Abstract
Helicoverpa armigera is one of the primary agricultural pests in the Old World, whereas H. zea is predominant in the New World. However, H. armigera was first documented in Brazil in 2013. Therefore, the geographical distribution, range of hosts, invasion source, and dispersal routes for H. armigera are poorly understood or unknown in Brazil. In this study, we used a phylogeographic analysis of natural H. armigera and H. zea populations to (1) assess the occurrence of both species on different hosts; (2) infer the demographic parameters and genetic structure; (3) determine the potential invasion and dispersal routes for H. armigera within the Brazilian territory; and (4) infer the geographical origin of H. armigera. We analyzed partial sequence data from the cytochrome c oxidase subunit I (COI) gene. We determined that H. armigera individuals were most prevalent on dicotyledonous hosts and that H. zea were most prevalent on maize crops, based on the samples collected between May 2012 and April 2013. The populations of both species showed signs of demographic expansion, and no genetic structure. The high genetic diversity and wide distribution of H. armigera in mid-2012 are consistent with an invasion period prior to the first reports of this species in the literature and/or multiple invasion events within the Brazilian territory. It was not possible to infer the invasion and dispersal routes of H. armigera with this dataset. However, joint analyses using sequences from the Old World indicated the presence of Chinese, Indian, and European lineages within the Brazilian populations of H. armigera. These results suggest that sustainable management plans for the control of H. armigera will be challenging considering the high genetic diversity, polyphagous feeding habits, and great potential mobility of this pest on numerous hosts, which favor the adaptation of this insect to diverse environments and control strategies.
Introduction
The Heliothinae (Lepidoptera: Noctuidae) subfamily has 381 described species, many of which are important agricultural pests from the Helicoverpa Hardwick and Heliothis Ochsenheimer genera [1]. The Helicoverpa genus contains two of the primary Heliothinae pest species: Helicoverpa armigera (Hübner) (Old World bollworm) and Helicoverpa zea (Boddie) (New World bollworm). Although the exact evolutionary relationship between H. armigera and H. zea remains uncertain, these insects are considered to be ‘twin’ or ‘sibling’ species, and they are able to copulate and produce fertile offspring under laboratory conditions [2]–[5]. Some hypotheses propose that H. zea evolved from a small portion of the larger H. armigera population (i.e., a “founder effect”) that reached the American continent approximately 1.5 million years ago, which is consistent with previous phylogeographic analyses of H. armigera and H. zea individuals [6], [7].
H. armigera is considered to be one of the most important agricultural pests in the world. This insect is widely distributed throughout Asia, Africa, Europe, and Australia, and it has been shown to attack more than 100 host species from 45 different plant families [8]–[10]. In contrast, H. zea is restricted to the American continent and is of lesser economic importance; it is a secondary pest of cotton, tomato, and, most significantly, maize crops [11]. However, the scenario in Brazil changed in 2013 when H. armigera individuals, which are considered to be A1 quarantine pests, were officially reported within the Brazilian territory [12]–[14]. This situation increased in severity due to the great dispersal ability of this insect as well as the steady reports from several regions of the world that described new H. armigera lineages showing tolerance/resistance to insecticides and genetically modified plants [15], [16]. It is estimated that H. armigera will cause a loss of more than US$2 billion to the 2013/14 Brazilian agriculture crop because of direct productivity losses and resources spent on phytosanitary products for soybean, cotton, and maize, which are the main crops of Brazilian agribusinesses. Therefore, H. armigera is now one of the most important pest species with respect to agriculture in Brazil [17].
High population densities of Helicoverpa spp. and the resulting economic damages to cultivated plants have been reported in different regions of Brazil, in particular in the Western state of Bahia [18]. Therefore, these reports suggest the existence of an invasion period prior to the first official report of H. armigera in Brazil. This atypical and confusing scenario was likely caused by the significant morphological similarities between H. zea and H. armigera [9], [19] and by major changes in pest management programs over recent years. In addition, these population changes may have been related to the release and increased cultivation of crops that express Bacillus thuringiensis (Bt) genes in Brazil.
Aside from the identification of H. armigera individuals within the Brazilian territory, many basic pieces of information concerning this species, including its geographical distribution, the types of hosts it attacks, its invasion source, and its dispersal routes, remain poorly understood or completely unknown. Therefore, we attempted to address some of these outstanding questions using a phylogeographic approach by analyzing genetic sequence data from a portion of the cytochrome c oxidase subunit I (COI) gene of Helicoverpa spp. specimens isolated from different hosts and regions of Brazil. This study was performed with the following goals in mind: (1) to confirm and evaluate the occurrence of H. armigera and H. zea individuals from different hosts and regions of Brazil; (2) to assess the demographic parameters and genetic structure of H. armigera and H. zea populations within the Brazilian territory, with a focus on the region, season, and host; (3) to assess the potential invasion (single or multiple) and dispersal routes for H. armigera within the Brazilian territory; and (4) to determine the geographical origin of the H. armigera populations present in Brazil. This information will be essential for understanding the genetic diversity and population dynamics of these pests as well as for guiding both immediate control strategies (legal and/or phytosanitary) and subsequent long-term integrated management programs for the Helicoverpa spp. complex in Brazil.
Results
Identification of Helicoverpa spp., hosts, and geographic locations
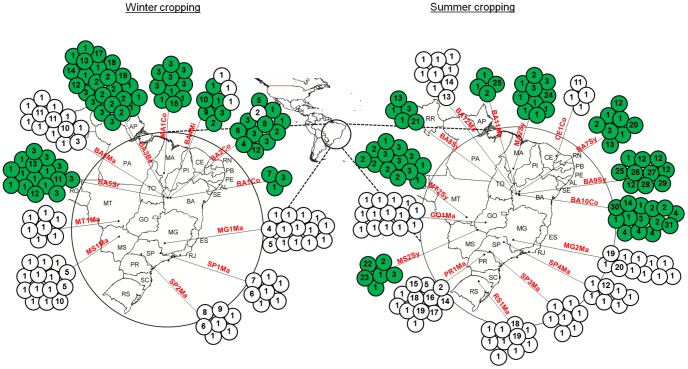
One hundred thirty-nine individuals from the 274 Helicoverpa spp. specimens initially sampled were identified as H. armigera (98–100% homology) and 134 individuals were identified as H. zea (98–100% homology) (GenBank Accession numbers KM274936–KM275209 are listed in Table 1). H. armigera was primarily found on soybean, bean, and cotton crops, and these insects were widely distributed throughout the Midwest and Northeast of Brazil during both crop periods (winter and summer) (Figure 1). H. armigera was also found on sorghum, millet, and maize crops. However, for maize, H. armigera individuals were only found at one site during the summer growing season in Northeastern Brazil (state of Bahia). H. armigera was not found on maize crops in the Midwest, Southeast, or South of Brazil. H. zea was primarily found on maize crops and was present in all sampled regions during both the winter and summer growing seasons. Of the winter crops, millet and cotton were exceptional in that they could simultaneously support H. zea and H. armigera (Figure 1). We found no correlations between specific H. armigera mitochondrial lineages (haplotypes) and specific hosts (Figure 1).
