Significance
Human mucosal surfaces contain a wide range of microorganisms. The biological effects of these organisms are largely unknown. Large-scale metagenomic sequencing is emerging as a method to identify novel microbes. Unexpectedly, we identified DNA sequences homologous to virus ATCV-1, an algal virus not previously known to infect humans, in oropharyngeal samples obtained from healthy adults. The presence of ATCV-1 was associated with a modest but measurable decrease in cognitive functioning. A relationship between ATCV-1 and cognitive functioning was confirmed in a mouse model, which also indicated that exposure to ATCV-1 resulted in changes in gene expression within the brain. Our study indicates that viruses in the environment not thought to infect humans can have biological effects.
Keywords: chlorovirus ATCV-1, infection, cognitive functioning, oropharyngeal virome, metagenomic sequencing
Abstract
Chloroviruses (family Phycodnaviridae) are large DNA viruses known to infect certain eukaryotic green algae and have not been previously shown to infect humans or to be part of the human virome. We unexpectedly found sequences homologous to the chlorovirus Acanthocystis turfacea chlorella virus 1 (ATCV-1) in a metagenomic analysis of DNA extracted from human oropharyngeal samples. These samples were obtained by throat swabs of adults without a psychiatric disorder or serious physical illness who were participating in a study that included measures of cognitive functioning. The presence of ATCV-1 DNA was confirmed by quantitative PCR with ATCV-1 DNA being documented in oropharyngeal samples obtained from 40 (43.5%) of 92 individuals. The presence of ATCV-1 DNA was not associated with demographic variables but was associated with a modest but statistically significant decrease in the performance on cognitive assessments of visual processing and visual motor speed. We further explored the effects of ATCV-1 in a mouse model. The inoculation of ATCV-1 into the intestinal tract of 9–11-wk-old mice resulted in a subsequent decrease in performance in several cognitive domains, including ones involving recognition memory and sensory-motor gating. ATCV-1 exposure in mice also resulted in the altered expression of genes within the hippocampus. These genes comprised pathways related to synaptic plasticity, learning, memory formation, and the immune response to viral exposure.
Mucosal sites of humans such as the oral pharynx and intestinal tract are not sterile but contain many microorganisms including bacteria, viruses, and fungi. It has recently become apparent that individuals differ in the composition of the microbial flora at their mucosal sites, characterized as the “microbiota” comprising the “microbiome.” Studies in both humans and animal models have shown that the microbiome can affect biological functions, including cognitive performance (1, 2). Bacterial and fungal components of the oral microbiome have been the subject of several prior studies. These studies generally rely on PCR-based amplification and subsequent analysis of ribosomal RNA sequences conserved among most species (e.g., refs. 3–6). Analysis of viral components in the microbiome (virome) has received less attention, largely because there are no universal target regions suitable for PCR amplification across different viral taxa. This problem can be overcome by whole genome sequencing in which viral particles are separated from the rest of the sample, sequenced, and then bioinformatically identified. Such analyses have identified bacteriophages as the most abundant component of the human oral microbiome (6–9). The virome can also be evaluated by sequencing unfractionated samples. This approach has the advantage of identifying viruses that might be in low concentrations or evade detection by standard fractionation methods (10–12).
In the process of analyzing whole genome sequences obtained from unfractionated samples of the oropharynx from healthy individuals participating in a study that involved the assessment of cognitive functioning, we unexpectedly discovered a substantial number of sequence reads very similar to virus Acanthocystis turfacea chlorella virus 1 (ATCV-1), a member of the genus Chlorovirus (family Phycodnaviridae). This family of algae-infecting viruses is common in aqueous environments but not previously thought to infect humans or animals or to inhabit human mucosal surfaces (13). Viruses that cross kingdoms are rare; however, some plant viruses can replicate in both their plant host as well as an invertebrate vector. However, there is one report indicating a possible algal-infecting virus associated with humans. In this report, cervico-vaginal secretion samples contained virus-like particles, and these samples inhibited the propagation of certain algal cultures, consistent with the presence of a virus capable of infecting algae (14).
The surprising discovery of this apparent human–ATCV-1 association led us to conduct further investigations in humans and in mice by using high throughput sequencing methods, PCR procedures, and immunological analyses. The presence of ATCV-1 genomes in the oropharynx of many individuals was evaluated in a Baltimore, Maryland-based cohort, and correlations were discovered between the presence of ATCV-1 and cognitive performance. These results prompted us to explore the ability of ATCV-1 to infect mice under experimental conditions and to study the effect of ATCV-1 infection on cognitive performance and brain gene expression.
Results
Detection of ATCV-1 DNA in Human Pharyngeal Samples.
