Significance
Synthetic biology envisages the creation of custom-based signaling by means of modular plug-and-play. This concept has primarily been realized in the construction of synthetic gene circuits. However, all real-time events in biology are processed by protein-based sensing and signal transducing systems; yet, the systematic bottom-up design of protein-based signaling systems remains elusive to date. Here we report a strategy for construction of modular protein switches based on artificially autoinhibited proteases whose activity can be modulated by specific proteolysis, ligand binding, or protein–protein interactions. We demonstrate that such protease-based ligand receptors or signal transducers can be assembled into different types of integrated signal sensing and amplification circuits that, in principle, can be connected to any biological process.
Keywords: synthetic biology, protein engineering, protein switches, proteases
Abstract
The bottom-up design of protein-based signaling networks is a key goal of synthetic biology; yet, it remains elusive due to our inability to tailor-make signal transducers and receptors that can be readily compiled into defined signaling networks. Here, we report a generic approach for the construction of protein-based molecular switches based on artficially autoinhibited proteases. Using structure-guided design and directed protein evolution, we created signal transducers based on artificially autoinhibited proteases that can be activated following site-specific proteolysis and also demonstrate the modular design of an allosterically regulated protease receptor following recombination with an affinity clamp peptide receptor. Notably, the receptor’s mode of action can be varied from >5-fold switch-OFF to >30-fold switch-ON solely by changing the length of the connecting linkers, demonstrating a high functional plasticity not previously observed in naturally occurring receptor systems. We also create an integrated signaling circuit based on two orthogonal autoinhibited protease units that can propagate and amplify molecular queues generated by the protease receptor. Finally, we present a generic two-component receptor architecture based on proximity-based activation of two autoinhibited proteases. Overall, the approach allows the design of protease-based signaling networks that, in principle, can be connected to any biological process.
A key objective in the emerging field of synthetic biology is to develop approaches for the systematic engineering of artificial signal transduction systems (1, 2), which is expected to advance our understanding of fundamental biological processes and create new biotechnological and medical applications (3). Engineering synthetic signaling systems have predominantly been realized with gene expression circuits that relay molecular queues either through transcription factors or functional nucleic acids (4–8). Here, rational engineering strategies are supported by the modular organization and function of transcription factors and regulatory DNA elements (4), as well as the predictability of base pairing interactions (8). However, transcription-based signaling circuits usually require hours to process and actuate a specific molecular queue (9). Further, the limited chemical diversity of nucleic acids compared with amino acids ultimately restricts their functionality. Thus, protein-based signaling networks that operate faster compared with gene-based signaling circuits are highly desirable.
The systematic engineering of protein-based modules that sense, process, and amplify defined molecular queues has provided a formidable challenge to date. At the molecular level, many biological response functions depend on allosterically regulated protein activities that couple an input to an output function solely through conformational changes. Engineering these has proven difficult with only a few successful designs reported to date (10). For instance, domain insertion strategies have been successfully applied to create a maltose-inducible version of β-lactamase (11) and rapamcyin-inducible mutants of Src, p38, and focal adhesion kinase (12, 13). However, domain insertion does not follow generic principles, and the successful designs either rely on extensive empirical optimization (11) or exploit distinct structural features in the input and output function domain (12, 13). Engineering allosteric receptors based on mutually exclusive binding interactions is considered to be more generic: e.g., the natural autoinhibition domains of neuronal Wiskott–Aldrich syndrome protein (N-WASP) could be replaced with artificial ones based on Src homology 3 (SH3) and PDZ peptide–ligand interactions (14, 15). Mutually exclusive binding interactions have also been used to create sensors based on pairs of interacting fluorescent or bioluminescent proteins and probes where ligand–receptor interactions modulate the efficiency of resonance energy transfer (RET) (16, 17). Similar sensors have been developed using reversibly associating split-luciferase halves (18) and β-lactamase fused to its inhibitor β-lactamase inhibitor protein (BLIP) where ligand–receptor interactions in the connecting linker modulate the activity of the reporter enzyme in response to a specific molecular queue (19, 20).
Beyond allosterically regulated proteins, biological signaling events can also be relayed through molecular proximity. In cellular contexts, this was exploited to create new phenotypes by rewiring existing signaling nodes either through artificial protein–protein interactions (PPIs) or recombination of signaling domains (21, 22). Molecular proximity is also exploited by many reporter systems that monitor distinct physiological states inside live cells or in complex biological samples. The most popular systems are based on reconstitution of split-protein fragments and fluorescent or bioluminescent proteins (or chemical probes) that report on bimolecular interactions (16). Due to the unfavorable biophysical properties of split-protein fragments, such systems are usually limited to applications in vivo or in cell-free systems. Conversely, the best reported fluorescent and bioluminescent RET-based probes have limited sensitivities with sensors detecting analytes in the mid-nanomolar range under optimal in vitro conditions (17).
