell Online. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal L, Scheel D, Rosahl S. The coi1-16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell Online. 2008;20:824–826. doi: 10.1105/tpc.107.056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macarisin D, Wisniewski ME, Bassett C, Thannhauser TW. Proteomic analysis of β-aminobutyric acid priming and abscisic acid – induction of drought resistance in crabapple (Malus pumila): effect on general metabolism, the phenylpropanoid pathway and cell wall enzymes. Plant Cell Environ. 2009;32:1612–1631. doi: 10.1111/j.1365-3040.2009.02025.x. [DOI] [Google Scholar]
- Kopischke M, Westphal L, Schneeberger K, Clark R, Ossowski S, Wewer V, Fuchs R, Landtag J, Hause G, Dörmann P. Impaired sterol ester synthesis alters the response of Arabidopsis thaliana to Phytophthora infestans. The Plant Journal. 2013;73(3):456–468. doi: 10.1111/tpj.12046. [DOI] [PubMed] [Google Scholar]
- Hendrix JW. Sterols in growth and reproduction of fungi. Annu Rev Phytopathol. 1970;8:111–130. doi: 10.1146/annurev.py.08.090170.000551. [DOI] [Google Scholar]
- Choi D, Bostock RM, Avdiushko S, Hildebrand DF. Lipid-derived signals that discriminate wound- and pathogen-responsive isoprenoid pathways in plants: methyl jasmonate and the fungal elicitor arachidonic acid induce different 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes and antimicrobial isoprenoids in Solanum tuberosum L. Proc Natl Acad Sci. 1994;91:2329–2333. doi: 10.1073/pnas.91.6.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjamos EC, Kucacute JA. Inhibition of steroid glycoalkaloid accumulation by arachidonic and eicosapentaenoic acids in potato. Science. 1982;217:542–544. doi: 10.1126/science.217.4559.542. [DOI] [PubMed] [Google Scholar]
- Choi D, Ward BL, Bostock RM. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell Online. 1992;4:1333–1344. doi: 10.1105/tpc.4.10.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krits P, Fogelman E, Ginzberg I. Potato steroidal glycoalkaloid levels and the expression of key isoprenoid metabolic genes. Planta. 2007;227:143–150. doi: 10.1007/s00425-007-0602-3. [DOI] [PubMed] [Google Scholar]
- Ros B, Thummler F, Wenzel G. Analysis of differentially expressed genes in a susceptible and moderately resistant potato cultivar upon Phytophthora infestans infection. Mol Plant Pathol. 2004;5:191–201. doi: 10.1111/j.1364-3703.2004.00221.x. [DOI] [PubMed] [Google Scholar]
- Stermer BA, Bostock RM. Involvement of 3-hydroxy-3-methylglutaryl coenzyme a reductase in the regulation of sesquiterpenoid phytoalexin synthesis in potato. Plant Physiol. 1987;84:404–408. doi: 10.1104/pp.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM, Kuc JA, Laine RA. Eicosapentaenoic and arachidonic acids from phytophthora infestans elicit fungitoxic sesquiterpenes in the potato. Science. 1981;212:67–69. doi: 10.1126/science.212.4490.67. [DOI] [PubMed] [Google Scholar]
- Bostock RM, Schaeffer DA, Hammerschmidt R. Comparison of elicitor activities of arachidonic acid, fatty acids and glucans from Phytophthora infestans in hypersensitivity expression in potato tuber. Physiol Mol Plant Pathol. 1986;29:349–360. doi: 10.1016/S0048-4059(86)80051-0. [DOI] [Google Scholar]
- Preisig CL, Kuć JA. Inhibition by salicylhydroxamic acid, BW755C, eicosatetraynoic acid, and disulfiram of hypersensitive resistance elicited by arachidonic acid or poly-l-lysine in potato tuber. Plant Physiol. 1987;84:891–894. doi: 10.1104/pp.84.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas J-L, Vailleau F, Davoine C, Ennar N, Agnel J-P, Tronchet M, Ponchet M, Blein J-P, Roby D, Triantaphylides C, Montillet J-L. The combined action of 9 lipoxygenase and galactolipase is sufficient to bring about programmed cell death during tobacco hypersensitive response. Plant Cell Environ. 2005;28:1367–1378. doi: 10.1111/j.1365-3040.2005.01369.x. [DOI] [Google Scholar]
