Abstract
Background
CD4+ memory T-cells are a major target for infection by HIV-1, whereby latent provirus can establish and endure suppressive antiretroviral therapies. Although HIV-1 subtype C strains (C-HIV) account for the majority of HIV-1 infections worldwide, the susceptibility of CD4+ memory T-cells to infection by CCR5- (R5) and CXCR4-using (X4) C-HIV is unknown. Here, we quantified the susceptibility of naïve and memory CD4+ T-cell subsets, including stem cell memory T-cells (TSCM), to infection by HIV-1 subtype C (C-HIV) strains from treatment-naïve subjects who progressed from chronic to advanced stages of disease whilst either maintaining CCR5-using (R5) viruses (subjects 1503 and 1854), or who experienced emergence of dominant CXCR4-using (X4) strains (subject 1109).
Findings
We show that R5 and X4 C-HIV viruses preferentially target memory and naïve CD4+ T-cell subsets, respectively. While TSCM were susceptible to infection by both R5 and X4 C-HIV viruses, the proportion of infected CD4+ T-cells that were TSCM was higher for R5 strains. Mutagenesis studies of subject 1109 viruses established the V3 region of env as the determinant underlying the preferential targeting of naïve CD4+ T-cells by emergent X4 C-HIV variants in this subject. In contrast, the tropism of R5 C-HIV viruses for CD4+ T-cell subsets was maintained from chronic to advanced stages of disease in subjects 1503 and 1854.
Conclusions
This study provides new insights into the natural history of tropism alterations for CD4+ T-cell subsets by C-HIV strains during progression from chronic to advanced stages of infection. Although not preferentially targeted, our data suggest that TSCM and other memory CD4+ T-cells are likely to be viral reservoirs in subjects with X4 C-HIV infection.
Electronic supplementary material
The online version of this article (doi:10.1186/s12977-014-0097-5) contains supplementary material, which is available to authorized users.
Keywords: HIV-1, Subtype C, T-cell, CD4+, TSCM
Findings
CD4+ T-cells are targets for infection by human immunodeficiency virus type 1 (HIV-1), whereby latent, replication-competent provirus can persist despite suppressive antiretroviral therapies [1]. Following exposure to antigen, naïve CD4+ T-cells (TN) proliferate and differentiate into antigen-specific effector cells or self-renewing memory T-cells [2,3]. Memory CD4+ T-cell subsets include long-lived central memory T-cells (TCM), transitional memory (TTM) and shorter-lived effector memory T-cells (TEM) [4,5]. Recently, a new memory T-cell subset was described in CD4+ and CD8+ T-cell populations in humans [6-8]. These cells, termed stem cell memory T cells (TSCM), account for approximately 2–4% of the CD4+ T-cell population in HIV-seronegative individuals, and are the longest-lived memory CD4+ T-cell subset [8,9]. Importantly, TSCM contribute to the persistent cellular HIV-1 reservoir [6].
HIV-1 entry into cells is mediated by the interaction between the viral envelope protein complex (Env) and cell-surface CD4, and then with a chemokine coreceptor, either CCR5 or CXCR4 (reviewed in [10]). The ability of HIV-1 to engage CCR5, CXCR4 or either coreceptor for entry is a major determinant of cellular tropism, and is used to classify viruses as R5, X4 or R5X4, respectively [11].
Although HIV-1 subtype C (C-HIV) strains account for >50% of HIV-1 infections worldwide and exhibit pathogenicity and tropism alterations that are distinct from other HIV-1 subtypes [12], at present there are few studies describing the susceptibility of CD4+ T-cell subsets to infection by clinical R5 C-HIV strains, and none concerning clinical X4 C-HIV isolates. Given recent reports that have described increased incidence of X4 C-HIV strains across multiple cohorts, particularly from patients with late-stage disease (up to 52%) [12-17], this lack of knowledge has implications for HIV/AIDS cure strategies, including the widely discussed use of histone deacetylase inhibitors to activate transcription of latent provirus and purge CD4+ memory T-cell reservoirs.