Table 1. Sampling sites for Helicoverpa armigera and Helicoverpa zea in Brazil, including the sites where these insects were sampled for this study, abbreviations, sample sizes for the mitochondrial genes (COI), crops sampled, geographic coordinates, dates sampled, and GenBank Accession.
| Sites (City, State) | Abbreviation (Site, Crop) | Crop | Sample size | Lat. (S) | Lon. (W) | Date | GenBank Accession | |
| H. armigera | H. zea | |||||||
| Winter cropping | ||||||||
| Barreiras, Bahia | BA1Co | Cotton | 3 | - | 12°08′54′′ | 44°59′33″ | 05.22.12 | KM274936–KM274938 |
| Luís E. Magalhães, Bahia | BA2Co | Cotton | 11 | 1 | 12°05′58″ | 45°47′54″ | 05.24.12 | KM274939–KM274950 |
| Balsas, Maranhão | MA1Co | Cotton | 10 | - | 07°31′59″ | 46°02′06″ | 06.23.12 | KM274987–KM274996 |
| Luís E. Magalhães, Bahia | BA3Be | Bean | 23 | - | 12°05′58″ | 45°47′54″ | 06.12.12 | KM274979–KM274986, KM275038–KM275052 |
| Luís E. Magalhães, Bahia | BA4Mi | Millet | 6 | 3 | 12°05′58″ | 45°47′54″ | 05.10.12 | KM274951–KM274959 |
| Luís E. Magalhães, Bahia | BA5Sr | Sorghum | 16 | - | 12°05′58″ | 45°47′54″ | 05.10.12 | KM274960–KM274975 |
| Capitólio, Minas Gerais | MG1Ma | Maize | - | 14 | 20°36′17″ | 46°04′19″ | 06.08.12 | KM274997–KM275010 |
| Luís E. Magalhães, Bahia | BA6Ma | Maize | - | 13 | 12°05′58″ | 45°47′54″ | 06.12.12 | KM274976–KM274978, KM275053–KM275062 |
| Itapira, São Paulo | SP1Ma | Maize | - | 7 | 22°26′11″ | 46°49′20″ | 06.12.12 | KM275011–KM275017 |
| Assis, São Paulo | SP2Ma | Maize | - | 7 | 22°39′40″ | 50°23′58″ | 06.15.12 | KM275018–KM275024 |
| São Gabriel do Oeste, Mato Grosso do Sul | MS1Ma | Maize | - | 13 | 19°23′37″ | 54°33′49″ | 06.27.12 | KM275025–KM275038 |
| Rondonópolis, Mato Grosso | MT1Ma | Maize | - | 7 | 16°28′17″ | 54°38′14″ | 08.01.12 | KM275063–KM275069 |
| Summer cropping | ||||||||
| Riachão das Neves, Bahia | BA7Sy | Soybean | 8 | - | 12°08′54″ | 44°59′33″ | 10.21.12 | KM275070–KM275077 |
| Luís E. Magalhães, Bahia | BA8Sy | Soybean | 5 | - | 12°05′58″ | 45°47′54″ | 10.31.12 | KM275078–KM275082 |
| Rondonópolis, Mato Grosso | MT2Sy | Soybean | 13 | - | 16°28′17″ | 54°38′14″ | 11.08.12 | KM275083–KM275092, KM275156–KM275158 |
| Chapadão do Sul, Mato Grosso do Sul | MS2Sy | Soybean | 6 | - | 18°46′44″ | 52°36′59″ | 11.29.12 | KM275097–KM275102 |
| Balsas, Maranhão | MA2Sy | Soybean | 10 | - | 07°31′59″ | 46°02′06″ | 01.06.13 | KM275103–KM275112 |
| São Desidério, Bahia | BA9Sy | Soybean | 10 | - | 12°21′08″ | 44°59′03″ | 01.15.13 | KM275127–KM275136 |
| Limoeiro do Norte, Ceará | CE1Co | Cotton | - | 4 | 05°08′56′′ | 38°05′52′′ | 10.08.12 | KM275093–KM275096 |
| São Desidério, Bahia | BA10Co | Cotton | 14 | - | 12°21′08″ | 44°59′03″ | 01.15.13 | KM275147–KM275155, KM275202–KM275206 |
| Cândido Mota, São Paulo | SP3Ma | Maize | - | 7 | 22°44′46″ | 50°23′15″ | 01.14.13 | KM275113–KM275119 |
| Jardinópolis, São Paulo | SP4Ma | Maize | - | 7 | 21°03′47″ | 47°45′05″ | 03.04.13 | KM275120–KM275126 |
| Barreiras, Bahia | BA11Ma | Maize | 4 | - | 11°33′33″ | 46°19′47″ | 02.21.13 | KM275137–KM275140 |
| Luís E. Magalhães, Bahia | BA12Ma | Maize | - | 9 | 12°05′58″ | 45°47′54″ | 03.28.13 | KM275141–KM275146, KM275207–KM275209 |
| Rolândia, Paraná | PR1Ma | Maize | - | 12 | 23°19′13′′ | 51°29′01′′ | 01.24.13 | KM275159–KM275170 |
| Passo Fundo, Rio Grande do Sul | RS1Ma | Maize | - | 10 | 28°16′08′′ | 52°37′15′′ | 01.30.13 | KM275171–KM275180 |
| Montividiu, Goiás | GO1Ma | Maize | - | 10 | 17°19′19′′ | 51°14′51′′ | 02.05.13 | KM275181–KM275190 |
| Capitólio, Minas Gerais | MG2Ma | Maize | - | 11 | 20°36′17″ | 46°04′19″ | 03.10.13 | KM275191–KM275201 |
| Total | 139 | 135 | ||||||
Figure 1. Geographic distributions of COI haplotypes of H. armigera and H. zea.
