Background: Salicylic acid-based inhibitors of signal transducer and activator of transcription 3 (STAT3) are predicted to interact with the Src homology 2 (SH2) domain, inhibiting dimerization.
Results: STAT3-inhibitor interactions were interrogated by hydrogen-deuterium exchange (HDX) mass spectrometry (MS).
Conclusion: HDX MS revealed local and global perturbations in STAT3 dynamics.
Significance: Understanding STAT3 inhibitor interactions with the SH2 domain is critical to inhibitor development.
Keywords: Allosteric Regulation, Hydrogen Exchange Mass Spectrometry, Hydrogen-Deuterium Exchange, Mass Spectrometry (MS), Protein Dynamic, Signal Transducers and Activators of Transcription 1 (STAT1), Dimerization Inhibitor, Signal Transducers and Activators of Transcription 3
Abstract
The activity of the transcription factor signal transducer and activator of transcription 3 (STAT3) is dysregulated in a number of hematological and solid malignancies. Development of pharmacological STAT3 Src homology 2 (SH2) domain interaction inhibitors holds great promise for cancer therapy, and a novel class of salicylic acid-based STAT3 dimerization inhibitors that includes orally bioavailable drug candidates has been recently developed. The compounds SF-1-066 and BP-1-102 are predicted to bind to the STAT3 SH2 domain. However, given the highly unstructured and dynamic nature of the SH2 domain, experimental confirmation of this prediction was elusive. We have interrogated the protein-ligand interaction of STAT3 with these small molecule inhibitors by means of time-resolved electrospray ionization hydrogen-deuterium exchange mass spectrometry. Analysis of site-specific evolution of deuterium uptake induced by the complexation of STAT3 with SF-1-066 or BP-1-102 under physiological conditions enabled the mapping of the in silico predicted inhibitor binding site to the STAT3 SH2 domain. The binding of both inhibitors to the SH2 domain resulted in significant local decreases in dynamics, consistent with solvent exclusion at the inhibitor binding site and increased rigidity of the inhibitor-complexed SH2 domain. Interestingly, inhibitor binding induced hot spots of allosteric perturbations outside of the SH2 domain, manifesting mainly as increased deuterium uptake, in regions of STAT3 important for DNA binding and nuclear localization.
Introduction
Signal transducer and activator of transcription 3 (STAT3), an Src homology 2 (SH2)4 domain-containing protein (molecular weight, 88,000), mediates intracellular signaling downstream of a number of cytokine and growth factor receptors and is involved in regulating cell proliferation, survival, differentiation, angiogenesis, cell migration, and inflammatory signaling (1). Dysregulated STAT3 activity promotes the progression of a multitude of hematological and solid malignancies (2). STAT3 is a transcription factor whose canonical activation pathway is modulated by tyrosine phosphorylation. Phosphorylation at Tyr-705 drives STAT3 homodimerization or heterodimerization with other STAT family members mediated by its SH2 domain (3), which binds the phosphotyrosine peptide with nanomolar affinity (4). Inhibition of SH2 domain function is thus a robust strategy to antagonize its biological activity by inhibiting STAT3 dimerization (5).
The SH2 domain is a structurally conserved feature of many intracellular signaling transducers and is capable of recognizing and binding to phosphorylated tyrosine residues presented in the context of specific protein sequences (6). Comprising minimal secondary structure, the SH2 domain and the phosphopeptide binding interface are highly unstructured and dynamic (7), a property that makes the development of small molecule inhibitors targeted at this important domain class a challenge. Gunning and co-workers (8, 9) have recently developed a number of pharmacological, orally bioavailable inhibitors of STAT3 based on the salicylic acid pharmacophore through a series of quantitative structure-activity relationship studies. Two such inhibitors, SF-1-066 and BP-1-102, demonstrate an IC50 of 35 and 19.7 μm, respectively, for the inhibition of STAT3 DNA binding activity in vitro and inhibit STAT3 dimerization (10). Additionally, BP-1-102 has demonstrated potent antitumor effects in vivo in human breast and lung cancer xenograft studies in mice (8). Given the demonstration that both SF-1-066 and BP-1-102 inhibit the binding of the phosphotyrosine peptide by the SH2 domain of STAT3 (8, 11), a proposed mechanism of action involves inhibitor binding to the SH2 domain that disrupts its function or otherwise blocks the binding of the phosphopeptide regulatory region. In silico docking studies identified a putative binding site for the two inhibitors comprising all three phosphopeptide-binding subpockets of the SH2 domain (10), supporting this mechanism. However, little experimental evidence exists corroborating that the putative binding site of SF-1-066 and BP-1-102 is indeed in the SH2 domain, and the mechanism of action of these molecules is not yet fully understood.
Although a number of x-ray crystal structures of STAT3 are available (12, 13), including that of the phosphorylated STAT3 dimer (14), derivation of x-ray crystal structures of STAT3 complexed with SF-1-066 and BP-1-102 has proven exceptionally challenging, and to date, no inhibitor-STAT3 structure has been resolved. Unlike these static images, dynamic representations of protein structures can offer unparalleled mechanistic insights into biological processes and interactions by depicting the time evolution of protein conformational ensembles (15). Hydrogen-deuterium exchange (HDX) mass spectrometry (MS) when coupled with microfluidic sample processing enables the interrogation of protein dynamics of the native protein structures in solution, permitting the in situ observation of enzyme-catalyzed reactions (16) and protein-ligand interactions (17). HDX implemented on a microfluidic device for rapid mixing, quenching, and proteolytic digestion of the protein analyte offers substantial advantages over other techniques, including easy implementation, unlimited protein analyte size, time scales compatible with protein breathing motions, and site-specific resolution of up to a few amino acids (18).
We have applied a time-resolved electrospray ionization MS HDX (TRESI-MS/HDX) approach to the study of protein-ligand interactions involving the highly unstructured SH2 ligand-binding domain in the context of the near full-length STAT3 protein. TRESI-MS/HDX is particularly useful for interrogating weakly structured protein regions where short deuterium labeling pulses are essential for accurately assessing the magnitudes of dynamic perturbations (18). Given that weakly structured regions experience moderate protection from exchange, conformational changes induced by ligand binding to such regions can be readily detected by TRESI-MS/HDX with a heightened degree of sensitivity. In the present work, TRESI-MS/HDX was applied to (i) experimentally identify the STAT3 binding site of the salicylic acid-based inhibitors SF-1-066 and BP-1-102 and (ii) probe the changes in protein dynamics induced in STAT3 upon complexation with these inhibitors.