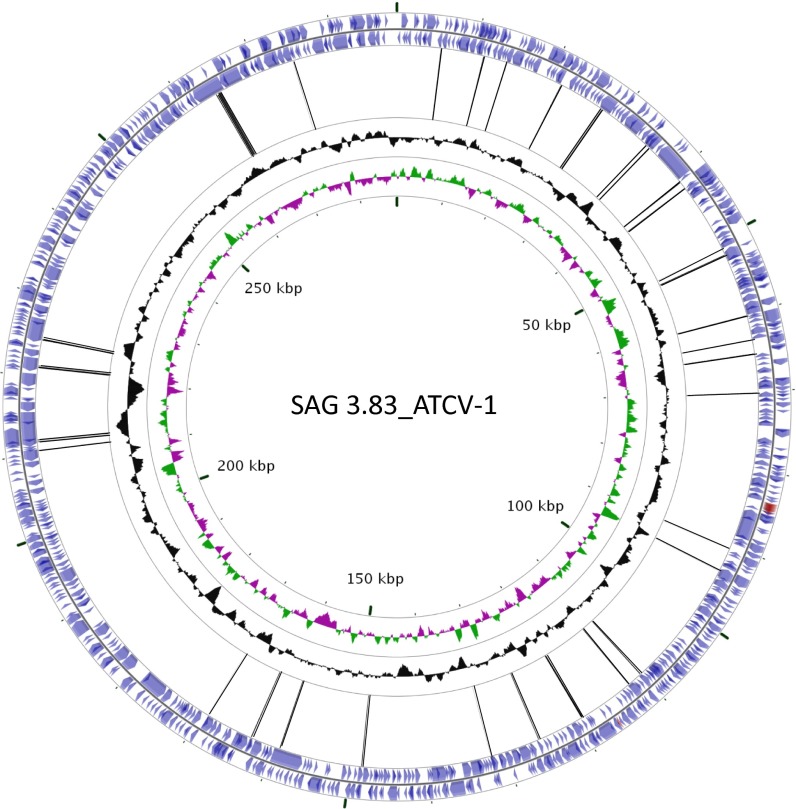
Metagenomic sequencing was performed on DNA extracted from oropharyngeal samples obtained from 33 adult individuals without a known psychiatric disorder or physical illness. The demographic characteristics of these individuals are reported in Table S1A. The viral fraction of these metagenomic analyses revealed a wide range of viruses consistent with human and bacterial components of the oropharyngeal cavity. However, there were an unexpected significant number of sequences that resembled chlorovirus ATCV-1. Individuals with and without detectable ATCV-1 sequences in their oropharyngeal samples did not differ significantly in terms of the demographic variables of age, sex, race, educational level, level of maternal education, cigarette smoking, basal metabolic index (BMI) score, a history of travel outside of North America, or place of birth. The sequence reads mapped to many ATCV-1 sites located throughout the viral genome (Fig. 1 and Table S2).
Fig. 1.
Chlorovirus ATCV-1 genome showing the gene block distributions [blue arrows, protein coding sequence (CDS); red arrows, tRNAs] on each strand of the genome. Histograms in black indicate the G+C distribution along the genome; colored histograms (green, magenta) indicate the GC skew of the genome. The most inner circle indicates the genome map position with the start position at “12 o’clock.” The viral genome is a linear dsDNA, but is represented here as a circle for convenience of presentation. Control throat swab deep sequencing consensus sequence reads are matched to ATCV-1, and two experiments (17 and 16 individuals per experiment) are represented by the black lines connecting the gene blocks. BLAST hits, 61; Query, ATCV-1; Subject, human throat swab chlorovirus consensus sequence reads (52).
A quantitative PCR (qPCR) procedure with a fluorescent-labeled probe (Taqman) was developed to allow throats of more individuals to be tested for ATCV-1 DNA. The assay relied on primers directed at ATCV-1 gene z100l. The sensitivity of the assay was ∼10 copies of target DNA based on standard curves generated from purified ATCV-1 DNA. The qPCR assay detected ATCV-1 DNA in all 10 individuals with at least two sequence reads homologous to ATCV-1 and in 12 of the 14 individuals who had one sequence read homologous to ATCV-1. The Taqman assay was negative when tested with either human DNA or extracts from buffer solutions. Furthermore, the assay was negative, with DNA extracted from either the ATCV-1 host Chlorella heliozoae or with DNA from two other chloroviruses, PBCV-1 and CVM-1, and their hosts Chlorella variabilis and Micractinium conductrix, respectively.
The Taqman assay was used on oropharyngeal samples from 92 individuals, including the 33 individuals tested above. Overall ATCV-1–like DNA was detected in 40 (43.5%) of the 92 samples. Individuals with or without detectable ATCV-1 DNA were similar in terms of demographic variables (Table S1B). No ATCV-1 DNA was detected in blood samples obtained from the study individuals by the qPCR assay.
Because the individuals in the study cohort were also participating in a study of cognitive functioning (15), we examined the association between detection of ATCV-1 DNA and performance on a battery of cognitive tests. A significant association occurred between the presence of oropharyngeal ATCV-1 DNA and a lower level of performance on the Trail Making Test Part A (Trails A), a test of visual motor speed (P < 0.002), as well as the total score of the Repeatable Battery for the Assessment of Neuropsychological Status (RBANS) (P < 0.014) (Table 1). Within the RBANS test, there were statistically significant differences between those who had detectable oropharyngeal ATCV-1 DNA and those who did not in the domains of delayed memory (P < 0.039) and attention (P < 0.011). These differences were independent of the covariates of age, sex, race, socioeconomic status, educational level, place of birth, and current cigarette smoking. On the other hand, no differences were observed between the presence/absence of ATCV-1 DNA and scores on the Wechsler Adult Intelligence Scale (WAIS) Information subtest, a test of general knowledge.
Table 1.
Association between ATCV-1 oropharyngeal DNA and performance on cognitive tests
| Cognitive Test | ATCV-1 DNA detected, n = 40 | ATCV-1 DNA not detected, n = 52 | Overall cohort, n = 93 | P value |
| Trails A, scaled score | 38.2 (12.4) | 46.7 (11.7) | 43.0 (12.7) | <0.002 |
| WAIS III, Information subtest, scaled score | 10.8 (2.7) | 10.8 (2.6) | 10.8 (2.6) | NS |
| RBANS | ||||
| Total Score | 81.3 (11.9) | 85.4 (11.5) | 83.6 (11.8) | <0.014 |
| Attention Index | 91.4 (17.5) | 98.5 (14.5) | 95.4 (16.2) | <0.011 |
| Delayed Memory Index | 85.2 (11.7) | 88.3 (9.9) | 87.0 (10.8) | <0.039 |
| Immediate Memory Index | 85.8 (15.5) | 89.3 (14.5) | 87.8 (14.9) | NS |
| Visuospatial/Constructional Index | 72.6 (9.1) | 74.4 (10.6) | 73.6 (9.9) | NS |
| Language Index | 93.3 (17.0) | 94.8 (17.0) | 94.2 (16.9) | NS |
Values listed are means (standard deviations). P values calculated by linear regression adjusted for age, sex, race, educational level, maternal education, cigarette smoking, and place of birth. NS indicates P > 0.1.
