Background: It is necessary to improve the antiviral response of IFN-α for chronic hepatitis B (CHB) patients.
Results: Hepatitis B virus (HBV) disrupted glucocorticoid-induced S-adenosylmethionine and methionine adenosyltransferase 1A (MAT1A) expression by hypermethylation within the MAT1A promoter.
Conclusion: Glucocorticoid-induced S-adenosylmethionine enhances the response of IFN-α by restoring STAT1 methylation in HBV-infected cells.
Significance: The combination therapy of glucocorticoids, S-adenosylmethionine, and IFN-α is possibly useful for CHB patients.
Keywords: Glucocorticoid, Hepatitis Virus, Interferon, Liver Cancer, S-Adenosylmethionine (AdoMet)
Abstract
Patients with chronic hepatitis B usually exhibit a low response to treatment with interferon α (IFN-α). An alternative approach to increase the response rate of IFN-α might be to immunologically stimulate the host with glucocorticoids (GCs) before treatment with IFN-α, but the underlying mechanism remains unclear. We hypothesized that the GCs enhance IFN signaling by inducing S-adenosylmethionine (AdoMet) when hepatitis B virus (HBV) replication was effectively suppressed by IFN-α. Here, we investigated the effect of GCs and IFN-α on AdoMet production and methionine adenosyltransferase 1A (MAT1A) expression in vitro. Furthermore, we determined whether post-transcriptional regulation is involved in HBV-repressed MAT1A expression and AdoMet production induced by dexamethasone (Dex). We found that AdoMet homeostasis was disrupted by Dex and that Dex directly regulated MAT1A expression by enhancing the binding of the glucocorticoid receptor (GR) to the glucocorticoid-response element (GRE) of the MAT1A promoter. HBV reduced AdoMet production by increasing methylation at GRE sites within the MAT1A promoter. The X protein of hepatitis B virus led to hypermethylation in the MAT1A promoter by recruiting DNA methyltransferase 1, and it inhibited GR binding to the GRE in the MAT1A promoter. Dex could increase an antiviral effect by inducing AdoMet production via a positive feedback loop when HBV is effectively suppressed by IFN-α, and the mechanism that involves Dex-induced AdoMet could increase STAT1 methylation rather than STAT1 phosphorylation. These findings provide a possible mechanism by which GC-induced AdoMet enhances the antiviral activity of IFN-α by restoring STAT1 methylation in HBV-infected cells.
Introduction
The hepatitis B virus (HBV)4 is the most common hepatitis virus, and it causes chronic infections in the human liver (1). Complete eradication of HBV is rarely achieved due to the persistence of its covalently closed circular DNA in host hepatocytes (2). One key component of the host antiviral responses is the interferon (IFN) system. The immunomodulatory agent interferon α (IFN-α) is known to reduce the quantity of covalently closed circular DNA, presumably by inducing T-cell cytotoxicity and lysis of infected hepatocytes, along with the production of cytokines for control of viral replication (3). However, patients with chronic hepatitis B (CHB) usually respond poorly to IFN-α treatment, and the underlying mechanism remains unclear (4). It is noteworthy that the HBV genome contains a specific DNA-binding site for the GR, and this HBV GR domain can be categorized as a functional glucocorticoid-response element (GRE). Treatment of CHB would benefit from an improved antiviral response to IFN-α.
An alternative approach to increase the efficacy and response rate observed with IFN might be to immunologically stimulate the host by withdrawing glucocorticoids (GCs) before treatment with IFN. In CHB infection, pulse GC treatment followed by abrupt withdrawal has been associated with an enhanced cellular immune response to hepatitis B, as indicated by a rise in alanine transaminase values and a transient reduction in markers of viral replication upon withdrawal of GCs (5). Pretreatment with GCs (“immunologic priming”) is believed to be synergistic when followed by treatment with IFN-α in a subgroup of patients (with low initial alanine transaminase values) (5, 6). Although there are different opinions concerning the rationale for a combination regimen of GCs and IFN-α, most studies suggest that sequential treatment with GCs and IFN-α for HBeAg-positive chronic hepatitis B may be more effective than IFN-α monotherapy in promoting the loss of hepatitis B “e” antigen and hepatitis B virus DNA (7). However, the antiviral mechanism of the combination regimen is unknown.
S-Adenosylmethionine (AdoMet), a principal biological methyl donor, is synthesized from methionine and ATP in a reaction catalyzed by methionine adenosyltransferase (8, 9). In mammals, there are two genes (MAT1A and MAT2A) that encode two homologous methionine adenosyltransferase catalytic subunits (10, 11). AdoMet biosynthesis is depressed in chronic liver disease (12). Preclinical studies indicate that this depression could exacerbate liver injury, and therefore, supplementation may be a useful therapy. AdoMet has been widely adopted worldwide as a therapy for chronic intrahepatic cholestasis liver disease (13). However, the effectiveness of AdoMet therapy in CHB has not been adequately addressed. It was previously reported that hepatic AdoMet synthetase activity is altered in adrenalectomized animals, suggesting a role for GCs in its regulation (14). This finding was further supported by a study indicating that GCs strongly up-regulate AdoMet synthetase, both in vivo and in hepatic cultured cells, and that they have a direct effect on enzyme gene expression (15). However, the mechanism by which AdoMet synthetase activity was up-regulated by GCs was not investigated in these studies. Interestingly, AdoMet synthetase activity has been reported to be significantly reduced in various liver disorders (16). Thus, it is tempting to speculate that at least some of the beneficial effects of GCs in chronic HBV-related liver diseases could be due to the direct stimulation of AdoMet synthetase, which in turn would increase the availability of AdoMet. In addition, AdoMet may improve IFN signaling and antiviral effects through increased methylation of STAT1, leading to enhanced STAT1-DNA binding and greater expression of interferon-stimulated genes (ISGs) (17). AdoMet represents the first IFN-sensitizing agent with in vivo efficacy. Thus, AdoMet appears to be a useful adjunct to IFN-based therapy, a factor that may be particularly important in the era of direct antivirals for HBV infection.
