Abstract
Background
Mitochondria are key players in the development and progression of heart failure (HF). Mitochondrial (mt) dysfunction leads to diminished energy production and increased cell death contributing to the progression of left ventricular (LV) failure. The fundamental mechanisms that underlie mt dysfunction in HF have not been fully elucidated.
Methods and Results
To characterize mt morphology, biogenesis and genomic integrity in human HF, we investigated LV tissue from non-failing (NF) hearts and end-stage ischemic (ICM) or dilated (DCM) cardiomyopathic hearts. Although mt dysfunction was present in both types of cardiomyopathy, mt were smaller and increased in number in DCM compared to ICM or NF hearts. Mt volume density and mtDNA copy number was increased by ~2-fold (P<0.001) in DCM hearts in comparison to ICM hearts. These changes were accompanied by an increase in the expression of mtDNA-encoded genes in DCM versus no change in ICM. mtDNA repair and antioxidant genes were reduced in failing hearts suggestive of a defective repair and protection system, which may account for the 4.1-fold increase in mtDNA deletion mutations in DCM (P<0.05 vs NF hearts, P<0.05 vs ICM).
Conclusions
In DCM, mt dysfunction is associated with mtDNA damage and deletions, which could be a consequence of mutating stress coupled with a PGC-1α-dependent stimulus for mt biogenesis. However, this maladaptive compensatory response contributes to additional oxidative damage. Thus, our findings support further investigations into novel mechanisms and therapeutic strategies for mt dysfunction in DCM.
Keywords: heart failure, mitochondrial biogenesis, mtDNA mutations, dilated cardiomyopathy, ischemic cardiomyopathy
Introduction
Mitochondria are the major site of energy production in the cell. Thus it is not surprising, that energy dependent tissues such as the heart are particularly sensitive to mt dysfunction. Accumulating evidence suggests that mt dysfunction reflected in the structure, function, and number of mitochondria within cardiac myocytes, leads to diminished energy production, loss of myocyte contractility, and increased cell death during the development of heart failure1, 2. However, despite extensive animal studies, the fundamental mechanisms behind mt dysfunction contributing to the development and progression of LV failure in humans have not been fully elucidated.
Each cardiac myocyte contains numerous mitochondria (50–100) and each mitochondria contains multiple copies of mtDNA (1–10 molecules/mitochondria)3. The abundance of mitochondria per cell is determined by the rate of mt biogenesis and cell division4. It has been reported that limited myocardial mt biogenesis occurs early in response to pathological stimuli but ultimately proliferation of mitochondria does not match the increased metabolic energy demand of the hypertrophied myocytes, which may contribute to the eventual decompensation of the heart5. The abundance of mitochondria per cell is tightly controlled by the activation of specific transcription factors and signaling pathways4, 6. Recent studies have revealed a central role of peroxisome proliferator-activated receptor γ coactivator (PGC)-1 alpha7 and c-Myc8 in mt biogenesis in the heart. Myc activation was shown to trigger mt biogenesis in adult heart by directly activating nuclear genes involved in mt replication and biogenesis, which was protective in response to ischemic stress8. Downregulation of PGC-1α and its target genes have been observed in number of rodent models of heart failure raising the intriguing possibility that impaired mt biogenesis can be a causal mechanism for mt dysfunction in HF9, 10. However, these models may not recapitulate the pathophysiology of human DCM. PGC-1α expression levels in human failing hearts has been more variable. Two studies examining the PGC-1α levels in human HF found a decrease in PGC-1α mRNA or protein levels11, 12. However, more recently it was reported PGC-1α protein levels are increased in failing human hearts. The difference in findings was attributed to the severity of HF as well as lack of age-matched control group used in previous studies13. All of these studies were done in cohorts of failing human hearts from mixed etiologies highlighting the need to further characterize mt biogenesis and related signaling pathways with respect to HF etiology.
Mt injury in failing heart can also be reflected by mtDNA damage, which is tightly associated with the expression of mt transcripts, proteins and mt function. Human mtDNA is a closed-circular, double-strand DNA molecule encompassing 16,569 bp and encoding 13 proteins that are essential for mt biogenesis and respiratory function14. Human mtDNA is more susceptible to oxidative damage and consequently acquires mutations at a higher rate than nuclear DNA15, 16 due to exposure to high levels of free radicals and reactive oxygen species (ROS) generated during respiration, lack of protective histones and limited capacity for repair of mtDNA damage (for review, see17, 18). Thus, increased oxidative stress may contribute to alterations in the abundance of mitochondria as well as the copy number and integrity of mtDNA under pathological conditions. Indeed, increased oxidative stress may play a critical role in regulating the decline in mt abundance and mtDNA copy number in stressed cells16. When the capacity of the antioxidant system is compromised, higher oxidative stress results in an increase in defective mitochondria and mutated mtDNA leading to a cyclic increase in ROS production and further oxidative damage19. Despite strong evidence suggestive of abnormal mt biogenesis and increased mutated mtDNA copy number in aging tissues19, the relative importance of this process in HF, particularly with respect to different HF etiologies, is not known.
We have characterized the changes in mt morphological dynamics, biogenesis, mtDNA content and damage in subjects with HF of differing etiologies. We examined LV tissue from NF and from end-stage ICM and DCM human hearts. Mitochondria were fragmented and increased in number in DCM failing hearts compared to ICM and NF hearts. There was increased mtDNA copy number in DCM accompanied by an increase in the expression of mtDNA-encoded genes. In contrast, these parameters were reduced in failing ICM hearts consistent with published report. The stimulus for abnormal mt biogenesis in end-stage DCM hearts appeared to be PGC-1α dependent. Importantly, mt abnormalities in DCM were associated with a defective mtDNA repair system and increase in mtDNA deletion mutations. Taken together, these observations suggest that mt dysfunction in failing human hearts may be etiology specific. The basis for these differences is uncertain but raise hope that a better understanding of the pathophysiologic mechanisms could lead to novel therapies to prevent HF progression.
Methods
Myocardial Samples from NF and Failing Hearts
The failing heart samples (n=16) were obtained from the left ventricular (LV) anterior wall during heart transplantation or implantation of an LV assist device. The non-failing heart samples (NF) (n=8) were obtained from the LV free wall and procured from National Disease Research Interchange (NDRI) and University of Pennsylvania. NF heart donors had no history of macroscopic or laboratory signs of cardiac diseases. The tissue collection was approved by the UCLA Institutional Review Board #11-001053 and #12-000207.
Histochemistry
Hematoxylin and eosin (H&E) staining was performed according to the rapid H&E technique for cryostat sections20. Cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) activity staining were performed as previously described21.