The odds ratios defining the association between the presence of oropharyngeal ATCV-1 DNA and low performance on the cognitive tests were significantly correlated. As depicted in Fig. 2, the presence of ATCV-1 oropharyngeal DNA was associated with low performance on Trails A with an odds ratio of 5.2 (95% confidence interval, 1.63–16.7; P < 0.005) and low performance on the RBANS Total Score with an odds ratio of 4.3 (95% confidence interval, 1.4–12.8; P < 0.01). Within the RBANS there was a strong association between oropharyngeal ATCV-1 DNA and low performance on the attention domain (odds ratio, 8.0; 95% confidence interval, 1.7–37.6; P < 0.008). These associations were independent of the covariates of age, sex, race, socioeconomic status, educational level, place of birth, and current cigarette smoking. No significant differences occurred with a low level of performance on the other RBANS domains or on the test of information (all P > 0.1).
Fig. 2.
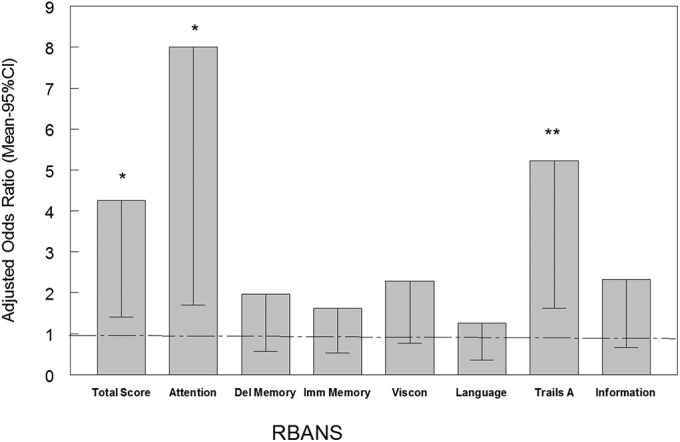
Odds of detecting ATCV-1 in the pharynx by percentile of score on cognitive testing. Bars represent the mean and 95% confidence interval odds of detecting ATCV-1 DNA in the oropharynx in individuals with the indicated test. The odds ratios are adjusted for the demographic variables of age, sex, race, maternal education, educational status, and place of birth in the United States. Trails A and Information are separate tests and not part of the RBANS. **P < 0.005, *P < 0.01, adjusted for the same covariates.
Effect of ATCV-1 on Mouse Behavior and Cognition.
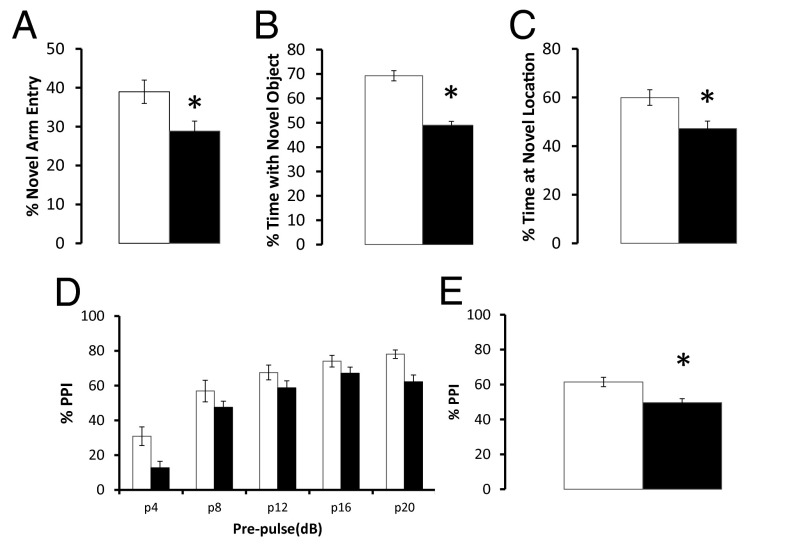
A series of behavior tests were performed on an equal number of male and female mice gavaged with either C. heliozoae alone (control, n = 20) or C. heliozoae infected with ATCV-1 at a multiplicity of infection of 10 per cell for 5 h (C. heliozoae/ATCV-1 exposed, n = 30). The behavior tests were started 6 wk postinoculation. An open field test and dark–light box were used to evaluate the effects of viral exposure on general locomotor activity and anxiety (16, 17). No significant group differences occurred in either test. The effect of ATCV-1 exposure on learning and memory was then tested using the Y-maze test to evaluate spatial recognition memory (17, 18). Mice exposed to ATCV-1 performed at a lower level on this test compared with control mice (Fig. 3A). ANOVA of the percentage of entries to the previously blocked arm showed a significant effect of group, F(1, 49) = 6.4, P = 0.015. This difference could not be explained by lower locomotor activity in ATCV-1–exposed mice, as both groups had similar numbers of total entrances over the observational period: 19.8 ± 1.45 for the control group and 19.5 ± 1.39 for the ATCV-1 group. The effects of ATCV-1 exposure on recognition of a novel object or a novel location were then evaluated. These tests evaluate different aspects of recognition memory in mice (19). No differences were observed in baseline exploratory activity of the objects during training between the ATCV-1–exposed and control group. In contrast, compared with control mice, ATCV-1–exposed mice spent significantly less time exploring the novel object, F(1, 48) = 62.3, P < 0.001 (Fig. 3B), or the novel location of the same object, F(1, 47) = 7.75, P = 0.008 (Fig. 3C). The passive avoidance test was used to evaluate memory to aversive stimuli in mice (20). No significant effect of virus exposure was observed in this test.