Since changes in HIV-1 cellular tropism may influence the size and composition of viral reservoirs, here we sought to quantify and compare the susceptibility of naïve and memory CD4+ T-cell subsets, including TSCM, to infection by clinical X4 and R5 C-HIV strains from patients who either maintained R5 viruses or experienced emergence of X4 strains during progression from chronic to advanced stages of C-HIV infection.
Experimental procedures
We utilized X4 (n = 2) and R5 (n = 15) Envs cloned from longitudinal plasma samples of three treatment-naïve C-HIV infected subjects (1503, 1854 and 1109) from the recently described MUSH cohort [12] who, over a 3-year sampling period, showed clinical and immunological evidence of progression from chronic to advanced stages of infection [12,17] (Table 1). Subjects 1503 and 1854 exclusively harboured R5 viruses in plasma samples provided at enrolment (E), 1-year later (I; intermediate) and 3-years after enrolment (F; final), whereas subject 1109 experienced a switch from dominant R5 viruses in enrolment and intermediate plasma samples to dominant X4 strains in the final plasma sample [12] (Table 1).
Table 1.
Subject 1503, 1854 and 1109 Envs
| Subject | Plasma sample | CD4 + T-cell count (cells/μL) | Viral Load (RNA copies/ml) | Env ID | Coreceptor usage |
|---|---|---|---|---|---|
| 1503 | E | 396 | 5.73 | 11 | R5 |
| 13 | R5 | ||||
| I | 168 | 5.69 | 7 | R5 | |
| 11 | R5 | ||||
| F | 259 | 5.71 | 7 | R5 | |
| 8 | R5 | ||||
| 1854 | E | 435 | 5.61 | 8 | R5 |
| 10 | R5 | ||||
| I | 279 | 6.57 | 4 | R5 | |
| 20 | R5 | ||||
| F | 272 | 5.67 | 4 | R5 | |
| 8 | R5 | ||||
| 1109 | E | 367 | 5.54 | 10 | R5 |
| 42 | R5 | ||||
| I | 402 | 5.59 | 3 | R5 | |
| F | 169 | 5.67 | 30 | X4 | |
| 39 | X4 | ||||
| - | - | - | F-30-E10V3 | R5 |
The CD4+ T-cell counts and plasma viral loads for these subjects have been reported previously, and are summarized again here to assist in the interpretation of the CD4+ T-cell subset tropism data [12]. Plasma viral load values are shown as log10. Envs were cloned directly from plasma samples provided at enrolment (E), 1-year later (I; intermediate) and 3-years after enrolment (F; final). Env coreceptor usage was determined by infecting NP2 or U87 cells expressing CD4 and either CCR5 or CXCR4 with Env-pseudotyped luciferase reporter viruses [12]. The 1109-F-30-E10V3 Env was generated by mutating 1109-F-30 to express the V3 region of 1109-E-10 [12,17]. Env ID, HIV-1 Env identification number as described in [12].
To determine the CD4+ T-cell subset tropism for each of these Envs, we utilized an in vitro multi-parameter flow cytometry-based infection assay, described in our recent studies [18,19] and in Additional file 1. Briefly, peripheral blood CD4+ T-cells isolated from four HIV-seronegative donors were infected with single-round Env-pseudotyped GFP reporter viruses, then HIV-1 infection was determined by GFP positivity and the distribution of infection among CD4+ T-cell subsets was determined using antibodies specific for cell-surface markers; TN (CD45RO−CCR7+CD27+), TSCM (CD45RO−CCR7+CD27+CD95+CD122+), TCM (CD45RO+CCR7+CD27+), TTM (CD45RO+CCR7−CD27+), TEM (CD45RO+CCR7−CD27−) and TEMRA (CD45RO−CCR7−CD27−) The fluorochrome labeled flow cytometry antibodies used in this study are described in Additional file 1: Table S1. Our T-cell subset gating strategy, which we have described previously [18], is illustrated in Additional file 1: Figure S1.
GFP reporter viruses pseudotyped with well-characterized T-cell tropic B-HIV Envs JR-CSF (R5), HXB2 (X4) and Macs1-Spleen12 (R5X4) were used as controls [19-22]. CD4+ T-cells were infected with 3000 infectious units of B- and C-HIV R5 viruses, and 1250 infectious units of B- and C-HIV R5X4 and X4 viruses, which we determined here (data not shown) and previously [18,19] to be virus inoculums that ensure infections within the linear range.