Mitochondrial Electron Transport Chain Activity Assays
Mitochondrial Complex II activity was determined by standard spectrophotometric enzyme assay as described22. Activity of Complex II was measured by changes in absorbance at 600 nm after reduction of artificial electron acceptor, 2,6-dichlorophenolindophenol (DCPIP). The absorbance at 600 nm was recorded continuously for 5 min. at every 30 sec. intervals. The relative Complex II activity was defined as the rate of change of absorbance per unit time of incubation.
Complex IV activity was measured using a microplate assay kit from Abcam according to the manufacturer’s procedure. Briefly, Complex IV was immunocaptured within the wells and activity was determined calorimetrically following the oxidation of reduced cytochrome c at 550 nm.
Transmission Electron Microscopy
Small pieces from the left myocardium were fixed in 2% glutaraldehyde, 2% paraformaldehyde at room temperature for 2h, followed by 24h at 4°C. Specimens were rinsed in 0.1 M phosphate buffer (PB), pH 7.4, fixed in 1% osmium tetroxide in PB (pH 7.4) at 4°C for 1h, dehydrated in graded ethanol followed by propylene oxide and embedded in Eponate 812 (TedPella Redding, CA). Ultra-thin sections (around 60–70 nm) from longitudinal parts were cut and contrasted with uranyl acetate followed by lead citrate and examined in a JEOL 10000X transmission electron microscope (Tokyo, Japan) at 80 kV.
Morphometric Analysis
Cardiac mt volume densities were determined from electron micrographs as described previously8. Data were expressed as volume density (volume of mitochondria [μm3] per cytoplasmic volume [μm3]).
Protein and RNA analysis
Western blots were performed on protein extracts from LV anterior wall samples, according to established protocols23. Total RNA was extracted from frozen tissue samples using Tri Reagent, Sigma and first strand cDNA were generated using Omniscript Reverse Transcriptase Kit (Qiagen) as per the manufacturer’s instructions. Primer sequences were designed with the OligoPerfect Designer software. Real-time quantitative PCR was conducted using ABI PRISM 7700 Sequence Detection System; (ABI, CA).
Oxyblot procedures
The Oxyblot Oxidized Protein Detection kit was purchased from Chemicon. Protein homogenates from LV anterior wall samples were derivatized with 1,3-dinitrophenylhydrazine (DNPH) for 15 min according to the manufacturer’s instructions and followed by electrophoresis SDS-polyacrylamide gel.
Mitochondrial DNA copy measurement
DNA was collected from LV anterior wall from NF, DCM and ICM hearts. Real-time PCR was performed for cytochrome c oxidase subunit 1 (COX1) and peroxisome proliferative activated receptor gamma coactivator-related (PPRC). Absolute COX1 DNA copies were normalized to a nuclear gene, PPRC1.
mtDNA Mutation Assays
The random mutation capture assays were performed as previously described21. Briefly, total DNA was digested with TaqI for 5 hr, with the addition of 100 units of TaqI every hour. DNA was then probed with primers flanking the TaqI restriction site in order to detect mtDNA genomes that contain a mutation in the TaqI restriction site. A second pair of primers was used to determine the amount of mtDNA genomes that was interrogated. The following primers were used for DNA amplification:
mtDNA control primer forward, ACAGTTTATGTAGCTTACCTCC;
mtDNA control primer reverse, TTGCTGCGTGCTTGATGCTTGT;
mtDNA deletion primer forward, GAACCAACACCTCTTTACAG;
mtDNA deletion primer reverse, CCTGCTAATGCTAGGCTGCC
Next generation sequencing was performed on Illumina GAIIx sequencer. Libraries of mtDNA were constructed according to the manufacturer’s instructions, and paired-end sequence reads of 75 nucleotides were obtained. To identify candidate deletions, each sequence read was trimmed to 70 nucleotides, and the 15 nucleotides at each end were mapped using Bowtie to the human mtDNA genome. Sequence reads with ends mapping to separate regions of the mtDNA genome were retrieved as candidate deletions. With this algorithm, deletions occurring within the central 40 nucleotide region of a sequence read could be identified. The candidate deletions were then manually verified. To minimize the false-positive rate, only deletions identified by four or more independent reads were included in the analysis.
SOD activity assay
To measure the MnSOD activity, mitochondria were isolated from frozen heart tissue using standard differential centrifugation in 10.0 mM Tris-HCl (pH 7.4), 10.0 mM Hepes, 250 mM sucrose, 0.5 mM EGTA and 0.02% protease inhibitor cocktail24. MnSOD activity was measured using an assay kit from Abcam according to the manufacturer’s instruction. The data were presented as the percentage inhibition of the rate of superoxide anion generation from xanthine oxidase.
Statistical Analysis
All data are presented as mean ± SEM. Results were compared by one-way ANOVA using Fisher protected least significant difference tests as post-hoc corrections for multiple testing. Significance was declared at a P value less than 0.05. After statistical analysis, mRNA expression data of genes were grouped together based on functional similarity for presentation purposes.
Results
Mitochondrial Functional Integrity and Respiratory Dysfunction in Failing Hearts
The characteristics of the subjects included in the ICM and DCM subgroups recorded at the time of sample collection are shown in Table 1. As a control, mRNA levels of brain natriuretic peptide (BNP) and alpha-myosin heavy chain (α-MHC) were determined by real-time PCR in all samples (Suppl. Fig. 1). As expected, upregulation of BNP and downregulation of α-MHC were observed in failing hearts confirming integrity of samples (Suppl. Fig. 1A & B).
Table 1.
Preimplantation clinical characteristics of the patients.
| NF | HEART FAILURE | ||
|---|---|---|---|
| ICM | DCM | ||
| NO. | 8 | 8 | 8 |
| Age in years, median (range) | 55 (50–80) | 53(40–60) | 57(40–75) |
| Gender, male/female | 4/4 | 6/2 | 6/2 |
| Ejection Fraction, (%) | 60±5* | 20±5 | 20±5 |
| Cardiac Index (L/min/m2) | No Data | 2.5±0.4 | 2.2±0.2 |
| PWP, mm Hg | No Data | 20±5 | 27±5 |
| Inotropic support, n | - | 5/8 | 7/8 |
| Diabetic, n | 0/4* | 5/8 | 1/8 |
| Renal disease, n | 0/4* | 1/8 | 3/8 |
| Hypertension, n | 0/4* | 5/8 | 1/8 |
| Smoking history, n | 5/8* | 1/8 | 0/8 |
Clinical data for 4 out of 8 NF hearts.
