Significance
We generated high-resolution multiorgan expression data showing that nearly half of all genes in the mouse genome oscillate with circadian rhythm somewhere in the body. Such widespread transcriptional oscillations have not been previously reported in mammals. Applying pathway analysis, we observed new clock-mediated spatiotemporal relationships. Moreover, we found a majority of best-selling drugs in the United States target circadian gene products. Many of these drugs have relatively short half-lives, and our data predict which may benefit from timed dosing.
Keywords: circadian, genomics, gene networks, noncoding RNA, chronotherapy
Abstract
To characterize the role of the circadian clock in mouse physiology and behavior, we used RNA-seq and DNA arrays to quantify the transcriptomes of 12 mouse organs over time. We found 43% of all protein coding genes showed circadian rhythms in transcription somewhere in the body, largely in an organ-specific manner. In most organs, we noticed the expression of many oscillating genes peaked during transcriptional “rush hours” preceding dawn and dusk. Looking at the genomic landscape of rhythmic genes, we saw that they clustered together, were longer, and had more spliceforms than nonoscillating genes. Systems-level analysis revealed intricate rhythmic orchestration of gene pathways throughout the body. We also found oscillations in the expression of more than 1,000 known and novel noncoding RNAs (ncRNAs). Supporting their potential role in mediating clock function, ncRNAs conserved between mouse and human showed rhythmic expression in similar proportions as protein coding genes. Importantly, we also found that the majority of best-selling drugs and World Health Organization essential medicines directly target the products of rhythmic genes. Many of these drugs have short half-lives and may benefit from timed dosage. In sum, this study highlights critical, systemic, and surprising roles of the mammalian circadian clock and provides a blueprint for advancement in chronotherapy.
Circadian rhythms are endogenous 24-h oscillations in behavior and biological processes found in all kingdoms of life. This internal clock allows an organism to adapt its physiology in anticipation of transitions between night and day. The circadian clock drives oscillations in a diverse set of biological processes, including sleep, locomotor activity, blood pressure, body temperature, and blood hormone levels (1, 2). Disruption of normal circadian rhythms leads to clinically relevant disorders including neurodegeneration and metabolic disorders (3, 4). In mammals, the molecular basis for these physiological rhythms arises from the interactions between two transcriptional/translational feedback loops (reviewed in ref. 5). Many members of the core clock regulate the expression of other transcripts. These clock-controlled genes mediate the molecular clock’s effect on downstream rhythms in physiology.
In an effort to map these connections between the core clock and the diverse biological processes it regulates, researchers have devoted significant time and effort to studying transcriptional rhythms (6–10). Although extremely informative, most circadian studies of this nature have analyzed one or two organs by using microarrays, and little work has been done to analyze either clock control at the organism level or regulation of the noncoding transcriptome. To address these gaps in our knowledge, we used RNA-sequencing (RNA-seq) and DNA arrays to profile the transcriptomes of 12 different mouse organs: adrenal gland, aorta, brainstem, brown fat, cerebellum, heart, hypothalamus, kidney, liver, lung, skeletal muscle, and white fat. We sampled organs every 6 h by RNA-seq and every 2 h by arrays, to develop an atlas of mouse biological space and time.
Using this resource, we examined the genomic characteristics of the rhythmic coding and noncoding transcriptomes, the differences between organs in timing (phase) and identity of oscillating transcripts, and the functional implications of rhythmic regulation in various biological pathways. Lastly, we explored the potential medical impact of circadian genes as drug targets and disease-associated genes. It is our hope that this work provides a rich dataset to the research community that could power many future studies.
Results
Genes and Noncoding Transcripts.
We defined a background set of 19,788 known protein-coding mouse genes and, for each organ, we used the JTK_CYCLE (11) algorithm to detect 24-h oscillations in transcript abundance. For this protein-coding gene analysis, we leveraged the high temporal resolution of the array data to accurately identify circadian genes. We set a 5% false-discovery rate (FDR) for detection, although the specific value of this cutoff did not affect the relative amount of rhythmic transcripts detected between organs (Fig. S1A). In this context, we defined the term “circadian gene” as any gene identified as cycling with a 24-h period by JTK_CYCLE, and passing the FDR cutoff listed above. We used the base-pair level RNA-seq data in a complimentary fashion to identify the expressed spliceforms of these circadian genes, and for our analysis of the noncoding transcriptome.
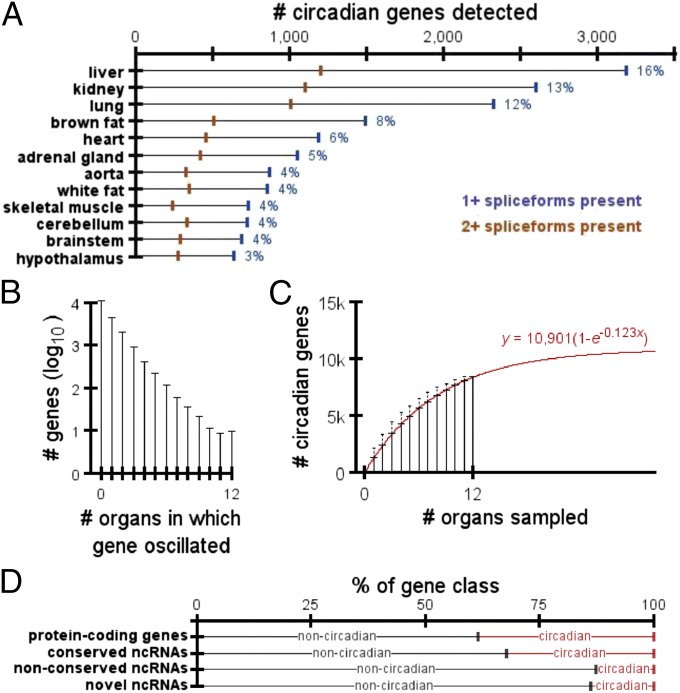
Following these analyses, we found liver had the most circadian genes (3,186), whereas hypothalamus had the fewest (642) (Fig. 1A). In fact, the three brain regions (cerebellum, brainstem, and hypothalamus) had the fewest circadian genes, collectively. Because of the technical difficulty of precisely sampling brain regions, we assume that heterogeneous mixtures of cell types within these complex organs may express different sets of genes, or may be out of phase with each other. This transcript/phase discrepancy within the same organ would make it difficult to accurately identify circadian genes in these brain regions. On average, 46% (SD = 0.036%) of circadian protein-coding genes expressed multiple spliceforms detected in the RNA-seq data.