R5 and X4 C-HIV Envs exhibit alternative CD4+ T-cell subset tropism profiles
Prior to infection we first showed that the proportion of each CD4+ T-cell subset (Additional file 1: Figure S2) and the proportion of cells expressing CCR5 and CXCR4 (Additional file 1: Table S2) was similar between donors, and was consistent with the findings of previous studies [10,18,19,23,24]. R5 C-HIV Envs mediated levels of CD4+ T-cell infection (mean 1% ± standard deviation 0.4% of infected CD4+ T-cells) similar to JR-CSF (1 ± 0.1%), and preferentially infected memory T-cells; TN (21.5 ± 8.9%), TSCM (3.3 ± 2.6%), TCM (24.7 ± 5.3%), TTM (20.1 ± 4.7%), TEM (14.9 ± 4.8%) and TEMRA (0.6 ± 0.4%) (Figure 1a). X4 C-HIV Envs mediated levels of CD4+ T-cell infection (3.5 ± 1.8%) similar to HXB2 (1.7 ± 0.9%), and preferentially infected naïve T-cells; TN (62.1 ± 12.1%), TSCM (1.1 ± 1.7%), TCM (16.9 ± 7.3%), TTM (13.5 ± 4.6%), TEM (5.7 ± 2.8%) and TEMRA (0.6 ± 0.7%), (Figure 1a). The preferential infection of memory and naive T-cells by R5 and X4 C-HIV Envs, respectively, could be influenced by the variation in the proportion of cells that express CCR5 and CXCR4 (Additional file 1: Table S2) as CD45RO− cells had a higher proportion of CD4+ T-cells expressing CXCR4 compared to CD45RO+ cells, which had higher proportions of CD4+ T-cells expressing CCR5. Together, these data suggest that, within these subjects, C-HIV tropism for CD4+ memory and naïve T-cells is constrained by Env coreceptor specificity.
Figure 1.
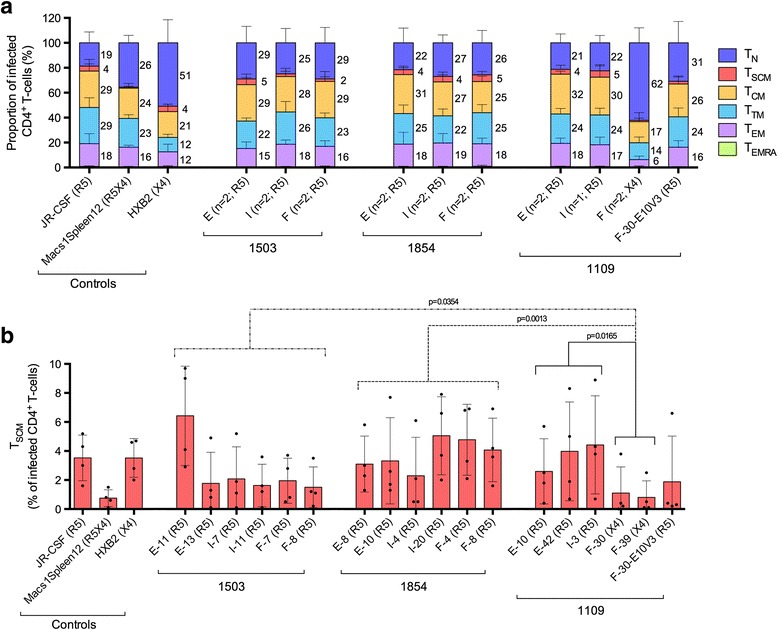
Proportion of CD4 + T-cell subsets infected by Env-pseudotyped GFP reporter viruses. (a) Values represent the percentage of infected CD4+ T-cells that belong to the indicated subset; naïve (TN, dark blue), stem cell memory (TSCM, red), central memory (TCM, yellow), transitional memory (TTM, light blue), effector memory (TEM, purple) and effector memory RA (TEMRA, green). Values were averaged across four HIV-seronegative donors. Error bars represent the standard deviation. CD4+ T-cells were infected with GFP reporter viruses pseudotyped with (i) subtype C HIV-1 Envs isolated from plasma samples of subjects 1503, 1854 and 1109 provided at enrolment (E), 1-year later (I; intermediate) and 3-years after enrolment (F; final), or (ii) subtype B HIV-1 control Envs. Text in parentheses indicates the number of Envs isolated per plasma sample and/or Env coreceptor usage; CCR5-specific (R5), CXCR4-specific (X4), uses CCR5 or CXCR4 (R5X4). (b) Values represent the percentage of infected CD4+ T-cells that are TSCM for each Env-pseudotyped GFP reporter virus per donor (individual dots). Columns display the mean and standard deviation. Statistical analyses were performed using Mann Whitney tests, p-values ≤0.05 were considered significant.