In this study, we propose that a combination regimen of GCs and IFN-α is associated with the GC induction of AdoMet production. Here, we investigated whether HBV, alone or in combination with Dex, could alter the alternative expression of MAT1A and AdoMet production, which was potentially associated with DNA methylation in the putative GRE of the MAT1A gene promoter in the examined hepatoma cells. We then analyzed the possible epigenetic mechanisms involved. Recent evidence suggests that HBV has evolved strategies to block the nuclear translocation of signal transducers and activators of transcription (STAT1) to limit IFN-α-induced cellular antiviral responses (18). Furthermore, we explored the effect of the GC-induced increase of AdoMet production on STAT1 methylation and phosphorylation.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Cell Culture
A 1474-bp promoter construct of the MAT1A gene, corresponding to the sequence from nt −1474 to 0 (relative to the transcriptional start site) of the 5′-flanking region of the human MAT1A gene, was generated from human genomic DNA by PCR using pMAT1A1.4Luc-F and pMAT1ALuc-R as forward and reverse primers carrying the MluI and XhoI sites at the 5′ and 3′ ends, respectively. The PCR product was cloned into the MluI and XhoI sites of the pGL3-Basic vector. The resulting construct was confirmed by DNA sequencing. The 5′-flanking deletion constructs of the MAT1A promoters, pMAT1A1.2Luc, pMAT1A0.9Luc, and pMAT1A0.8Luc, were similarly generated by PCR, using the pMAT1A1.4Luc construct as a template. The forward primers were pMAT1A1.2Luc-F1, pMAT1A0.9Luc-F2, and pMAT1A0.8Luc-F3. Site-directed deletion of two GRE sites was performed by 30 cycles of PCR using the pMAT1A1.4Luc or pMAT1A0.9Luc construct as a template and the appropriate primers. In the pMAT1A1.4ΔLuc1 plasmid, MAT1A-GRE1 (nt −876 to −862) was deleted. In the pMAT1A1.4ΔLuc2 plasmid, MAT1A-GRE2 (nt −1022 to −1008) was deleted. In the pMAT1A1.4ΔLuc3 plasmid, both MAT1A-GRE1 (nt −876 to −862) and MAT1A-GRE2 (nt −1022 to −1008) were deleted. In the pMAT1A0.9ΔLuc plasmid, MAT1A-GRE1 (nt −876 to −862) was deleted. Four site-directed mutations were constructed by PCR using pMAT1A-1.4Luc as a template. Four CpG sites were mutated separately from C to A. Ligation was verified by sequence analysis. The PCR primer sequences are shown in Table 1.
TABLE 1.
DNA sequences of primers used in the study
qRT-PCR is quantitative RT-PCR; F is forward; R is reverse.
| Primer name | Sequence (5′ to 3′) | bp |
|---|---|---|
| qRT-PCR | ||
| MAT1A-F | AGAGTGCTTGTCCAGGTT | 97 |
| MAT1A-R | GCTCTCGCTCTGTCTTCT | |
| GRE1-F | TCAGAATACAGGTGCGTGCT | 74 |
| GRE1-R | CTGCGTCTCATCTGGATTGGT | |
| GRE2-F | ATTCCCCATTGTTCCTTGGGT | 70 |
| GRE2-R | TGTACTAAATGACAGCGTCTCACA | |
| HBV-GRE-F | CTGGCCAAAATTCGCAGTCC | 84 |
| HBV-GRE-R | GACACATCCAGCGATAGCCA | |
| GAPDH-F | CAAGTTCAACGGCACAGTCA | 100 |
| GAPDH-R | CCATTTGATGTTAGCGGGAT | |
| Truncation | ||
| pMAT1A1.4Luc-F | CGCACGCGTTTCCAGAAGAGGTCACCTTAATACT | 1474 |
| pMAT1A1.2Luc-F1 | CGCACGCGTAGTCCAAGCTTTGATGCACAAGGTT | 1274 |
| pMAT1A0.9Luc-F2 | CGCACGCGTAAACTGGACTTTGATAATTTCCCTG | 974 |
| pMAT1A0.8Luc-F3 | CTCACGCGTACCTCCCCAGATAGATACTTTTTAA | 874 |
| pMAT1ALuc-R | ATACTCGAGCTTCTCACACTTCTCCACTCACGCT | |
| Mutation | ||
| del−876 to −862-F4 | TAAAAAAAAAACACACACACACACTGTAACCTCCCCAGATAGATAC | |
| del−876 to −862-R2 | GTATCTATCTGGGGAGGTTACAGTGTGTGTGTGTGTTTTTTTTTTA | |
| del−1022 to −1008-F5 | TCAAAGCAAAAAATAATGCAAGAGTTATGTTACATGTATAGAACTGAATATGTAGATG | |
| del−1022 to −1008-R3 | CATCTACATATTCAGTTCTATACATGTAACATAACTCTTGCATTATTTTTTGCTTTGA | |
| MUT CpG1-F | CAGCTGGATCTGAGAGTGTGAGAAGCTGTCATTTAG | |
| MUT CpG1-R | CTAAATGACAGCTTCTCACACTCTCAGATCCAGCTG | |
| MUT CpG2-F | CAAAAAATAATGCAAGAGTTATGTGAACAAGATGTTTATTACATGTATAGAACTGAATA | |
| MUT CpG2-R | TATTCAGTTCTATACATGTAATAAACATCTTGTTCACATAACTCTTGCATTATTTTTTG | |
| MUT CpG3-F | GTATAGAACTGAATATGTAGATGTTTATAATACGGAAGCTTTGGGAAA | |
| MUT CpG3-R | TTTCCCAAAGCTTCCGTATTATAAACATCTACATATTCAGTTCTATAC | |
| MUT CpG4-F | GTTTCTGTTAGCAGAAACAAGTGGACTCAAAGCTTTTCC | |
| MUT CpG4-R | GGAAAAGCTTTGAGTCCACTTGTTTCTGCTAACAGAAAC | |
| ChIP | ||
| chip-GRE1F | CACATTGTTCTCTGTAACCTCCCCA | 254 |
| chip-GRE1R | TCCCCTGCGTCTCATCTGGATTGGT | |
| chip-GRE2F | ATTCCCCATTGTTCCTTG | 154 |
| chip-GRE2R | AACATCGTGTTCACATAACT | |
| chip-HBV-GRE-F | CTCGTGTTACAGGCGGGGTTTTTCT | 206 |
| chip-HBV-GRE-R | ATAAAACGCCGCAGACACATCCAGC | |
| MSP | ||
| MAT1A-F | AGGAAGAGAGTTGTATTTTGAGTTAGTTGTGATGTT | |
| MAT1A-R | CAGTAATACGACTCACTATAGGGAGAAGGCTCAATTCCCCATTATTCCTTAAATCT | |
Cell lines, including the human normal liver cell L02 and the hepatoma cell lines Huh7, Hep3B, and HepG2, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), where they were characterized by mycoplasma detection, DNA fingerprinting, isozyme detection, and determination of cell viability. The HepG2.2.15 cell line was derived from HepG2 cells and stably expresses HBV (Genotype D, Serotype ayw, U95551), which was used as an HBV replication model (19–21). The stable cell lines were maintained in DMEM containing 400 μg/ml G418. The plasmid pCMV-HBV-1.3, which expresses HBV (genotype C, serotype adr, FJ899793), was a gift from Dr. Ying Zhu (State Key Laboratory of Virology, College of Life Sciences, Wuhan University, China). All cells were cultured in the recommended media supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and streptomycin at 37 °C in an incubator with 5% CO2.