EXPERIMENTAL PROCEDURES
Reagents
SF-1-066 and BP-1-102 were synthesized as described previously (10) and were dissolved in DMSO. Deuterium oxide (D2O; 99.99%), ammonium acetate (99.99%), acetic acid (99.7%), and pepsin-agarose beads were purchased form Sigma-Aldrich. Dialysis cassettes (30,000 molecular weight cutoff) were purchased from Fisher Scientific. Vivaspin 20 (30,000 molecular weight cutoff) columns were purchased from GE Healthcare.
Cloning, Expression, and Purification of the STAT3 Fusion Protein
Plasmid containing mouse His-tagged STAT3 was provided by Dr. Rob C. Laister (Toronto, Ontario, Canada). STAT3, residues 127–688 (100% sequence identity on the protein level between mouse and human), was excised with NdeI and BamHI and subcloned directly into the pMAL-c5X plasmid digested with the same restriction enzymes to generate an N-terminal maltose-binding protein (MBP)-tagged fusion of STAT3. The resulting pMAL-c5X-STAT3 plasmid was transformed into Escherichia coli BL21(DE3) for inducible expression. The resulting MBP-human STAT3 fusion was purified by affinity chromatography on amylose resin (New England BioLabs Inc., Ipswitch, MA). Purified STAT3 was further concentrated to 80 μm using the Vivaspin 20 filtration column and buffer-exchanged into 100 mm ammonium acetate, pH 7.4 using a 30,000 molecular weight cutoff dialysis cassette. Purity was confirmed by SDS-PAGE and absorbance at 280 nm to be above 95%.
Microfluidic Device Construction
The microfluidic device for the HDX workup was constructed as described previously (18). Briefly, a standard poly(methyl methacrylate) block (8.9 × 3.8 × 0.6 cm) was laser-ablated to incorporate two input channels for introduction of reactants, a proteolytic digestion chamber, and a third output channel using the VersaLaser engraver (Universal Laser, Scottsdale, AZ). The rapid mixing device, described previously (19), was embedded into the first input channel to supply the protein solution and D2O for labeling. The second channel supplied the quenching solution (5% acetic acid, pH 2.4). A proteolytic digestion chamber was edged in a rectangular shape (30 × 5 × 0.05 mm) onto the device and filled with pepsin-agarose beads (average diameter, 20–50 μm). The output channel fed directly into the MS. A blank poly(methyl methacrylate) block was used as a cover to seal the device, lined with a silicon-rubber gasket for a liquid-tight seal. A custom-built pressure clamp was used to secure the device seal (LAC Machine and Tooling Ltd., Toronto, Ontario, Canada). Metal capillaries were melted into the etched poly(methyl methacrylate) chip to connect automated supply syringes to the device and provide an outlet for direct coupling to the MS. Reactants were supplied into the device with gas-tight Hamilton syringes through polytetrafluoroethylene tubing using automated infusion pumps (Harvard Apparatus, Holliston, MA).
Hydrogen-Deuterium Exchange Mass Spectrometry
HDX experiments were carried out at 20.5 °C in triplicate with an 80 μm solution of STAT3 in 100 mm ammonium acetate buffer, pH 7.4. STAT3 was preincubated with 400 μm SF-1-066, 400 μm BP-1-102, or a DMSO-only blank control for 2 h on ice. The DMSO-only blank was used to account for the potential impact of residual DMSO (5%, v/v) from inhibitor solutions on the HDX reaction.
The microfluidic device was directly coupled to the front end of a modified QStar Elite hybrid quadrupole time-of-flight mass spectrometer (AB Sciex, Framingham, MA). A bypass switch was introduced to simulate the presence of a commercial ESI source. STAT3 solutions were infused through a polyamide-coated glass capillary into the rapid mixing device at a flow rate of 1 μl min−1. Deuterium was supplied through the outer metal capillary at a flow rate of 3 μl min−1. The resulting maximum deuteration was 75%. Deuteration pulse length was controlled by the withdrawal of the inner capillary and was calculated by measuring the position of the inner capillary in reference to the outer capillary in the rapid mixing device. Pulse lengths ranged from 179 ms up to 7.56 s. 5% acetic acid solution, pH 2.4 at 20.5 °C, was supplied to quench the exchange through the second input channel at a flow rate of 15 μl min−1. ESI-MS acquisition was carried out in a positive ion mode with an electrospray voltage of +4700 to +4900 V, 60-V declustering potential, and 250-V focusing potential. Spectra were acquired over the range of 300–1600 m/z with a scanning rate of 1 s−1.
In Silico Docking of SF-1-066 and BP-1-102 to STAT3 SH2 Domain Crystal Structure
Structures of SF-1-066 and BP-1-102 were docked into the STAT3 SH2 domain structure (derived from Protein Data Bank code 1BG1) using the GOLD software package (Cambridge Crystallographic Data Centre, Cambridge, UK) as described previously (9).
Data and Statistical Analyses
All MS spectra analyses were performed using mMass software, version 5.5 (20). Peptic peptide identification was performed using a FindPept tool on the ExPASy proteomic server (Swiss Institute of Bioinformatics, Basel, Switzerland) and confirmed by collision-induced dissociation where needed. Deuterium incorporation was computed using software for isotopic distribution analysis developed in house and was normalized to the maximum deuterium uptake of 75%. HDX data were fitted using single exponential non-linear regression and normalized in SigmaPlot (Systat Software, San Jose, CA). Protein structures were rendered using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC).