One hundred and thirty nine and 135 COI haplotypes were analyzed for these species, respectively. The samples were separated into two temporal groups (winter crops and summer crops). Each circle represents the haplotypes identified in a given population; a number within a circle denotes the COI haplotypes identified in that population. Colored circles refer to H. armigera specimens, and white circles refer to H. zea specimens. The abbreviations refer to the sampled locations and crops (Table 1).
Dataset assembly, haplotypes, and demographic analysis
Following alignment and editing, we were unable to identify indels or stop codons in the sequences from either species. However, using the most common haplotype for each species as a reference, eight non-synonymous substitutions were observed in 17 H. armigera individuals, and four non-synonymous substitutions were observed in eight H. zea individuals. However, considering the relatively high mutation rate reported for the COI gene in the Helicoverpa genus [20], as well the absence of indels and stop codons, it is unlikely that these sequences represent numts (nuclear mitochondrial DNA).
Twenty-six polymorphic sites were found among the 139 H. armigera individuals sampled, which yielded 31 haplotypes with a haplotype diversity (Hd) of 0.821 and a nucleotide diversity (Pi) of 0.0028. Sequence analysis of the 134 sampled H. zea individuals identified 19 polymorphic sites, which yielded 20 haplotypes with an Hd of 0.420 and a Pi of 0.0011 (Table 2). No significant differences in Hd or Pi were found for either species when the individuals were separated by growing season according to the sampled crops (Table 2). The results from Tajima's D test were only not significant for H. armigera individuals (p = 0.07) sampled on summer crops; however, Fu's Fs test was significant (p<0.01). The Tajima's D and Fu's Fs test results for both H. armigera and H. zea were negative and significant when the individuals were tested as a single group and when the individuals were split into groups based on the crop on which they were sampled (summer or winter; temporally). These results indicate an excess of low frequency polymorphisms and are consistent with either population expansion or purifying selection (Table 2). In addition, the model of sudden expansion [21] did not reject the hypothesis of expansion demographics for H. armigera (SSD = 0.0012, p = 0.48; Raggedness = 0.0433, p = 0.61) or H. zea (SSD = 0.0002, p = 0.90; Raggedness = 0.1492, p = 0.72).
Table 2. Number of individuals, haplotype designation, and genetic diversity for the sampled populations grouped according to geographical origin.
| Group | N. Individuals (samples) | N. haplotypes | Distribution of Haplotypes (n) | Haplotype Diversity (Hd) | Nucleotide diversity (Pi) | Tajima's D test (p value) | Fu's Fs test (p value) |
| H. armigera | |||||||
| Pooled | 139 (14) | 31 | - | 0.821 | 0.0028 | −1.729 (<0.01) | −26.361 (<0.01) |
| Winter cropping | 69 (6) | 19 | H1(22); H2(9); H3(21); H4(1); H5(1); H6(1); H7(1); H8(1); H9(1); H10(1); H11(1); H12(2); H13(2); H14(1); H15(1); H16(1); H17(1); H18(1); H19(1). | 0.805 | 0.0028 | −1.608 ( = 0.03) | −11.891 (<0.01) |
| Summer cropping | 70 (8) | 19 | H1(22); H2(13); H3(11); H4(4); H12(4); H13(2); H14(1); H20(1); H21(1); H22(1); H23(1); H24(1); H25(2); H26(1); H27(1); H28(1); H29(1); H30(1); H31(1). | 0.835 | 0.0028 | −1.353 ( = 0.07) | −11.254 (<0.01) |
| H. zea | |||||||
| Pooled | 135 (16) | 20 | - | 0.420 | 0.0011 | −2.190 (<0.01) | −22.912 (<0.01) |
| Winter cropping | 65 (8) | 11 | H1(50); H2(1); H3(1); H4(1); H5(3); H6(2); H7(1); H8(1); H9(1); H10(2); H11(2). | 0.408 | 0.0009 | −2.156 (<0.01) | −9.735 (<0.01) |
| Summer cropping | 70 (8) | 13 | H1(53); H2(1); H5(1); H11(1); H12(1); H13(1); H14(2); H15(1); H16(1); H17(1); H18(2); H19(3); H20(2). | 0.427 | 0.0012 | −1.967 (<0.01) | −10.411 (<0.01) |
Statistical analysis of population structure
The results of the analysis of molecular variance (AMOVA) with two hierarchical levels showed that the greatest amount of total variation was accounted for by differences among individuals within populations: 92.89% for H. armigera (ΦST = 0.071) and 94.22% for H. zea (ΦST = 0.058) (Table S1). For the AMOVA with three hierarchical levels for H. armigera, the largest percentage of variation occurred within populations, separating individuals into groups by time (winter and summer crops; 93.17%, ΦCT = 0.006; ΦSC = 0.074; ΦST = 0.068), host group (mono- and dicotyledonous; 99.24%, ΦCT = −0.01; ΦSC = 0.018; ΦST = 0.007), and each host type (crop; 93.19%, ΦCT = −0.042; ΦSC = 0.105; ΦST = 0.068) (Table S1). The group separation for H. armigera was not significant for any of the three tested groups (p>0.10). The AMOVA with three hierarchical levels divided the H. zea individuals into groups by time (winter and summer crops), which showed a larger variation within populations (93.76%, ΦCT = 0.010; ΦSC = 0.052; ΦST = 0.062); the group division was not significant (p>0.10) (Table S1).
Network analysis and Bayesian phylogeny
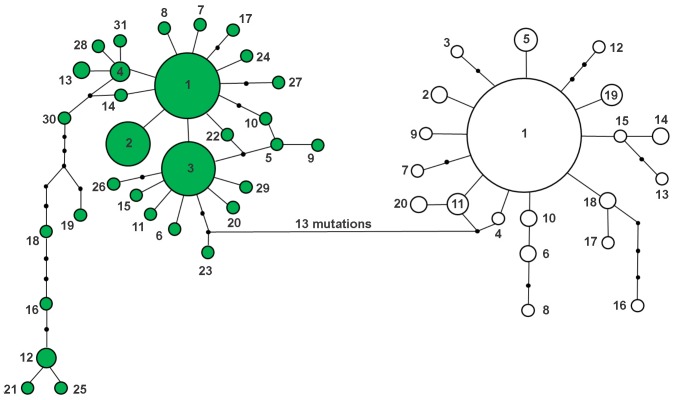
Analysis of the genetic connections between the Helicoverpa spp. represented in the haplotype network revealed a close genetic relation between H. armigera and H. zea, which were separated by only 13 mutational steps (Figure 2). By separately analyzing the connections between the genetic haplotypes of each species, we inferred the existence of two predominant maternal lineages for H. armigera: H1 (31.65%) and H3 (23.02%), which were located at the center of the haplotype network. The other haplotypes of H. armigera, with the exception of haplotype H2 (15.83%), all had frequencies below 5%. Haplotypes H19, H18, H16, H12, H21, and H25 formed an outer cluster within the haplotype network of H. armigera (Figure 2). The haplotype network for H. zea revealed a genetic haplotype relationship with a single central high-frequency lineage (H1 = 76.30%) surrounded by low-frequency haplotypes (<5%) (Figure 2).