Quantitative qRT-PCR Analysis
For the analysis of mRNA levels, total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Quantification of total RNA was performed with a NanodropTM spectrophotometer (Thermo Scientific) at 260 and 280 nm. cDNA was synthesized using a cDNA synthesis kit (Toyobo, Japan). For the analysis of production levels in ChIP assays, the enriched DNA fragments in ChIPs were quantified with quantitative RT-PCR. Amplification was performed with the iQ5 quantitative PCR system (Bio-Rad) and SYBR® Green Master Mix (Toyobo, Japan). GAPDH was used for normalization of the relative expression. Relative mRNA levels were determined using the 2−ΔΔCT method. The gene-specific primers are listed in Table 1.
Transfection and Luciferase Reporter Assays
All transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. For the luciferase assay, the cells were transiently co-transfected with the pRL-TK plasmid (Promega) containing the Renilla luciferase gene, which was used for internal normalization, and various constructs containing different lengths of the MAT1A 5′-flanking region. After 48 h post-transfection, cell lysates were prepared, and the luciferase activity was measured using a luciferase assay system (Promega). The cell lysates (10 μl) and luciferase assay substrates (50 μl) (Promega) were mixed, and the fluorescence intensity was detected using a luminometer (Promega). The assays were performed in triplicate and expressed as the means ± S.E. relative to the vector control as percent values. All transfection experiments were performed at least three times.
Fluorescent Immunocytochemistry
Hepatoma cells were fixed with 4% paraformaldehyde for 20 min and then permeabilized with 0.5% Triton X-100 for 10 min. The fixed cells were blocked with 3% fetal calf serum for 30 min. For specific detection of GR protein, the cells were incubated with polyclonal rabbit anti-rat GR antibody (1:100, Cell Signaling Technology) overnight at 4 °C. The samples were incubated with Cy3-labeled goat anti-rabbit IgG (1:100, eBioscience) in PBS for 1 h at 37 °C. Then the cells were washed twice with PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min. The images were analyzed using fluorescence microscopy (NIKON Eclipse 80i, Japan). Identical illumination and camera settings were used within each dataset.
Immunohistochemistry
Fifty cases (25 pairs) of HBV-associated hepatocellular carcinoma (HCC) were collected from Zhongnan Hospital, Wuhan University, between January 2011 and October 2012. Chemotherapy and radiation therapy were not administered before tumor excision. Both the tumors and corresponding peritumoral noncancerous tissues were selected for each case. All patients tested positive for HBV surface antigen in their serum. Matched normal human liver tissues were obtained from liver trauma patients undergoing partial hepatectomy. Written informed consent was obtained from each patient.
Both HCC tissues and adjacent nontumor liver tissues were sectioned to produce 4-μm thick slices. The tissue sections were deparaffinized in xylene and rehydrated with ethanol, and the endogenous peroxidase was inactivated with 0.3% hydrogen peroxide for 10 min. All of the procedures were performed using an UltraSensitiveTM S-P kit (Maixinbio, China) according to the manufacturer's instructions. The negative control sections were incubated in PBS without the antibody under the same experimental conditions. The total immunostaining score was calculated as the sum of the positive percentage and the staining intensity of the stained cells, which ranged from 0 to 6. The percent positivity was scored as 0 (0–25%), 1 (26–50%), 2 (51–75%), and 3 (≥75%). The staining intensity was scored as 0 (no staining), 1 (weakly stained), 2 (moderately stained), and 3 (strongly stained). The results of the immunohistochemical staining were scored by two experienced pathologists, who were blinded to the clinical data. A negative expression of protein was defined as a total score ≤3, and a positive expression was defined as a total score ≥4.
Immunoblotting
For the detection of protein, cytoplasm and nuclear protein extracts were prepared from cells treated with vehicle or dexamethasone. The protein concentration of each sample was determined using a NanodropTM spectrophotometer (Thermo Scientific). Protein (100 μg) from each sample was examined by SDS-PAGE (4% stacking and 10% separating gels) and then transferred overnight onto PVDF membranes (Millipore). The membranes were immunoblotted with the following: polyclonal rabbit anti-human MAT1A antibody (1:200, Abgent); X protein of hepatitis B virus (HBx) antibody (1:1000, Abcam); GR antibody (1:1000, Cell Signaling Technology); DNMT3A antibody (1:500, Abgent); DNMT1 antibody (1:500, Abgent); GAPDH antibody (1:1000, Santa Cruz Biotechnology); lamin B2 antibody (1:1000, Cell Signaling Technology); STAT1 antibody (1:1000, Cell Signaling Technology); or p-STAT1 (Tyr-701) antibody (1:1000, Cell Signaling Technology) overnight. The blots were then incubated with peroxidase-conjugated goat anti-rabbit antibody or goat anti-mouse antibody (1:4000, Millipore) for 1 h. The PVDF membranes were subsequently subjected to immunoblotting analysis using an ECL immunoblotting kit according to the manufacturer's recommended protocol (Beyotime Institute of Biotechnology, China).
Quantification of the AdoMet and AdoHcy Levels by HPLC
For the assay, 1 × 105 cells were mixed with 200 μl of solvent consisting of 50% A and 50% B and stored at −20 °C for 10 min. After sonication, the samples were centrifuged at 2000 × g for 15 min at 4 °C and filtered through 0.45-μm Millex-HV filters (Millipore). The AdoMet and AdoHcy levels were determined by reversed-phase HPLC. The contents were quantified using a Dionex Ultimate 3000 system. The compounds were separated on a reversed-phase UltimateTMAQ-C18 column (5 μm, 4.6 × 250 mm; Welch, China) connected to a guard column (5 μm; Scienhome, China) at 26 °C. The two mobile phases consisted of 5 mm ammonium formate and 0.2% (v/v) formic acid aqueous solution, pH 3.0 (Buffer A), and HPLC-grade methanol (TEDIA) (Buffer B). HPLC-grade ammonium formate, formic acid, AdoMet, and AdoHcy standards were purchased from Sigma. The wavelength for detection was 254 nm. The column was equilibrated with 80% A and 20% B. The flow rate was 0.5 ml/min. The sample injection volume was 10 μl. The data were acquired and processed using Chromeleon software (Dionex).
Coimmunoprecipitation
The primary antibody (antibody to methyl- and dimethylarginine) was incubated with a 25% slurry of protein A/G-agarose beads (Santa Cruz Biotechnology) in IP buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton, 1 mm EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin) with gentle agitation for 2 h at room temperature. The beads were washed three times with 1 ml of IP buffer and then incubated with cell lysates for 2 h at room temperature. The beads were then washed five times with 1 ml of IP buffer and resuspended in Laemmli sample buffer. The beads were boiled for 2 min and then centrifuged. The resulting samples were analyzed by immunoblotting procedures as described above (Western blot with anti-STAT1 antibody).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using an EZ-ChIP kit (Millipore) according to the manufacturer's instructions. Briefly, the cells were transfected with relevant plasmids and then cross-linked using 1% formaldehyde at 37 °C for 10 min. The cells were washed twice with ice-cold PBS and resuspended in 1 ml of lysis buffer. DNA was sheared to reduce the DNA length to between 200 and 1000 bp by sonication 9 times for 10 s each time, using an Ultrasonic Processor VCX 600 (Sonics and Materials, Newtown, CT) at 30% power. The recovered supernatants were incubated with an antibody directed against GR (5 μg, Cell Signaling Technology) or an isotype control IgG for 4 h in the presence of ChIP Dilution Buffer (900 μl) and ChIP blocked protein G-agarose (60 μl). The immunoprecipitated DNA was retrieved from the beads with a 1% SDS and 1.1 m NaHCO3 solution maintained at 65 °C overnight. The DNA was then purified using a PCR purification kit (Axygen), and PCR was performed on the extracted DNA using specific primers (Table 1).