RESULTS
TRESI-MS Analyses of STAT3 and STAT3·Inhibitor Complexes
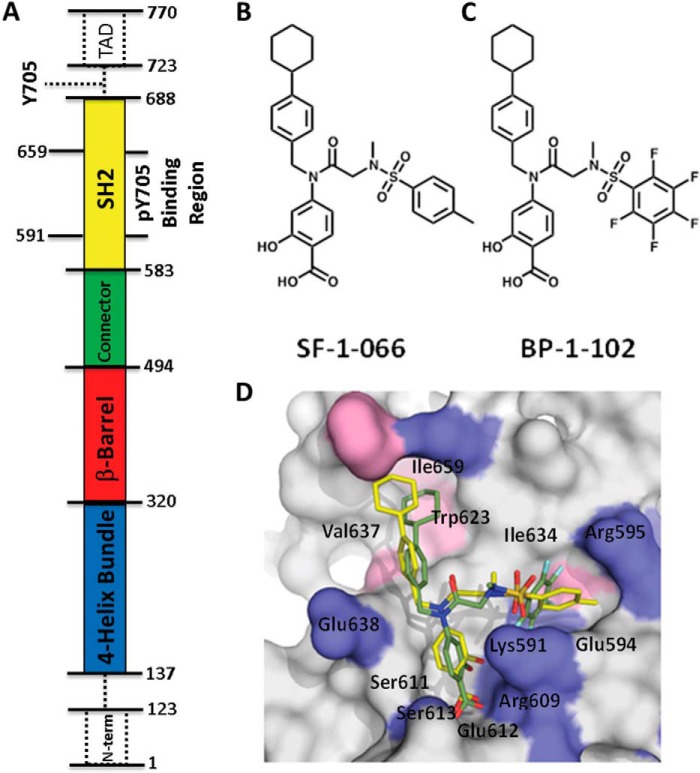
A slightly truncated construct of STAT3 (Fig. 1A) was preincubated with a 400 μm concentration or a 1:5 molar excess of SF-1-066 or BP-1-102 inhibitor (inhibitor structures are shown in Fig. 1, B and C). This concentration was expected to have saturated STAT3 given the experimentally determined Kd values for SF-1-066 and BP-1-102 of 2.74 μm and 504 nm, respectively (8, 11). The STAT3 construct utilized in this study is devoid of the Tyr-705 necessary for STAT3 dimerization (13), ensuring that the protein remains as a monomer in solution and that any dynamic changes observed are not as a result of intermolecular interactions. The construct does retain the full SH2 domain containing the Tyr(P) binding site, however. Fig. 1D shows how SF-1-066 (yellow) and BP-1-102 (green) are predicted to bind to the SH2 domain of STAT3 based on in silico docking simulations. In these models, residues Lys-591, Arg-609, Ser-611, and Ser-613 are predicted to make direct contact with the Tyr(P)-mimetic portion of the scaffold, ortho-hydroxybenzoic acid, via hydrogen bonding and electrostatic interactions in the Tyr(P)-binding subpocket. The hydrophobic cyclohexylbenzyl moiety is predicted to form van der Waals contacts with the distal, hydrophobic subpocket of the SH2 domain lined by residues Trp-623, Val-637, and Ile-659. The sulfonamide-substituted group of the inhibitors is predicted to access an additional subpocket of the SH2 domain and is predicted to interact with Lys-591, Glu-594, Arg-595, and Ile-634 (8, 10).
FIGURE 1.
Domain structure of STAT3 and structures of the salicylic acid-based inhibitors with in silico predicted binding configuration. A, the domain structure of STAT3 is shown oriented as in later figures. Dotted lines and boxes indicate regions not included in the construct, including Tyr-705, which upon phosphorylation is the reciprocal binding partner of the SH2 domain in STAT3 homodimerization, and the transcriptional activator domain (TAD). B, SF-1-066 structure. C, BP-1-102 structure. D, representation of the results of in silico docking of SF-1066 (yellow) and BP-1-102 (green) inhibitors to the phosphotyrosine peptide-binding pocket of the STAT3 SH2 domain. Key contact residues at the SH2 domain surface are indicated.
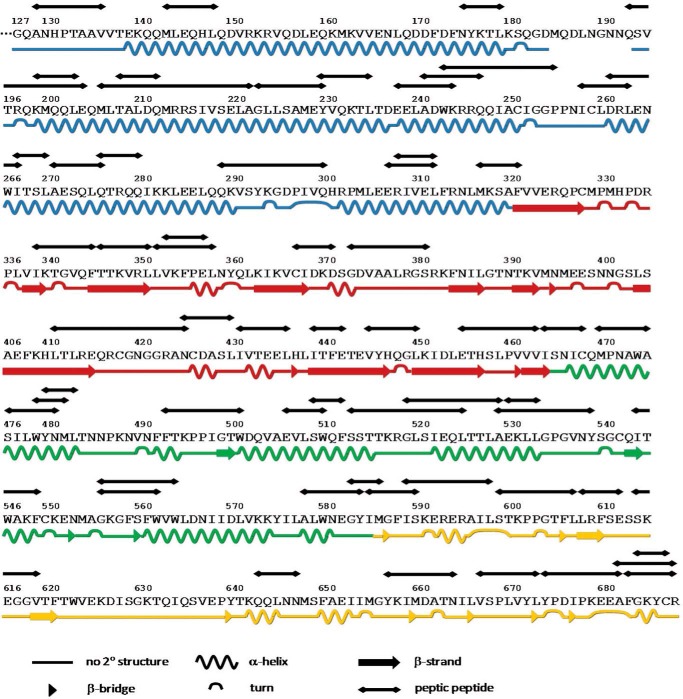
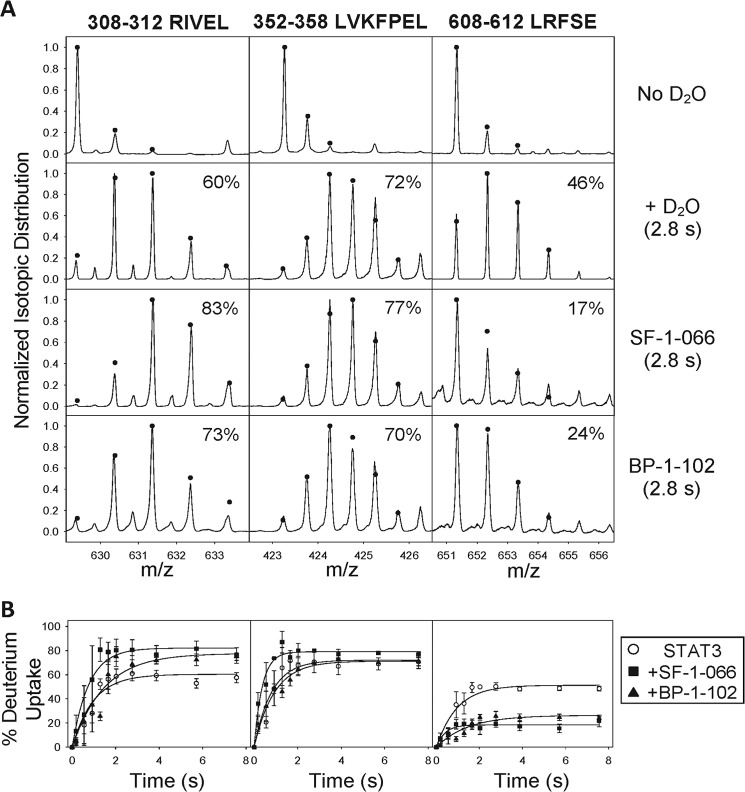
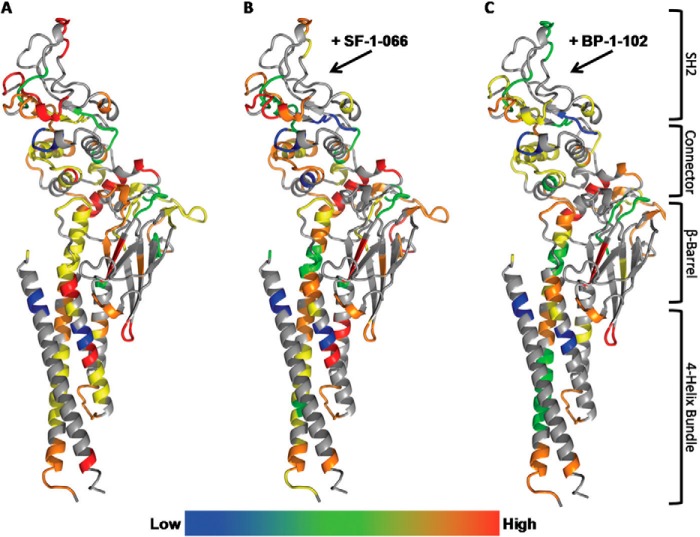