Fig. 3.
Behavioral effects of oral ATCV-1 exposure. Mice were orally infected with C. heliozoae alone (open bars) or with ATCV-1–infected C. heliozoae (solid bars) as described in the text. (A) Spatial recognition memory; the y axis displays the percentage of the previously blocked (i.e., novel) arm entries; *P = 0.015 measured by one-way ANOVA. (B) Novel object recognition; the y axis depicts the percentage of time spent exploring the novel object; *P < 0.001 measured by one-way ANOVA. (C) Place recognition memory recognition; the y axis depicts the percentage of time spent exploring the new location of the familiar object; *P < 0.008 measured by one-way ANOVA. (D) Impaired PPI; mice were exposed to presentation of pulse alone (120 dB) and prepulse–pulse combinations across different prepulse intensities; for example, p4 indicates pairing of the prepulse (4 dB above the background noise of 70 dB) with the pulse alone (120 dB) (see the text for more details); the y axis displays the percentage of PPI. (E) Impaired average PPI; the y axis displays the percentage of PPI; *P < 0.015 measured by post hoc test.
The effects of ATCV-1 exposure on the acoustic startle and its prepulse inhibition (PPI) were assessed. These are translational measures of sensorimotor gating that are impaired in patients with neurological and psychiatric illnesses (21). No group difference was found in the baseline acoustic startle response, and both groups exhibited comparable amplitudes (in mV): 193.1 ± 8.4 for control mice and 212.4 ± 8.6 for ATCV-1–exposed mice. However, ATCV-1 exposure impaired PPI in mice. Two-way repeated measures ANOVA with treatment as a between-subject factor and PPI as a within-subjects factor indicated a significant group effect, F(1, 45) = 6.9, P = 0.015, and a significant effect of type of prepulse, F(4, 234) = 98.2, P < 0.001 (Fig. 3D). No Group × Prepulse interaction was found. Post hoc test results revealed that compared with control mice the ATCV-exposed group had significantly reduced PPI averaged across all prepulses (P < 0.05) (Fig. 3E).
Hippocampus Gene Expression in ATCV-1–Exposed Mice.
Gene expression in the hippocampus of mice was evaluated to compare ATCV-1 exposure (26 wk after ATCV-1 exposure) and control mice. The hippocampus was selected because it contains pathways essential for learning, memory, and behavior (22, 23). Exposure to ATCV-1 was associated with a significant up-regulation or down-regulation of 1,285 individual genes (Fig. S1), which could be constructed into 34 functional pathways that met the inclusion criteria (Table S3). Pathways with multiple components relevant to the response of the mice to ATCV-1 exposure and the development of cognitive changes following infection include pathways related to dopamine receptor signaling (Fig. S2), cyclin-dependent kinase 5 (CDK5) signaling (Fig. S3), antigen presentation (Fig. S4), immune cell adhesion (Figs. S5 and S6), and eukaryotic initiation factor 2 (EIF2) (Fig. S7) with color coding legend (Fig. S8).
Antibodies to ATCV-1 in Exposed Mice.
Following the completion of the behavioral studies, there were a total of 47 mice from which serum could be obtained for antibody testing ∼6 mo following oral inoculation. This set included 28 of the 30 mice that had been inoculated with ATCV-1–infected C. heliozoae (15 females, 13 males; 2 males died before testing) and 19 mice inoculated with C. heliozoae alone (10 males, 9 females; 1 female died before testing). We found detectable antibodies to ATCV-1 by enzyme immunoassay (ELISA) in 10 of the 28 mice exposed to ATCV-1 (5 males and 5 females) but in none of the 19 mice exposed to C. heliozoae alone (P < 0.0033, Fisher’s exact test).
The presence of antibodies to ATCV-1 proteins was examined by Western blot in 12 available blood samples from mice exposed to ATCV-1 in this study. Of five tested samples that were positive by ELISA, four also reacted to multiple ATCV-1 proteins but not to C. heliozoae proteins (Fig. S9). One of the predominant proteins recognized was tentatively identified as the ATCV-1 major capsid protein. No reaction to ATCV-1 proteins occurred in Western blots with the seven samples that were seronegative with ELISAs. Sera obtained from mice exposed to C. heliozoae in the absence of ATCV-1 did not react with ATCV-1 proteins.
Discussion
Metagenomic sequencing of DNA extracted from human oropharyngeal samples identified sequences homologous to chlorovirus ATCV-1 in 14 of 33 (42.4%) individuals from a cohort of adults without a known psychiatric disorder or physical illness who were living in an urban area in the United States. DNA sequences found in study individuals mapped to many sites on the ATCV-1 genome and included many viral genes (Fig. 1). The finding of ATCV-1 sequences by metagenomic sequencing was confirmed by a sequence-specific (gene z100l) qPCR assay that detected ATCV-1 sequences in oropharyngeal samples from 40 of 92 (43.5%) individuals from the same study cohort. There were no differences between individuals, with or without ATCV-1 sequences, with respect to demographic variables, such as age, sex, race, socioeconomic status, cigarette smoking, travel history, or place of birth.