Env determinants underlying the linkage between C-HIV coreceptor specificity and CD4+ T-cell tropism alterations
To better understand the underlying Env determinants of C-HIV tropism for CD4+ T-cell subsets, we quantified the susceptibility of CD4+ T-cells isolated from peripheral blood of four HIV-seronegative donors to infection by GFP reporter viruses pseudotyped with X4 1109-F-30 Env that was mutated to express the V3 region of the R5 1109-E-10 Env (referred to as 1109-F-30-E10V3) (Additional file 1: Table S3). Previously we utilized this Env to define the critical C-HIV V3 alterations necessary to confer CXCR4 usage [12,18]. In these studies, we confirmed that either an Ile314-Gly315 insertion or an Arg318Pro alteration in the V3 loop crown (GPGQ) motif was sufficient to confer an R5X4 phenotype, and that both of these alterations together conferred a strictly X4 phenotype. Here, the R5 1109-F-30-E10V3 Env displayed a CD4+ T-cell subset tropism profile that was similar to enrolment and intermediate 1109 R5 Envs, preferentially infecting memory CD4+ T-cells; TN (30.9 ± 17.2%), TSCM (2.1 ± 3.6%), TCM (26.4 ± 6.5%), TTM (24.4 ± 7.1%), TEM (15.9 ± 5.2%) and TEMRA (0.4 ± 0.4%) (Figure 1a). These findings establish (i) the V3 loop as a key determinant of CD4+ T-cell subset tropism for viruses from subject 1109, and (ii) the linkage between C-HIV coreceptor specificity and CD4+ T-cell tropism alterations.
V3 determinants of 1109-F-30 tropism for CD4+ T-cell subsets are shared by the majority of CXCR4-using C-HIV Envs
Our analyses of X4 C-HIV Envs were limited to just subject 1109 because this was the only individual within our longitudinal cohort to experience the emergence of CXCR4-using strains [12]. To gauge the likelihood that our findings would translate to other CXCR4-using C-HIV strains, we analyzed every previously published R5 (n = 1653) and CXCR4-using (n = 203) C-HIV Env sequence for expression of the defined 1109-F-30 V3 determinants of CXCR4-usage and tropism for CD4+ T-cell subsets, namely the two amino acid insertion at positions 314–315 and/or the GPGQ crown alteration. Additional file 1: Table S4 shows that of the CXCR4-using V3 sequences, 27.1% contained a 314–315 insertion, of which 81.9% contained Ile314-Gly315 (36.5% overall), 62.6% exhibited a GPGQ alteration, of which 55.1% exhibited GRGQ (58.1% overall), and 23.2% contained both a 314–315 insertion and a GPGQ alteration, of which 84.9% exhibited both Ile314-Gly315 and GRGQ (32.4% overall). These proportions were even higher when assessing just strictly X4 C-HIV Envs, where 44.6% contained a 314–315 insertion, of which 81.8% contained Ile314-Gly315 (36.5% overall), 83.8% exhibited a GPGQ alteration, of which 69.3% exhibited GRGQ (58.1% overall), and 39.2% contained both a 314–315 insertion and a GPGQ alteration, of which 82.7% exhibited both Ile314-Gly315 and GRGQ (32.4% overall). Conversely, none of the R5 V3 sequences contained a 314–315 insertion and only 1.9% exhibited a GPGQ alteration (none of which were GRGQ). Although further functional studies are required to establish the CD4+ T-cell subset tropism properties of diverse CXCR4-using C-HIV strains, these sequence analyses suggest that a substantial proportion of previously reported X4 C-HIV Envs will likely exhibit a similar profile to that of the X4 Envs from subject 1109.