Fig. 1.
Breakdown of circadian genes and ncRNAs. (A) Number of protein-coding genes in each organ with circadian expression. Blue marks indicate the number of genes with at least one spliceform detected by RNA-seq. Orange marks indicate the number of genes with at least two spliceforms detected by RNA-seq. Blue numbers to the right of each bar list the percentage of protein-coding genes with rhythmic expression in each organ. (B) Distribution of the number of organs in which a protein-coding gene oscillated. (C) Average total number of circadian genes detected as a function of the number of organs sampled. Error bars represent SD. Best-fit model has been overlayed in red. (D) Percentages of each transcript class that did vs. did not oscillate in at least one organ.
Transcript abundance for 43% of protein-coding genes oscillated in at least one organ (Fig. 1B). Only 10 genes oscillated in all organs: Arntl, Dbp, Nr1d1, Nr1d2, Per1, Per2, and Per3 (core clock factors), and Usp2, Tsc22d3, and Tspan4. Although the organs we analyzed provide a broad sampling across the entire organism, there are still many more to study that may contain additional circadian genes. The average number of total circadian genes, y, detected by randomly sampling x organs was closely modeled by the exponential function y = a(1−e-bx), where e is Euler’s number and the coefficients a (asymptote) and b (rate of asymptotic approach) equal 10,901 and 0.123, respectively (R2 > 0.99; Fig. 1C). This estimate remains unchanged if we exclude the potentially noisy, heterogeneous organs discussed above (Fig. S1B). In other words, as we continue to sample additional organs, we predict ∼10,901 mouse protein-coding genes (55% of the background set) will show circadian oscillations somewhere in the body.
To study the noncoding transcriptome, we used NONCODE to define a background set of 1,016 mouse-human conserved noncoding RNAs (ncRNAs) (Fig. S2A and SI Methods). We found 32% of conserved ncRNAs oscillated (a similar proportion compared with protein-coding genes), whereas nonconserved ncRNAs were less likely to oscillate (Fig. 1D). This result suggests our set of conserved ncRNAs may be functionally relevant. Unlike protein-coding genes, no individual ncRNA oscillated in more than five organs. This observation is unsurprising, given that ncRNA expression is known to be organ-specific (12). We also found 712 of 5,154 unannotated, spliced noncoding transcripts (SI Methods) had rhythmic expression. Eighty percent of these aligned to the human genome (BLASTN; E < 10−10, sequence identity >70%), indicating they are conserved between human and mouse.
These conserved, clock-regulated ncRNAs covered a diverse set of functional classes (Fig. S2B). We found 30 were antisense to protein-coding genes, half of which were themselves circadian. There was no general phase relationship between sense and antisense ncRNAs. For example, in the liver, both Galt (galactose-1-phosphate uridylyltransferase) and an overlapping antisense ncRNA oscillated in phase with each other (Fig. S3 A–D). We also identified host genes for 39 circadian miRNAs and four small nucleolar RNA (snoRNA) host genes: Cbwd1, Snhg7, Snhg11, and Snhg12. Because snoRNAs were recently shown to have light-driven oscillations in Drosophila brains (13), these findings provide further evidence of the clock’s potential to influence ribosome biogenesis (14). We also found 74 conserved lincRNAs with circadian oscillations, the majority of which were Riken transcripts with no known function. Finally, we also found 1,979 genes with unannotated antisense transcripts, 187 of which showed sense and antisense oscillations in the same organ. Of these antisense transcripts, 43 oscillated at least 8 h out of phase with their sense transcripts. Genes with antiphase, antisense oscillators included Arntl and Per2 (Fig. S3 E–H). A known Per2 antisense transcript (9, 10) oscillated in four organs, the most of any antisense transcript, providing further evidence of its functional relevance. Taken together, our data reflect a vast and diverse set of transcripts regulated by the clock at the organism level.
Gene Parameters.