As noted in the introduction, ATCV-1 is a member of the genus Chlorovirus, family Phycodnaviridae. Viruses in the phycodnavirus family, together with those in the Poxviridae, Iridoviridae, Ascoviridae, Asfarviridae, Mimiviridae, and Marseilleviridae, are proposed to have a common evolutionary ancestor and are often referred to as nucleocytoplasmic large DNA viruses (24–26). Chloroviruses have large dsDNA genomes (290–370 kb) that encode up to 410 proteins and many tRNAs (13). Chloroviruses infect certain unicellular, eukaryotic, exsymbiotic chlorella-like green algae, called zoochlorellae. Zoochlorellae are associated with the protozoan Paramecium bursaria, the coelenterate Hydra viridis, the heliozoon A. turfacea, and other freshwater and marine invertebrates and protozoans (27, 28). Three such zoochlorellae are Chlorella NC64A (renamed C. variabilis), Chlorella Pbi (renamed M. conductrix), and C. heliozoae, the host for ATCV-1. ATCV-1 is a representative of a group of viruses that infect C. heliozoae and collectively are referred to as SAG viruses.
Genomes from 41 chloroviruses infecting these three hosts have been sequenced, assembled, and annotated (29). Collectively, the 41 viruses encode genes from 632 protein families, whereas any given virus only has 330–410 protein-encoding genes. Thus, the genomic diversity among these viruses is large. Furthermore, the viruses encode some unusual proteins that might influence brain function including potassium ion and aquaglyceroporin channels, potassium and calcium transporters, polyamine metabolic enzymes, a histone methyltransferase, and numerous sugar enzymes including several glycosyltransferases.
Chloroviruses are common in inland waters throughout the world with titers as high as thousands of plaque-forming units (PFUs) per milliliter of indigenous water, although titers are typically 1–100 PFUs/mL. The viruses cannot infect zoochlorellae when they are in their symbiotic phase, and we have no evidence that the zoochlorellae grow free of their hosts in indigenous waters.
To our knowledge, this is the first report of chlorovirus gene sequences in the oral pharynx of humans. There have been several recent reports of oral viromes, but ATCV-1 or other chloroviruses were not mentioned. There are two explanations for this apparent absence. First, in one report the authors specifically looked for known human viral pathogens (30), and so they would have missed the chloroviruses. Second, in three other reports, the samples were filtered through both 0.45-µm and 0.2-µm filters; the material that passed through both filters was used to extract DNA. These researchers were primarily evaluating bacteriophages (7, 8, 31). The icosahedral-shaped chloroviruses are 190 nm in diameter and have a 34-nm spike structure protruding from one unique vertex (32). Therefore, it is likely that ATCV-1 and most chloroviruses would be trapped on a 0.2-µm filter.
The fact that the individuals in our study population were participating in a project, which included measures of cognitive functioning, allowed us to examine the association of detectable ATCV-1 DNA in the pharynx and performance on a range of cognitive tests. Surprisingly, the presence of ATCV-1 DNA in the oropharynx was associated with modest but statistically significant decreases in performance on tests including Trails A and RBANS Attention, both of which measure visual processing and visual motor speed.
The association between the level of ATCV-1 and decreased performance on cognitive tests was independent of demographic variables (Tables S1A and S1B). Also, there was no association between the level of oropharyngeal ATCV-1 DNA and WAIS Information, indicating that the association between ATCV-1 DNA and the other cognitive domains was not due to level of general knowledge or educational background. Studies of associations between environmental factors and cognitive performance in humans must always be interpreted with caution because it is possible that exposure to a single infectious agent might be associated with unmeasured exposures, such as other infectious agents, heavy metals, pollutants, or other environmental factors that also can be associated with alterations in cognitive functioning (33).
For this reason, we developed a mouse model for assessing behavior and cognitive functioning following ATCV-1 exposure by the oral route. As a group, mice inoculated by the oral route showed signs of impairment in new object or new location recognition memory and sensory-motor gating as measured by PPI several months after a single viral exposure (Fig. 3).
Exposure of mice to ATCV-1 also resulted in changes in gene expression within the hippocampus, the region of the brain most associated with spatial memory and navigation (22, 34, 35). Of particular interest in light of the cognitive assays were alterations in the Cdk5 pathway (Fig. S3) because this pathway is central to learning and memory formation (36). Also of note were differences in expression of genes in the dopamine pathway (Fig. S2) because perturbation of the dopamine system impairs novel object recognition and PPI in rodents (37, 38), as do alterations in the pathway of EIF2 because this factor is central to the control of long-term synaptic plasticity and memory storage (39).
Although it is difficult to directly relate the cognitive tests used in our human study with memory tests in mice, it is striking that exposure to ATCV-1 in both humans and mice was associated with decrements in performance on tasks calling for visual spatial abilities (Figs. 2 and 3). More detailed cognitive assessment of humans and mice exposed to ATCV-1 might better define these associations and the relationship between exposure to ATCV-1 and cognitive functioning.
There are several questions relating to ATCV-1 exposure in humans that remain to be addressed. One concerns the source of the acquisition of ATCV-1 in the virome. ATCV-1–like viruses are common in inland waters such as those around Baltimore, so exposure to these water sources would be relatively common. The factors involved in the acquisition of ATCV-1 in the oropharynx following exposure will be the subject of additional investigations. ATCV-1 is the reference genome for the SAG virus group, but we have previously sequenced 12 other SAG viruses and there are 2–3 distinct clades (29). Therefore, the exact nature of SAG viruses capable of colonizing the human oropharynx and other mucosal sites is also an important subject for future studies.