Figure 2. Haplotype network based COI sequences from H. armigera and H. zea samples collected in Brazil.
Partial mtDNA COI (658 bp) sequences from H. armigera (colored circles) and H. zea (white circles) were analyzed from samples collected in Brazil. Each haplotype is represented by a circle and is identified by a number from 1–31. The H. armigera and H. zea COI haplotypes are shown as described in Table 2. The numbers of nucleotide substitutions between the haplotypes are indicated by black circles. The total number of nucleotide substitutions separating the H. armigera specimens from the H. zea specimens is shown.
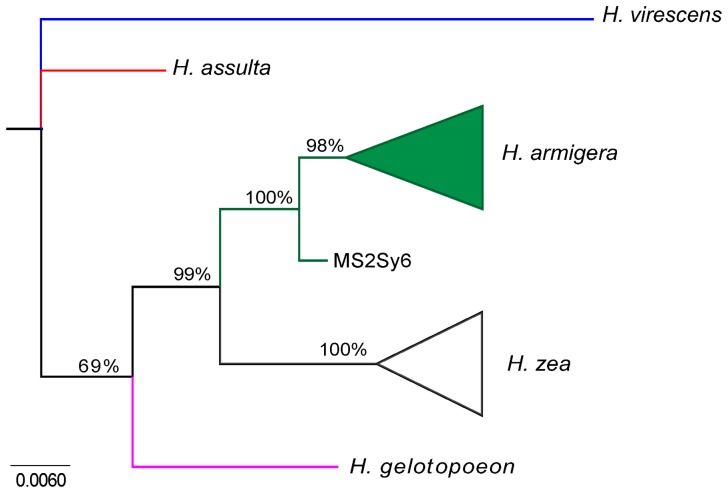
The optimal nucleotide substitution model identified by the MODELTEST 2.3 software program was the GTR+I+G model (Generalized time reversible + Proportion of invariable sites + Gamma distribution model). The estimated model parameters were based on empirical base frequencies (A = 0.3092, C = 0.1463, G = 0.1312, and T = 0.4133), with the proportion of invariable sites (I) set to 0.7393 and the gamma distribution shape parameter set to 0.5778. The consensus tree generated by the Bayesian analysis divided the Helicoverpa spp. specimens sampled in Brazil into two monophyletic clades (H. armigera and H. zea) with an associated probability of 99% (Figure 3; Figure S1). The probabilities separating the H. zea individuals into groups within this species were not significant. A single H. armigera individual (MS2Sy6) was separated from the other individuals with an associated probability of 98%. Finally, Helicoverpa gelotopoeon showed a closer phylogenetic relationship to H. armigera and H. zea compared with H. assulta (Figure 3; Figure S1).
Figure 3. Bayesian phylogenetic tree of H. armigera and H. zea individuals sampled in Brazil.
This phylogenetic tree is based on partial COI haplotype sequences and includes H. assulta and H. gelotopoeon sequences. Numbers near the interior branches indicate posterior probability (×100) values. The outgroup used was Heliothis virescens. H. armigera COI haplotypes and Genbank Accession numbers can be found in Table S2.
Network analysis: Brazilian vs. Old World Helicoverpa armigera
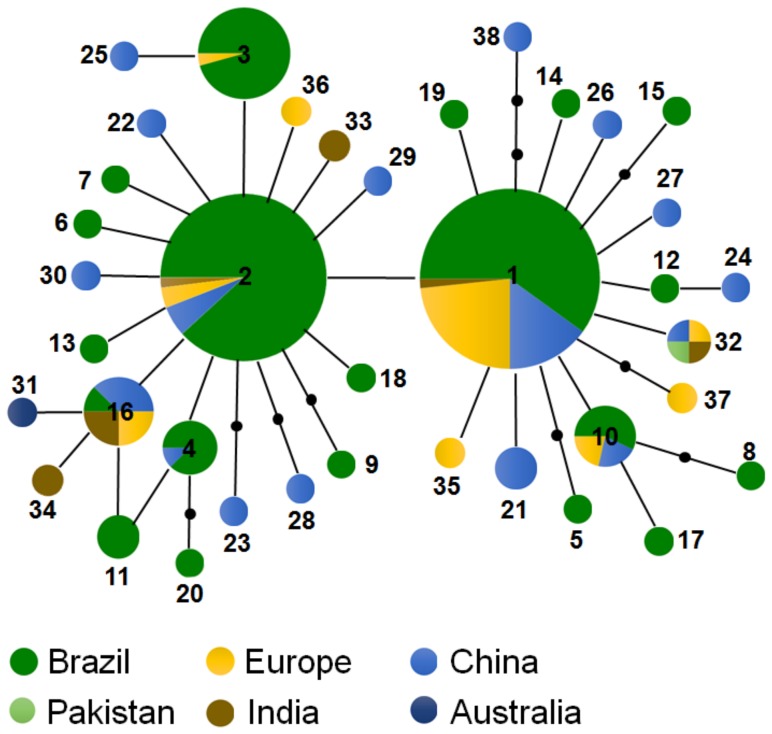
The haplotype network constructed using the edited sequences collected in Brazil, along with numerous Old World sequences, identified 38 distinct haplotypes (Figure 4). H1 (28%) and H2 (24%), which are widely distributed throughout Brazil, Europe, and China, were the most frequent haplotypes and occupied the central region of the haplotype network. All other haplotypes, with the exception of H3 and H10, showed frequencies below 5%. Finally, the majority of haplotypes with low frequencies represented by singletons were located at the network extremities (Figure 4).
Figure 4. Haplotype network based COI sequences from H. armigera samples from Brazil and Old World specimens.