TRESI-HDX was carried out on a microfluidic device that facilitates variable HDX labeling times from 179 ms to 7.56 s with on-chip digestion and electrospray ionization. Digestion of STAT3 was reasonably efficient in our system, resulting in sequence coverage of 70% corresponding to 62 unique peptides and an average spatial resolution of 7.3 amino acids (Fig. 2). Typical HDX MS data are depicted in Fig. 3A for STAT3 peptides that experienced no change in deuteration, increased deuteration, or decreased deuteration as a result of complexation with SF-1-066 and BP-1-102 STAT3 inhibitors. Kinetic profiles depicting the change in peptide deuteration as a function of labeling time are shown in Fig. 3B. Kinetic profiles of all 62 peptides are depicted in supplemental Fig. S1. Data were normalized fit to a single exponential, and the amplitude parameter was used to calculate deuterium uptake. In Fig. 4, deuterium uptake is mapped onto the crystal structure of STAT3 (Protein Data Bank code 1BG1), giving a broad picture of the dynamic nature of the free SH2 domain and the changes in dynamics that occur upon complexation with inhibitors (Fig. 4, A–C).
FIGURE 2.
Primary protein sequence and secondary structure assignment (based on the Dictionary of Protein Secondary Structure assignment (34)) of the human STAT3 (Protein Data Bank code 1BG1) segment of the MBP-STAT3 fusion protein. All 62 peptic peptide resolved and analyzed in the HDX experiments are indicated above the primary sequence. Total STAT3 sequence coverage is 70%.
FIGURE 3.
Analysis of site-specific HDX. A, representative spectra of peptic peptides derived from STAT3 for non-deuterated STAT3 (top row), STAT3 labeled with a 2.8-s HDX pulse (second row), and STAT3 complexed with SF-1-066 (third row) or BP-1-102 (bottom row) labeled by a 2.8-s HDX pulse (bottom row). Raw spectra depict the observed shifts in isotopic distribution following HDX. Peptides that exhibited an increase (left column), a decrease (right column), or no change (middle column) in deuterium uptake are represented. Percent deuterium uptake is indicated on each spectrum. B, normalized HDX kinetic plots of representative peptides in A for the free STAT3 (open circles) and STAT3 complexed with SF-1-066 (filled squares) or BP-1-102 (filled triangles). Data represent an average of triplicate runs (error bars represent S.E.).
FIGURE 4.
Average deuterium uptake mapped and color-coded onto the crystal structure of STAT3 (Protein Data Bank code 1BG1) for free STAT3 (A), STAT3 complexed with SF-1-066 (B), and STAT3 complexed with BP1–102 (C). STAT3 protein is depicted as a stick representation colored by element. Low (blue shading) represents 0–20% deuterium uptake followed by green (20–40%), yellow (40–60%), orange (60–80%), and then High (red shading) representing 80–100% deuterium uptake. The domain organization of STAT3 is mapped on the right. The arrows point to the region demonstrated to bind the inhibitors in this study.