Another set of questions concerns the mechanisms by which ATCV-1 might be associated with alterations in cognitive functioning. Although the exact mechanisms of the behavioral changes in the exposed host remain unclear, the observed cognitive deficits are unlikely to be related to sickness behavior, as no overt signs of malaise were noted in exposed mice. Similar to a number of other microbial infections, we think that both direct and indirect effects of pathogens could play a role (e.g., refs. 40–42). The finding of alterations in several pathways involved in antigen processing and immune cell functioning in the hippocampus of mice exposed to ATCV-1 (Table S3 and Figs. S4–S6) suggests that immune mechanisms may be involved, as have been documented in other biological systems (43). We found evidence of an immune response to ATCV-1 in about 35% of mice exposed to ATCV-1 when measured 6 mo following a single exposure. Thus, our serological and gene expression data implicate immune response to ACTV-1 as a mechanism underlying the cognitive deficits. It is conceivable that immune activation produced secretion of proinflammatory cytokines that affected neuronal functioning, leading to behavioral abnormalities. In this context, both shared and unique profiles of cytokine up-regulation have been shown for various microbial infections (Borna virus vs. Toxoplasma), and it is plausible that differential neurobehavioral outcomes of different microbial infections may be at least in part explained by unique “signatures” of cytokine expression (44).
Earlier blood samples were not obtained from this cohort of mice to avoid affecting the behavioral tests. Further studies of the kinetics of ATCV-1 infection and the immune response to infection are thus warranted. Our studies document that ATCV-1 is part of the human virome and is associated with cognitive changes in humans and experimentally infected animals. An increased understanding of the role of ATCV-1 and related viruses may lead to a new understanding of the role of the oropharyngeal virome in human health and cognition.
Materials and Methods
Studies in Humans.
Study population.
The study cohort consisted of 92 individuals living in the Baltimore, Maryland metropolitan area who did not have current or past psychiatric disorders and who did not have a serious medical illness that would be likely to affect cognitive performance. The overall characterization of the study population including the methods of recruitment and measures used for their characterization was previously described (45). Control individuals were enrolled after they were screened to rule out the presence of current or past psychiatric disorders with the Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition Axis I Disorders–Nonpatient Edition (46). Participants also met the following criteria: proficient in English, no history of i.v. substance abuse, absence of mental retardation, absence of HIV infection, absence of a serious medical disorder that would affect cognitive functioning, and no indication of alcohol or substance use disorder. Demographic data including age, self-reported race, level of highest educational attainment, level of maternal education, and current use of cigarettes were obtained from all participants. All of the participants provided written informed consent following explanation of the study goals and procedures. The study was approved by the Institutional Review Boards of Johns Hopkins University and Sheppard Pratt Hospital.
Clinical samples.
Oropharyngeal samples were obtained by swabbing the back of the throat using sterile cotton swabs (BBL Culture Swabs, Becton Dickinson). On the day of collection, the swabs were transported from the collection site to the processing laboratory at room temperature and then frozen until processed further, as described below.
Cognitive testing.
All of the participants underwent a battery of cognitive tests, as previously described (45). These included the RBANS (47), Trails A (48), and the Information subtest of the WAIS III (49). Details of these tests are described in SI Materials and Methods.
Sample Processing.
DNA was extracted from throat swabs using Qiagen’s Gentra Puregene Buccal Cell Kit. The collection brush heads from the swab ends were excised and incubated at 65 °C overnight in the kit cell lysis solution. The manufacturer’s instructions were followed with some minor changes to the protocol as follows: (i) During the isopropyl alcohol precipitation, 2 µL of 5 mg/mL linear acrylamide (Life Technologies), a chemically synthesized reagent, was substituted for the glycogen carrier to minimize possible contamination by reagents extracted from natural sources; (ii) following this precipitation step, incubation on ice was added for 15 min; (iii) centrifugation following the 70% (vol/vol) ethanol wash was extended to 5 min from 1 min; and (iv) final elution of DNA was reduced from 100 µL to 30 µL.
Metagenomic Sequencing.
DNA samples from 33 individuals (two independent experiments with 17 and 16 individuals) were analyzed by metagenomic sequencing. The demographic information on these individuals is reported in Table S1A. The method used for sequencing is detailed in SI Materials and Methods.
qPCR Analysis.
Oropharyngeal samples from a larger set of individuals (n = 92) were tested by a qPCR system. These individuals included the 33 individuals evaluated by the initial metagenomic sequencing method. The demographic information related to these individuals is presented in Table S1B. The method of sample collection and DNA extraction was identical to that used in the metagenomic sequencing. The qPCR was performed using the 5′–3′ exonuclease activity of Thermus aquaticus polymerase (Taqman) (50). Target primers were directed at the portion of the genome encoding ATCV-1 protein Z100L. This target region was selected because the primers did not have appreciable homology with any other viruses, bacteria, or eukaryotic organisms.
The method used for the Taqman assay was as follows: A 20× assay mix was made using forward primer 5′-GCA ATT CCG ATA GTA ATG GTC A-3′, reverse primer 5′- CTT GTT TGG CCT TTC ACA AA-3′, and probe /56-FAM/AG TAA ACC CAC ACC CTT TGG TAG CCA /36-TAMSp/ containing 18-µM primers and a 4-µM probe. A 20-µL reaction volume was used using 10 µL of 2× Gene Expression Master Mix (Applied Biosystems) and 1 µL of the 20× assay mix with the remaining volume consisting of input DNA and sterile water. The qPCR reaction profile consisted of one cycle of 10 min at 95 °C followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C. Reactions were performed in a Stratagene Mx3005P Thermocycler (Agilent Technology). Results were quantified with a standard curve generated from the testing of 10-fold dilutions of a plasmid created to contain the target. Testing of this standard curve indicated that 10 copies of ATCV-1 DNA could be reliably detected in the qPCR reaction. A sample, which contained ≥10 copies of ATCV-1 genomic DNA, was considered to contain ATCV-1 DNA. DNA extracted from related chloroviruses PBCV-1 and CVM-1 (13, 29) and human DNA did not produce products with this assay.
Statistical Analysis of Virome Studies.