Thus far, all synthetic signal recognition or relay units represent single stage architectures, which imposes limits on their sensitivity and the potential for subsequent propagation and processing of the signal. Similarly, reporter systems only incorporate limited types of readouts that, to date, have largely been constrained by the availability of functional modulators (i.e., notably inhibitors) for the reporter enzymes. To overcome these limitations, we developed a generic approach for the engineering protease-based signal transducers and receptors based on artificially engineered autoinhibited proteases. We demonstrate that protease-based signal transducers and receptors can be assembled into signal sensing and amplification circuits. Importantly, we derive and discuss key theoretical and empirical design guidelines that should greatly facilitate the engineering of synthetic protease-based signaling systems for molecular diagnostics or cellular engineering.
Results
Rationale of Synthetic Protease-Based Signaling.
To create a generic approach for the bottom-up construction of protein-based signaling systems to custom specification, we chose proteases as elementary signal transducing units. The reason for this choice is threefold. First, proteases comprise a large family of structurally related enzymes with specific and frequently nonoverlapping substrate specificities, which provides a rich repertoire of potentially orthogonal signal transducers. By virtue of their catalytic activity, proteases are also able to amplify a biomolecular response that is not dependent on cofactors or external sources of energy, and thus significantly reduces the complexity of protease-based signaling systems. In addition, peptide bond cleavage causes drastic changes in the physical properties of a protein that can be detected rapidly in multiple ways. Peptide bond cleavage is also a very common modulator of protein function that facilitates the development of protease-responsive effectors.
Although protease-based signaling systems are common in nature (e.g., caspases in apoptosis and clotting factors in the blood coagulation cascade), elements of these cascades cannot be readily adopted for the construction of synthetic signaling systems. First and foremost, the substrate specificities of naturally occurring transducer proteases are deeply intertwined with the regulation and actuation of their endogenous signaling networks. At the same time, changing their substrate specificity to orthogonalize them provides a formidable protein engineering challenge. The same applies to the complex structural transitions that underlie the allosteric activation of transducer proteases. Therefore, we devised a strategy for the de novo creation of artificially autoinhibited proteases. In this approach, a protease of choice is extended with a competitive autoinhibitor (AI) that blocks its active site in the basal state (Fig. 1A). Depending on the functional elements in the connecting linker, the activity of the autoinhibited protease can be allosterically regulated by modulating access of the competitive inhibitor to the active site by, for instance, (i) incorporating into the linker cleavage sites for an activating protease, thus creating a protease-inducible transducer; or (ii) replacing a linker with a ligand-binding domain capable of a conformational transition, thus creating a reversibly regulated, ligand-responsive receptor protease.
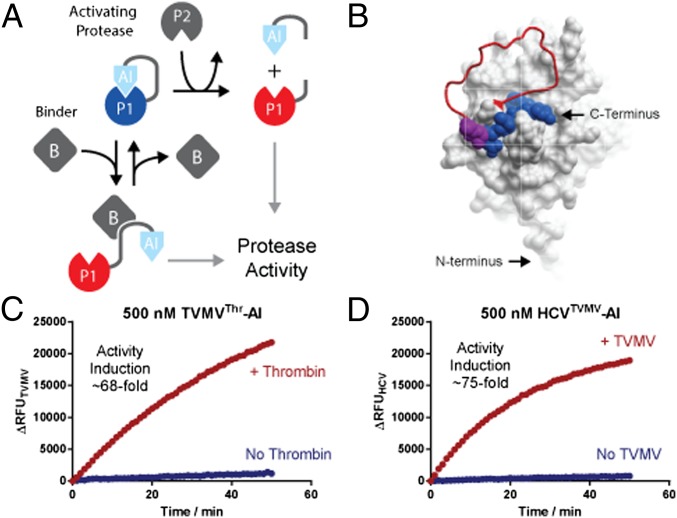
Fig. 1.
Principle design of protease-based signal transducers and ligand sensors. (A) An elementary signal transducer is created by connecting a protease domain (P1) to an autoinhibitory (AI) domain that binds and blocks the active site of the protease. Proteases are activated by dislodging the bound inhibitor from the active site through a conformational rearrangement of the linker. In the simplest case, such a rearrangement constitutes cleavage by an activating protease (P2), which separates the AI domain from the transducer protease and thus relieves autoinhibition to activate the transducer protease. Alternatively, a conformational rearrangement can be mediated through a ligand (B) binding event stabilizing the AI-domain away from the active site. (B) Surface representation of TVMV protease (Protein Data Bank ID code 3MMG) carrying the C-terminal extension (shown as red worm) containing a cleavage site for thrombin and a product peptide bound to the active site (shown in blue space filing representation). The position P5 and P6 of the substrate peptide that was varied in the library are shown as space filling magenta alanines. (C and D) Time-resolved traces of protease activities of TVMV- and HCV-based signal transducers in the presence or absence of thrombin and TVMV, respectively. Protease activities were assayed using 500 nM protease and 5 µM of quenched fluorescent substrate peptide.