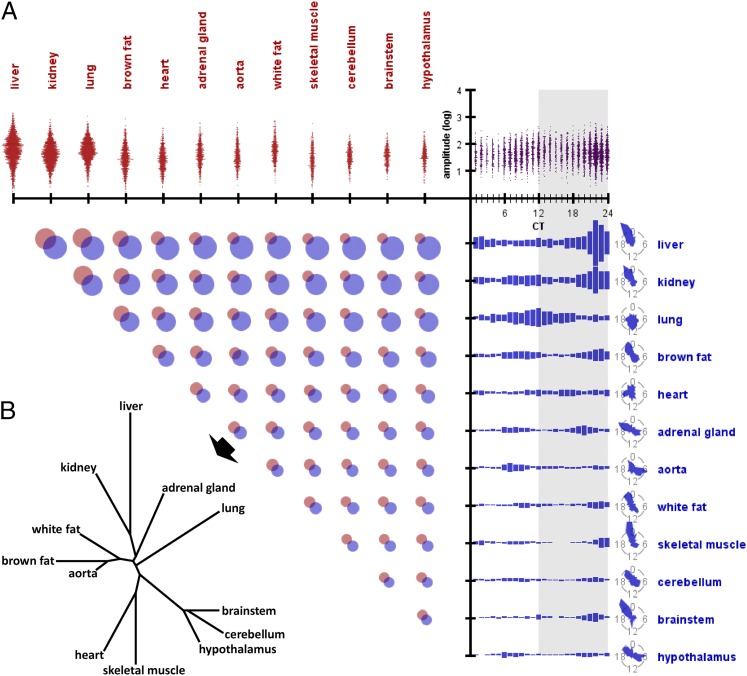
Our data agree with the finding from previous multiorgan studies that the vast majority of circadian-gene expression is organ-specific (6, 7), with little overlap of circadian-gene identity between organs (Fig. 2A). In most organs, expression of circadian genes peaked in the hours preceding subjective dusk or dawn, often in a bimodal fashion. Heart and lung were notable exceptions, with phase distributions that diverged substantially from other organs. Moreover, those circadian genes with expression peaks clustered around subjective dusk or dawn also tended to have the highest average oscillation amplitude, compared to genes with expression peaks at other times of day. Taken together, these data suggest that the body may experience daily “rush hours” of transcription at these critical times. Using the average phase difference between the shared circadian genes of any two organs as a distance metric, we were able to construct an ontogenic tree that recovered recognizable organ lineage (Fig. 2B) (15). Thus, developmentally related organs tended to share genes that oscillate synchronously.
Fig. 2.
Parameters of circadian genes across organs. (A) Relationships among organ, oscillation amplitude, and oscillation phase of circadian genes. (Upper Left) Histograms of amplitudes within each organ (number of circadian genes within each amplitude bin is shown on the horizontal axis, grouped by organ). (Upper Right) Histograms of amplitudes within each phase, across all organs. (Lower Right) Histograms of phases within each organ, with summary radial diagrams (number of circadian genes within each phase bin is shown on the vertical axis, grouped by organ). Larger versions of these radial diagrams are included in Fig. S1C for clarity. (Lower Left) Venn diagrams of the identities of the genes that oscillated within a given pair of organs. (B) Ontogenic tree constructed using the average phase differences between each organ pair’s shared circadian genes as the distance metric. Shared genes correspond to the overlapping regions from Venn diagrams in A.
Having examined their oscillation patterns, we looked for genomic characteristics common to rhythmically expressed genes. Circadian genes clustered physically in the genome (Fig. S4A and SI Methods). Their lengths tended to be longer than nonrhythmic genes (Mann–Whitney u test P << 10−15; Fig. S4B). This trend was maintained at the level of 5′UTR, CDS, and 3′UTR (Fig. S4 C–E). These results are in agreement with previous findings about oscillating liver transcripts (16). By using gapped, junction-spanning reads to discriminate between expressed spliceforms, we found circadian genes had more spliceforms than noncircadian genes (Mann–Whitney u test P << 10−15; Fig. S4 F–H). Furthermore, we found that the spliceforms expressed by circadian genes, including the identity of the dominant spliceform, tended to differ across organs more than for noncircadian genes. These findings are consistent with the idea that the circadian genes have more regulatory capacity than noncircadian genes.
Remarkably, 1,400 genes were phase-shifted with respect to themselves by at least 6 h between two organs, with 131 genes completely antiphased (Fig. S4I). For example, at dusk, the transcript levels of Vegfa (vascular endothelial growth factor) peaked in brown fat but reached a nadir in heart. To our knowledge, such drastic phase discrepancies of individual genes between organs have not been reported. The mechanisms for these phenomena are unclear, because the genes did not share any obvious transcription-factor or miRNA-binding motifs. The core clock genes oscillated synchronously, with the peak phases of a given gene falling within 3 h of each other across all organs (Fig. S5). Several core clock genes showed 1- to 2-h phase advances and delays in skeletal muscle and cerebellum, respectively, compared with other organs. However, these cases were in the minority, and given the limitations in our ability to precisely resolve small (<2 h) phase differences from data with a 2-h resolution, their significance remains unclear. This finding indicates that the antiphased patterns observed in genes like Vegfa are not due to phase differences between the core clocks of each organ. Rather, these phenomena are due to additional, organ-specific levels of timing regulation positioned between the core clock and these output genes.
Pathways.
Given the high temporal and spatial resolution of our study, we were able to examine ways in which time and space influenced biological pathways. We used the Reactome (17) database as a basis for our pathway network and found many pathways enriched for circadian genes both within and across organs (Fig. S6).