Electrophoresis Mobility Shift Assay (EMSA)
Nuclear extracts were prepared from the cells treated as described above. EMSA was performed using a nonradioactive EMSA kit following the manufacturer's instructions (Pierce). The sequence of the GRE1 probe was P1, 5′-CACACACACACACATTGTTCTCTGTA-3′. The GRE2 probe was P2, 5′-GAGTTATGTGAACACGATGTTTATTACATG-3′, and the HBV-GRE probe was P3, 5′-CCAACCTCCTTGTCCTCCAATTTGTCCTGGT-3′. The 5′ end of the oligonucleotides was biotin-labeled. Ten micrograms of crude nuclear protein were incubated for 20 min at room temperature in a 15-μl binding reaction system, including 1.5 μl of 10× binding buffer, 1.5 μl of poly(dI-dC) (1.0 μg/μl), and double-distilled H2O to a final volume of 15 μl. Then 0.6 μl of Bio-GRE1 probe or Bio-GRE2 probe or Bio-HBV-GRE probe (500 fm) was added, and the reaction was incubated for 20 min at room temperature. Where indicated, 2 μl of specific unlabeled competitor oligonucleotide was added before the labeled probe to the ×100× competing system and incubated for 20 min. Protein-DNA complexes were resolved by electrophoresis at 4 °C on a 6.5% polyacrylamide gel and subjected to autoradiography. Electrophoresis was conducted on a 6.5% nondenaturing polyacrylamide gel at 175 V in 0.25× TBE (1× TBE is 89 mm Tris-HCl, 89 mm boric acid, and 5 mm EDTA, pH 8.0) at 4 °C for 1 h. For the supershift experiments, purified polyclonal antibody directed against GR (4 μg, Cell Signaling Technology) or IgG was incubated with protein-DNA complexes on ice for 20 min. The gels were placed on the bonding membrane, and the proteins were transferred at 394 mA in 0.5× TBE at room temperature for 40 min. Then the membrane was cross-linked in a UV cross-linking apparatus for 10 min (immobilization), blocked, streptavidin-HRP labeled, washed, and then equilibrated. Images were obtained using an Imager apparatus (Alpha Innotech, San Leandro, CA).
Quantitative Methylation Changes of MAT1A Promoter
To perform bisulfite conversion of the genomic DNA, an Epitect bisulfite kit (Qiagen AG, Basel, Switzerland) was used according to the manufacturer's protocol. We designed primers for the MAT1A promoter to cover the regions with the CpG sites (Table 1). The selected amplicon was located in the core regulatory regions of the promoter, which covered the two GREs. The primers were designed using MethPrimer. For PCR amplification, a T7 promoter tag was added to the reverse primer, and a 10-mer tag sequence was added to the forward primer to balance the PCR primer length. The following PCR conditions were used for the amplification of the bisulfite-treated genomic DNA: one cycle, 94 °C for 4 min; 45 cycles, 94 °C for 20 s; 56 °C for 30 s; 72 °C for 1 min; and one cycle, 74 °C for 3 min. Unincorporated dinucleotide triphosphates were removed by shrimp alkaline phosphatase (Sequenom, San Diego) treatment. Typically, 2 μl of the PCR product was then directly used as a template for the transcription reaction. Twenty units of T7 R&DNATM polymerase (Epicenter, Madison, WI) were used to incorporate dTTP in the transcripts. Ribonucleotides were used at 1 mmol/liter and the dNTP substrate at 2.5 mmol/liter. In the same step, RNase A (Sequenom, San Diego) was added to cleave the in vitro transcripts (T-cleavage assay). The samples were diluted with double distilled H2O to a final volume of 7 μl. Then the samples were incubated at 37 °C for 3 h. Next, 20 μl of double distilled H2O was added to each sample. Phosphate backbone conditioning was achieved by adding 6 mg of Clean Resin (Sequenom, San Diego) before performing MALDI-TOF MS analysis. A total of 12 nl of the RNase A-treated product was robotically dispensed onto a silicon matrix of preloaded chips (SpectroCHIP; Sequenom, San Diego), and the mass spectra were collected using a MassARRAY Compact MALDI-TOF (Sequenom, San Diego). The methylation ratios of the spectra were generated using EpiTYPER software version 1.0 (Sequenom, San Diego).
Statistical Analysis
Measurement data are presented as the mean ± S.D. or the mean ± S.E., and count data were analyzed using a χ2 or Fisher's exact test. Data that satisfied the normal distribution criterion were used in the group design of the t test for the statistical analysis. The differences among the groups were tested by a one-way analysis of variance followed by a post hoc test (LSD). A value of p < 0.05 was considered significant. All statistical analyses were performed using professional statistical software (SPSS 15.0 for Windows, SPSS Inc., Chicago).