Developing Protease-Based Signal Transducers.
To create a protease-based elementary signal transducing unit, we set out to develop a protease-inducible autoinhibited version of the NIa protease from tobacco vein mottling virus (TVMV). Members of the NIa protease family have naturally evolved to operate in the complex environment of the eukaryotic cytosol and display stringent substrate specificities toward their native seven-amino-acid-long cleavage sites in the viral polyprotein (23). This stringent substrate specificity fulfills the requirement for a high degree of orthogonality, which is crucial for making the behavior of synthetic signaling systems predictable in cellular context (4). In addition, NIa proteases including TVMV are well characterized biochemically (24) and structurally (25).
As genetically encodable active site inhibitors of NIa proteases are not available, the AI domains were developed de novo. Briefly, the N-terminal cleavage product of the optimal TVMV peptide substrate ETVRFQ (24) served as a lead sequence for the creation of a competitive active site inhibitor. The lead peptide was appended to the C terminus of TVMV by a connecting linker encoding a cleavage site for thrombin flanked by Gly- and Ser-rich sequences. The linker covered a distance of 18 Å between the N terminus of the active site-bound peptide product and the last structured residue E217 in TVMV (25). The resulting mutant was recombinately produced in Escherichia coli, and its proteolytic activity could be induced approximately fivefold following thrombin cleavage (Figs. S1 and S2A). To improve the performance of the module, an activity-based optimization screen was performed to improve binding of the AI domain to the protease. To this end, additional hydrophobic interactions between P5 and P6 in the AI domain and T214 and V216 in the adjacent β-sheet β13 (25) of TVMV were screened (SI Materials and Methods). The activity of the resulting mutant slightly improved to ∼13-fold (Fig. S2B); yet, kinetic analysis suggested that residual background activity primarily arose from proteases lacking an AI domain as opposed to insufficiently strong competitive inhibition (Fig. S2C). A second screen was therefore devised to identify dipeptide motifs that were capable of binding across the P1-P1′ junction of the TVMV active site without being cleaved and enable the introduction of a C-terminal His6 affinity purification tag to purify full-length proteins (SI Materials and Methods). This screen yielded an autoinhibited mutant of TVMV with an AI domain EYVRFAP whose activity was induced ∼68-fold following thrombin cleavage (Fig. 1C), whereas, on its own, the AI domain inhibited TVMV with a Ki of 196 ± 27 μM, indicating that even a weak inhibitor could function as an effective AI domain (Fig. S2E). This result can be rationalized with a simple mathematical model that guides the design and configuration of autoinhibited protease modules (SI Materials and Methods and Fig. S3). Notably, the induction ratio is linearly proportional to the effective, intramolecular concentration of the inhibitor, which, in practice, depends on the proximity of the N and C termini to the active site, as well as the structure and length of the connecting linkers. At the same time, an AI domain must not bind its transducer protease too tightly to minimize continuing competitive inhibition after cleavage. As a rule of thumb, the Ki of a potential inhibitor should be at least two- to fivefold greater than the operating concentration of the transducer protease to achieve at least 66–83% of the maximum induction ratio (Fig. S3). Both criteria are satisfied in the case of the newly developed TVMV-based signal transducer.
To confirm the general applicability of the approach and to create another autoinhibited protease module that can be used to relay and amplify protease-based signals, we developed a protease-based signal transducer from hepatitis C virus (HCV) NS3 serine protease. Conveniently, noncleavable peptides capable of bridging the P1-P1′ junction of the HCV’s active site have been reported previously (26). Applying the design principles used for developing TVMV-based signal transducers, we engineered a TVMV-inducible autoinhibited signal transducer based on HCV (Fig. 1D). The HCV activity of the resulting transducer could be induced >75-fold following cleavage by TVMV (Fig. 1D). Crucially, TVMV and HCV proteases are naturally orthogonal (Fig. S4) and can be readily assembled into functional signaling cascades without undesirable cross-activation. At the same time, we would like to highlight that reaction conditions were chosen to favor high induction ratios by applying below KM substrate concentrations and comparatively low concentrations of the protease-based signal transducer; these are easily controlled in vitro, but in cells may ultimately require optimization e.g., either by using stronger inhibitors or fine-tuning expression levels to regulate the concentration of the substrate or the protease-based signal transducers.
Protease-Based Ligand Sensors.