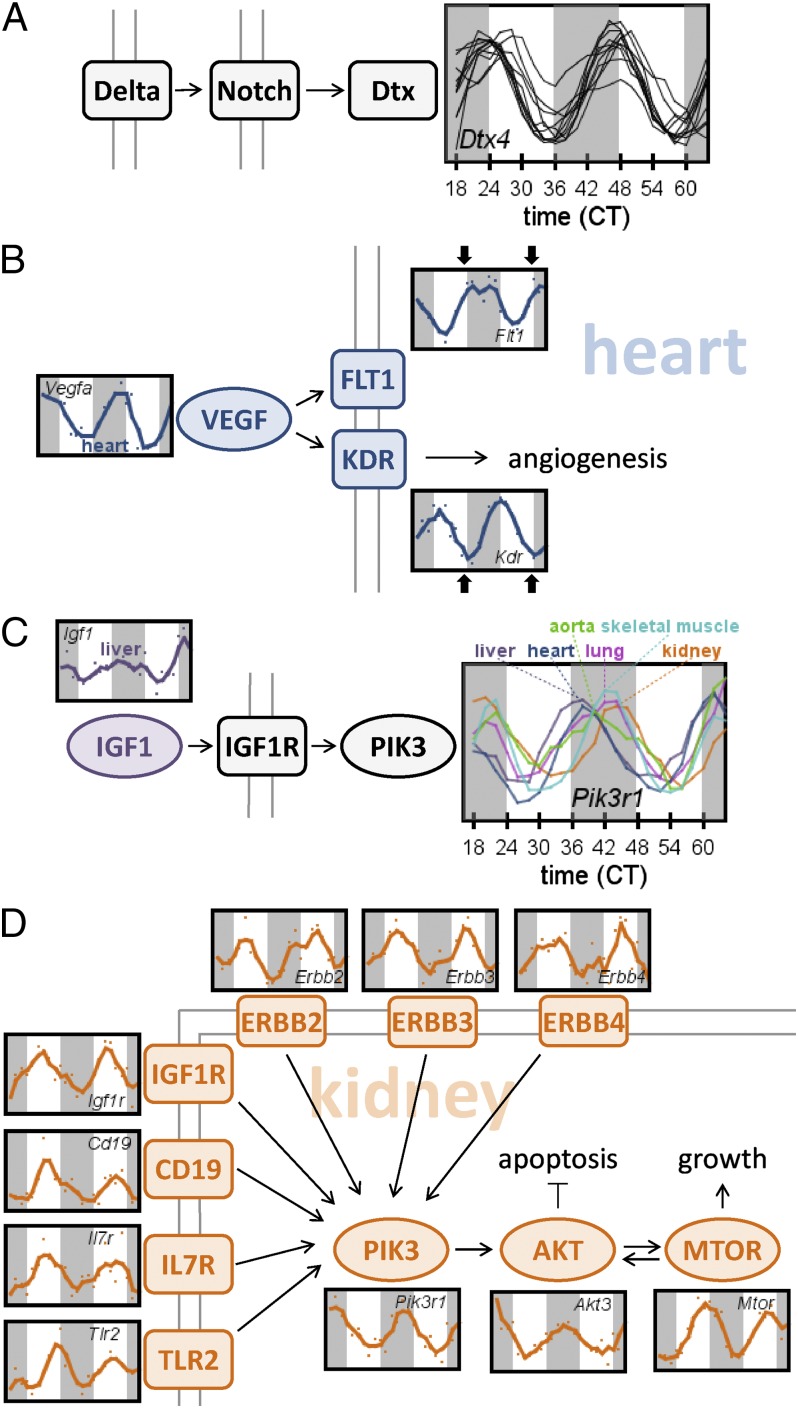
Several genes oscillated synchronously across all organs, like the core clock genes. For example, Dtx4, a Notch pathway E3 ubiquitin ligase, oscillated in phase with Arntl in all organs (Fig. 3A). We also noted that genes with “opposite” functions (e.g., activators vs. repressors) often had opposite phases. For example, members of the initial vascular endothelial growth factor (VEGF) signaling cascade oscillated in the heart (Fig. 3B). These VEGF pathway members included the primary circulating ligand, Vegfa, and its two principal membrane-bound receptors, Flt1 and Kdr. This cascade regulates angiogenesis, with critical roles in development, cancer, and diabetes (18). At dusk, expression of Vegfa and Kdr in the heart was low, whereas Flt1 was high. KDR is thought to mediate most of the known cellular responses to VEGF signaling, whereas FLT1 is thought to be a decoy receptor (19). Thus, the rhythmic timing of these receptors appears to reflect function, in that FLT1 (the decoy) is present when KDR is not and vice versa.
Fig. 3.
Exploring pathways across biological space and time. (A) Expression of the deltex gene Dtx4 in all organs superimposed. (B) Example of pathway components’ timing reflecting function: expression profiles from the heart, for Vegfa and its two receptors Kdr and Flt1. Black arrows highlight times at which Flt1 and Kdr are anti-phased. (C) Example of systemic pathway orchestration segregating in time and space: expression profile of Igf1 in the liver, compared with its downstream target Pik3 in several organs. (D) Example of widespread pathway component synchronization within the same space (organ): expression profiles from the kidney for multiple signaling receptors that activate the PIK3-AKT-MTOR pathway.
Whereas members of some systemic pathways, such as the core circadian clock, were expressed in phase across organs, many were not. For instance, expression of the insulin-like growth factor Igf1 oscillated in the liver, peaking in the early subjective night (Fig. 3C). Because the liver produces nearly all of the circulating IGF1 (20), IGF signaling throughout the entire body is likely under clock influence. IGF1 is one of the most potent natural activators of the PIK3-AKT-MTOR pathway, which stimulates growth, inhibits apoptosis, and has a well-known role in cancer (21). However, peak expression of Pik3r1, which encodes the regulatory subunit for PIK3, did not occur at the same time across all organs. Instead, there was a steady progression throughout the night spanning nearly 10 h, as it peaked first in liver, then heart, followed by aorta, lung, skeletal muscle, and finally in kidney (Fig. 3C). Because the core clocks of these organs were in phase with each other, as mentioned earlier, the timing differences of Pik3r1 are most likely driven by some unknown, organ-specific mechanism situated between the core clock pathway and Pik3r1.