Partial mtDNA COI (590 bp) sequences from this species were analyzed. Thirty-eight haplotypes were identified from 212 individuals sampled from China (n = 35), Thailand (n = 1), Australia (n = 1), Pakistan (n = 2), Europe (n = 28), India (n = 6), and Brazil (n = 139). H. armigera COI haplotypes are shown as described in Table S2. Each circle represents a haplotype and its number. The colors represent the frequency of each haplotype in the country/continent, with dark green (Brazil), light green (Pakistan), yellow (Europe), brown (India), light blue (China), and dark blue (Australia).
Discussion
Our results indicate a widespread distribution for H. armigera throughout the Midwest and Northeast of Brazil on a variety of crops, particularly dicotyledons, beans, soybeans, and cotton as well as, to a lesser extent, millet, sorghum, and maize. This pest was not found on maize crops in the Midwest, Southeast, or South of Brazil, despite the fact that these crops were initially identified as sources of H. armigera in this system. H. armigera individuals associated with maize crops were only found at a single sampling site in the Northeast (state of Bahia) during February 2013. In contrast, H. zea individuals were essentially found only on maize crops, with the exception of a few individuals collected from millet and cotton crops, where H. zea individuals were found alongside H. armigera individuals. Before the documentation of H. armigera in Brazil in 2013, we had hypothesized that major source of Helicoverpa spp. attacking different host plant was maize crops. However, our findings showed that targeting the control of H. armigera on maize crops may not be effective because H. zea was the predominant species in this host plant. The possibility of the formation of hybrid individuals between these two species, which has been reported under laboratory conditions [3], [4], needs to be investigated under field conditions to improve our pest management programs.
Demographic analyses using neutrality tests and a Mismatch Distribution Analysis indicated an expansion of the H. armigera and H. zea populations within the Brazilian territory. Population expansions were also consistent with the Haplotype network structure, which was characteristic of species undergoing processes of demographic expansion [22]. Brazilian H. armigera individuals showed two primary maternal lineages, whereas H. zea showed a single primary lineage, all of which were surrounded by numerous lower-frequency haplotypes. Therefore, these central high-frequency haplotypes represent the ancestral haplotypes, with the low-frequency haplotypes more recently derived [23]. Furthermore, signs of the H. armigera population expansion are likely because of the recent introduction of this pest into Brazil. Following the founder event, during which a portion of the overall genetic diversity of the species was introduced to Brazil, the H. armigera population further propagated. According to Nibouche et al. [24], H. armigera can migrate as far as 2,000 km, which likely facilitated the colonization of a variety of crops. The migration and colonization of crop areas by a small group of individuals can cause bottleneck effects, which, combined with plague population-suppression strategies (e.g., insecticide use that kills all but a small portion of the population), can lead to the types of demographic expansions observed for H. zea and H. armigera in Brazil [25]–[27]. In addition, the expansion of maize, soybean, and cotton crops into the North and Northeast of Brazil over the previous decade may also be responsible, in part, for the demographic expansion of these species, specifically H. zea. Additionally, assuming that not all COI variation is neutral, Helicoverpa spp. populations could be suffering selection, especially considering that populations have colonized new environments recently. However, further studies using a larger number of molecular markers from nuclear and mitochondrial genome regions would answer these questions. The H. armigera and H. zea population genetics were not structured according to space, time (winter and summer crops), or host (crops). Unstructured genetic networks have been reported for other populations of these two pest species in other parts of the world, which were based on several molecular markers, including mtDNA, allozymes, and microsatellites [7], [24], [25], [28], [29], [30]. Both species showed wide spatial haplotype distributions, and no genetic relationships were identified using a haplotype network analysis or an AMOVA. This scenario may be because these populations have a polyphagous feeding habit and migratory characteristics.
The unstructured population of H. armigera and the wide distribution of the two ancestral maternal lineages within the Brazilian territory did not allow us to infer any hypothetical invasion or dispersal routes for this species within the region. However, we noted that the haplotype and nucleotide diversities found for H. armigera in Brazil are similar to or greater than those reported for natural H. armigera populations in the Old World [7], [20]. For example, one outer branch of the H. armigera haplotype network, formed by haplotypes H19, H18, H16, H12, H21, and H25, is noteworthy for having the greatest genetic distance from the central haplotypes (H1 and H3), and these haplotypes have yet to be identified in Old World populations [7], [20]. In addition, joint analysis of the haplotypes from Brazil and the Old World yielded an overall structure that was similar to the haplotype network obtained only from the Brazilian individuals. In particular, the two most frequent haplotypes were identified throughout Brazil, Europe, China, and India, whereas the majority of the singletons were from Brazil and China. The cited literature, along with our results that showed a wide geographic distribution for H. armigera during the first half of 2012, support the hypothesis of an invasion period prior to the first reports of this species in Brazil. Alternatively, these findings are also consistent with a more recent invasion that involved a large gene pool, multiple invasion events, or some combination of these events.
The low genetic divergence observed between H. armigera and H. zea in the haplotype network analysis and the Bayesian phylogeny confirms the close genetic relatedness of these two species. Therefore, the reported co-occurrence of these species in time and space, as well as on the same hosts (as described here), could allow for the formation of hybrid individuals, which has been reported under laboratory conditions [3], [4]. Although the existence of hybrids in the wild remains unconfirmed, this scenario is of significant concern. In particular, recombination or introgression phenomena between H. armigera, which is reportedly resistant to control methods, and H. zea, which has adapted to the environmental conditions of the American continent, may enable gene transfer and fixation in some individuals. Therefore, hybridization may enable the selection of breeds with enhanced hybrid vigor and the ability to rapidly adapt to current management and suppression methods.
The population studies described in this study indicate a recent demographic expansion and a high mitochondrial genetic diversity for H. armigera and H. zea in Brazil. Therefore, the sustainable management of H. armigera will likely become a significant challenge for Brazilian entomology in the coming years, especially considering the polyphagous feeding habit, the great dispersal ability, and the numerous reports of resistance to insecticides and Bt crops for this insect [8], [24], [31]–[35]. This scenario requires immediate attention, as there is an imminent risk of H. armigera expanding throughout the American territory and perhaps reaching agricultural areas in Central and North America. However, it was not possible to trace the invasion and dispersal routes of H. armigera in the Brazilian territory. Nevertheless, the hypotheses of an invasion period prior to the first reports in the literature and/or an invasion that involved a diverse gene pool are both consistent with the observed high incidence and rapid adaptation of H. armigera in the Brazilian territory. Our confirmation that the predominant maternal lineages in the Brazilian territory are the same compared with those in Europe and Asia may represent a starting point to guide H. armigera management programs. Indeed, control strategies have a greater chance of success when reliable information is gathered in the regions where the pests, their hosts, and their natural enemies have co-evolved over a significant period of time.