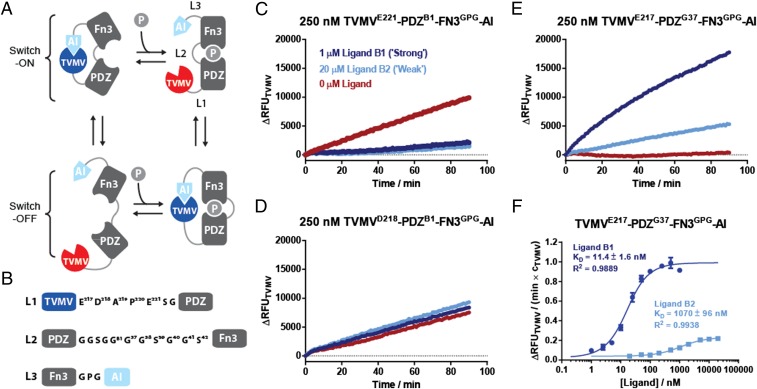
Allosterically regulated receptors are ubiquitous in biology and key to the function of signaling systems. We hence set out to create a ligand activated allosteric protease receptor based on our TVMV signal transducer. To this end, we sought to incorporate a receptor scaffold that undergoes a large conformational transition on ligand binding between the TVMV’s catalytic and AI domains. As a model receptor, we chose an “affinity clamp”—an artificial two-domain receptor composed of a circularly permutated Erbin PDZ domain connected by a flexible serine-glycine linker to an engineered fibronectin type III (FN3) domain (27, 28). Peptide-based ligands RGSIDTWV (B1) and PQPSDTWV (B2) bind the affinity clamp with Kd values of 0.6 and 5 nM, respectively, by inducing association of the PDZ and FN3 domains and structuring of the connecting linker (27, 28). To create a peptide-responsive version of TVMV, we replaced the protease-cleavable linker in the AI-TVMV module with the ePDZ-b1 version of the affinity clamp (27). As shown in Fig. 2A, this arrangement of functional domains allows in principle four major conformational states, two of which would switch TVMV-based protease activity either ON or OFF on ligand binding. We hypothesized that these four conformational states can be selectively accessed by varying the length and the structure of the linkers L1, L2, and L3 that connect the more rigid functional domains (Fig. 2B). To test this idea, we generated a range of linker truncations and elongations: as a starting point, a short GPG motif was chosen for linker L3 (connecting FN3 to the AI domain) while progressively shortening linker L1 (connecting TVMV to PDZ). This truncation screen yielded a series of allosteric protease receptors including mutant TVMVE221-PDZB1-FN3GPG-AI whose activity was suppressed by ligand binding (Fig. 2C). Further shortening linker L1 by three amino acids yielded a protease TVMVD218-PDZB1-FN3GPG-AI that was unresponsive to either ligand B1 or B2 (Fig. 2D). A single amino acid deletion in L1 then yielded a mutant TVMVE217-PDZB1-FN3GPG-AI that responded with 30-fold induction of activity to the addition of ligand B1 (Fig. S5A). Inserting an additional glycine residue in L2 yielded the most effective switch-ON mutant TVMVE217-PDZG37-FN3GPG-AI that responded to the addition of ligand B1 with a 37-fold activity induction (Fig. 2D) while reaching ∼34% activity compared with the TVMV-based signal transducer, suggesting that the affinity clamp remains in a partially autoinhibited state (Fig. S5D). The affinity of the chimeric receptor for B1 and B2 was reduced compared with the parental ePDZ-b1 binder ∼20- and 200-fold, respectively (Table S1; mutant TVMVE217-PDZB1-FN3GPG-AI), suggesting that binding of the AI domain to TVMV directly competes with binding of the FN3 domain to the ligand–PDZ complex (27). At the same time, different linker compositions only had a negligible effect on induction ratios and apparent Kds for switch-ON receptors, giving further support to this assumption (Fig. S5 and Table S1). Here, increasing L2 linker length likely balances steric constraints with the entropic penalty associated with linker structuring for the two competing binding interactions in a similar fashion. Furthermore, increasing L2 has a smaller effect on the apparent Kd in the protease-based receptor compared with in the affinity clamp on its own where a GGSGG insertion has previously been shown to result in a fivefold reduction in binding affinity (28). This reduced reduction in binding affinity is likely because binding of TVMV to the AI domain constrains the affinity clamp in a conformation that is unfavorable for PDZ ligand binding; thus, increasing the length of linker L2 is not associated with the same entropic penalty in the protease-based receptor as to the original affinity clamp.
Fig. 2.