Some pathways known to function systemically were only rhythmic in a single organ. For example, IGF1’s principal membrane-bound receptor, IGF1R, is present in numerous organs. However, Igf1r expression oscillated only in kidney. In addition to Igf1r, many other membrane-bound receptors that activate the PIK3-AKT-MTOR cascade were also rhythmically expressed only in kidney (Fig. 3D). These receptors included Erbb2, Erbb3, and Erbb4 (tyrosine kinase receptors), Tlr2 (toll-like receptor), Cd19 (antigen receptor), and Il7r (cytokine/interleukin receptor). These receptors were all notably in phase with one another, all having peak expression in the subjective midday. Thus, there is kidney-specific clock regulation of PIK3-AKT-MTOR signaling that is distinct from, and in addition to, the already clock-regulated IGF1 signal coming from the liver.
Drug Targets and Disease.
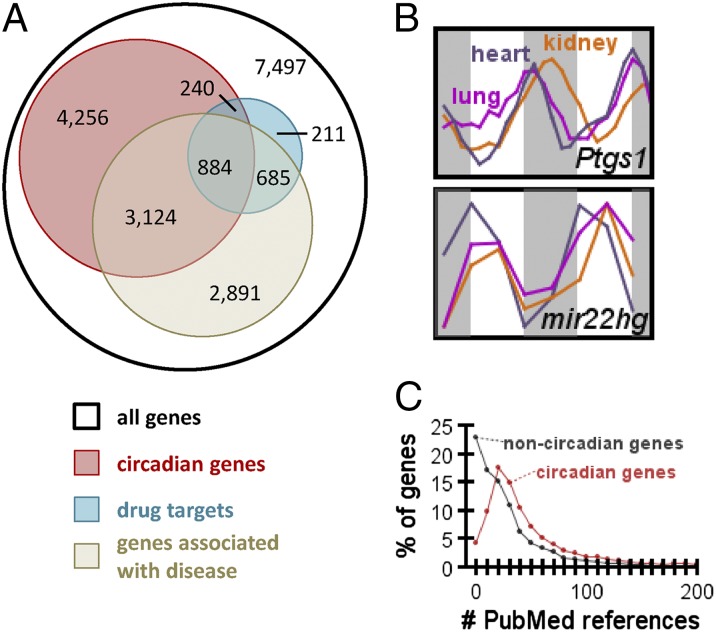
Timing is an important but underappreciated factor in drug efficacy. For example, short half-life statins work best when taken before bedtime, as cholesterol synthesis peaks when we sleep (22). To find new opportunities for prospective chronotherapy, we investigated which of the best-selling and commonly taken drugs target genes with rhythmic expression (association between circadian genes and drug targets by Pearson’s χ2 test, P << 10−15; Fig. 4A). By “drug targets,” we are referring to genes with products directly bound and functionally affected by a given drug. Notably, 56 of the top 100 best-selling drugs in the United States, including all top 7, target the product of a circadian gene (Dataset S1). Nearly half of these drugs have half-lives less than 6 h (Table 1), suggesting the potential impact time of administration could have on their action. Most of these drugs have not been associated with circadian rhythms and are not dosed with consideration for body time. Furthermore, 119 of the World Health Organization’s list of essential medicines target a circadian gene, including many of the most common and well known targets (Dataset S2). For example, Ptgs1 (cyclooxygenase-1, alias Cox1), the primary target of low dose aspirin therapy used in secondary prevention of heart attacks (23), oscillated in the heart, lung, and kidney (Fig. 4B). Given that aspirin has a short half-life and that heart attacks have a circadian rhythm (2), dosing aspirin at an optimal time of the day has great potential. Consistent with this observation, clinical reports have suggested nighttime administration of low dose aspirin may be important for its cardio-protective effects (24). Our data suggest a mechanism for Ptgs1’s circadian regulation as well. Mir22 is a microRNA predicted to target PTGS1, and its host transcript oscillated antiphase to Ptgs1 in the heart, lung, and kidney. This miRNA may therefore regulate Ptgs1 function. To test this hypothesis, we transfected mir22 mimics into NIH 3T3 cells and knocked down endogenous quantities of PTGS1 protein by ∼50% (Fig. S7). We also observed a slight, nonsignificant decrease in Ptgs1 mRNA levels in these same samples. These data suggest that mir22 operates on PTGS1 predominantly at the posttranscriptional level, although it remains possible that Ptgs1 is a transcriptional target of the clock through other mechanisms.
Fig. 4.
Circadian disease genes and drug targets. (A) Overlap between circadian genes, known disease-associated genes, and drug targets. Sources for disease genes and drug targets are included in SI Methods. (B) Example of a common drug having an oscillatory gene target: expression profiles for the aspirin target Ptgs1 from heart, lung, and kidney. Traces from these organs for the mir22 host gene, predicted to target Ptgs1, are also shown. (C) Number of PubMed references for circadian vs. noncircadian genes.
Table 1.