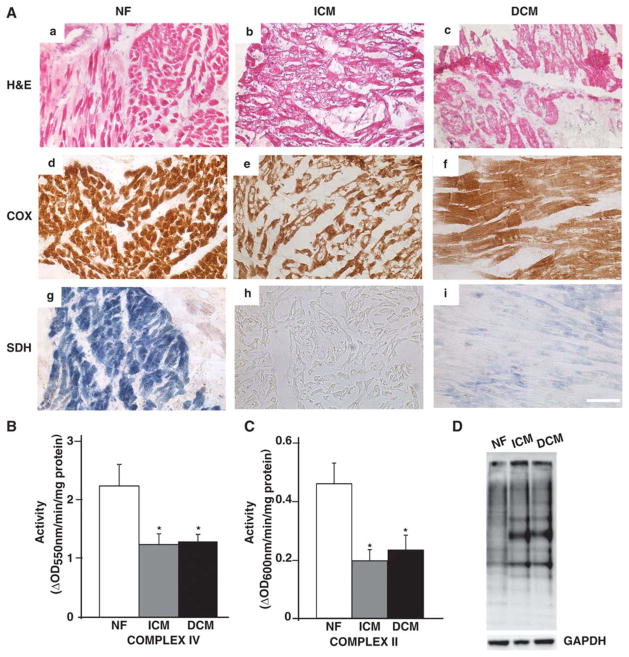
To determine the functional integrity of the mitochondria, end-stage DCM and ICM hearts were examined for activity of enzymes of the electron transport chain, SDH and COX (Fig. 1A). Histological analysis revealed cardiac myocyte degeneration and cytoplasmic vacuolation in both ischemic and dilated cardiomyopathic hearts (Fig. 1A-b & c), which was associated with enlarged cross-sectional area of individual myocytes, particularly in DCM hearts (Fig. 1A-c). Histochemical staining for COX and SDH activities was performed to examine qualitative differences in these electron transport activities in ICM and DCM. This demonstrated decreased staining of COX (Fig. 1A e&f) and SDH in both ICM and DCM hearts (Fig. 1A-h & i) in comparison to NF hearts (Fig. 1A-d & g). To quantitate the observed decrease in electron transport chain activities, spectrophotometric enzyme assays was performed for complex IV and II on mt rich fractions isolated from NF, end-stage DCM and ICM hearts. As shown in Fig. 1 B&C, both ICM and DCM failing hearts demonstrated decrease in complex IV (Fig. 1B; P<0.05 vs NF hearts) and complex II (Fig. 1C; P<0.05 vs NF hearts) activities by almost 50% consistent with severe dysfunction of mt respiratory chain during HF in humans. The mt respiratory dysfunction in failing hearts prompted us to investigate whether increased levels of oxidation products could be detected in these failing human hearts. Analysis of protein oxidative stress was done by derivatizing carbonyl groups on oxidized proteins from NF and failing hearts with DNPH on oxyblot using immunoblotting. Representative example of oxyblot shown in Fig. 1D revealed increase in oxidized proteins in failing hearts.
Figure 1. Mitochondrial functional integrity and respiratory dysfunction in failing hearts.
(A) H&E (a–c), COX (d–f) and SDH (g–i) stained frozen sections from NF, ICM and DCM left ventricular myocardium (n=3/group) showing cardiac myocyte degeneration and reduced functional state of electron transport chain complexes IV and II. Scale bar represents 50μm. Enzymatic activity of electron transport chain complexes (B) II and (C) IV on mt rich fractions obtained from NF, ICM and DCM hearts (*P<0.05 vs NF; n=8/group). (D) Representative micrograph of measurement of oxidized proteins in the failing hearts using oxyblot.
Mitochondrial Morphology and Biogenesis in Failing Hearts
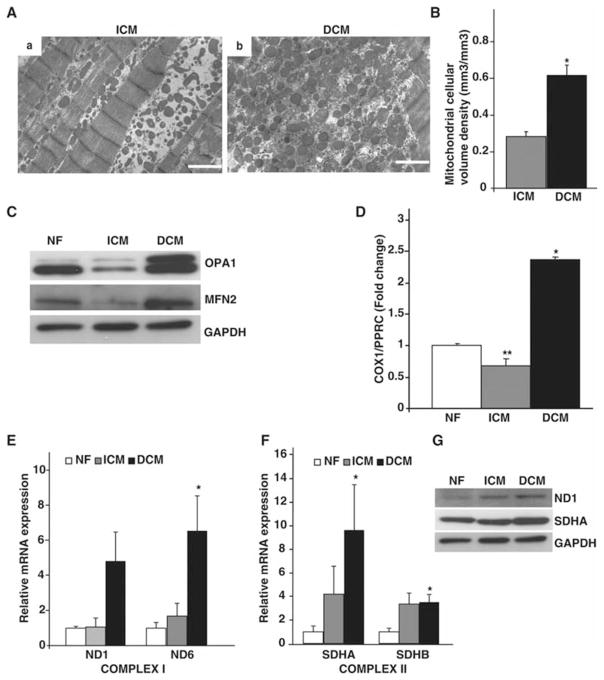
To determine whether mt morphology was also altered in end-stage human HF, electron microscopy was performed on myocardial sections from ICM (Fig. 2A-a) and DCM (Fig. 2A-b) hearts. Mitochondria were more numerous but with abnormal morphology in DCM. The marked mt proliferation was accompanied by myofibrillar displacement and loss in DCM sections. Quantitative morphometric analysis confirmed a 2.2-fold increase in mt volume density in DCM hearts (Fig. 2B; P<0.001 vs ICM hearts). In contrast, electron micrographs of ICM myocardium depicted fewer mitochondria with fragmented morphology (Fig. 2A-a). To determine if the observed morphological changes in failing hearts was caused by an imbalance in mt dynamics, we analyzed total ventricular lysates from NF, ICM and DCM failing hearts for mt fusion and fission proteins. As shown in Fig. 2C, mt fusion proteins such as MFN2, and OPA1 were increased in DCM hearts but decreased in ICM hearts. These observations suggest an imbalance in mt fusion and fission in end-stage HF.
Figure 2. Mitochondrial morphology and biogenesis in failing hearts.
(A) Transmission electron microscopy performed on heart sections from (a) ICM and (b) DCM myocardium demonstrating abnormal mt biogenesis in failing hearts. Scale bar represents 1μm. (B) Quantitative morphometric measurement of mt cellular volume density (μm3/μm3) based on analysis of electron micrographs from ICM and DCM ventricles (*P<0.001 vs NF; n=4/group). (C) Total ventricular protein lysates from indicated phenotypes were probed with antibodies against mt fusion proteins (OPA1 and MFN2) along with GAPDH as a loading control (n=4/group). (D) Quantitative real-time PCR on the mt gene COX1, along with the nuclear gene PPRC as an internal control from NF, ICM and DCM hearts showing increase in mtDNA content in DCM hearts. (*P<0.001, **P<0.05 vs NF; n=8/group). (E) Quantification of mt encoded Complex I ND1 and ND6 genes on NF, ICM and DCM hearts. (n=8/group, *P<0.05 vs NF). (F) Quantification of nuclear encoded Complex II SDHA and SDHB genes on NF, ICM and DCM hearts. (*P<0.05 vs NF). (G) Total ventricular protein lysates from indicated phenotypes probed with antibodies against electron transport Complex I and II proteins (ND1 and SDHA) along with GAPDH as a loading control (n=4/group).