Construction of protease-based peptide receptors. (A) The fusion of an affinity clamp with an autoinhibited TVMV can occupy four major conformational states, which include subequilibria that respond to the addition of a ligand in either switch-ON or switch-OFF fashions. (B) Summary of linker compositions L1, L2, and L3 that connect the different functional elements. (C–E) Time-resolved protease activity measurements of key mutants that respond to the addition of ligand in switch-OFF, neutral, and switch-ON fashions. (F) Apparent Kds for the stronger and weaker binding ligand peptides determined by plotting the gradient of TVMV-based activity against the ligand concentration. Protease activities are normalized over the concentration of the ligand-sensing TVMV protease. Protease activities were assayed with 5 μM TVMV substrate peptide.
An Integrated Signal Sensing and Amplification Circuit.
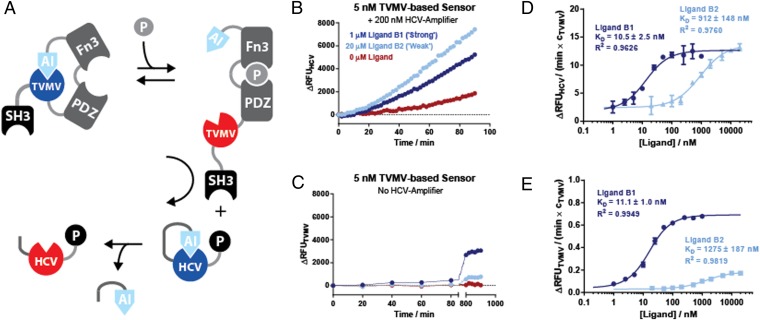
We next sought to connect the TVMV-based allosteric receptor with an HCV-based signal amplifier to form an integrated signal sensing and amplification circuit (Fig. 3A). To ensure efficient signal relay between the TVMV-based signal sensor and the HCV-based signal transducer, the first SH3 domain of the Crk adaptor protein (29) was added to the N terminus of a TVMV-based ligand sensor, whereas its cognate peptide ligand PPPPLPPKRRR (30) was fused to the C terminus of the HCV-based signal transducer. In this way, the sensitivity of the assay could be improved between one and two orders of magnitude as equivalent responses could be recorded in a shorter period using less protein compared with the TVMV-based ligand sensor on its own (Fig. 3C) or the TVMV-based ligand sensor in an untagged, two-stage amplification system (Fig. S6A).
Fig. 3.
(A) Construction of an integrated signal sensing and amplification circuit from an allosterically regulated protease sensor TVMVE217-PDZG38-FN3GPG-AI and an HCV-based signal transducer. Efficient signal transmission between the sensor and the transducer is ensured by protein–peptide interactions between an SH3 domain and its cognate peptide ligand fused to the modules. (B) Activation of HCV protease activity in response to affinity clamp ligand peptides B1 and B2 in an integrated signaling circuit. (C) A control experiment where the protease activity of the TVMV affinity clamp chimera was measured directly in the presence or absence of the two peptide ligands B1 and B2. (D) Determination of apparent Kd values for B1 and B2 using experimental setup shown in B with different ligand concentrations. The rate of HCV activity in the exponential phase (normalized over the concentration of TVMV) was plotted against the concentration of the ligand peptide and fitted to Eq. S2. (E) As in D but directly measuring the activity of the TVMV-based receptor from experiment shown in C. RFU units generated through the cleavage of TVMV- and HCV-specific peptide protease substrates in the two different systems scale by a factor of 1.8. Protease activities in D and E are normalized over the concentration of the ligand sensing TVMV protease. Proteases were assayed with 5 μM quenched fluorescent substrate peptide.
In addition, the relative responsiveness to ligand binding changed in the two-stage system: first, no differences in the induction ratio of the TVMV-based sensor to different strength ligands were evident in the two-stage system as the weaker binding ligand B2 activated the HCV-based signal amplifier slightly faster compared with the stronger binding ligand B1 (Fig. 3B and Fig. S6B). Considering the ligand binding kinetics differ only by an eightfold faster off-rate for ligand B2 (28), it appears that processing of a TVMV cleavage site within an HCV-based signal amplifier (as opposed to a much smaller TVMV-based peptide substrate) imposes additional kinetic constraints. Those may include displacement of the AI domain from the TVMV active site and isomerization of the affinity clamp following dissociation of the FN3-domain or the peptide ligand to enable access of the TVMV-based ligand sensor to the cleavage site in the HCV-based signal amplifier. Furthermore, nonspecific activation was observed even in the absence of ligand in both the SH3-tagged (Fig. 3B) and the untagged two-stage systems (Fig. S6A). As noise propagation is an inherent feature of signal amplification, further optimization and modeling of the system to obtain optimal signal-to-nose ratios will be required.