- Cacas J-L, Marmey P, Montillet J-L, Sayegh-Alhamdia M, Jalloul A, Rojas-Mendoza A, Clérivet A, Nicole M. A novel patatin-like protein from cotton plant, GhPat1, is co-expressed with GhLox1 during Xanthomonas campestris-mediated hypersensitive cell death. Plant Cell Rep. 2009;28:155–164. doi: 10.1007/s00299-008-0622-x. [DOI] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Kollehn DO, Fenske R, Finnegan PM, O’Brien PA, Hardy GE, Lambers H. Acclimation responses of Arabidopsis thaliana to sustained phosphite treatments. J Exp Bot. 2013;64:1731–1743. doi: 10.1093/jxb/ert037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta. 2010;231:1237–1249. doi: 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- Lazzarato L, Trebbi G, Pagnucco C, Franchin C, Torrigiani P, Betti L. Exogenous spermidine, arsenic and beta-aminobutyric acid modulate tobacco resistance to tobacco mosaic virus, and affect local and systemic glucosylsalicylic acid levels and arginine decarboxylase gene expression in tobacco leaves. J Plant Physiol. 2009;166:90–100. doi: 10.1016/j.jplph.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Halim VA, Eschen-Lippold L, Altmann S, Birschwilks M, Scheel D, Rosahl S. Salicylic acid is important for basal defense of solanum tuberosum against Phytophthora infestans. Mol Plant-Microbe Interact. 2007;20:1346–1352. doi: 10.1094/MPMI-20-11-1346. [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20:228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Kuflu K, Romero MC, Gijzen M. A soybean seed protein with carboxylate-binding activity. J Exp Bot. 2005;56:2335–2344. doi: 10.1093/jxb/eri226. [DOI] [PubMed] [Google Scholar]
- Mika A, Minibayeva F, Beckett R, Lüthje S. Possible functions of extracellular peroxidases in stress-induced generation and detoxification of active oxygen species. Phytochem Rev. 2004;3:173–193. [Google Scholar]
- Baysal Ö, Gȕrsoy YZ, Örnek H, Duru A. Induction of oxidants in tomato leaves treated with DL-b-Amino butyric acid (BABA) and infected with Clavibacter michiganensis ssp. Eur J Plant Pathol. 2005;112:361–369. doi: 10.1007/s10658-005-6234-1. [DOI] [Google Scholar]
- Almasia NI, Bazzini AA, Hopp HE, Vazquez-Rovere C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol Plant Pathol. 2008;9:329–338. doi: 10.1111/j.1364-3703.2008.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A, Moreno M, Madueno F, Molina A, Garcia-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant-Microbe Interact. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- Nahirnak V, Almasia NI, Fernandez PV, Hopp HE, Estevez JM, Carrari F, Vazquez-Rovere C. Potato snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol. 2012;158:252–263. doi: 10.1104/pp.111.186544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri FP, Lobato MC, Gonzalez Altamiranda E, Daleo GR, Huarte M, Guevara MG, Andreu AB. BABA effects on the behaviour of potato cultivars infected by Phytophthora infestans and Fusarium solani. Eur J Plant Pathol. 2009;123:47–56. doi: 10.1007/s10658-008-9340-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of number of Gene Ontology terms. The number GO terms per probe of the Agilent JHI Solanum tuberosum 60 k v1 microarray annotated using either Parallel-OrthoMCL across 26 plant genomes or the Solanaceae Genomics Resource annotation.
Complete list of enriched GO terms. Significant differentially expressed genes after 10 mM BABA treatment were analysed by GOEast and GO terms with p >0.1 were considered significant.
Significant differentially expressed transcripts. A full list of significant differentially expressed transcripts (adj. p-value <0.05) in potato leaves following treatment with 10 mM BABA. Samples were analysed two days after treatment of the leaf canopy with BABA.
Text file 1. Extended gene ontology terms for microarray probes. Extended gene ontology terms for the custom-made Agilent expression array (JHI Solanum tuberosum 60 k v1). The file is formatted for use in GOEast.
Text file 2. Extended gene ontology terms for PGSC gene models v.3.4. Extended gene ontology terms for the PGSC gene models. The file is formatted for use in GOEast.