TSCM are susceptible to infection by CCR5- and CXCR4-using clinical C-HIV viruses
Recently, a new subset of CD4+ T-cells, TSCM, was found to contribute significantly to the viral reservoir during HIV-1 disease progression [6]. Here, we found that the proportion of CD4+ T-cells infected by X4 C-HIV Envs from subject 1109 that were TSCM (1 ± 1.4%) was significantly less (by Mann Whitney test) than that of R5 C-HIV strains from subject 1109 (3.7 ± 2.9%, p = 0.0165), 1503 (2.6 ± 2.6%, p = 0.0354) and 1854 (3.8 ± 2.4%, p = 0.0013) (Figure 1b). Unexpectedly, the proportion of TSCM infected by the R5 1109-F-30-E10V3 Env (1.9 ± 3.2%) was statistically similar to both R5 and X4 C-HIV isolates from subject 1109, suggesting that additional mutations in the 1109-F-30 Env are required to confer TSCM entry levels alike subject 1109 R5 Envs.
These results suggest that TSCM are more susceptible to infection by R5 C-HIV clinical isolates than X4 strains. Future studies involving replication-competent viruses are required to more accurately quantify the susceptibility of TSCM to infection by C-HIV in vivo. While it is important to note that TSCM represent a small proportion of the total circulating CD4+ T-cells and their level of infection was relatively low, demonstration of their susceptibility to infection by both R5 and X4 clinical C-HIV viruses provides new information that may help inform the design and implementation of therapies that target the TSCM HIV-1 reservoir, such as anti-cancer drugs that block stem cell-specific molecular pathways expressed by TSCM cells [25].
The susceptibility of CD4+ T-cell subsets to CCR5-mediated infection is maintained during progression from chronic and advanced C-HIV infection
To determine whether C-HIV CD4+ T-cell subset tropism is altered during progression from chronic to advanced C-HIV infection, we performed intra-patient statistical analysis to compare the proportion of CD4+ T-cell subsets infected by Envs cloned from enrolment, intermediate and final time point plasma samples (Figure 1a). Statistical significance (by Mann Whitney test) was only observed for subject 1109 between Envs from the final time point and Envs from enrolment (E) or intermediate (I) time points, where the emergence of X4 strains was associated with a significant increase in the proportion of infected CD4+ T-cells that were TN (E p = 0.0002, I p = 0.004), and a significant decrease in the proportion of infected TCM (E p = 0.0002, I p = 0.0444), TTM (E p = 0.0002, I p = 0.004), TEM (E p = 0.0003, I p = 0.004) and TSCM (E p = 0.0263, I p = 0.0202).
Interestingly, these results suggest that as subjects 1503 and 1854 progressed from chronic to advanced C-HIV infection over a 3 year period [12] (Table 1), circulating R5 viruses maintained their tropism profiles for CD4+ T-cell subsets. While additional studies involving more C-HIV Envs isolated over longer periods of time are required to confirm these findings, our data support those from a study by Buzon et al. [6], which showed that the susceptibility of TSCM to infection by R5 subtype B HIV-1 is maintained during disease progression, and those from a study by Chomont et al. [1], which showed that HIV-1 can persist in TCM and TTM of subjects who exhibit reconstitution of CD4+ T-cells during suppressive antiretroviral therapy.
Conclusions
These findings provide new insights into the natural history of tropism alterations for CD4+ T-cell subsets by R5 and X4 C-HIV strains during progression from chronic to advanced stages of infection. Specifically, we showed that R5 C-HIV strains maintain their preference for infecting memory CD4+ T-cell subsets during later stages of C-HIV infection, whereas emergent X4 C-HIV strains preferentially target TN. Furthermore, by demonstrating the susceptibility of TSCM to infection by C-HIV, our results highlight the potential for this subset of CD4+ T-cells to serve as a long-lived viral reservoir in individuals harboring CCR5- and CXCR4-using C-HIV viruses.