The demographic correlates for the presence of ATCV-1 DNA were calculated by χ2 analysis for categorical variables such as sex, race, cigarette smoking, history of travel outside of North America, and place of birth and by two-way analysis of variance for continuous variables such as age, level of education, BMI score, and maternal education, the latter being used as a marker of socioeconomic status (51). Individual performances on the cognitive tests described above were further compared with the performance of individuals who did or did not have detectable ATCV-1 genomes in their pharynx using linear regression models incorporating the covariates of age, sex, race, educational level, maternal education, cigarette smoking, and place of birth. Logistic regression models were used to calculate the odds ratios, which define the association between the presence of ATCV-1 DNA in the throat with low performance on the cognitive tests as defined above, using the same covariates that were used in the linear regression models. To have adequate statistical power, analyses of cognitive functioning were only performed on the larger cohort (n = 92), on whom ATCV-1 DNA was detected by the qPCR method described above.
Mouse Model of Infection.
Animal model studies were performed to determine the effect of ATCV-1 exposure via the oral route on cognition and other behaviors. The conditions of viral growth and mouse inoculation were determined by a set of preliminary experiments. All protocols were approved by the Animal Care and Use Committee at Johns Hopkins University.
A total of 50 C57BL/6 male and female mice (Charles River Laboratories) were used to evaluate the effect of ATCV-1 exposure on cognition and behavior. Mice were housed five per cage (28.3 cm length × 17.4 cm width × 13 cm height) unless separated due to fighting. Animals had free access to food and water at all times. Mice were inoculated at 9–11 wk of age, as described below.
Exposure to ATCV-1.
C. heliozoae host algae were either uninfected (C. heliozoae control) or infected with ATCV-1 at a multiplicity of infection of 10 PFU per cell for 5 h (C. heliozoae/ATCV-1), pelleted (3,800 × g × 5 min, 4 °C), and then resuspended in 0.4× PBS. Mice were gavaged with 0.2 mL of either C. heliozoae/ATCV-1 (n = 30) or C. heliozoae control preparations containing ∼4 × 107 cells (n = 20), with both groups equally divided between males and females.
The tests used to monitor the behavior of the mice exposed to ATCV-1, as well as the procedures used for RNA extraction, microarray analysis, data normalization and statistical analysis for microarray transcriptomics, pathway analysis, and measurement of antibodies to ATCV-1 and related chloroviruses in mouse blood samples are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Fabrice Casseus, Joshua Crawford, Ross McFarland, and ChunXia Yang for their excellent technical assistance with the mice studies. This work was supported by grants from the Stanley Medical Research Institute, National Institute of Mental Health P50 Silvio O. Conte Center at Johns Hopkins (MH-94268), National Science Foundation–Experimental Program to Stimulate Competitive Research Grant EPS-1004094, and a P20-RR15635 grant from the Centers of Biomedical Research Excellence program of the National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418895111/-/DCSupplemental.
References
- 1.Al-Asmakh M, Anuar F, Zadjali F, Rafter J, Pettersson S. Gut microbial communities modulating brain development and function. Gut Microbes. 2012;3(4):366–373. doi: 10.4161/gmic.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davari S, Talaei SA, Alaei H, Salami M. Probiotics treatment improves diabetes-induced impairment of synaptic activity and cognitive function: Behavioral and electrophysiological proofs for microbiome-gut-brain axis. Neuroscience. 2013;240:287–296. doi: 10.1016/j.neuroscience.2013.02.055. [DOI] [PubMed] [Google Scholar]
- 3.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19(4):636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6(5):915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pride DT, Salzman J, Relman DA. Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses. Environ Microbiol. 2012;14(9):2564–2576. doi: 10.1111/j.1462-2920.2012.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robles-Sikisaka R, et al. Association between living environment and human oral viral ecology. ISME J. 2013;7(9):1710–1724. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wommack KE, et al. VIROME: A standard operating procedure for analysis of viral metagenome sequences. Stand Genomic Sci. 2012;6(3):427–439. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solonenko SA, et al. Sequencing platform and library preparation choices impact viral metagenomes. BMC Genomics. 2013;14:320. doi: 10.1186/1471-2164-14-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wylie KM, Weinstock GM, Storch GA. Virome genomics: A tool for defining the human virome. Curr Opin Microbiol. 2013;16(4):479–484. doi: 10.1016/j.mib.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Etten JL, Dunigan DD. Chloroviruses: Not your everyday plant virus. Trends Plant Sci. 2012;17(1):1–8. doi: 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stepanova OA, Solovyova YV, Solovyov AV. Results of algae viruses search in human clinical material. Ukrainica Bioorganica Acta. 2011;2:53–56. [Google Scholar]
- 15.Dickerson F, et al. Association between cytomegalovirus antibody levels and cognitive functioning in non-elderly adults. PLoS ONE. 2014;9(5):e95510. doi: 10.1371/journal.pone.0095510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourin M, Hascoët M. The mouse light/dark box test. Eur J Pharmacol. 2003;463(1-3):55–65. doi: 10.1016/s0014-2999(03)01274-3. [DOI] [PubMed] [Google Scholar]