RESULTS
AdoMet Homeostasis Was Disrupted by Pharmacologic Concentrations of Glucocorticoids through Inducing MAT1A Expression
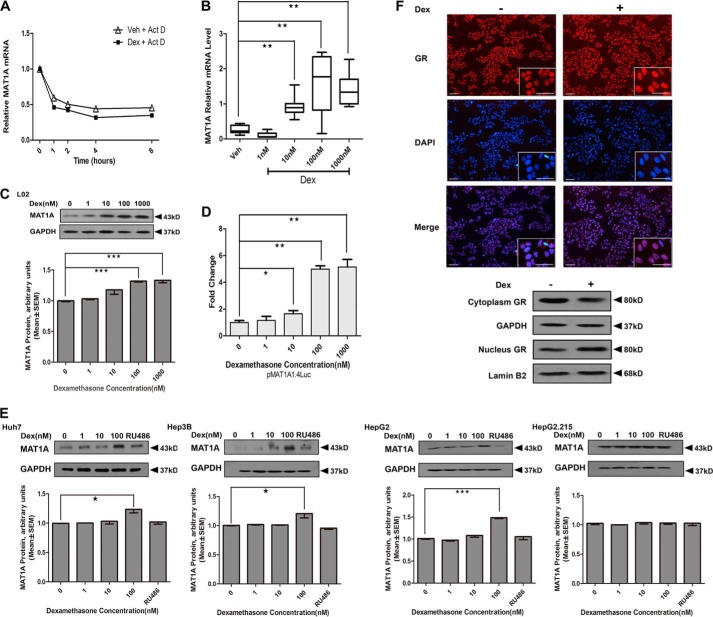
To determine the effects of GCs on AdoMet and AdoHcy, we treated different liver cells with Dex. Dex was chosen in our studies because it is similar to GCs and has been used extensively in humans. We observed that the levels of AdoMet and the ratio of AdoMet/AdoHcy were markedly increased in Dex-treated cells, including normal hepatic L02 cells and HepG2 cells. Next, we determined the specificity of Dex in the activation of AdoMet production. We treated these cells with RU486 (an antagonist of GR) before supplying Dex. The results indicated that RU486 can counteract the stimulatory effect of Dex on AdoMet production, which indicates that GCs induced AdoMet production by binding to a specific receptor, GR (Table 2). Because AdoMet production was dependent on MAT1A expression, to elucidate the mechanism of the GC-induced increase of AdoMet production, we analyzed the effect of Dex on MAT1A expression. The results showed that the half-life of MAT1A mRNA was identical, whereas the absolute level of MAT1A mRNA was higher in Dex-treated L02 cells compared with vehicle-treated cells (Fig. 1, A and B), which suggested that Dex did not affect the stability of the MAT1A mRNA. Furthermore, we found that MAT1A protein was significantly induced when Dex was supplied at doses of 100 and 1000 nm (Fig. 1C). We analyzed the possible mechanisms involved by investigating the effects of Dex on the MAT1A promoter activity. The luciferase activity of MAT1A was significantly increased in a dose-dependent manner in the Dex-treated cells (Fig. 1D). These results were confirmed in other hepatoma cell lines, including Huh7, Hep3B, and HepG2. However, MAT1A expression was blocked dramatically with RU486 treatment in the aforementioned cells (Fig. 1E). The results showed that GCs induced MAT1A expression by binding to the GR. Next, we analyzed the GR localization in hepatoma cells. We observed an increased amount of GR importation to the nucleus in response to ligand binding in different hepatoma cells. The level of GR increased in the nucleus and decreased in the cytoplasm of the Dex-treated cells compared with the vehicle-treated cells (Fig. 1F). These results demonstrated that the GR participated in Dex-induced MAT1A expression via translocation to the nucleus.
TABLE 2.
Changes of AdoMet, AdoHcy, and the AdoMet/AdoHcy ratio in L02, HepG2, and HepG2.2.15 per 105 cells treated with different concentrations of Dex and RU486
Results represent the mean ± S.E. from four to five separate determinations.
| Cell lines | Treatment | Concentration | AdoMet | AdoHcy | AdoMet/AdoHcy |
|---|---|---|---|---|---|
| ng | ng | ||||
| L02 | Dex (nm) | 0 | 4.13 ± 0.18 | 2.18 ± 0.14 | 1.89 ± 0.13 |
| 1 | 5.51 ± 0.11a | 2.40 ± 0.12 | 2.40 ± 0.15a | ||
| 10 | 8.03 ± 0.19a | 2.48 ± 0.15 | 3.24 ± 0.14a | ||
| 100 | 9.37 ± 0.17a | 2.60 ± 0.17 | 3.60 ± 0.11a | ||
| RU486 (nm) | 100 | 3.78 ± 0.13 | 2.12 ± 0.03 | 1.79 ± 0.13 | |
| HepG2 | Dex (nm) | 0 | 3.57 ± 0.15 | 1.93 ± 0.11 | 1.85 ± 0.13 |
| 1 | 5.30 ± 0.17a | 2.10 ± 0.16 | 2.53 ± 0.16a | ||
| 10 | 7.24 ± 0.11a | 2.21 ± 0.19 | 3.28 ± 0.17a | ||
| 100 | 8.87 ± 0.14a | 2.43 ± 0.37 | 3.66 ± 0.21a | ||
| RU486 (nm) | 100 | 3.47 ± 0.12 | 1.99 ± 0.09 | 1.75 ± 0.11 | |
| HepG2.2.15 | Dex (nm) | 0 | 3.17 ± 0.07 | 1.74 ± 0.06 | 1.82 ± 0.07 |
| 1 | 3.09 ± 0.04 | 1.77 ± 0.12 | 1.75 ± 0.08 | ||
| 10 | 3.17 ± 0.08 | 1.75 ± 0.05 | 1.81 ± 0.06 | ||
| 100 | 3.19 ± 0.02 | 1.69 ± 0.04 | 1.89 ± 0.03 | ||
| RU486 (nm) | 100 | 2.97 ± 0.11 | 1.65 ± 0.09 | 1.80 ± 0.10 |
a p < 0.05 versus Dex 0 nm by unpaired Student's t test.
FIGURE 1.
Effect of Dex on MAT1A promoter activity and expression. A, analysis of MAT1A mRNA stability in L02 cells. Each level of Dex-treated and -untreated MAT1A mRNA before actinomycin D (Act D) treatment was considered as 1, and the relative levels were calculated. B and C, MAT1A mRNA and MAT1A protein were examined after L02 cells were treated with vehicle (Veh) or the indicated concentration of Dex for 24 h. D, effect of Dex concentration on the luciferase activity in L02 cells transfected with pMAT1A1.4Luc. E, MAT1A protein levels were detected in Huh7, Hep3B, HepG2 and HepG2.2.15 cells after treatment with the vehicle or Dex with or without RU486 for 24 h. The inset shows the representative immunoblots of different concentration points. *, p < 0.05; **, p < 0.01 and ***, p < 0.001. F, GR localization was investigated in the aforementioned cells treated with Dex for 12 h and then fixed, and endogenous GR was labeled (red). DNA was counterstained with DAPI (blue). GR protein levels and distributions were detected in the cytoplasm and nucleus, respectively. GAPDH or lamin B2 was used as a loading control. Scale bar, 50 μm. Shown is a representative of results from five independent experiments.
Role of the GRE in the Stimulatory Effect of GCs on the MAT1A Alternative Promoter Activity
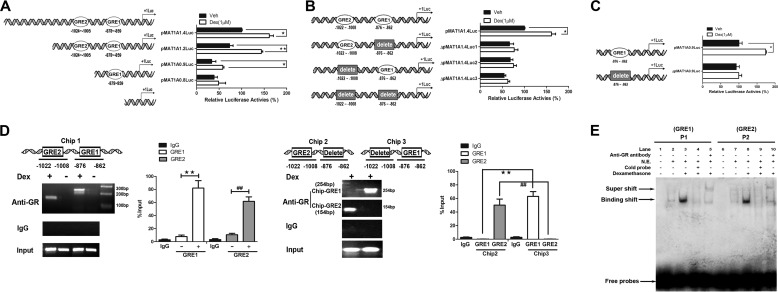
To further explore the mechanism of the effect of GCs on MAT1A expression, we investigated the role of the cis-regulatory elements of the MAT1A promoter in response to Dex regulation. When a series of truncated MAT1A promoter mutants was generated, we found that the Dex-induced increase of MAT1A promoter activity was inhibited by a deletion from nt −1474 to −874 (Fig. 2A), which suggested that the sequence between nt −1474 and −874 is critical for the activation of MAT1A by Dex. Analyses of the cis-regulatory elements of the MAT1A promoter revealed two GR-binding sites in this region, including MAT1A-GRE1 (nt −876 to −862) and MAT1A-GRE2 (nt −1022 to −1008). To evaluate the roles of these GREs in the activation of the MAT1A promoter by Dex, experiments involving deletion and site-directed mutagenesis at positions GRE1 and GRE2 were conducted (Fig. 2, B and C). The results showed that the luciferase activity in cells transfected with pMAT1A1.4Luc or pMAT1A0.9Luc was significantly induced by Dex, but the actual luciferase activity units of pMAT1A0.9Luc was less than 50% compared with that of pMAT1A1.4Luc. However, the induction of Dex on pMAT1A1.4Luc or pMAT1A0.9Luc was disrupted when the GRE1 or GRE2 site was deleted or mutated. These results suggested that GREs were required for the activation of MAT1A expression mediated by Dex.