Materials and Methods
Sampling procedures
Permit access to collect material used in our research at various crop sites was granted by respective growers. GPS coordinates of each location are listed in Table 1.
Brazilian agriculture has shown successive and overlapping crops in space and time, and these crops can be largely separated into two harvest groups that are primarily characterized by their rainfall needs. In particular, winter crops are grown between May and September and require low rainfall, whereas summer crops are grown between October and April and require high rainfall. Our initial sampling design was directed at understanding the H. zea population dynamics and primarily involved maize fields. However, attacks on soybean, cotton, bean, sorghum, and millet crops were also reported between May 2012 and April 2013 (Brazilian agricultural year). Therefore, we directed our sampling efforts towards a variety of crops and regions throughout Brazil. We also focused on the Western region of Bahia State, Brazil, which was the site of numerous Helicoverpa spp. attacks, to determine whether maize crops were the main source of H. zea in the Brazilian agricultural system. A total of 274 Helicoverpa caterpillars were collected at 19 sampling sites from six different crops (Table 1). In the absence of morphological characters or nuclear markers to reliably distinguish between H. zea and H. armigera, species identification was carried out using the sequence fragment of COI mitochondrial gene by comparing with H. zea and H. armigera species barcodes [7], [18], [19], [39] and determining homology with BlastN tool.
DNA extraction, PCR amplification, and gene sequencing
Genomic DNA was isolated from the thorax of each adult using an Invisorb Spin Tissue Kit (STRATEC Molecular, Berlin, Germany), according to the manufacturer's protocol. A fragment of the COI mitochondrial gene was amplified by polymerase chain reaction (PCR) with the primers LCO(F) (5′ - GGT CAA CAA ATC ATA AAG ATA TTG G - 3′) and HCO(R) (5′ - TAA ACT TCA GGG TGA CCA AAA AAT CA - 3′) [36]. Amplification reactions were performed using 10 ng genomic DNA, 50 mM MgCl2, 0.003 mg.mL−1 BSA, 6.25 mM dNTPs, 10 pmol each primer, 1 U Taq DNA Polymerase (Life Technologies, Carlsbad, CA, USA), and 10% 10× Taq Buffer in a final volume of 25 µL. The PCR program consisted of an initial denaturation step at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 30 s, and polymerization at 72°C for 1.5 min, with a final extension step at 72°C for 10 min. Following amplification, the aliquots were visually inspected using agarose gel (1.5% w/v) electrophoresis. The amplicons were purified by ethanol precipitation, and a second round of amplification was performed using the Big Dye Terminator v3.1 Cycle Sequencing system (Applied Biosystems, Foster City, CA, USA), which was followed by further purification. DNA sequencing was performed using the ABI3500xl automated genetic analyzer (Applied Biosystems, Foster City, CA, USA) at the State University of Campinas (Universidade Estadual de Campinas, Campinas, São Paulo, Brazil).
Dataset assembly, haplotypes, and demographic analysis
All sequences were manually edited using the Chromas Lite version 2.01 [37] software program and were aligned using the ClustalW tool from the BioEdit version 7.0 [38] software program. After editing and aligning the COI sequences, we determined the 658 bp consensus sequence, which was then posteriorly compared with the H. zea and H. armigera species barcodes [41] to determine homology using the BlastN tool, which is available online at NCBI [40].
The MEGA version 4 [41] software program was used to inspect the COI sequences from each species individually for the presence of numts [42]. In particular, we searched for the following numt signatures: (i) insertions/deletions (indels); (ii) stop codons leading to premature protein termination; and (iii) increased rates of non-synonymous mutations. The presence of signatures (i) and (ii) was considered sufficient to regard a sequence as a COI numt. In the presence of signatures (i) or (ii), signature (iii) was used to confirm the sequence as a numt. The presence of signature (iii) alone was not considered sufficient to define a sequence as a numt.
Haplotype and nucleotide diversity parameters for each species were estimated using the DnaSP version 5 [43] software program. Neutrality tests using Tajima's D [44] and Fu's Fs [45] were performed using the Arlequin version 3.1 [46] software program, and significance was determined using 1,000 random samples in coalescent simulations. Based on the recommendations in the Arlequin manual, we activated the “Infer from distance matrix” option under “Haplotype definition”, and the Fu's Fs statistical values were considered to be significant at a level of 5% only when the P-value was below 0.02. The diversity estimates and neutrality tests were performed using all sampled individuals from each species, which were divided into winter-crop and summer-crop groups. A Mismatch Distribution Analysis using a spatial expansion model [21] was also performed using the Arlequin version 3.1 software program, and significance was determined using 1,000 bootstrap replicates. We used the goodness-of-fit of the observed mismatch distribution to the expected distribution from the spatial expansion model and the sum of square deviations (SSD) as a test statistic (P-value support).
Population structure analysis
Using Arlequin 3.1, we also performed an AMOVA at the two- and three-hierarchy levels [47]. For the three-hierarchy AMOVA, we first separated the samples depending on whether they were collected on winter or summer crops and then further divided them by host plant (monocotyledonae or dicotyledonae).
Network analysis and Bayesian phylogenies
Genetic differences and connections among Helicoverpa spp. haplotypes were determined by constructing a maximum parsimony network [48] using the TCS 1.21 software program [49]. To resolve ambiguities present in the haplotype network, we used the criteria of coalescence theory and population geography proposed by Crandall and Templeton [23].
We used the distance matrix option in the PAUP *4.0 software program to calculate the inter- and intra-species genetic distances, which were inferred using the nucleotide substitution model and the Akaike Information Criteria [50] selected by MODELTEST 2 [51]. The MrBayes v3.2 software program [52] was used to estimate Bayesian phylogenies. In particular, the Bayesian analysis was performed with 10 million generations using one cold and three heated chains. Helicoverpa assulta (Guenée) (GenBank Accession number: EU768937), H. gelotopoeon Dyar (EU768938), and H. virescens (IN799050) sequences were included as outgroups for the Bayesian analysis. We obtained a 50%-majority-rule consensus tree with posterior probabilities that were equal to the bipartition frequencies.