Mapping the Inhibitor Binding Site in STAT3
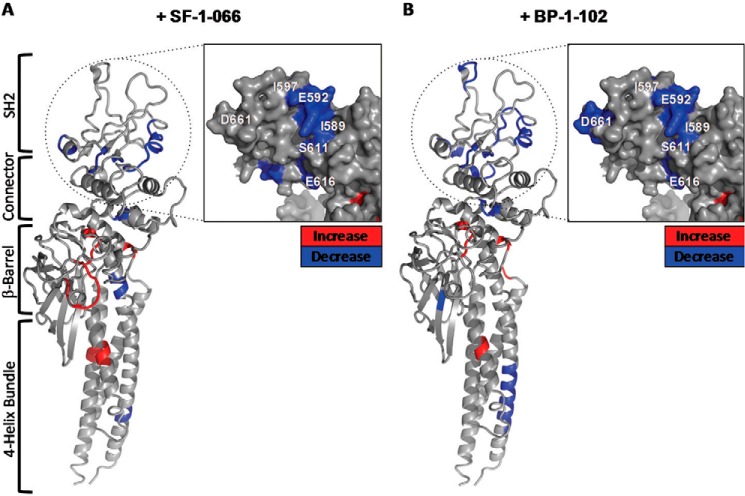
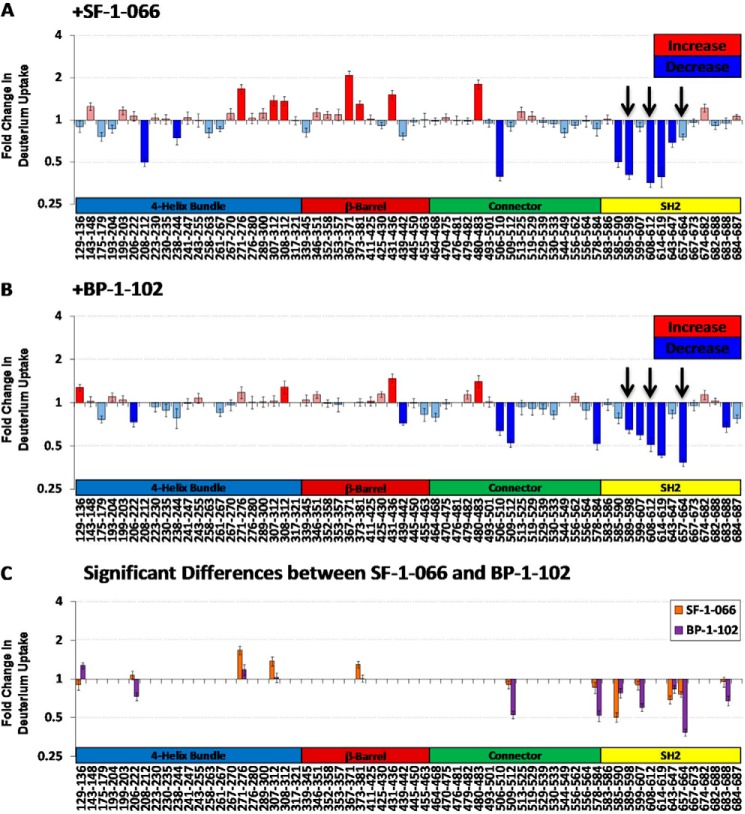
Mapping the -fold change in deuterium uptake for inhibitor-bound versus free STAT3 to the crystal structure (Fig. 5, A and B) enables the visualization of regions exhibiting pronounced changes in local backbone dynamics. Decreases in deuterium uptake may indicate the steric exclusion of solvent molecules or alternatively reorganization of the local backbone hydrogen bonding network, resulting in a net increase in hydrogen bonding. Increases in deuterium uptake typically result from a disruption of secondary structure or increased local accessibility to solvent. The STAT3 SH2 domain (Fig. 5, A and B, insets) exclusively experienced significant, large decreases in deuterium uptake upon complexation with both SF-1-066 and BP-1-102 inhibitors in five and six unique peptides, respectively. Fig. 6, A and B, map along the STAT3 primary sequence for each peptic peptide the -fold change in deuterium uptake in the inhibitor-complexed STAT3 relative to STAT3 without inhibitor for SF-1-066 and BP-1-102, respectively. Fig. 6, A and B, also clearly highlight the clustering of large decreases in deuterium uptake within the STAT3 SH2 domain upon complexation of both inhibitors.
FIGURE 5.
-Fold changes in deuterium uptake from free STAT3 are shown for STAT3 complexed with SF-1-066 (A) and STAT3 complexed with BP1–102 (B) are shown. Hot spots of differential deuterium uptake mapped onto the crystal structure of STAT3 (Protein Data Bank code 1BG1) are color-coded as decreases (blue shading) or increases (red shading) in uptake. Insets show a magnified view of the SH2 domain depicted from the face of the predicted inhibitor-binding pocket. The domain organization of STAT3 is mapped on the left.
FIGURE 6.
-Fold changes in deuterium uptake shown for all peptic peptides comprising the STAT3 primary sequence coverage in the HDX experiments. -Fold changes in deuterium uptake from free STAT3 are shown for STAT3 complexed with SF-1-066 (A) and STAT3 complexed with BP1–102 (B). Arrows denote regions of the binding pocket in closest contact with the inhibitors in the in silico model. C, significant differences in relative deuterium uptake between the STAT3·SF-1-066 (orange) and STAT3·BP-1-102 (purple) complexes are highlighted. Data represent an average of triplicate runs (error bars represent S.E.).
This decrease is pronounced in those regions of the Tyr(P)-binding pocket that are in closest proximity to inhibitor functional groups in the in silico model. Peptides ISKERERAIL (589–598), LRFSE (608–612), and YKIMDATN (657–664) for instance that line the surface of the putative inhibitor-binding pocket all exhibited a 2–3-fold reduction in deuterium uptake when inhibitors were present (Fig. 6, A and B, arrows). This observation is consistent with the occlusion of the SH2 phosphotyrosine-binding subpockets where the inhibitors are predicted to bind, resulting in decreased solvent accessibility.
Allosteric Changes Induced in STAT3 by the SH2 Dimerization Inhibitors
Multiple peptides from outside of the SH2 domain exhibited pronounced changes in relative deuterium uptake upon complexation with inhibitor (Figs. 5, A and B, and 6, A and B). The majority of these conformational disturbances manifested as significant increases in deuterium uptake and were largely localized to regions of α-helical secondary structure. The profile of allosteric differences induced by SF-1-066 versus BP-1-102 was somewhat different. Fig. 6C highlights the regions that exhibited relative changes in HDX that were significantly different between the SF-1-066·STAT3 (orange) and BP-1-102·STAT3 (purple) complexes.
DISCUSSION
This study highlights the utility of structural mass spectrometry based on TRESI-MS/HDX in probing protein-ligand interactions and understanding dynamic changes and mechanisms associated with ligand binding. The “bottom-up” HDX approach as implemented here enables one to generate accurate, subsecond time scale kinetic profiles of site-specific exchange in the native protein structures. This structural technique is particularly useful for interrogating protein domains with minimal secondary structure or a high degree of intrinsic disorder. Although NMR has been used to probe disordered proteins to a degree, it is highly limited by protein size in applications involving protein-ligand interactions (15, 21).