To test the linearity of the response in the integrated signal sensing and amplification circuit, we titrated it with increasing concentrations of the ligand peptides (Fig. 3D). Plotting the activity of HCV in the exponential phase against the ligand concentration showed a dose-dependent response that could be fit to the value of 11 ± 1.0 nM for B1 and 921 ± 187 nM for B2 (Fig. 3D and Fig. S7 A–D). These affinities are comparable to the apparent Kd determined for the stand-alone sensor (Fig. 3E). To obtain a quantitative measure for signal amplification, we compared the protease signal generated (output) at the different ligand concentrations (input) in the single and two-stage system while normalizing over the concentration of the primary TVMV-based ligand sensor and taking into account 1.8-fold lower quantum yield of the HCV substrate peptide (Fig. S4C). Regarding absolute signal gain, we estimate signal amplification factors of >40- and >150-fold for the B1 and B2 ligands, respectively, over time scales where the signal is proportional to the bound ligand (Fig. 3D, Fig. S7 E and F, and SI Materials and Methods). Conversely, regarding signal-to-noise ratios, signal amplification drops to 0.3 for ligand B1 while remaining constant at 1.3 for ligand B2 (Fig. S7 G and H). Over prolonged time periods, signal-to-noise ratios are further reduced as all of the HCV-based signal transducer is eventually cleaved even in the absence of ligand, leading to continuous generation of an HCV-based protease signal.
Protease-Based Proximity Sensors.
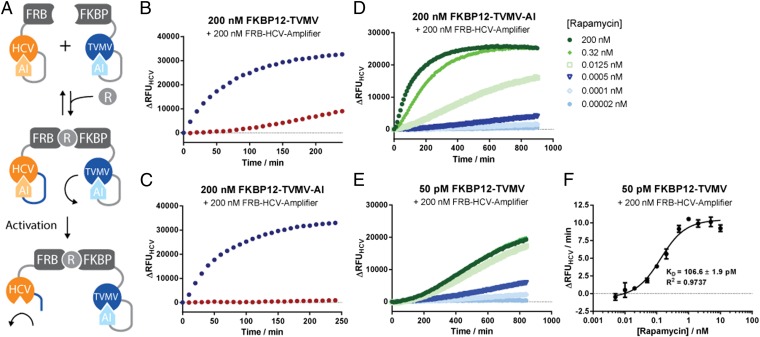
The observed PPI-enhanced coupling of autoinhibited protease modules in a signal amplification cascade (Fig. 3A) prompted us to explore further utilities of this architecture. We conjectured that ligand-induced molecular proximity can drive proteolytic activation of autoinhibited protease modules and thus be exploited to create protease-based receptors. Activation of such receptors would be governed by changes in intermolecular distances rather than a conformational change. To validate this concept, we fused our autoinhibited TVMV- and HCV-based signal transducers to FKBP12 and FRB domains, which selectively associate with each other in the presence of rapamycin (31) (Fig. 4A). We tested a configuration where HCV was connected to AI via a TVMV cleavable linker, whereas TVMV was connected to AI via a thrombin cleavage sequence. The latter enabled us to test the role of TVMV’s AI domain in the activation of the rapamycin receptor. We then monitored the activity of HCV in the absence and presence of rapamycin and in the presence and the absence of thrombin. As can be seen in Fig. 4 B and C, the addition of rapamycin induced HCV activity both in the presence and the absence of thrombin, indicating that molecular proximity was the main driving force in the system’s activation. However, the preactivation with thrombin resulted in higher background activity at the chosen concentrations (Fig. 4 B and C). Titration experiments demonstrated that we could confidently detect concentrations of rapamycin below 0.5 pM using 200 nM of the TVMV- and HCV-based signal transducers each (Fig. 4D). Furthermore, we could faithfully reproduce the Kd for rapamycin binding to FKBP12 with 107 ± 2 pM and Kd = 117 ± 3 pM for the autoinhibited and uninhibited TVMV-based signal transducers, respectively (Fig. 4 E and F and Fig. S7 I and J). Although the detection of rapamycin at subpicomolar concentrations required an extended measurement (Fig. 4E), we expect that connecting the receptor system to further signal amplification modules will significantly reduce the response time. Overall, we successfully created a two-component receptor architecture that does not rely on conformational changes but uses molecular clustering following ligand binding. From an engineering perspective, this greatly facilitates sensor design as it does not involve optimization of intramolecular interactions or linker compositions, potentially allowing the detection of any molecule for which specific binders can be obtained.
Fig. 4.
(A) Principle design of autoinhibited protease-based proximity sensors. (A) TVMV-inducible mutant of HCV was fused to FRB and a thrombin-inducible mutant of TVMV was fused to FKBP12. In the presence of rapamycin, TVMV colocalizes with HCV resulting in rapid cleavage of the latter. (B and C) Time-resolved traces of protease activities of HCV activity of the sensors in the absence (red) or presence (blue) of rapamycin. Note that background activation is significantly reduced in the presence of the AI domain (D and E). Titration of rapamycin to the two-component sensor induces HCV activation at subpicomolar concentrations. (F) Quantification of Kds for rapamycin for uninhibited TVMV at different ligand concentrations. Proteases were assayed with 5 μM quenched fluorescent substrate peptide.