Network analysis: Brazil vs. Old World
Seventy-three sequences from a variety of Old World sites that were present in GenBank were included with the 139 H. armigera sequences we collected in Brazil. In particular, 73 sequences were obtained from specimens collected in China (N = 35) [GenBank Accession numbers GQ892840 - GQ892855, GQ995232 - GQ995244 [20], HQ132369 (Yang, 2010), JX392415, and JX392497 (not published)], Thailand (1) [(EU768935)], Australia (1) [(EU768936) [5]], Pakistan (2) [(JN988529 and JN988530) (not published)], Europe (28) [(FN907979, FN907980, FN907988, FN907989, FN907996 - FN907999, FN908000 - FN908003, FN908005, FN908006, FN908011, FN908013 - FN908018, FN908023, FN908026, GU654969, GU686757, GU686955, and JF415782) (not published)] and India (6) [(HM854928-HM854932 and JX32104) (not published)] (Table S2). This new data set was edited and aligned as follows. The sequences were different lengths; thus, the editing and alignment processes generated a total of 212 sequences 590 bp in length, excluding indels. The sequences from individuals collected in Brazil, which were previously analyzed using a fragment length of 658 bp, as entered into GenBank (see Table 1), were edited by removing the first 36 bp and the last 32 bp. Using the TCS 1.21 software program [49], we subjected this data set to haplotype network analysis using a maximum parsimony network [48] to investigate the genetic connections between haplotypes from Brazil and the Old World as well as to infer the origins of maternal lineages within H. armigera populations in Brazil.
Supporting Information
Bayesian phylogenetic tree of H. armigera and H. zea individuals sampled in Brazil. This phylogenetic tree is based on partial COI haplotype sequences and includes H. assulta and H. gelotopoeon sequences. Numbers near the interior branches indicate the posterior probability (×1,000) values. The outgroup used was Heliothis virescens. H. armigera COI haplotypes and Genbank Accession numbers can be found in Table S2.
(TIF)
Hierarchical analysis of molecular variance (AMOVA), for population genetics structure of Helicoverpa armigera and H. zea with a mithocondrial (COI) region marker.
(DOCX)
Global Helicoverpa armigera including the Brazilian H. armigera haplotypes, and relevant GenBank Accession numbers. Numbers of individuals sequenced from each locality are indicated in parentheses.
(DOCX)
Acknowledgments
We thank Celito Breda, Diego Miranda, Fábio Wazne, Germison Tomquelski, José Wilson de Souza, Marcos Michelotto, Milton Ide, Paulo Saran, Pedro Brugnera, Pedro Matana Junior, Rodrigo Franciscatti, Rodrigo Sorgatto, Rubem Staudt, Sérgio de Azevedo, and SGS Gravena (SISBIO License #18018-1) for helping to collect insect samples in different Brazilian regions. We also thank Jaqueline Campos for technical assistance and Prof. José Baldin Pinheiro for providing laboratory space and equipment.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Sequences were deposited in NCBI and GenBank Accession numbers KM274936–KM275201 are listed in Table 1.
Funding Statement
This work was partially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Grant 308150/2009-0) and Comitê Brasileiro de Ação a Resistência a Inseticidas (IRAC-BR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pogue MG (2013) Revised status of Chloridea Duncan and (Westwood), 1841, for the Heliothis virescens species group (Lepidoptera: Noctuidae: Heliothinae) based on morphology and three genes. Syst Entomol 38: 523–542. [Google Scholar]
- 2. Mitter C, Poole RW, Matthews M (1993) Biosystematics of the Heliothinae (Lepidoptera: Noctuidae). Annu Rev Entomol 38: 207–225. [Google Scholar]
- 3. Laster ML, Hardee DD (1995) Intermating compatibility between north american Helicoverpa zea and Heliothis armigera (Lepidoptera: Noctuidae) from Russia. J Econ Entomol 88: 77–80. [Google Scholar]
- 4. Laster ML, Sheng CF (1995) Search for hybrid sterility for Helicoverpa zea in crosses between the north american H. zea and H. armigera (Lepidoptera: Noctuidae) from China. J Econ Entomol 88: 1288–1291. [Google Scholar]
- 5. Cho S, Mitchell A, Mitter C, Regier J, Matthews M, et al. (2008) Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Syst Entomol 33: 581–594. [Google Scholar]
- 6. Mallet J, Korman A, Heckel D, King P (1993) Biochemical genetics of Heliothis and Helicoverpa (Lepidoptera: Noctuidae) and evidence for a founder event in Helicoverpa zea . Ann Entomol Soc Am 86: 189–197. [Google Scholar]
- 7. Behere GT, Tay WT, Russell DA, Heckel DG, Appleton BR, et al. (2007) Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea . BMC Evol Biol 7: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fitt GP (1989) The ecology of Heliothis species in relation to agroecosystems. Annu Rev Entomol 34: 17–52. [Google Scholar]
- 9. Pogue M (2004) A new synonym of Helicoverpa zea (Boddie) and differentiation of adult males of H. zea and H. armigera (Hübner) (Lepidoptera: Noctuidae: Heliothinae). Ann Entomol Soc Am 97: 1222–1226. [Google Scholar]
- 10. Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ (2008) Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321: 1676–1678. [DOI] [PubMed] [Google Scholar]
- 11. Degrande PE, Omoto C (2013) Estancar prejuízos. Cultivar Grandes Culturas Abril 32–35. [Google Scholar]
- 12. Czepack C, Albernaz KC, Vivan LM, Guimarães HO, Carvalhais T (2013) Primeiro registro de ocorrência de Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) no Brasil. Pesq Agropec Trop 43: 110–113. [Google Scholar]
- 13. Specht A, Sosa-Gómez DR, Paula-Moraes SV de, Yano SAC (2013) Identificação morfológica e molecular de Helicoverpa armigera (Lepidoptera: Noctuidae) e ampliação de seu registro de ocorrência no Brasil. Pesq Agropec Bras 48: 689–692. [Google Scholar]
- 14.Agropec Consultoria (2013) Pragas quarentenárias: consulta a dados sobre pragas quarentenárias presentes e ausentes no Brasil. Available: http://spp.defesaagropecuaria.com/. Accessed 2014 Jan 29.