Here we applied the microfluidics-integrated TRESI-MS/HDX workflow to derive averaged dynamic structures of free and inhibitor-bound STAT3. Our examination of the dynamic changes in this protein largely confirmed in silico predictions with respect to the inhibitor binding regions as well as revealed additional regions inside the STAT3 SH2 domain that exhibit strong protection from exchange due to the interaction with the inhibitors. SF-1-066 and BP-1-102 represent a novel, state-of-the-art class of salicylic acid-based SH2-targeted STAT3 inhibitors with low micromolar and nanomolar range affinity for STAT3, respectively; however, no experimental evidence directly describes their interaction with the SH2 domain (8, 10).
It has been proposed that these inhibitors bind to the SH2 domain of STAT3 and preclude the binding of its ligand, the phospho-Tyr705-containing peptide of another STAT3 molecule, which in turn inhibits the ability of STAT3 to dimerize. This mechanism is supported by the observation that both SF-1-066 and BP-1-102 can inhibit the binding of phospho-Tyr-705-containing peptide to STAT3 in vitro (8, 11) and inhibit the biological activity of STAT3 both in vitro and in vivo. It is unclear whether the ability of these compounds to inhibit phosphopeptide binding is a consequence of their direct complexation with the SH2 domain and competitive inhibition as predicted by in silico docking studies (10) or a consequence of their interaction with other regions of the STAT3 protein. Our data for the first time provide an experimental proof that the SF-1-066 and BP-1-102 compounds bind exclusively to the SH2 domain on the STAT3 molecule and complete the evidence set supporting the originally proposed mechanism for their biological activity. Additionally, our data reveal a series of increases in HDX outside the STAT3 SH2 domain due to inhibitor binding, suggesting allosteric effects that are potentially destabilizing to the protein.
Peptides ISKERERAIL (589–598), LRFSE (608–612), and YKIMDATN (657–664), which exhibited significant decreases in deuterium uptake upon inhibitor binding, map to the surface of the inhibitor-binding pocket predicted by docking studies (Fig. 6, A and B). The side chain of Arg-609, mapping near the peptide LRFSE (608–612), was predicted to hydrogen bond with the salicylic acid moiety common to both inhibitors, greatly stabilizing the binding energy of the molecules (8, 22). This is consistent with large decreases in backbone deuteration observed in the LRFSE (608–612) peptide given the tight predicted occupancy of this subpocket by the salicylic acid moiety for both inhibitors.
The peptide YKIMDATN (657–664), lining the cyclohexylbenzyl-binding subpocket, only experienced a significant decrease in deuterium uptake in the case of the higher affinity inhibitor BP-1-102 (Fig. 6, A and B). The binding of the cyclohexylbenzyl moiety to the predicted subpocket is dominated by relatively weak van der Waals interactions (10). This observation with respect to the YKIMDATN (657–664) peptide, which lines the deepest end of the cyclohexylbenzyl subpocket, is consistent with the tighter binding of the cyclohexylbenzyl moiety of BP-1-102 compared with SF-1-066 as predicted by in silico docking (Fig. 1D). The only structural difference between the two inhibitors is the substituent at the sulfonamide end of the molecule, a tosyl sulfonamide for SF-1-066 and a pentafluorobenzyl sulfonamide for BP-1-102. Despite this, in the peptide lining the relevant subpocket, ISKERERAIL (589–598), a significant but similar decrease in deuteration was observed that is not reflective of any differential effect on the backbone amides in this region.
Other regions of the SH2 domain not immediately lining the surface of the predicted inhibitor-binding site also exhibited significant decreases in deuterium uptake upon inhibitor binding. Some of these segments interact directly with specific regions of the binding pocket. For example, the peptide SKEGGV (614–619) is located in proximity to LRFSE (608–612), which lines the salicylic acid moiety-binding subpocket, with the backbone amide of the Ser-611 making a hydrogen bond with the backbone carbonyl of Gly-618. Both inhibitors induce a significant decrease in deuterium uptake in the SKEGGV (614–619) peptide.
Overall, the higher affinity inhibitor, BP-1-102, induced more decreases in deuterium uptake compared with SF-1-066 (Fig. 6C). This and other observations of significant decreases in deuterium uptake in the SH2 domain suggest that either (i) the inhibitors are able to bind to regions of the SH2 domain outside of the in silico predicted binding pocket or (ii) the binding of the inhibitors to the predicted binding pocket results in an overall stabilization of and possibly the induction of additional secondary structure in a large portion of the SH2 domain. We tend to favor the latter possibility given the lack of clustered HDX decreases elsewhere in the molecule outside of the SH2 domain that would point to nonspecific binding by the inhibitors to STAT3.
Our data suggest that the core of the STAT3 SH2 domain represents a highly dynamic network potentially able to transmit local perturbations in dynamics to distal regions of the domain. Indeed, studies of dynamics of SH2 domains of other proteins reveal a general, global reduction or stabilization in dynamics in a large portion of the SH2 domain upon peptide ligand binding, resulting in increased rigidity that propagates throughout the entire domain (23, 24). Overall, the clustering of significant decreases in deuterium uptake in the SH2 domain of inhibitor-complexed STAT3 provides experimental evidence supporting the SH2 domain being the target site of the salicylic acid-based inhibitors SF-1-066 and BP-1-102. This finding corroborates in silico docking predictions as well as other experimental evidence. In fluorescence polarization assays with STAT3 and a fluorescently labeled phosphotyrosine peptide (5-carboxyfluorescein-GpYLPQTV-NH2 where pY is phosphotyrosine) that mimics the natural SH2 domain target substrate, both SF-1-066 and BP-1-102 inhibitors competed strongly with substrate binding with an IC50 of 20 and 4.1 μm, respectively (8, 11).
Interestingly, both inhibitors induced a burst of dynamic changes, mainly manifesting as significant relative increases in deuterium uptake, outside of the SH2 domain of STAT3 (Fig. 6, A and B). These allosteric changes induced in the inhibitor-complexed structures propagate into STAT3 domains and regions involved in DNA binding (β-barrel and connector domains) and nuclear localization of the protein (four-helix bundle and β-barrel domains) (14, 25). Interestingly, the majority of allosteric changes mapped to (or near) regions important to the DNA binding activity of STAT3 (DNA-binding domain, residues 406–514) (26). The backbone amide of Val-432 for instance is directly involved in DNA binding, and Glu-435 forms a hydrogen bond that is important for maintaining the structural rigidity of one of the DNA binding loops (14). We observed a significant increase in deuterium uptake in the peptide IVTEEL (431–436) with both inhibitors. This suggests the possibility that the salicylic acid-based inhibitors SF-1-066 and BP-1-102 could also be acting via an allosteric mechanism to modulate non-canonical DNA binding of STAT3, a prediction that will need to be explored experimentally with the use of STAT3 mutants.