FIGURE 2.
Functional characterization of the GRE in the MAT1A promoter. HepG2 cells were transfected with pMAT1A-1.4 Luc, pMAT1A-1.2 Luc, pMAT1A-0.9 Luc, and pMAT1A-0.8 Luc for 24 h, followed by treatment with Dex for another 24 h. *, p < 0.05 and **, p < 0.01. A–C, effects of truncation, site-directed mutation, and deletion mutation in the GRE sequence on the MAT1A promoter activity were analyzed. D, role of Dex in the binding of the GR to the GREs of the MAT1A promoter was determined by ChIP assays. The productions of Chip-GRE1 and Chip-GRE2 were quantified by qPCR. **, p < 0.01, and ##, p < 0.01. E, analyses of the effect of Dex on the binding of the GR to GRE1 (P1) and GRE2 (P2) in the MAT1A promoter by EMSA. Productions of Chip-GRE1 and Chip-GRE2 were quantified by quantitative PCR. Shown is a representative result from three independent experiments. Veh, vehicle.
To explore the interactions between the GRE sites and the GR, ChIP assays were performed. The results showed that PCR products were only produced from DNA isolated from the Dex-treated cells (Fig. 2D, Chip1). Then we deleted the two GRE sites separately and found that only PCR products of GRE1 were produced when GRE2 was deleted, and only PCR products of GRE2 were produced when GRE1 was deleted after being treated with Dex (Fig. 2D, Chip2 and Chip3). The results showed that the production of Chip-GRE1 (254 bp) was higher than that of Chip-GRE2 (154 bp), which suggested that there was more binding of GR protein to the GRE1 site than to the GRE2 site. These findings indicated that the two complete GRE sequences are functional in the context of the MAT1A promoter by binding with GR.
To determine the specific binding of the GR to the GRE sites in the MAT1A promoter, EMSAs were performed. We observed one faint band in the absence of Dex, indicating the presence of a protein-DNA complex (binding shift band) (Fig. 2E, 2nd lane); however, the binding shift band was enhanced in the presence of Dex (Fig. 2E, 3rd lane). Moreover, the binding shift band was eliminated in the presence of a cold probe (Fig. 2E, 4th lane). In addition, a specific protein-DNA complex (supershift band) was detected in the presence of an anti-GR antibody (Fig. 2E, 5th lane). Similar results were observed when the GRE2 probe (P2) was used (Fig. 2E, 6th to 10th lanes).
HBV Down-regulated MAT1A Expression by Up-regulating DNMT1 but Not DNMT3A
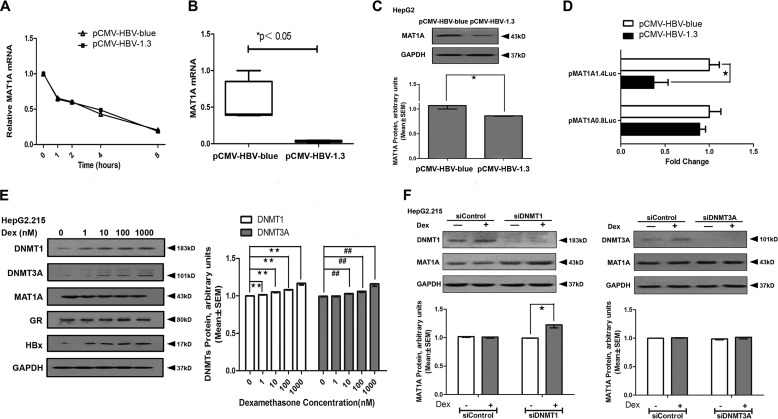
Although AdoMet production and MAT1A expression were induced by Dex, we found that the levels of AdoMet and the ratio of AdoMet/AdoHcy were not altered in HepG2.2.15 cells that were stably transfected with HBV after Dex treatment (Table 2). Furthermore, Dex also failed to induce MAT1A expression in HepG2.2.15 (Fig. 1E). These results suggested that the effect of Dex on MAT1A expression may be disrupted by HBV. It has been reported that HBx plays a crucial role in hepatocarcinogenesis by inducing aberrant epigenetic modifications (23). To verify the role of HBV and HBx in the regulation of MAT1A expression, we studied whether post-transcriptional regulation is involved. We observed that the half-life of MAT1A mRNA was identical, whereas the absolute level of MAT1A mRNA was lower in pCMV-HBV1.3-transfected HepG2 cells compared with the mock-transfected cells (Fig. 3, A and B), which suggested that HBV did not affect the stability of MAT1A mRNA. We also found that the levels of the MAT1A protein (Fig. 3C) were lower in HepG2 cells transfected with pCMV-HBV1.3 than with mock-transfected cells. To determine the effects of HBV on luciferase activity, HepG2 cells were transiently transfected with pMAT1A-1.4Luc or pMAT1A-0.8Luc. There was a significant reduction of luciferase activity in pMAT1A-1.4Luc when the cells were transfected with pCMV-HBV1.3 compared with the mock vector (Fig. 3D). This suggests that HBV suppressed MAT1A promoter activity by way of the sequence between nt −1474 and −874, which was critical for the activation of MAT1A by Dex. However, Dex failed to induce MAT1A expression, but DNMT1 and DNMT3A were induced in a dose-dependent manner in HepG2.2.15 cells (Fig. 3E). Furthermore, we found that MAT1A expression was inducible by Dex when DNMT1 was knocked down with siDNMT1 (5′-AGATTTGTCCTTGGAGAACGG-3′), whereas MAT1A expression was not induced by Dex when DNMT3A was knocked down with siDNMT3A (5′-AGAAGTGTACACGGACATGTG-3′) (Fig. 3F). These results suggested that Dex-induced MAT1A expression was disrupted by HBV, perhaps due to HBx recruiting of DNMT1 to increase methylation at the putative GRE of the MAT1A promoter.
FIGURE 3.