- 15. Yang Y, Li Y, Wu Y (2013) Current status of insecticide resistance in Helicoverpa armigera after 15 years of Bt cotton planting in China. J Econ Entomol 106: 375–381. [DOI] [PubMed] [Google Scholar]
- 16. Martin T, Ochou GO, Djihinto A, Traore D, Togola M, et al. (2005) Controlling an insecticide-resistant bollworm in West Africa. Agric Ecosyst Environ 107: 409–411. [Google Scholar]
- 17.MAPA (2014) Combate à praga Helicoverpa armigera. Brasilia: MAPA. [Google Scholar]
- 18. Tay WT, Soria MF, Walsh T, Thomazoni D, Silvie P, et al. (2013) A brave new world for an old world pest: Helicoverpa armigera (Lepidoptera: Noctuidae) in Brazil. Plos One 8: e80134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Behere GT, Tay WT, Russell DA, Batterham P (2008) Molecular markers to discriminate among four pest species of Helicoverpa (Lepidoptera: Noctuidae). Bull Entomol Res 98: 599–603. [DOI] [PubMed] [Google Scholar]
- 20. Li QQ, Li DY, Ye H, Liu XF, Shi W, et al. (2011) Using COI gene sequence to barcode two morphologically alike species: the cotton bollworm and the oriental tobacco budworm (Lepidoptera: Noctuidae). Mol Biol Rep 38: 5107–5113. [DOI] [PubMed] [Google Scholar]
- 21. Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 22. Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity 103: 285–298. [DOI] [PubMed] [Google Scholar]
- 23. Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nibouche S, Bues R, Toubon JF, Poitout S (1998) Allozyme polymorphism in the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae): comparison of African and European populations. Heredity 80: 438–445. [Google Scholar]
- 25. Endersby NM, Hoffmann AA, McKechnie SW, Weeks AR (2007) Is there genetic structure in populations of Helicoverpa armigera from Australia? Entomol Exp Appl 122: 253–263. [Google Scholar]
- 26. Albernaz KC, Silva-Brandao KL, Fresia P, Consoli FL, Omoto C (2012) Genetic variability and demographic history of Heliothis virescens (Lepidoptera: Noctuidae) populations from Brazil inferred by mtDNA sequences. Bull Entomol Res 102: 333–343. [DOI] [PubMed] [Google Scholar]
- 27. Domingues FA, Silva-Brandão KL, Abreu AG, Perera OP, Blanco CA, et al. (2012) Genetic structure and gene flow among Brazilian populations of Heliothis virescens (Lepidoptera: Noctuidae). J Econ Entomol 105: 2136–2146. [DOI] [PubMed] [Google Scholar]
- 28. Zhou XF, Faktor O, Applebaum SW, Coll M (2000) Population structure of the pestiferous moth Helicoverpa armigera in the Eastern Mediterranean using RAPD analysis. Heredity 85: 251–256. [DOI] [PubMed] [Google Scholar]
- 29. Han Q, Caprio MA (2002) Temporal and spatial patterns of allelic frequencies in cotton bollworm (Lepidoptera: noctuidae). Environ Entomol 31: 462–468. [Google Scholar]
- 30. Asokan R, Nagesha S, Manamohan M, Krishnakumar N, Mahadevaswamy H, et al. (2012) Molecular diversity of Helicoverpa armigera Hübner (Noctuidae: Lepidoptera) in India. Orient Insects 46: 130–143. [Google Scholar]
- 31. Gunning RV, Dang HT, Kemp FC, Nicholson IC, Moores GD (2005) New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl Environ Microbiol 71: 2558–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X, Liang Z, Siddiqui ZA, Gong Y, Yu Z, et al. (2009) Efficient screening and breeding of Bacillus thuringiensis subsp. kurstaki for high toxicity against Spodoptera exigua and Heliothis armigera . J Ind Microbiol Biotechnol 36: 815–820. [DOI] [PubMed] [Google Scholar]
- 33. Martin T, Ochou GO, Hala-N′Klo F, Vassal J-M, Vaissayre M (2000) Pyrethroid resistance in the cotton bollworm, Helicoverpa armigera (Hübner), in West Africa. Pest Manag Sci 56: 549–554. [Google Scholar]
- 34. Achaleke J, Brevault T (2010) Inheritance and stability of pyrethroid resistance in the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) in Central Africa. Pest Manag Sci 66: 137–141. [DOI] [PubMed] [Google Scholar]
- 35. Nair R, Kalia V, Aggarwal KK, Gujar GT (2013) Variation in the cadherin gene sequence of Cry1Ac susceptible and resistant Helicoverpa armigera (Lepidoptera: Noctuidae) and the identification of mutant alleles in resistant strains. Curr Sci 104: 215. [Google Scholar]
- 36. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–299. [PubMed] [Google Scholar]
- 37.Technelysium Pty Ltd (1998–2005) Chromas lite version 2.01. Available: http://www.technelysium.com.au/chromas_lite.html.
- 38. Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series [Google Scholar]
- 39.Encyclopedia of Life. Available: http://www.eol.org. Accessed 2013 July 4.
- 40.Matten T (2002) The BLAST Sequence Analysis Tool. In: McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US). [Google Scholar]
- 41. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 42. Lopez JV, Yuhki N, Masuda R, Modi W, O′Brien SJ (1994) Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39: 174–190. [DOI] [PubMed] [Google Scholar]
- 43. Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 44. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Excoffier L, Laval G, Schneider S (2005) Arlequin v. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 47. Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to humam mitochondrial - DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- 50. Akaike H (1974) A new look at the statistical model identification. IEEE T Automat Contr 19: 716–723. [Google Scholar]
- 51.Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Uppsala University: Evolutionary Biology Centre. [Google Scholar]
- 52. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian phylogenetic tree of H. armigera and H. zea individuals sampled in Brazil. This phylogenetic tree is based on partial COI haplotype sequences and includes H. assulta and H. gelotopoeon sequences. Numbers near the interior branches indicate the posterior probability (×1,000) values. The outgroup used was Heliothis virescens. H. armigera COI haplotypes and Genbank Accession numbers can be found in Table S2.
(TIF)
Hierarchical analysis of molecular variance (AMOVA), for population genetics structure of Helicoverpa armigera and H. zea with a mithocondrial (COI) region marker.
(DOCX)
Global Helicoverpa armigera including the Brazilian H. armigera haplotypes, and relevant GenBank Accession numbers. Numbers of individuals sequenced from each locality are indicated in parentheses.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Sequences were deposited in NCBI and GenBank Accession numbers KM274936–KM275201 are listed in Table 1.