Since mt morphological and functional alterations are often associated with changes in mtDNA18, 25, we examined total mtDNA content. Total DNA (genomic and mtDNA) isolated from NF, ICM and DCM ventricles was assayed for levels of the mtDNA-encoded gene, COX1 and compared to the nuclear-encoded gene PPRC (Fig. 2D). The observed increase in mt number in DCM failing hearts was associated with a 2.41-fold increase in mtDNA (P<0.001 vs NF hearts). In contrast, ICM hearts displayed a 32% reduction in mtDNA content when compared to NF hearts (P<0.05). To rule out the possibility that the observed increased mtDNA was from a relative increase in the non-cardiac myocyte population in the failing hearts, expression levels of fibroblast marker DDR2 (Discoidin domain receptor 2) was determined from total ventricular RNA from NF, ICM and DCM hearts. There was no significant difference in expression of DDR2 in failing hearts when compared to NF hearts (Suppl. Fig. 2).
To investigate the basis for the mt dysfunction (Fig. 1) we assessed expression levels of a panel of genes encoded by mtDNA (Complex I genes ND1 and ND6) and nuclear DNA (nDNA) encoded genes (Complex II genes SDHA and SDHB) in failing hearts. As shown in Fig. 2E, DCM hearts demonstrated increased expression of both mtDNA encoded subunits ND1 and ND6 (Fig. 2E; 4.8 and 6.5-fold respectively; P<0.05 vs NF hearts), whereas ICM hearts had no significant change in expression of both mtDNA encoded genes (Fig. 2E). Interestingly, expression levels of nDNA encoded genes SDHA and SDHB were upregulated in both HF models compared to NF hearts (Fig. 2F). These changes in gene expression were associated with a corresponding increase in expression of mt encoded Complex I protein, ND1 and nuclear encoded Complex II protein, SDHA in DCM hearts (Fig 2G). Taken together, these results demonstrate that mtDNA content and mtDNA-encoded genes are upregulated in DCM hearts and conversely, not significantly changed in ICM failing hearts.
Impaired Mitochondrial Biogenesis in Heart Failure
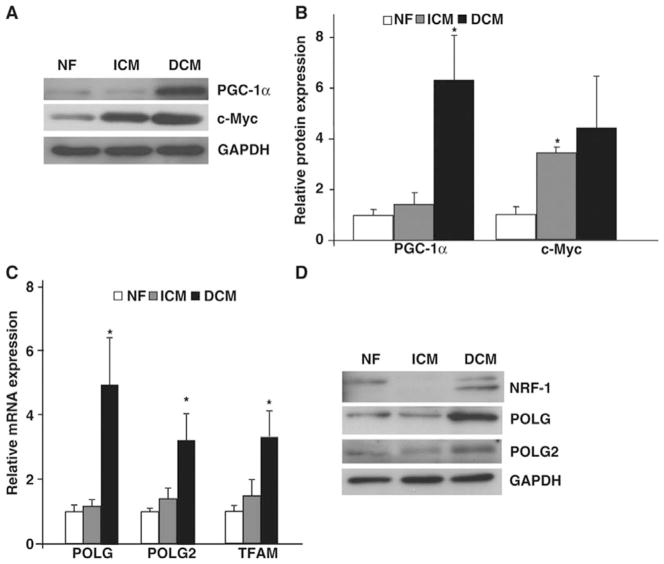
To determine the mechanisms underlying abnormal mt biogenesis in failing hearts, we evaluated the expression levels of key transcriptional regulators of mt biogenesis in heart, PGC-1α26 and c-Myc8 in NF, ICM and DCM hearts (Fig. 3 A & B). As shown in Fig. 3A, DCM hearts showed a 6-fold increase in protein levels (Fig. 3B; P<0.05 vs NF hearts) whereas protein levels of PGC-1α did not change significantly in ICM failing hearts. Protein levels of c-Myc were increased in both ICM and DCM (Fig. 3B), suggesting the important factor mediating the difference in mt biogenesis in the two etiologies was PGC-1α.
Figure 3. Mitochondrial biogenesis in failing hearts.
Total ventricular protein from NF, ICM and DCM ventricles assayed by (A) immunoblot and (B) quantified for mt biogenesis regulators PGC-1α and c-Myc. (*P<0.05 vs NF; (n=4/group). (C) Real-time PCR from NF and failing ventricles for mt biogenesis genes POLG, POLG2 and TFAM. (*P<0.05, **P<0.05 vs NF; n=8/group). (D) Total ventricular lysates from NF, ICM and DCM probed with antibodies against NRF-1, POLG and POLG2 along with GAPDH as a loading control (n=4/group).
Since increased mt biogenesis in DCM failing hearts was associated with elevated levels of PGC-1α, we RNA for analyzed total ventricular downstream targets of PGC-1α such as nuclear respiratory factor 1 (NRF-1), mtDNA polymerase catalytic subunits POLG and POLG2 and mt transcription factor A (TFAM). As shown in Fig. 3C, there was a 4.9-fold increase in POLG (P<0.05), 3.25-fold (P<0.05) in POLG2 and 3.34-fold increase in TFAM (P<0.05) compared to NF hearts, while ICM hearts demonstrated no significant changes in the expression of these genes. These changes in expression of mt biogenesis regulatory genes were associated with a corresponding increase in protein expression of NRF-1, POLG and POLG2 in DCM hearts with no significant changes in ICM hearts when compared to NF hearts (Fig. 3D). Thus, our results suggest that there is PGC-1α-associated abnormal mt biogenesis in DCM failing hearts in contrast to no significant change of mtDNA replication and biogenesis in ICM failing hearts.