Effect of HBV on MAT1A promoter activity and expression. A, analysis of MAT1A mRNA stability in HepG2 cells transfected with HBV. Each level of Dex-treated and -untreated MAT1A mRNA before actinomycin D treatment was considered as 1, and the relative levels were calculated. B and C, MAT1A mRNA and MAT1A protein were examined after HepG2 cells were transfected with HBV for 24 h. The inset shows the representative immunoblots of MAT1A with different transfections. D, effect of HBV on luciferase activity in HepG2 cells transfected with pMAT1A1.4Luc. *, p < 0.05. E, DNMT1, DNMT3A, MAT1A, GR, HBx, and GAPDH protein levels were detected after HepG2.2.15 cell treatment with vehicle or Dex for 24 h. The inset shows representative immunoblots of DNMT1 and DNMT3A at different concentrations. **, p < 0.01; ##, p < 0.01. F, DNMT1, DNMT3A, MAT1A, and GAPDH protein levels were detected after HepG2.2.15 cells were transfected with siControl, siDNMT1, or siDNMT3A and treated with vehicle or Dex (100 nm) for 24 h. The inset shows the representative immunoblots of MAT1A with different treatments. *, p < 0.05. Shown is a representative result from three independent experiments.
HBV Could Suppress the Dex-induced Increase of MAT1A Expression by Promoting DNA Hypermethylation of the MAT1A Promoter
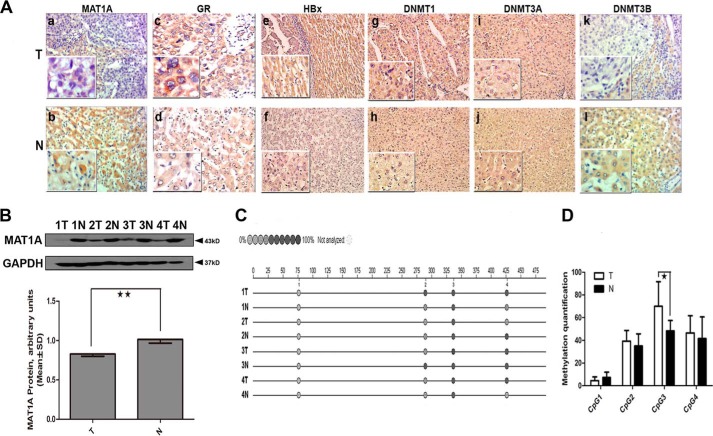
To study HBV suppression of Dex-induced MAT1A expression in vivo, we tested the expressions of HBx and DNMT in HBV-associated HCC tissues, and we searched for a possible linker role for DNA methylation in the Dex-dependent interaction of the GR, the MAT1A promoter, and HBx. As shown in Fig. 4A, HBx had a higher expression in HCC tissue, which was consistent with our previous findings (22); in addition, DNMT1 had a higher level of expression, whereas DNMT3B had a lower level of expression in HCC tissues compared with adjacent nontumor tissues. Interestingly, there is a positive correlation between HBx expression and DNMT1 expression, and a negative correlation between HBx expression and DNMT3B expression in liver tumor tissues (Table 3). As shown in Fig. 4B, the protein level of MAT1A was significantly decreased by 17.82% (0.83 ± 0.06 versus 1.01 ± 0.09, p = 0.015) in the HCC tissues compared with adjacent nontumor tissues. Previous studies have reported that HBx expression increased total DNMT activities by up-regulating DNMT1 and DNMT3A and selectively promoting regional hypermethylation of specific tumor suppressor genes. HBx also induced global hypomethylation by down-regulating DNMT3B (23). As mentioned earlier, we found that HBx could recruit DNMT1 to increase methylation at the putative GRE of the MAT1A promoter (Fig. 3). Therefore, we speculated that HBx might promote regional hypermethylation by up-regulating DNMT1 and lead to repressed MAT1A expression. Next, we investigated the methylation profile of CpG sites in the promoter sequence of MAT1A in four pairs of liver tissues. We found that the rates of methylation of CpG sites of the MAT1A promoter were higher in HBV-associated HCC tissues than in adjacent nontumor tissues (Fig. 4, C and D).
FIGURE 4.
Determination of MAT1A, GR, HBx, and DNMTs expression and methylation profiles in the MAT1A promoter in HBV-associated HCC tissues. A, representative results of immunohistochemistry analyses. Panels a and b, MAT1A; panels c and d, GR; panels e and f, HBx; panels g and h, DNMT1; panels i and j, DNMT3A; panels k and l, DNMAT3B. B, four adjacent paired HBV-associated HCC tissues (T) and peritumoral noncancerous tissues (N) were selected for immunoblotting analyses using antibodies to MAT1A and GAPDH proteins. The inset shows representative immunoblots of different tissues. **, p < 0.01. C and D, methylation profile of CpG sites for promoter sequence of MAT1A. *, p < 0.05. The color of the circles is related to the percent of methylation in each CpG site. Shown is a representative result from four independent experiments.
TABLE 3.
Correlation of HBx protein expression with DNMT1, DNMT3A, DNMT3B, MAT1A, GR protein expression, and patients' clinicopathologic characteristics in hepatocellular carcinomas and noncancerous tissues
The correlations between the protein expression and tissue types were analyzed using a χ2 or Fisher's exact test.
| Characteristic | HBx expression HCC tissues |
Correlation | p value | HBx expression noncancerous tissues |
Correlation | p value | |||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | ||||||
| DNMT1 expression | Negative | 14 | 3 | 0.557 | 0.010a | 19 | 2 | 0.600 | 0.016a |
| Positive | 2 | 6 | 1 | 3 | |||||
| DNMT3A expression | Negative | 5 | 5 | 0.870 | 11 | 4 | 0.615 | ||
| Positive | 8 | 7 | 9 | 1 | |||||
| DNMT3B expression | Negative | 1 | 12 | −0.923 | 0.000a | 3 | 1 | 1.000 | |
| Positive | 12 | 0 | 17 | 4 | |||||
| MAT1A expression | Negative | 18 | 2 | 0.656 | 0.005a | 7 | 3 | 0.500 | 0.034a |
| Positive | 1 | 4 | 3 | 12 | |||||
| GR expression | Negative | 4 | 3 | 1.000 | 8 | 8 | 0.428 | ||
| Positive | 8 | 9 | 7 | 3 | |||||
| Sex | Male | 10 | 10 | 1.000 | 16 | 4 | 1.000 | ||
| Female | 3 | 2 | 4 | 1 | |||||
| Liver cirrhosis | No | 6 | 2 | 0.557 | 0.010a | 5 | 3 | 0.673 | |
| Yes | 3 | 14 | 8 | 9 | |||||
| AFP (ng/ml) | ≤ 200 | 3 | 0 | 0.538 | 0.024a | 2 | 1 | 0.543 | |
| >200 | 5 | 17 | 8 | 14 | |||||
a p < 0.05 was considered significant.