Drugs of the top-100 best-seller list that target circadian genes and have half-life < 6h
| Rank | Sales, $ | Trade name | Indications | Circadian-gene targets | Organs in which targets oscillate |
| 2 | 1.46 b | Nexium | Gastritis, GERD, Esophagitis | Atp4a | L |
| 5 | 1.28 b | Advair Diskus | Asthma, Chronic obstructive pulmonary di... | Serpina6, Pgr, Nr3c2, Adrb2, Pla2g4a | Lu, H, L, K, S, A |
| 11 | 794 m | Rituxan | Rheumatoid arthritis, Non-Hodgkin's lymp... | Fcgr2b, Ms4a1, Fcgr3 | L, K, S |
| 20 | 538 m | Diovan | Hypertension, Heart failure | Slc22a6, Agtr1a, Slco1b2, Car4, Kcnma... | H, AG, L, K, S |
| 27 | 431 m | Vyvanse | Attention deficit hyperactivity disorder | Adra1b | L |
| 32 | 392 m | Tamiflu | Influenza | Neu2, Neu1, Ces1g, Slc22a8, Slc15a1, ... | Lu, L, BF, K, C |
| 33 | 383 m | Ritalin | Attention deficit hyperactivity disorder | Slc6a4 | AG, K |
| 37 | 348 m | AndroGel | Hypogonadism | Slc22a4, Slc22a3, Ar, Cyp1a1, Cyp2b10... | Lu, H, BS, WF, AG... |
| 38 | 346 m | Lidoderm | Pain | Slc22a5, Cyp2b10, Egfr, Abcb1a | Lu, H, AG, BF, L,... |
| 44 | 304 m | Seroquel XR | Bipolar disorder, Major depressive disor... | Htr2c, Htr1b, Htr2a, Chrm2, Drd4, Adr... | Lu, H, BS, WF, AG... |
| 45 | 289 m | Viagra | Erectile dysfunction | Cyp1a1, Pde6g, Abcc5, Abcc10, Pde5a, ... | Lu, H, BS, WF, AG... |
| 47 | 281 m | Niaspan | Hyperlipidemia | Slco2b1, Slc22a5, Qprt, Slc16a1 | Lu, H, BS, AG, WF... |
| 48 | 279 m | Humalog | Diabetes mellitus T2 | Igf1r | K |
| 49 | 274 m | Alimta | Mesothelioma, Nonsmall cell lung cancer | Tyms, Atic, Gart, Slc29a1 | Lu, H, BS, BF, L,... |
| 54 | 267 m | Combivent | Asthma, Chronic obstructive pulmonary di... | Slc22a5, Slc22a4, Chrm2, Adrb1, Adrb2 | Lu, H, BS, BF, K,... |
| 56 | 262 m | ProAir HFA | Asthma, Chronic obstructive pulmonary di... | Adrb1, Adrb2 | Lu, K, S |
| 62 | 240 m | Janumet | Diabetes mellitus T2 | Slc47a1, Slc22a2, Prkab1, Abcb1a, Dpp4 | H, BS, AG, Hy, L,... |
| 66 | 236 m | Toprol XL | Hypertension, Heart failure | Slc22a2, Adrb1, Adrb2, Abcb1a | Lu, H, AG, BF, L,... |
| 71 | 220 m | Vytorin | Hyperlipidemia | Hmgcr, Cyp2b10, Soat1, Abcc2, Anpep, ... | Lu, H, BS, AG, BF... |
| 78 | 209 m | Aciphex | Gastritis, GERD, Esophagitis | Cyp1a1, Atp4a, Abcg2 | Lu, H, BS, WF, L,... |
| 90 | 189 m | Lunesta | Insomnia | Ptgs1, Tspo, Gabra3 | Lu, H, AG, K |
| 98 | 173 m | Prilosec | Gastritis, GERD, Esophagitis | Cyp1a1, Atp4a, Abcg2, Cyp1b1, Abcb1a | Lu, H, BS, WF, AG... |
| 99 | 171 m | Focalin XR | Attention deficit hyperactivity disorder | Slc6a4 | AG, K |
Rank and sales are based on USA 2013 Q1 data from Drugs.com. A, aorta; AG, adrenal gland; BF, brown fat; BS, brainstem; C, cerebellum; H, heart; Hy, hypothalamus; K, kidney; L, liver; Lu, lung; S, skeletal muscle; WF, white fat.
Beyond drug targets, circadian genes were also enriched among disease-associated genes (Pearson’s χ2 test, P << 10−15; Fig. 4A) and were highly studied in biomedical research. They received significantly more PubMed citations than nonoscillating genes (Mann–Whitney u test, P << 10−15; Fig. 4C). Furthermore, oscillating genes were also associated with nearly every major disease funded by National Institutes of Health at significantly higher rates than expected by chance (Fig. S8). Cancer, diabetes mellitus type 2, Alzheimer’s disease, schizophrenia, Down’s syndrome, obesity, and coronary artery disease were most strongly associated with circadian genes. For example, many of these oscillating genes are involved in neurodegeneration, including Fus, Tdp43, alpha synuclein, gamma synuclein, Atxn1, Atxn2, Atxn3, Atxn7, Atxn10, Psen1, and Psen2. These genes are mutated in frontotemporal dementia, ALS, Parkinson’s disease, spinocerebellar ataxia, and Alzheimer’s disease. They were predominantly rhythmic outside of the brain in peripheral organs (Psen2 had nearly fourfold amplitude in liver and peaked at subjective day, when mice are going to sleep). We speculate that promoters for these genes may have evolved sensitivity to global changes in redox state, which varies between day and night (25). Lending credence to the association between clocks and neurodegeneration are two clinical observations: Many patients with neurodegeneration-linked dementia display “sundowning” (behavioral problems in the early evening), and most patients with neurodegeneration eventually develop circadian sleep disorders (3).