Mitochondrial DNA Damage in Heart Failure
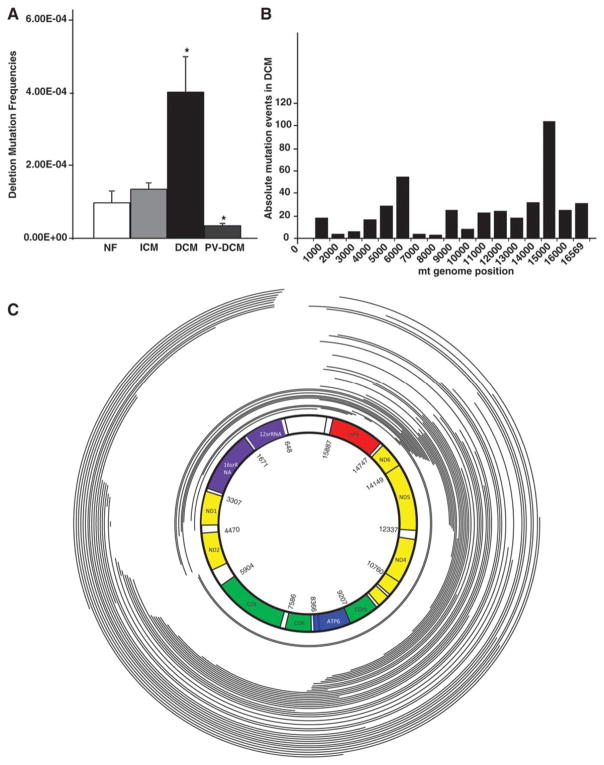
To determine if increase in mtDNA in DCM failing hearts is associated with mtDNA damage, we analysed mtDNA for deletion mutations. The prevalence of mtDNA deletions is inversely correlated with mt function in heart and skeletal muscle21. mtDNA deletion frequencies in the common deletion site in the major arc of the mt genome were determined using the random mutation capture (RMC) assay21. As shown in Fig. 4A, DCM failing hearts showed a 4.1-fold increase in frequency of mtDNA deletions (P<0.05 vs NF hearts). The trend for increased mtDNA deletions in ICM hearts was not significant when compared to NF hearts. To determine if the deletion mutations in DCM failing hearts were acquired during development of heart failure, we analyzed heart samples from DCM subjects who had been supported with left ventricular assist devices (LVAD) for an average of 2 months (Fig. 4A). LVAD support is associated with reverse remodeling and correction of mt defects in end-stage failing hearts (for review see27). mtDNA deletions were significantly reduced in LVAD-supported DCM hearts when compared to DCM failing hearts (Fig. 4A; P<0.05 vs DCM). These data suggest that deletion mutations observed in mtDNA of DCM failing hearts accumulate with the pathophysiologic signals in the failing ventricle.
Figure 4. Mitochondrial DNA deletion mutations in failing and in reverse remodeled hearts.
(A) Deletions were PCR-amplified and quantified using RMC assay on DNA obtained from NF, ICM, DCM and Post-VAD supported hearts demonstrating increased frequency of mtDNA mutations in DCM hearts (*P<0.05, vs normal; n=8/group). (B) Histogram showing frequency of deletion mutations observed across the mt genome in one of the DCM failing heart. (C) Schematic representation of the human mtDNA-deletion mutations obtained from an end-stage DCM failing heart. mtDNA deletion mutations were determined by Solexa sequencing of mtDNA and breakpoints were determined by DNA sequence analysis. Arcs represent the deleted regions of the genome.
To confirm and better characterize the increased mtDNA deletions in an unbiased manner, we performed next generation sequencing of mtDNA from NF and DCM failing hearts. For each sample, we obtained 5×105 to 1.1×106 mtDNA reads spanning 75 base pairs resulting in a 3,500–4,000-fold mt genome coverage. Both ends of each sequence read were mapped onto the mtDNA genome, and sequences with ends that mapped to different locations were identified as deletions. To minimize the chances of artifactual deletions, we only considered deletions that were identified by four or more independent sequence reads (Table 2). Two hundred and thirteen distinct deletions were identified by four or more independent sequence reads in DCM hearts representing deletions in 0.05% of the mt genomes. Although all DCM hearts exhibited mtDNA deletions, we observed deletions in only one NF heart representing a deletion rate of 0.003% in NF hearts (Table 2). Supplementary Fig. 3 depicts positions, sizes, frequencies and affected genes for each deletion for all 6 DCM subjects used in the study. We did not observe a correlation between the age of the subject and the frequency of mtDNA deletions; however the sample size was small and the stimulus for these mutations in failing myocardium may be much greater than that occurring with normal aging (Table 2). The histogram for one representative end-stage DCM failing heart is shown in Fig. 4B. It demonstrates that most of the mtDNA deletions were concentrated in the mt major arc. Fig. 4C is a schematic representation of the spectrum of deletion mutations observed in the same DCM failing heart demonstrating that most of the mtDNA deletion mutations were in the major arc of mt genome. These data suggest that the mt dysfunction is associated with mtDNA damage and deletions specifically in DCM hearts, which could be a consequence of mutating stress coupled with a PGC-1α-dependent stimulus for mt biogenesis.
Table 2.
Deletions of mtDNA found in NF and DCM patients by High-Throughput Sequencing.
| Myocardial sample | Age and sex of the patient | # Distinct deletions | Independent reads | Deletion Frequency (%) |
|---|---|---|---|---|
| NF 1 | 54 M | 90 | 448 | 0.01 |
| NF 2 | 86 M | n.d. | n.d. | n.d. |
| NF 3 | 84 M | n.d. | n.d. | n.d. |
| DCM 24 | 31 M | 73 | 353 | 0.03 |
| DCM 33 | 63 M | 11 | 45 | 0.01 |
| DCM 28 | 74 M | 55 | 246 | 0.02 |
| DCM 15 | 62 M | 61 | 265 | 0.03 |
| DCM 46 | 49 M | 4 | 16 | 0.003 |
| DCM 44 | 51 M | 9 | 132 | 0.2 |
Distinct deletions is column A, independent reads is sum of column E and deletion frequency is percentage of sum of column H over column I in Fig. S3.