The vast majority of allosteric changes induced by inhibitor binding (Fig. 6, A and B) mapped to regions predicted to have α-helical secondary structure (Fig. 2). Increases in HDX in α-helical regions most likely result from the loss or remodeling of secondary structure, affecting the hydrogen bonding network. STAT3 complexed with SF-1-066 exhibited more allosteric effects (Fig. 6C) compared with the tighter bound BP-1-102 complex, which may reflect the need for a degree of conformational freedom in the SH2 domain to efficiently “transmit” the allosteric signal. Nevertheless, this observation is qualitative and speculative and would necessitate further investigation. Allosteric changes were not observed in any peptides derived from the MBP protein that was fused via a polyasparagine linker to the N terminus of the STAT3 in our construct (data not shown). This confirms that changes induced by the binding of SF-1-066 and BP-1-102 inhibitors to STAT3 outside the SH2 domain are not a result of experimental error.
Allosteric modulation is an important mechanism of communication between distinct domains within the protein, and exactly how the information is transmitted between domains is an area of active investigation (27). The ability of the SH2 domain to induce conformational changes in other domains in the same protein has been demonstrated for the C-terminal Src kinase (Csk) (28), a protein-tyrosine kinase that phosphorylates other kinases of the Src family. In Csk, the binding of the SH2 domain target peptide, Csk-binding protein, appears to induce conformational changes that are transmitted through the SH2 linker to the active site and other regions of the protein. Conversely, binding of Csk nucleotide substrates to the kinase domain was shown to impact HDX protection in other regions of the protein, including the SH2 domain, depending on the nature of the nucleotide present (29). Our study adds to the work of others examining allosteric modulation by HDX MS (30–32), highlighting the vast utility of this structural technique in probing the evolution of allosteric changes in intact proteins under physiological conditions.
In summary, in silico predicted interactions of SF-1-066 and BP-1-102 inhibitors with regions of the STAT3 SH2 domain were experimentally confirmed by analyzing changes in site-specific deuterium uptake upon STAT3 complexation with the inhibitors. Showing a large contiguous decrease in deuterium uptake around the putative SH2 domain-binding site, our data support a mechanism of inhibition driven by “pacification” of the SH2 domain through complexation. This compliments prior biochemical evidence that points to disruption of STAT3 dimerization as the primary inhibitory mechanism. However, inhibitor binding was also found to induce substantial changes in the dynamics of the DNA-binding and nuclear localization domains, which could reflect additional modes of inhibition targeting non-canonical activation pathways (12, 33). Our study highlights the incremental benefits of dynamic structure analysis to understanding protein-ligand interactions via the examination of conformational ensembles rather than static representations of the protein complex structures.
Supplementary Material
Acknowledgment
We thank Dr. Rob C. Laister, Princess Margaret Cancer Center (Toronto, Ontario, Canada), for generously providing the STAT3 cDNA.
This work was supported in part by National Sciences and Engineering Research Council (NSERC) Discovery Grant 257588 and an Ontario Ministry of Research and Innovation early researcher award (to D. J. W.).

This article contains supplemental Fig. S1.
- SH2
- Src homology 2
- HDX
- hydrogen-deuterium exchange
- TRESI
- time-resolved electrospray ionization
- MBP
- maltose-binding protein
- Csk
- C-terminal Src kinase.
REFERENCES
- 1. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bromberg J., Darnell J. E., Jr. (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19, 2468–2473 [DOI] [PubMed] [Google Scholar]
- 3. Haan S., Hemmann U., Hassiepen U., Schaper F., Schneider-Mergener J., Wollmer A., Heinrich P. C., Grötzinger J. (1999) Characterization and binding specificity of the monomeric STAT3-SH2 domain. J. Biol. Chem. 274, 1342–1348 [DOI] [PubMed] [Google Scholar]
- 4. Ladbury J. E., Lemmon M. A., Zhou M., Green J., Botfield M. C., Schlessinger J. (1995) Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl. Acad. Sci. U.S.A. 92, 3199–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Page B. D., Ball D. P., Gunning P. T. (2011) Signal transducer and activator of transcription 3 inhibitors: a patent review. Expert Opin. Ther. Pat. 21, 65–83 [DOI] [PubMed] [Google Scholar]
- 6. Moran M. F., Koch C. A., Anderson D., Ellis C., England L., Martin G. S., Pawson T. (1990) Src homology region 2 domains direct protein-protein interactions in signal transduction. Proc. Natl. Acad. Sci. U.S.A. 87, 8622–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finerty P. J., Jr., Mittermaier A. K., Muhandiram R., Kay L. E., Forman-Kay J. D. (2005) NMR dynamics-derived insights into the binding properties of a peptide interacting with an SH2 domain. Biochemistry 44, 694–703 [DOI] [PubMed] [Google Scholar]
- 8. Zhang X., Yue P., Page B. D., Li T., Zhao W., Namanja A. T., Paladino D., Zhao J., Chen Y., Gunning P. T., Turkson J. (2012) Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. U.S.A. 109, 9623–9628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fletcher S., Page B. D., Zhang X., Yue P., Li Z. H., Sharmeen S., Singh J., Zhao W., Schimmer A. D., Trudel S., Turkson J., Gunning P. T. (2011) Antagonism of the Stat3-Stat3 protein dimer with salicylic acid based small molecules. ChemMedChem 6, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Page B. D., Fletcher S., Yue P., Li Z., Zhang X., Sharmeen S., Datti A., Wrana J. L., Trudel S., Schimmer A. D., Turkson J., Gunning P. T. (2011) Identification of a non-phosphorylated, cell permeable, small molecule ligand for the Stat3 SH2 domain. Bioorg. Med. Chem. Lett. 21, 5605–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X., Yue P., Fletcher S., Zhao W., Gunning P. T., Turkson J. (2010) A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem. Pharmacol. 79, 1398–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nkansah E., Shah R., Collie G. W., Parkinson G. N., Palmer J., Rahman K. M., Bui T. T., Drake A. F., Husby J., Neidle S., Zinzalla G., Thurston D. E., Wilderspin A. F. (2013) Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and x-ray crystallography. FEBS Lett. 587, 833–839 [DOI] [PubMed] [Google Scholar]