- 17.Pletnikov MV, et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13(2):173–186, 115. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- 18.Melnikova T, et al. Cycloxygenase-2 activity promotes cognitive deficits but not increased amyloid burden in a model of Alzheimer’s disease in a sex-dimorphic pattern. Neuroscience. 2006;141(3):1149–1162. doi: 10.1016/j.neuroscience.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Antunes M, Biala G. The novel object recognition memory: Neurobiology, test procedure, and its modifications. Cogn Process. 2012;13(2):93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tayebi Meybodi K, Vakili Zarch A, Zarrindast MR, Djahanguiri B. Effects of ultra-low doses of morphine, naloxone and ethanol on morphine state-dependent memory of passive avoidance in mice. Behav Pharmacol. 2005;16(3):139–145. doi: 10.1097/00008877-200505000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Swerdlow NR, Braff DL, Taaid N, Geyer MA. Assessing the validity of an animal model of deficient sensorimotor gating in schizophrenic patients. Arch Gen Psychiatry. 1994;51(2):139–154. doi: 10.1001/archpsyc.1994.03950020063007. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, King JA, Burgess N, O’Keefe J. How vision and movement combine in the hippocampal place code. Proc Natl Acad Sci USA. 2013;110(1):378–383. doi: 10.1073/pnas.1215834110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsien JZ, et al. On initial brain activity mapping of episodic and semantic memory code in the hippocampus. Neurobiol Learn Mem. 2013;105:200–210. doi: 10.1016/j.nlm.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75(23):11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117(1):156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L. The systematics of Zoochlorella revisited employing an integrative approach. Environ Microbiol. 2011;13(2):350–364. doi: 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- 28.Esteban GF, Fenchel T, Finlay BJ. Mixotrophy in ciliates. Protist. 2010;161(5):621–641. doi: 10.1016/j.protis.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Jeanniard A, et al. Towards defining the chloroviruses: A genomic journey through a genus of large DNA viruses. BMC Genomics. 2013;14:158. doi: 10.1186/1471-2164-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura S, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS ONE. 2009;4(1):e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willner D, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherrier MV, et al. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc Natl Acad Sci USA. 2009;106(27):11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Lewis G. Environmental toxicity and poor cognitive outcomes in children and adults. J Environ Health. 2014;76(6):130–138. [PMC free article] [PubMed] [Google Scholar]
- 34.Cabral HO, et al. Oscillatory dynamics and place field maps reflect hippocampal ensemble processing of sequence and place memory under NMDA receptor control. Neuron. 2014;81(2):402–415. doi: 10.1016/j.neuron.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Lyon L, Saksida LM, Bussey TJ. Spontaneous object recognition and its relevance to schizophrenia: A review of findings from pharmacological, genetic, lesion and developmental rodent models. Psychopharmacology (Berl) 2012;220(4):647–672. doi: 10.1007/s00213-011-2536-5. [DOI] [PubMed] [Google Scholar]
- 36.Shah K, Lahiri DK. Cdk5 activity in the brain—Multiple paths of regulation. J Cell Sci. 2014;127(Pt 11):2391–2400. doi: 10.1242/jcs.147553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Buuse M. Modeling the positive symptoms of schizophrenia in genetically modified mice: Pharmacology and methodology aspects. Schizophr Bull. 2010;36(2):246–270. doi: 10.1093/schbul/sbp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valsamis B, Schmid S. Habituation and prepulse inhibition of acoustic startle in rodents. J Vis Exp. 2011;55(55):e3446. doi: 10.3791/3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa-Mattioli M, Sonenberg N. Translational control of long-term synaptic plasticity and memory storage by eIF2alpha. Crit Rev Neurobiol. 2006;18(1-2):187–195. doi: 10.1615/critrevneurobiol.v18.i1-2.190. [DOI] [PubMed] [Google Scholar]
- 40.Yolken RH, Torrey EF. Viruses, schizophrenia, and bipolar disorder. Clin Microbiol Rev. 1995;8(1):131–145. doi: 10.1128/cmr.8.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pletnikov MV, Moran TH, Carbone KM. Borna disease virus infection of the neonatal rat: Developmental brain injury model of autism spectrum disorders. Front Biosci. 2002;7:d593–d607. doi: 10.2741/A797. [DOI] [PubMed] [Google Scholar]
- 42.Kannan G, Pletnikov MV. Toxoplasma gondii and cognitive deficits in schizophrenia: An animal model perspective. Schizophr Bull. 2012;38(6):1155–1161. doi: 10.1093/schbul/sbs079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohman RA, Rhodes JS. Neurogenesis, inflammation and behavior. Brain Behav Immun. 2013;27(1):22–32. doi: 10.1016/j.bbi.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart AM, et al. 2014. Cytokine and endocrine parameters in mouse chronic social defeat: Implications for translational ‘cross-domain’ modeling of stress-related brain disorders. Behav Brain Res, pii: S0166-4328(14)00549-X, 10.1016/j.bbr.2014.08.037.
- 45.Dickerson F, et al. Association between cognitive functioning, exposure to Herpes Simplex Virus type 1, and the COMT Val158Met genetic polymorphism in adults without a psychiatric disorder. Brain Behav Immun. 2008;22(7):1103–1107. doi: 10.1016/j.bbi.2008.04.156. [DOI] [PubMed] [Google Scholar]
- 46.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. (SCID-I/P) Biometrics Research, New York State Psychiatric Institute; New York: 2002. [Google Scholar]
- 47.Randolph C. RBANS Manual—Repeatable Battery for the Assessment of Neuropsychological Status. Psychological Corporation; San Antonio, TX: 1998. [Google Scholar]
- 48.Reitan RM. 1979. Manual for Administration of Neuropsychological Test Batteries for Adults and Children (Reitan Neuropsychology Laboratory, Tucson, AZ)
- 49.Wechsler D. Wechsler Adult Intelligence Scale III. Psychological Corporation; San Antonio, TX: 1997. [Google Scholar]
- 50.Bergallo M, et al. Real time PCR TaqMan assays for detection of polyomaviruses KIV and WUV in clinical samples. J Virol Methods. 2009;162(1-2):69–74. doi: 10.1016/j.jviromet.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elo IT. Social class differentials in health and mortality: Patterns and explanations. Comparative perspective. Annu Rev Sociol. 2009;35:553–572. [Google Scholar]
- 52.Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics. 2012;13:202. doi: 10.1186/1471-2164-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.