Mitochondrial DNA Repair in Failing Hearts
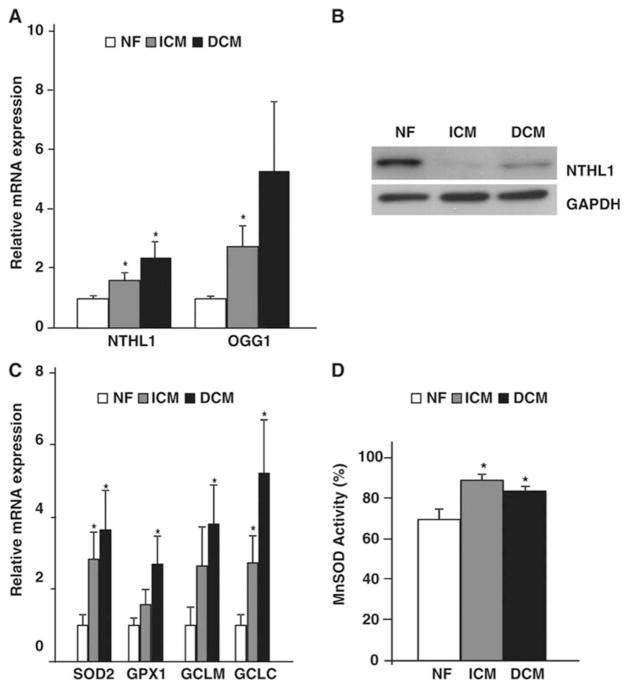
To evaluate the status of antioxidants and DNA repair mechanisms we examined ventricular RNA. Base excision repair (BER) genes such as human endonuclease III (NTHL11) and 8-oxoG DNA glycosylase (OGG1) were upregulated in both HF etiologies. There was an increase in NTHL11 expression by 1.62 and 2.38-fold, and OGG1 by 2.75 and 5.3-fold in ICM and DCM hearts, respectively, (Fig. 5A; P<0.05 vs NF hearts). Expression levels of a panel of antioxidant genes such as manganese superoxide dismutase (SOD2), glutathione peroxidase-1 (GPX1), γ-glutamyl-cysteine synthetase light subunit (GCLM) and γ-glutamyl-cysteine synthetase heavy subunit (GCLC) in failing hearts were also elevated. Significant increases in mRNA levels for SOD2 (2.83 and 3.64-fold), GPX1 (1.5 and 2.69-fold), GCLM (2.65 and 3.8-fold) and GCLC (2.74 and 5.2-fold) were found in ICM and DCM failing hearts (Fig. 5 C; P<0.05 vs NF hearts). A small, but significant increase in activity of MnSOD was also observed in mitochondria isolated from NF, ICM and DCM hearts (Fig. 5D). Interestingly, despite elevations of BER gene expression, protein levels for NTHL1 were reduced in failing hearts (Fig. 5B).
Figure 5. Mitochondrial DNA repair in failing hearts.
Total ventricular RNA (A) and protein (B) from NF, ICM and DCM ventricles assayed by real-time PCR and immunoblot for mt DNA repair genes NTHL1 and OGG1. (*P<0.05 vs NF; n=8/group). (C) Real-time PCR from NF and failing ventricles for mitochondrial anti-oxidant genes SOD2, GPX1, GCLM and GCLC. (*P<0.05 vs NF; n=8/group. (D) Total ventricular protein lysates from NF, ICM and DCM determined for MnSOD activity (n=8/group).
Discussion
Although the underlying injuries and stressors that lead to DCM or ICM are diverse, it has been assumed the pathophysiology, particularly with respect to cardiac mt dysfunction, is similar between these etiologies. This may be related to the fact that the mechanisms responsible for the mt dysfunction in human hearts are poorly understood and the animal models used to elucidate mechanisms may not reflect the true pathophysiology of human DCM. In the current study we performed a detailed morphological and molecular analysis of end-stage failing human hearts to characterize mt morphological integrity, proliferation, mtDNA content and damage in ICM and DCM. Our data demonstrates that DCM is associated with an increase in the number of fragmented mitochondria, mtDNA copy number and expression of mtDNA-encoded genes. In contrast, end-stage ICM hearts show a reduction in the number of mt, with corresponding decrease in mtDNA. This abnormal mt proliferation in DCM hearts appears to be PGC-1α-dependent and interestingly, is associated with an increase in mtDNA deletions. We speculate that this increase in mutated mtDNA in DCM hearts is a consequence of a stimulus for mt biogenesis in the setting of increased oxidative stress and a defective mtDNA repair system. Although certain limitations of the current study such as low sample size with a male predominance and increase in metabolic factors such as diabetes and hypertension in ICM, cannot be disregarded, however, this study was intended to be a pilot study to provide important insight into the mechanisms underlying human HF.
The electron transport chain (ETC) is an essential component of ATP production, and is embedded in the inner mt membrane. Defects in the individual complexes have been documented in HF (for review, see28). Decreased ETC enzyme activity occurs in all HF patients independent of the etiology29. Consistent with this, both DCM and ICM were associated with a decrease in activity of COX and SDH, subunits of Complex IV and II, respectively, indicative of mt respiratory chain dysfunction in HF, which was also corroborated with demonstration of increased oxidation of protein in these failing hearts. The impairment of ETC was accompanied by defects in the mt dynamics in failing hearts. Although mt were more numerous with smaller and fragmented morphology in both DCM and ICM hearts suggesting a predominance of mt fission, in DCM there was concomitant mt biogenesis leading to an increase in total mt volume in contrast to ICM hearts, which displayed a significant reduction in mt volume. Defects in mt organization and the presence of abnormally small and fragmented mitochondria have been previously observed in end-stage DCM, myocardial hibernation and ventricular associated congenital heart disease30. There was also an increase in mt fusion proteins such as OPA1 and MFN2 in DCM hearts; however, both were decreased in ICM hearts. Decreased protein levels of OPA1 have also been previously reported in human ICM, however, with increased MFN2. In contrast, in DCM OPA1 protein levels were unchanged, but MFN2 was increased31. The significance and contribution of abnormal mt dynamics in HF remains to be determined.
Alterations in intracellular levels of ROS are often associated with changes in mt abundance, mtDNA copy number and expression of respiratory genes (for review,16). Our data demonstrated increased mt proliferation in DCM hearts associated with increased mtDNA copy number and expression of mtDNA-encoded genes, while ICM hearts displayed a reduction in mtDNA. Our observations in ICM are similar to those made in mouse models of pathological remodeling after myocardial infarction, where the decline of mt function was associated with a reduction in mtDNA copy number32. Our findings in human DCM hearts parallel those reported in aging human tissues33, 34 and in response to DNA-damaging agents35 where increased mtDNA is associated with oxidative damage. The increase in mt mass and mtDNA in these studies has been proposed to be a compensatory mechanism against oxidative damage to mtDNA and respiratory chain components16. However, it should be noted that mtDNA estimations done in HF patients in the current study were compared with age matched NF hearts, thus implying, that age cannot account for the increase in mtDNA content in DCM hearts. Interestingly, Karamanlidis et al. recently showed a decrease in mtDNA, with an increase in PGC-1α, in failing hearts13. This unexpected finding might be due to use of ventricular samples from mixed etiology in their study. Our data from DCM hearts suggests that mt biogenesis was PGC-1α associated since Myc levels were similar in DCM and ICM and PGC-1α targets, NRF-1, mtDNA transcription factor, TFAM and mtDNA polymerase catalytic subunits POLG and POLG2 required for mtDNA replication were induced in DCM. Thus, under oxidative stress, there is a coordinated nuclear and mt genome activation to induce mt biogenesis presumably to compensate for mt dysfunction. In skeletal muscle, mt proliferation has been shown to partly compensate for the respiratory dysfunction by maintaining overall ATP production36. However, in cardiac muscle, induction of mt biogenesis has been proposed to be a maladaptive response. In fact, cardiac-specific induction of PGC-1α in mice results in cardiac dysfunction with morphologic features of myocyte mt proliferation and myofibrillar disorganization and loss37.