Discussion
We report a generally applicable approach for construction of signal sensors, transducers, and amplifiers based on highly specific proteases. We developed a modular design strategy where transducer proteases and AI domains are connected through linkers containing different types of functional units modulating the activity of the resulting autoinhibited enzyme. This approach enables rapid mix-and-match development of functional modules as evidenced by the sensors developed in this study. Demonstration that AI domains require only weak affinities for the active site and can be easily derived from the protease’s substrate peptides makes construction of additional autoinhibited protease modules straightforward. The same AI domains were used to create an allosterically regulated receptor protease based on an artificial peptide receptor known as an affinity clamp. We demonstrate that, through adjustments of linker length, both ON- and OFF-switches could be generated using this approach. Notably, affinity-clamping is proposed to be a modular and generic architecture for the creation of allosteric protein binders (27) that can be used for the development of allosterically regulated protease receptors. The high degree of orthogonality of the developed modules combined with the simplicity of engineering connectivity also allowed us to create two component receptors and signal relays. The system’s modularity, tunable substrate KMs, and control over the kinetic parameters via local concentrations enabled rapid, systems level optimization.
The signal generated by protease-based signal sensors, transducers, and proximity sensors can be connected to practically any type of output, e.g., activation of natural or artificially engineered zymogens (32, 33), including previously developed protease-inducible reporter systems such as BLIP–β-lactamase fusion proteins (19, 20). Because only modest affinities and specificities of protein or peptide binders are required for the construction of artificial zymogens, their development is likely to be straightforward, and neither depends on specific structural features (32) or on significant structural rearrangements (33). In vivo artificial zymogens or incorporation of appropriate cleavage sites into cellular proteins could enable actuation of such artificial signaling circuits through potentially any protein-mediated activity. Such a molecular queue can be a binding event or a chemical modification mediated by the enzymatic activity of a protein kinase, phosphatase, or other modifying enzyme. Efficient reverse reactions can also be introduced, for example, by exploiting the N-end rule pathway, ultimately enabling more complex signaling motives with negative feedback (34, 35). We anticipate that this approach will find application in the design of orthogonal, tunable, and highly sensitive systems both in vitro and in vivo.
Materials and Methods
General cloning procedures and the amino acid sequences of all TVMV- and HCV-based signal transducers and ligand receptors are listed in SI Materials and Methods. Library design and library screening procedures that were used to engineer an autoinhibited protease module based on TVMV are outlined in SI Materials and Methods. All TVMV- and HCV-based signal transducers and ligand receptors were fused to maltose binding protein, expressed recombinantly in Escherichia coli, and purified by Ni affinity chromatography as described in SI Materials and Methods. The activity of different TVMV- and HCV-based signal transducers and ligand receptors was assayed in vitro by fluorescence spectroscopy using purified proteins and fluorescently quenched peptide substrates according to procedures outlined in SI Materials and Methods. A list of fluorescently quenched peptide substrates used to assay TVMV- and HCV-based protease activities and the peptide ligands for the affinity clamp are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Prof. Matt Trau for stimulating discussions and Marinna Nilsson and Masuda Nabi for excellent technical assistance. This work was supported in part by Australian Research Council (ARC) Discovery Project Grant DP1094080, ARC Future Fellowship Grant FT0991611, National Health and Medical Research Council (NHMRC) Project Grant 569652, and NHMRC Program Grant APP1037320 (to K.A.). This study was funded in part by Movember through Prostate Cancer Foundation of Australia’s Research Program.
Footnotes
Conflict of interest statement: V.S. and K.A. are coinventors on a Patent Cooperation Treaty (PCT) patent application that covers aspects of the protease-based biosensor technology described in this publication.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405220111/-/DCSupplemental.