Acknowledgements
We thank J. Sodroski and D.F.J. Purcell for supplying pSVIII-HXB2 and pNL4-3Env-GFP plasmids, respectively. This study was supported by grants from (i) the Australian National Health and Medical Research Council (NHMRC) to PRG and MJC (1022066), (ii) the National Institutes of Health (NIH) to PRG and MJC (R21 MH100594), and (iii) the Delaney AIDS Research Enterprise (DARE) to MJC (U19 AI096109). KC is supported by an Australian Postgraduate Award from the University of Melbourne. PRG is supported by an Australian Research Council (ARC) Future Fellowship (FT2). The authors gratefully acknowledge the contribution to this work by the Victorian Operational Infrastructure Support Program, received by the Burnet Institute.
Abbreviations
- (HIV-1)
Human immunodeficiency virus type 1
- (AIDS)
Acquired immunodeficiency syndrome
- (B-HIV)
Subtype B HIV-1
- (C-HIV)
Subtype C HIV-1
- (TN)
Naïve CD4+ T-cell
- (TSCM)
Stem cell memory CD4+ T-cell
- (TCM)
Central memory CD4+ T-cell
- (TTM)
Transitional memory CD4+ T-cell
- (TEM)
Effector memory CD4+ T-cell
- (TEMRA)
Effector memory RA CD4+ T-cell
- (Env)
HIV-1 envelope protein
- (R5)
CCR5-specific HIV-1
- (X4)
CXCR4-specific HIV-1
- (R5X4)
CCR5- or CXCR4-using HIV-1
- (GFP)
Green fluorescent protein
- (E)
Plasma samples provided at enrolment
- (I)
1-year later
- (F)
3-years after enrolment
Additional file
Extended description of Materials, Methods and Results, including the CD4 + T-cell gating strategy, per patient CD4 + T-cell subset distributions, CD4 + T-cell subset CCR5 and CXCR4 expression levels, subject 1109 V3 sequence analysis, and V3 sequence accession numbers.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KC and JKF performed the experiments. KC, JKF and PRG designed the experiments. KC, JKF, GP and MJC analysed the data. KC, JKF and PRG wrote the manuscript. All authors helped edit the manuscript. All authors read and approved the final manuscript.
Contributor Information
Kieran Cashin, Email: kieran@burnet.edu.au.
Geza Paukovics, Email: paukovic@burnet.edu.au.
Martin R Jakobsen, Email: mrj@immunology.au.dk.
Lars Østergaard, Email: larsoest@rm.dk.
Melissa J Churchill, Email: churchil@burnet.edu.au.
Paul R Gorry, Email: gorry@burnet.edu.au.
Jacqueline K Flynn, Email: jflynn@burnet.edu.au.
References
- 1.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fritsch RD, Shen X, Sims GP, Hathcock KS, Hodes RJ, Lipsky PE. Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27. J Immunol. 2005;175:6489–6497. doi: 10.4049/jimmunol.175.10.6489. [DOI] [PubMed] [Google Scholar]
- 3.Luckey CJ, Weaver CT. Stem-cell-like qualities of immune memory; CD4+ T cells join the party. Cell Stem Cell. 2012;10:107–108. doi: 10.1016/j.stem.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 5.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 6.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lugli E, Gattinoni L, Roberto A, Mavilio D, Price DA, Restifo NP, Roederer M. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat Protoc. 2013;8:33–42. doi: 10.1038/nprot.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, Wang E, Douek DC, Price DA, June CH, Marincola FM, Roederer M, Restifo NP. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lugli E, Dominguez MH, Gattinoni L, Chattopadhyay PK, Bolton DL, Song K, Klatt NR, Brenchley JM, Vaccari M, Gostick E, Price DA, Waldmann TA, Restifo NP, Franchini G, Roederer M. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest. 2013;123:594–599. doi: 10.1172/JCI66327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorry PR, Ancuta P. Coreceptors and HIV-1 pathogenesis. Curr HIV/AIDS Rep. 2011;8:45–53. doi: 10.1007/s11904-010-0069-x. [DOI] [PubMed] [Google Scholar]