In the current study, we found that mt respiratory dysfunction, abnormal mt proliferation and biogenesis in failing DCM hearts were associated with an increased frequency of mtDNA deletion mutations. Since the frequency of mtDNA deletions was found to be significantly reduced with LVAD-support in DCM failing hearts, this data suggests that the accumulation of mtDNA deletions are acquired during the development of heart failure as opposed to part of a more global mt syndrome. Although, the exact cause of accumulation of mtDNA deletions in DCM hearts is unknown, mtDNA deletion mutations and mt abnormalities have been previously described in idiopathic DCM in humans38, 39. It has been proposed, based on the size of mtDNA deletion mutations, that deletion-containing mt genomes have a replicative advantage40. Support for the replicative advantage of smaller genomes comes from re-population kinetic studies, where it was shown that partially deleted mtDNA molecules re-populate mitochondria faster than full-length genomes41. Although, the deletion mutation frequency observed in DCM failing hearts was quite low to affect the mt function, it is interesting to note that it has been shown that deletion mutations in cardiac myocytes accumulate clonally in aging heart42. mtDNA deletions were shown to accumulate to high fractions in certain cells within the cardiac tissue while most cells remain deletion free, which unfortunately could not be demonstrated in the our study as 4-fold mutation rate observed was in total ground up LV tissues. However, focal accumulation of mtDNA deletions beyond a certain threshold could lead to drop out of cardiac myocytes causing respiratory chain abnormalities and further production of free radicals exacerbating the HF syndrome. Whether the rates of mtDNA deletions we observed in DCM hearts have pathophysiologic consequences or are simply a marker for the enhance mtDNA synthesis remains to be determined. DCM is the most challenging form of heart failure, since it lacks any molecular diagnostic assays or effective therapies. In fact, accumulation of mtDNA deletions could potentially serve as a marker for the enhance mtDNA synthesis, mitochondrial genomic instability or mtDNA damage in DCM failing hearts and are areas of mitochondrial biology that should be looked in detail in future.
In our study, mtDNA damage in both models of HF was not associated with induction of antioxidant enzymes such as SOD2, GPX1 and GCL or BER enzymes, NTHL1 and OGG1, despite a marked up-regulation of these genes at the RNA level. Similar results have been previously reported in human HF43, leading us to speculate that end-stage HF is unable to respond to oxidative stress by adequately increasing the antioxidant response and mtDNA repair mechanisms. The best characterized DNA repair pathway in mitochondria has been BER44. However, accumulation of mtDNA deletions has been linked to decline in BER activities. The accumulation of mtDNA deletions in aging cells is associated with decreased mitochondrial BER glycosylases expression levels45. Additionally, mice deficient for NEIL1, a BER glyocsylase, accumulate mtDNA deletions compared to wild-type mice46, suggesting an important role of mitochondrial BER in prevention of mtDNA damage and deletions.
In summary, our findings suggest while HF of both etiologies is associated with mt dysfunction they exhibit divergent phenotypes with respect to mt biogenesis. DCM is associated with enhanced mt biogenesis and mtDNA deletions. mtDNA deletions can cause mt dysfunction, resulting in an inability of the heart muscle to maintain adequate energy production and continuing to cause oxidative stress. Although we cannot resolve whether the rate of mtDNA deletion we observed contributed to the mt dysfunction, it does suggest a unique pathophysiology for the development of DCM. A better understanding of mechanisms underlying human HF could lead to etiology specific therapies in HF, something that up to now has been lacking.
Supplementary Material
SHORT COMMENTARY.
Although the underlying injuries and stressors that lead to dilated cardiomyopathy (DCM) or ischemic cardiomyopathy (ICM) are diverse, it has been assumed the pathophysiology, particularly with respect to cardiac mitochondrial (mt) dysfunction, is similar between these etiologies. This may be related to the fact that the mechanisms responsible for the mt dysfunction in human hearts are poorly understood and the animal models used to elucidate mechanisms may not reflect the true pathophysiology of human DCM. In the current study we performed a detailed morphological and molecular analysis of end-stage failing human hearts to characterize mt morphological integrity, proliferation, mtDNA content and damage. Our data demonstrates that DCM is associated with an increase in the number of fragmented mitochondria, mtDNA copy number and expression of mtDNA-encoded genes. This abnormal mt proliferation in DCM hearts appears to be PGC-1α-dependent and interestingly, is associated with an increase in mtDNA deletions. We speculate that this increase in mutated mtDNA in DCM hearts is a consequence of a stimulus for mt biogenesis in the setting of increased oxidative stress and a defective mtDNA repair system.
In summary, our findings suggest DCM is associated with more mtDNA deletions than ICM, which can cause mt dysfunction, resulting in an inability of the heart muscle to maintain adequate energy production and continuing to cause oxidative stress. This suggests a unique pathophysiology for the development of DCM and thus a potential novel target for etiology specific therapies in HF, something that up to now has been lacking.
Acknowledgments
We would like to thank Dr. Alan Garfinkel for his expert assistance with statistical analysis. Next generation sequencing was performed at Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech.
Funding Sources: This work was supported by the NIH (HL70748 and HL080111 to W.R.M. and HL070079, HL103205 and HL098954 to Y.W.).