Discussion
In this study, we used RNA sequencing and DNA microarrays to characterize circadian oscillations in transcript expression across 12 mouse organs. We found that the RNA abundance of 43% of mouse protein-coding genes cycle in at least one organ. Based on these results, we project that more than half of the mouse protein-coding genome is rhythmic somewhere in the body. This observation is similar to the proportion of liver genes encoding proteins detected by mass spectrometry, which also showed transcriptional rhythms (26). There is precedence for such large-scale rhythms in unicellular and plant studies (27, 28), and previous work has suggested the same may be true in the mammalian system (29). We found the majority of these transcriptional rhythms are organ-specific. This characteristic, in addition to our high sampling resolution, explains why we found more rhythmic genes across 12 organs than previous studies (including those from our laboratory) that focused on only a few organs. Furthermore, the organ specificity of these transcriptional rhythms indicates that although the molecular clock is active throughout the body, it regulates biological processes quite differently in each organ. Again, this observation is in agreement with existing literature (6, 7).
The major exception to this finding is the set of core clock genes, as these genes oscillated in phase across all 12 organs (Fig. S5). While external cues such as restricted feeding or jet lag can phase shift these peripheral oscillators with respect to one another (30, 31), our observation agrees with the notion that peripheral clocks are largely synchronized in a healthy organism. Taken as a whole, our data can be used to address questions about the regulatory mechanisms for clock-controlled genes and these analyses are the subject of ongoing work.
Additionally, we found a functionally diverse set of ncRNAs with rhythmic expression. Those ncRNAs conserved between human and mouse oscillated in the same proportion as protein-coding genes, suggesting their functional importance. Although some of these rhythmic ncRNAs have recognized functions, like snoRNA and miRNA host genes, little is known about the majority. The oscillations of these ncRNAs may prove advantageous for functional studies, e.g., linking a cycling miRNA to its predicted target genes by comparing their cycles.
Recent studies have found additional layers of complexity in hepatic rhythms by examining DNA binding patterns of core clock genes, changes in epigenetic marks, and oscillations in the proteome (9, 10, 26, 32). By providing a window into every stage of the transcriptional/translational oscillations in the liver (open chromatin, transcription factor binding, transcription, protein accumulation), one could use these complementary datasets to model how the interplay among DNA, RNA, and protein results in rhythmic output in liver biology. However, these proteomic and chromatin-immunoprecipitation assays are still quite challenging to perform, especially for organs with more limited material than the liver. By applying the model developed in liver to our data, one could make organism-level predictions about the rhythmic characteristics of epigenetic marks and proteins.
The field of chronotherapeutics has appreciated the system-level effects of circadian biology for quite some time. At its core, this field aims to understand how time of day influences the metabolism, efficacy, toxicity, and off-target effects of therapeutics (1). Consider the case of statins (reviewed in ref. 33), a class of drug that lowers cholesterol by inhibiting HMGCR (HMG-CoA reductase). HMGCR is the rate-limiting enzyme in cholesterol biosynthesis and its activity peaks during the night. Statins with short half-lives showed maximal efficacy when taken in the evening (when their target gene was most active). Longer-acting statins show no changes in efficacy based on the time of administration. The flexibility offered by these latter statins may serve to increase patient compliance and, ultimately, improve their health outcomes. This example is one among many, demonstrating the ability of chronotherapeutic practices to positively impact drug treatment.
Nevertheless, the influence of time of administration on the majority of pharmaceuticals on the market today has not been extensively studied, and circadian effects are not a routine aspect of drug efficacy and safety trials. Our data indicate that circadian genes are highly associated with diseases, and many of the most commonly-used medications in the world target circadian genes. Furthermore, our data indicate the following: (i) the majority of the top-selling drugs on the market have circadian targets, and (ii) a substantial fraction have half-lives less than 6 h (Dataset S3). These data allow for prospective chronotherapeutic studies, because they indicate which drugs are sensitive to time-of-day administration, and when and where they may do so. This is illustrated by the example we provide in the results, where we hypothesize that circadian oscillations in expression of the aspirin target gene, Ptgs1, are responsible for rhythms in aspirin’s cardio-protective effects. More broadly, these data will be a great resource for the field, and we invite the reader to explore this dataset through our web interface (bioinf.itmat.upenn.edu/circa).
Methods
Animal Preparation and Organ Collection.
Mice were prepared as previously described (8). Briefly, 6-wk-old male C57/BL6 mice were acquired from Jackson Labs, entrained to a 12h:12h light:dark schedule for 1 wk, then released into constant darkness. Starting at CT18 postrelease, three mice were killed in the darkness every 2 h, for 48 h. Specimens from the following organs were quickly excised and snap-frozen in liquid nitrogen: aorta, adrenal gland, brainstem, brown fat (anterior dorsum adipose), cerebellum, heart, hypothalamus, kidney, liver, lung, skeletal muscle (gastrocnemius), and white fat (epididymal adipose). Food and water were supplied ad libitum at all stages before killing. All procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Microarray Data.
Organ samples were homogenized in Invitrogen TRIzol reagent by using a Qiagen Tissuelyser. RNA was extracted by using Qiagen RNeasy columns as per manufacturer’s protocol, then pooled from three mice for each organ and time point. The reason for pooling was to average out both biological variance between individual animals and technical variance between individual dissections. RNA abundances were quantified by using Affymetrix MoGene 1.0 ST arrays and normalized by using Affymetrix Expression Console software (RMA). Probesets on the Affymetrix MoGene 1.0 ST array were cross-referenced to best-matching gene symbols by using Ensembl BioMart software, then filtered for known protein-coding status. The resulting 19,788 genes formed the protein-coding background set.