References
- 1.Wang YH, Wei KY, Smolke CD. Synthetic biology: Advancing the design of diverse genetic systems. Annu Rev Chem Biomol Eng. 2013;4:69–102. doi: 10.1146/annurev-chembioeng-061312-103351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng AA, Lu TK. Synthetic biology: An emerging engineering discipline. Annu Rev Biomed Eng. 2012;14:155–178. doi: 10.1146/annurev-bioeng-071811-150118. [DOI] [PubMed] [Google Scholar]
- 3.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nat Rev Genet. 2012;13(1):21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slusarczyk AL, Lin A, Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet. 2012;13(6):406–420. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 5.Galloway KE, Franco E, Smolke CD. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341(6152):1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proc Natl Acad Sci USA. 2012;109(15):5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Briggs N, McLain JR, Ellington AD. Stacking nonenzymatic circuits for high signal gain. Proc Natl Acad Sci USA. 2013;110(14):5386–5391. doi: 10.1073/pnas.1222807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padirac A, Fujii T, Rondelez Y. Nucleic acids for the rational design of reaction circuits. Curr Opin Biotechnol. 2013;24(4):575–580. doi: 10.1016/j.copbio.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Olson EJ, Tabor JJ. Post-translational tools expand the scope of synthetic biology. Curr Opin Chem Biol. 2012;16(3-4):300–306. doi: 10.1016/j.cbpa.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Golynskiy MV, Koay MS, Vinkenborg JL, Merkx M. Engineering protein switches: Sensors, regulators, and spare parts for biology and biotechnology. ChemBioChem. 2011;12(3):353–361. doi: 10.1002/cbic.201000642. [DOI] [PubMed] [Google Scholar]
- 11.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci USA. 2005;102(32):11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagliyan O, et al. Rational design of a ligand-controlled protein conformational switch. Proc Natl Acad Sci USA. 2013;110(17):6800–6804. doi: 10.1073/pnas.1218319110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28(7):743–747. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301(5641):1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 15.Dueber JE, Mirsky EA, Lim WA. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol. 2007;25(6):660–662. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- 16.Ibraheem A, Campbell RE. Designs and applications of fluorescent protein-based biosensors. Curr Opin Chem Biol. 2010;14(1):30–36. doi: 10.1016/j.cbpa.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 17.Griss R, et al. Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring. Nat Chem Biol. 2014;10(7):598–603. doi: 10.1038/nchembio.1554. [DOI] [PubMed] [Google Scholar]
- 18.Binkowski B, Fan F, Wood K. Engineered luciferases for molecular sensing in living cells. Curr Opin Biotechnol. 2009;20(1):14–18. doi: 10.1016/j.copbio.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Banala S, Aper SJ, Schalk W, Merkx M. Switchable reporter enzymes based on mutually exclusive domain interactions allow antibody detection directly in solution. ACS Chem Biol. 2013;8(10):2127–2132. doi: 10.1021/cb400406x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nirantar SR, Yeo KS, Chee S, Lane DP, Ghadessy FJ. A generic scaffold for conversion of peptide ligands into homogenous biosensors. Biosens Bioelectron. 2013;47:421–428. doi: 10.1016/j.bios.2013.03.049. [DOI] [PubMed] [Google Scholar]
- 21.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: The logic and plasticity of eukaryotic protein circuitry. Curr Opin Struct Biol. 2004;14(6):690–699. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328(5976):368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrington JC, Dougherty WG. A viral cleavage site cassette: Identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc Natl Acad Sci USA. 1988;85(10):3391–3395. doi: 10.1073/pnas.85.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tözsér J, et al. Comparison of the substrate specificity of two potyvirus proteases. FEBS J. 2005;272(2):514–523. doi: 10.1111/j.1742-4658.2004.04493.x. [DOI] [PubMed] [Google Scholar]
- 25.Sun P, Austin BP, Tozser J, Waugh DS. Structural determinants of tobacco vein mottling virus protease substrate specificity. Protein Sci. 2010;19(11):2240–2251. doi: 10.1002/pro.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingallinella P, et al. Optimization of the P’-region of peptide inhibitors of hepatitis C virus NS3/4A protease. Biochemistry. 2000;39(42):12898–12906. doi: 10.1021/bi001590g. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc Natl Acad Sci USA. 2008;105(18):6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Makabe K, Biancalana M, Koide A, Koide S. Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution. J Mol Biol. 2009;392(5):1221–1231. doi: 10.1016/j.jmb.2009.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14(3):242–251. doi: 10.1016/j.ymben.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Posern G, et al. Development of highly selective SH3 binding peptides for Crk and CRKL which disrupt Crk-complexes with DOCK180, SoS and C3G. Oncogene. 1998;16(15):1903–1912. doi: 10.1038/sj.onc.1201714. [DOI] [PubMed] [Google Scholar]
- 31.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127(13):4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 32.Plainkum P, Fuchs SM, Wiyakrutta S, Raines RT. Creation of a zymogen. Nat Struct Biol. 2003;10(2):115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitrea DM, Parsons LS, Loh SN. Engineering an artificial zymogen by alternate frame protein folding. Proc Natl Acad Sci USA. 2010;107(7):2824–2829. doi: 10.1073/pnas.0907668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taxis C, Knop M. TIPI: TEV protease-mediated induction of protein instability. Methods Mol Biol. 2012;832:611–626. doi: 10.1007/978-1-61779-474-2_43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.