- 11.Berger EA, Doms RW, Fenyo EM, Korber BT, Littman DR, Moore JP, Sattentau QJ, Schuitemaker H, Sodroski J, Weiss RA. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 12.Jakobsen MR, Cashin K, Roche M, Sterjovski J, Ellett A, Borm K, Flynn J, Erikstrup C, Gouillou M, Gray LR, Saksena NK, Wang B, Purcell DF, Kallestrup P, Zinyama-Gutsire R, Gomo E, Ullum H, Østergaard L, Lee B, Ramsland PA, Churchill MJ, Gorry PR. Longitudinal analysis of CCR5 and CXCR4 usage in a cohort of antiretroviral therapy-naive subjects with progressive HIV-1 subtype C infection. PLoS One. 2013;8:e65950. doi: 10.1371/journal.pone.0065950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connell BJ, Michler K, Capovilla A, Venter WD, Stevens WS, Papathanasopoulos MA. Emergence of X4 usage among HIV-1 subtype C: evidence for an evolving epidemic in South Africa. AIDS. 2008;22:896–899. doi: 10.1097/QAD.0b013e3282f57f7a. [DOI] [PubMed] [Google Scholar]
- 14.Graf T, Pinto AR. The increasing prevalence of HIV-1 subtype C in Southern Brazil and its dispersion through the continent. Virology. 2013;435:170–178. doi: 10.1016/j.virol.2012.08.048. [DOI] [PubMed] [Google Scholar]
- 15.Kassaye S, Johnston E, McColgan B, Kantor R, Zijenah L, Katzenstein D. Envelope coreceptor tropism, drug resistance, and viral evolution among subtype C HIV-1-infected individuals receiving nonsuppressive antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;50:9–18. doi: 10.1097/QAI.0b013e31818ffdff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michler K, Connell BJ, Venter WD, Stevens WS, Capovilla A, Papathanasopoulos MA. Genotypic characterization and comparison of full-length envelope glycoproteins from South African HIV type 1 subtype C primary isolates that utilize CCR5 and/or CXCR4. AIDS Res Hum Retroviruses. 2008;24:743–751. doi: 10.1089/aid.2007.0304. [DOI] [PubMed] [Google Scholar]
- 17.Cashin K, Jakobsen MR, Sterjovski J, Roche M, Ellett A, Flynn JK, Borm K, Gouillou M, Churchill MJ, Gorry PR. Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection. Retrovirology. 2013;10:98. doi: 10.1186/1742-4690-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn JK, Paukovics G, Cashin K, Borm K, Ellett A, Roche M, Jakobsen MR, Churchill MJ, Gorry PR. Quantifying susceptibility of CD4+ stem memory T-cells to infection by laboratory adapted and clinical HIV-1 strains. Viruses. 2014;6:709–726. doi: 10.3390/v6020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flynn JK, Paukovics G, Moore MS, Ellett A, Gray LR, Duncan R, Salimi H, Jubb B, Westby M, Purcell DF, Lewin SR, Lee B, Churchill MJ, Gorry PR, Roche M. The magnitude of HIV-1 resistance to the CCR5 antagonist maraviroc may impart a differential alteration in HIV-1 tropism for macrophages and T-cell subsets. Virology. 2013;442:51–58. doi: 10.1016/j.virol.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 20.De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 21.Cashin K, Roche M, Sterjovski J, Ellett A, Gray LR, Cunningham AL, Ramsland PA, Churchill MJ, Gorry PR. Alternative coreceptor requirements for efficient CCR5- and CXCR4-mediated HIV-1 entry into macrophages. J Virol. 2011;85:10699–10709. doi: 10.1128/JVI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray L, Roche M, Churchill MJ, Sterjovski J, Ellett A, Poumbourios P, Sherieff S, Wang B, Saksena N, Purcell DF, Wesselingh S, Cunningham AL, Brew BJ, Gabuzda D, Gorry PR. Tissue-specific sequence alterations in the human immunodeficiency virus type 1 envelope favoring CCR5 usage contribute to persistence of dual-tropic virus in the brain. J Virol. 2009;83:5430–5441. doi: 10.1128/JVI.02648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaff JM, Wilen CB, Harrison JE, Demarest JF, Lee B, Doms RW, Tilton JC. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J Virol. 2010;84:6505–6514. doi: 10.1128/JVI.00374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]