RNA-seq Data.
RNA samples from CT22, CT28, CT34, CT40, CT46, CT52, CT58, and CT64 were pooled for each organ, as described above (96 total pools). These RNA pools were converted into Illumina sequencing libraries by using Illumina TruSeq Stranded mRNA HT Sample Preparation Kits as per manufacturer’s protocol. Briefly, 1 μg of total RNA was polyA-selected, fragmented by metal-ion hydrolysis, and converted into double-stranded cDNA by using Invitrogen SuperScript II. The cDNA fragments were subjected to end-repair, adenylation, ligation of Illumina sequencing adapters, and PCR amplification. Libraries were pooled into groups of six and sequenced in one Illumina HiSeq 2000 lane by using the 100-bp paired-end chemistry (16 lanes total). Details on alignment and quantification are included in SI Methods. Annotation information for conserved ncRNAs and antisense transcripts included in Datasets S4 and S5.
Oscillation Detection.
The JTK_CYCLE (11) package for R was used, with parameters set to fit time-series data to exactly 24-h periodic waveforms. Significance was bounded by q < 0.05 for array data sampled at 2 h and by P < 0.05 for sequencing data sampled at 6 h.
Supplementary Material
Acknowledgments
We thank Jeanne Geskes, Trey Sato, George Paschos, Julie Baggs, Takeshige Kunieda, Zivjena Vucetic, Lei Yin, Boning Li, and Kathy Totoki for help with the circadian time point collection; the Penn Genome Frontiers Institute sequencing core for library construction and sequencing; the Penn Molecular Profiling Facility for preparation and running of the microarrays; and Katharina Hayer and the Institute for Translational Medicine and Therapeutics bioinformatics core for assistance with data analysis. This work is supported by the National Heart, Lung, and Blood Institute (5-R01-HL097800-04) and by the Defense Advanced Research Planning Agency [12-DARPA-1068 (to John Harer, Duke University)]. J.B.H. is supported by the National Institute of Neurological Disorders and Stroke (1R01NS054794-06) and by the Penn Genome Frontiers Institute under a grant with the Pennsylvania Department of Health. R.Z. is supported by National Human Genome Research Institute Grant T32HG000046. H.I.B. is supported by NIH Heart, Lung, and Blood Institute Grant T32 5T32HL007953-13 (to Dr. Allan Pack, University of Pennsylvania).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54652).
See Commentary on page 15869.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408886111/-/DCSupplemental.
References
- 1.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 2.Curtis AM, Fitzgerald GA. Central and peripheral clocks in cardiovascular and metabolic function. Ann Med. 2006;38(8):552–559. doi: 10.1080/07853890600995010. [DOI] [PubMed] [Google Scholar]
- 3.Hastings MH, Goedert M. Circadian clocks and neurodegenerative diseases: Time to aggregate? Curr Opin Neurobiol. 2013;23(5):880–887. doi: 10.1016/j.conb.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Storch K-F, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 8.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16(6):833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24(4):616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22(7):1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jouffe C, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11(1):e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar R, et al. LifeMap Discovery™: The embryonic development, stem cells, and regenerative medicine research portal. PLoS ONE. 2013;8(7):e66629. doi: 10.1371/journal.pone.0066629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G, et al. Gene and genome parameters of mammalian liver circadian genes (LCGs) PLoS ONE. 2012;7(10):e46961. doi: 10.1371/journal.pone.0046961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database issue):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folkman J. Angiogenesis: An organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6(4):273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 19.Zygmunt T, et al. Semaphorin-PlexinD1 signaling limits angiogenic potential via the VEGF decoy receptor sFlt1. Dev Cell. 2011;21(2):301–314. doi: 10.1016/j.devcel.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sjögren K, et al. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci USA. 1999;96(12):7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franke TF. PI3K/Akt: Getting it right matters. Oncogene. 2008;27(50):6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 22.Miettinen TA. Diurnal variation of cholesterol precursors squalene and methyl sterols in human plasma lipoproteins. J Lipid Res. 1982;23(3):466–473. [PubMed] [Google Scholar]
- 23.Antithrombotic Trialists’ Collaboration Collaborative meta-analysis of randomised trials of antiplatelet therapy for prevention of death, myocardial infarction, and stroke in high risk patients. BMJ. 2002;324(7329):71–86. doi: 10.1136/bmj.324.7329.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermida RC, et al. Differing administration time-dependent effects of aspirin on blood pressure in dipper and non-dipper hypertensives. Hypertension. 2005;46(4):1060–1068. doi: 10.1161/01.HYP.0000172623.36098.4e. [DOI] [PubMed] [Google Scholar]
- 25.Musiek ES, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123(12):5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauvoisin D, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci USA. 2014;111(1):167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9(12):1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 28.Michael TP, McClung CR. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 2003;132(2):629–639. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ptitsyn AA, Gimble JM. True or false: All genes are rhythmic. Ann Med. 2011;43(1):1–12. doi: 10.3109/07853890.2010.538078. [DOI] [PubMed] [Google Scholar]
- 30.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 31.Vosko AM, Colwell CS, Avidan AY. Jet lag syndrome: Circadian organization, pathophysiology, and management strategies. Nat Sci Sleep. 2010;2:187–198. doi: 10.2147/NSS.S6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schachter M. Chemical, pharmacokinetic and pharmacodynamic properties of statins: An update. Fundam Clin Pharmacol. 2005;19(1):117–125. doi: 10.1111/j.1472-8206.2004.00299.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.