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Capetanaki Y. Desmin cytoskeleton: A potential regulator of muscle mitochondrial behavior and function. Trends Cardiovasc Med. 2002;12:339–348. doi: 10.1016/s1050-1738(02)00184-6. [DOI] [PubMed] [Google Scholar]
- 2.Marin-Garcia J, Goldenthal MJ. Mitochondrial centrality in heart failure. Heart Fail Rev. 2008;13:137–150. doi: 10.1007/s10741-007-9079-1. [DOI] [PubMed] [Google Scholar]
- 3.Marin-Garcia J, Goldenthal MJ. the mitochondrial organelle and the heart. Rev Esp Cardiol. 2002;55:1293–1310. doi: 10.1016/s0300-8932(02)76802-4. [DOI] [PubMed] [Google Scholar]
- 4.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 5.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: A question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyes CD, Hood DA. Origins and consequences of mitochondrial variation in vertebrate muscle. Annu Rev Physiol. 2003;65:177–201. doi: 10.1146/annurev.physiol.65.092101.142705. [DOI] [PubMed] [Google Scholar]
- 7.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the pgc-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, Wang Y, Jin ES, Jeffrey FM, Portman M, Maclellan WR. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking ppar-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnier A, Fortin D, Delomenie C, Momken I, Veksler V, Ventura-Clapier R. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J Physiol. 2003;551:491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebastiani M, Giordano C, Nediani C, Travaglini C, Borchi E, Zani M, Feccia M, Mancini M, Petrozza V, Cossarizza A, Gallo P, Taylor RW, d’Amati G. Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J Am Coll Cardiol. 2007;50:1362–1369. doi: 10.1016/j.jacc.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 12.Garnier A, Zoll J, Fortin D, N’Guessan B, Lefebvre F, Geny B, Mettauer B, Veksler V, Ventura-Clapier R. Control by circulating factors of mitochondrial function and transcription cascade in heart failure: A role for endothelin-1 and angiotensin ii. Circ Heart Fail. 2009;2:342–350. doi: 10.1161/CIRCHEARTFAILURE.108.812099. [DOI] [PubMed] [Google Scholar]
- 13.Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res. 2010;106:1541–1548. doi: 10.1161/CIRCRESAHA.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 15.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Tsutsui H. Oxidative stress in heart failure: The role of mitochondria. Intern Med. 2001;40:1177–1182. doi: 10.2169/internalmedicine.40.1177. [DOI] [PubMed] [Google Scholar]
- 18.Tsutsui H, Kinugawa S, Matsushima S. Mitochondrial oxidative stress and dysfunction in myocardial remodelling. Cardiovasc Res. 2009;81:449–456. doi: 10.1093/cvr/cvn280. [DOI] [PubMed] [Google Scholar]
- 19.Lee HC, Wei YH. Mitochondrial role in life and death of the cell. J Biomed Sci. 2000;7:2–15. doi: 10.1007/BF02255913. [DOI] [PubMed] [Google Scholar]
- 20.Sheehan DC. Theory and practice of histotechnology. 1980:143–144. [Google Scholar]
- 21.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 22.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 23.MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel rb- and p300-binding protein inhibits transactivation by myod. Mol Cell Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rustin P, Chretien D, Bourgeron T, Gerard B, Rotig A, Saudubray JM, Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 25.Rosca MG, Hoppel CL. Mitochondria in heart failure. Cardiovasc Res. 2010;88:40–50. doi: 10.1093/cvr/cvq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators pgc-1alpha and pgc-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambardekar AV, Buttrick PM. Reverse remodeling with left ventricular assist devices: A review of clinical, cellular, and molecular effects. Circ Heart Fail. 2011;4:224–233. doi: 10.1161/CIRCHEARTFAILURE.110.959684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casademont J, Miro O. Electron transport chain defects in heart failure. Heart Fail Rev. 2002;7:131–139. doi: 10.1023/a:1015372407647. [DOI] [PubMed] [Google Scholar]
- 29.Jarreta D, Orus J, Barrientos A, Miro O, Roig E, Heras M, Moraes CT, Cardellach F, Casademont J. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc Res. 2000;45:860–865. doi: 10.1016/s0008-6363(99)00388-0. [DOI] [PubMed] [Google Scholar]
- 30.Hom J, Yu T, Yoon Y, Porter G, Sheu SS. Regulation of mitochondrial fission by intracellular ca2+ in rat ventricular myocytes. Biochim Biophys Acta. 2010;1797:913–921. doi: 10.1016/j.bbabio.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial opa1, apoptosis, and heart failure. Cardiovasc Res. 2009;84:91–99. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 2001;88:529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 33.Barrientos A, Casademont J, Cardellach F, Estivill X, Urbano-Marquez A, Nunes V. Reduced steady-state levels of mitochondrial rna and increased mitochondrial DNA amount in human brain with aging. Brain Res Mol Brain Res. 1997;52:284–289. doi: 10.1016/s0169-328x(97)00278-7. [DOI] [PubMed] [Google Scholar]
- 34.Pesce V, Cormio A, Fracasso F, Vecchiet J, Felzani G, Lezza AM, Cantatore P, Gadaleta MN. Age-related mitochondrial genotypic and phenotypic alterations in human skeletal muscle. Free Radic Biol Med. 2001;30:1223–1233. doi: 10.1016/s0891-5849(01)00517-2. [DOI] [PubMed] [Google Scholar]
- 35.Masayesva BG, Mambo E, Taylor RJ, Goloubeva OG, Zhou S, Cohen Y, Minhas K, Koch W, Sciubba J, Alberg AJ, Sidransky D, Califano J. Mitochondrial DNA content increase in response to cigarette smoking. Cancer Epidemiol Biomarkers Prev. 2006;15:19–24. doi: 10.1158/1055-9965.EPI-05-0210. [DOI] [PubMed] [Google Scholar]
- 36.Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci U S A. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 38.Arbustini E, Diegoli M, Fasani R, Grasso M, Morbini P, Banchieri N, Bellini O, Dal Bello B, Pilotto A, Magrini G, Campana C, Fortina P, Gavazzi A, Narula J, Vigano M. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am J Pathol. 1998;153:1501–1510. doi: 10.1016/S0002-9440(10)65738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marin-Garcia J, Goldenthal MJ, Ananthakrishnan R, Pierpont ME, Fricker FJ, Lipshultz SE, Perez-Atayde A. Specific mitochondrial DNA deletions in idiopathic dilated cardiomyopathy. Cardiovasc Res. 1996;31:306–313. [PubMed] [Google Scholar]
- 40.McKenzie D, Bua E, McKiernan S, Cao Z, Aiken JM. Mitochondrial DNA deletion mutations: A causal role in sarcopenia. Eur J Biochem. 2002;269:2010–2015. doi: 10.1046/j.1432-1033.2002.02867.x. [DOI] [PubMed] [Google Scholar]
- 41.Moraes CT, Kenyon L, Hao H. Mechanisms of human mitochondrial DNA maintenance: The determining role of primary sequence and length over function. Mol Biol Cell. 1999;10:3345–3356. doi: 10.1091/mbc.10.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khrapko K, Bodyak N, Thilly WG, van Orsouw NJ, Zhang X, Coller HA, Perls TT, Upton M, Vijg J, Wei JY. Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions. Nucleic Acids Res. 1999;27:2434–2441. doi: 10.1093/nar/27.11.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sam F, Kerstetter DL, Pimental DR, Mulukutla S, Tabaee A, Bristow MR, Colucci WS, Sawyer DB. Increased reactive oxygen species production and functional alterations in antioxidant enzymes in human failing myocardium. J Card Fail. 2005;11:473–480. doi: 10.1016/j.cardfail.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: The long and short of it. Cellular and molecular life sciences : CMLS. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen B, Zhong Y, Peng W, Sun Y, Hu YJ, Yang Y, Kong WJ. Increased mitochondrial DNA damage and decreased base excision repair in the auditory cortex of d-galactose-induced aging rats. Mol Biol Rep. 2011;38:3635–3642. doi: 10.1007/s11033-010-0476-5. [DOI] [PubMed] [Google Scholar]
- 46.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the neil1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.