- 13. Ren Z., Mao X., Mertens C., Krishnaraj R., Qin J., Mandal P. K., Romanowski M. J., McMurray J. S., Chen X. (2008) Crystal structure of unphosphorylated STAT3 core fragment. Biochem. Biophys. Res. Commun. 374, 1–5 [DOI] [PubMed] [Google Scholar]
- 14. Becker S., Groner B., Müller C. W. (1998) Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394, 145–151 [DOI] [PubMed] [Google Scholar]
- 15. Resetca D., Wilson D. J. (2013) Characterizing rapid, activity-linked conformational transitions in proteins via sub-second hydrogen deuterium exchange mass spectrometry. FEBS J. 280, 5616–5625 [DOI] [PubMed] [Google Scholar]
- 16. Liuni P., Jeganathan A., Wilson D. J. (2012) Conformer selection and intensified dynamics during catalytic turnover in chymotrypsin. Angew. Chem. Int. Ed. Engl. 51, 9666–9669 [DOI] [PubMed] [Google Scholar]
- 17. Rob T., Gill P. K., Golemi-Kotra D., Wilson D. J. (2013) An electrospray ms-coupled microfluidic device for sub-second hydrogen/deuterium exchange pulse-labelling reveals allosteric effects in enzyme inhibition. Lab. Chip 13, 2528–2532 [DOI] [PubMed] [Google Scholar]
- 18. Rob T., Liuni P., Gill P. K., Zhu S., Balachandran N., Berti P. J., Wilson D. J. (2012) Measuring dynamics in weakly structured regions of proteins using microfluidics-enabled subsecond H/D exchange mass spectrometry. Anal. Chem. 84, 3771–3779 [DOI] [PubMed] [Google Scholar]
- 19. Wilson D. J., Konermann L. (2003) A capillary mixer with adjustable reaction chamber volume for millisecond time-resolved studies by electrospray mass spectrometry. Anal. Chem. 75, 6408–6414 [DOI] [PubMed] [Google Scholar]
- 20. Strohalm M., Kavan D., Novák P., Volný M., Havlícek V. (2010) mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 82, 4648–4651 [DOI] [PubMed] [Google Scholar]
- 21. Montelione G. T., Zheng D., Huang Y. J., Gunsalus K. C., Szyperski T. (2000) Protein NMR spectroscopy in structural genomics. Nat. Struct. Biol. 7, (suppl.) 982–985 [DOI] [PubMed] [Google Scholar]
- 22. Fletcher S., Singh J., Zhang X., Yue P., Page B. D., Sharmeen S., Shahani V. M., Zhao W., Schimmer A. D., Turkson J., Gunning P. T. (2009) Disruption of transcriptionally active Stat3 dimers with non-phosphorylated, salicylic acid-based small molecules: potent in vitro and tumor cell activities. Chembiochem 10, 1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Engen J. R., Gmeiner W. H., Smithgall T. E., Smith D. L. (1999) Hydrogen exchange shows peptide binding stabilizes motions in Hck SH2. Biochemistry 38, 8926–8935 [DOI] [PubMed] [Google Scholar]
- 24. Shoelson S. E., Sivaraja M., Williams K. P., Hu P., Schlessinger J., Weiss M. A. (1993) Specific phosphopeptide binding regulates a conformational change in the PI 3-kinase SH2 domain associated with enzyme activation. EMBO J. 12, 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma J., Zhang T., Novotny-Diermayr V., Tan A. L., Cao X. (2003) A novel sequence in the coiled-coil domain of Stat3 essential for its nuclear translocation. J. Biol. Chem. 278, 29252–29260 [DOI] [PubMed] [Google Scholar]
- 26. Horvath C. M., Wen Z., Darnell J. E., Jr. (1995) A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9, 984–994 [DOI] [PubMed] [Google Scholar]
- 27. Ma B., Tsai C. J., Haliloğlu T., Nussinov R. (2011) Dynamic allostery: linkers are not merely flexible. Structure 19, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong L., Lieser S. A., Miyashita O., Miller M., Tasken K., Onuchic J. N., Adams J. A., Woods V. L., Jr., Jennings P. A. (2005) Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J. Mol. Biol. 351, 131–143 [DOI] [PubMed] [Google Scholar]
- 29. Hamuro Y., Wong L., Shaffer J., Kim J. S., Stranz D. D., Jennings P. A., Woods V. L., Jr., Adams J. A. (2002) Phosphorylation driven motions in the COOH-terminal Src kinase, CSK, revealed through enhanced hydrogen-deuterium exchange and mass spectrometry (DXMS). J. Mol. Biol. 323, 871–881 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J., Adrián F. J., Jahnke W., Cowan-Jacob S. W., Li A. G., Iacob R. E., Sim T., Powers J., Dierks C., Sun F., Guo G. R., Ding Q., Okram B., Choi Y., Wojciechowski A., Deng X., Liu G., Fendrich G., Strauss A., Vajpai N., Grzesiek S., Tuntland T., Liu Y., Bursulaya B., Azam M., Manley P. W., Engen J. R., Daley G. Q., Warmuth M., Gray N. S. (2010) Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463, 501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iacob R. E., Pene-Dumitrescu T., Zhang J., Gray N. S., Smithgall T. E., Engen J. R. (2009) Conformational disturbance in Abl kinase upon mutation and deregulation. Proc. Natl. Acad. Sci. U.S.A. 106, 1386–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Panjarian S., Iacob R. E., Chen S., Wales T. E., Engen J. R., Smithgall T. E. (2013) Enhanced SH3/linker interaction overcomes Abl kinase activation by gatekeeper and myristic acid binding pocket mutations and increases sensitivity to small molecule inhibitors. J. Biol. Chem. 288, 6116–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inghirami G., Chiarle R., Simmons W. J., Piva R., Schlessinger K., Levy D. E. (2005) New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle 4, 1131–1133 [DOI] [PubMed] [Google Scholar]
- 34. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.