HBV Inhibited MAT1A Expression by Site-specific Hypermethylation within the GRE in the MAT1A Promoter
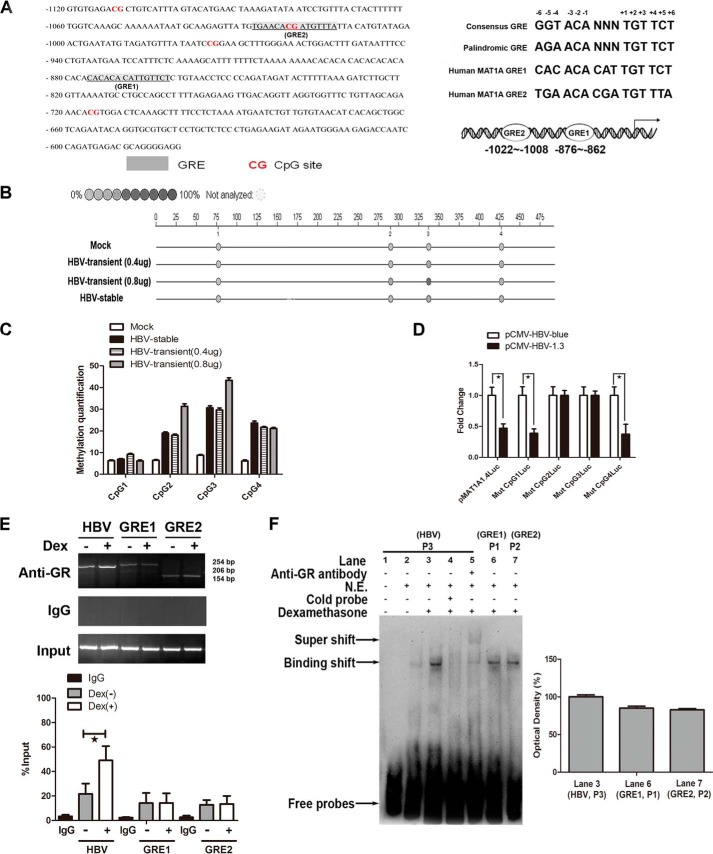
To clarify the role of HBV in aberrant epigenetic modifications at the putative GRE of the MAT1A promoter, we located two putative GR-binding sites in the GRE1 (nt −876 to −862) and GRE2 (nt −1022 to −1008) in the human MAT1A promoter. Five bases are necessary for maximal GRE function: −3, −2, +2, +3, and +5 (24). Of these five bases, the MAT1A-GRE1 sequence (5′-CACACACATTGTTCT-3′) contains the five optimal bases. However, the MAT1A-GRE2 sequence (5′-TGAACACGATGTTTA-3′) has only one different base (+5), where a C is substituted for a T (Fig. 5A). Thus, the MAT1A-GRE2 contains all but one of the nucleotides, which is required for full functional activity. This may be the primary reason for more binding of the GR protein to the GRE1 site than the GRE2 site. To demonstrate HBV-induced aberrant epigenetic modifications at the putative GRE in the MAT1A promoter, CpG methylation was tested by a MALDI-TOF mass array (Fig. 5B). The analysis of the DNA fragments of the MAT1A promoter, containing CpGs between nt −1120 and −620, revealed an increased methylation density in the 2nd and 3rd CpGs with increasing concentrations of transfected with pCMV-HBV1.3 (Fig. 5C). It was interesting to note that there was no significant reduction of luciferase activity when the CpG2 and CpG3 sites were mutated (Fig. 5D). These CpGs overlap with the GREs, which are important determinants for the induction of MAT1A expression, and the methylation of these CpG sites by HBV significantly reduced the activity of the MAT1A promoter.
FIGURE 5.
Effect of HBV on the methylation profile of CpGs and competition with the GR for binding to the consensus GRE in the MAT1A promoter. A, putative GRE-binding sites in the 5′-flanking region of the MAT1A gene are underlined. The human MAT1A-GRE1 and MAT1A-GRE2 were compared with the consensus GRE and the palindromic GRE. B, color of the circles is related to the percent of methylation in each CpG site. C, effect of HBV on the methylation profile of the CpG sites for the MAT1A promoter sequence. D, effect of HBV on the relative luciferase activity of the MAT1A promoter when four CpG sites were mutated in a wild-type pMAT1A-1.4Luc plasmid. *, p < 0.05. E, GR-binding profiles were examined by ChIP assays in HepG2.2.15 cells. Productions of Chip-GRE1, Chip-GRE2, and Chip-HBV were quantified by qPCR. *, p < 0.05. F, analyses of the effect of Dex on the binding of the GR to GRE of HBV (P3), and GRE1 (P1) and GRE2 (P2) of the MAT1A promoter by EMSA. Shown is a representative result from three independent experiments. N.E., nuclear extraction.
It is noteworthy that the HBV genome contains a specific DNA-binding site for the GR, and this HBV GR domain can be categorized as a functional GRE. Therefore, we further examined GR-binding profiles in HepG2.2.15 cells using ChIP analyses (Fig. 5E). The results indicated that the GR preferred to bind to the DNA sequence of HBV rather than to the promoter of MAT1A. To confirm that HBV was able to compete with MAT1A in binding to the GR at the GRE site, EMSAs were performed (Fig. 5F). We observed that the intensity of the band in lane 3 was stronger than that in lane 6 or lane 7 (Fig. 5F). The results indicated that there was more nuclear protein binding to the HBV probe than to the MAT1A promoter probe (GRE1 and GRE2 probes) after treatment with Dex. Taken together, all these results demonstrated that Dex-induced MAT1A gene expression was inhibited by HBV through site-specific hypermethylation at the GRE within the MAT1A promoter in hepatoma cells.
IFN-α Could Restore HBV-suppressed MAT1A Expression through an Antiviral Pathway
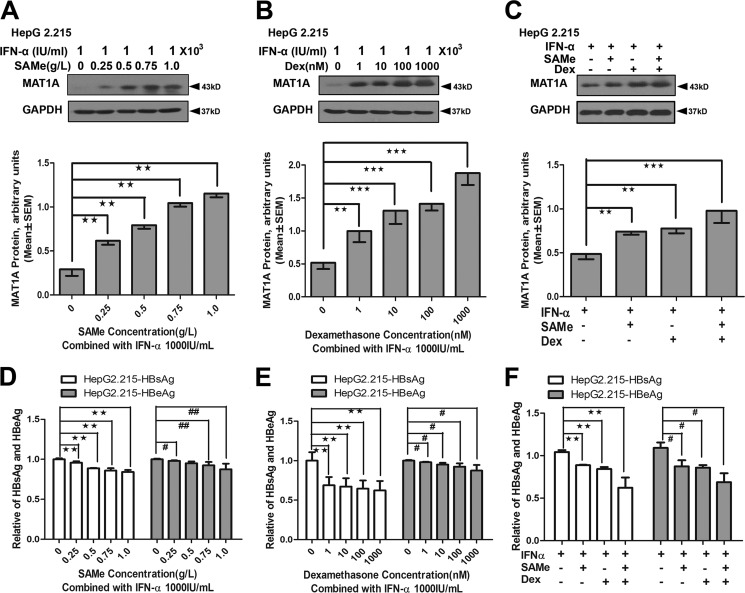
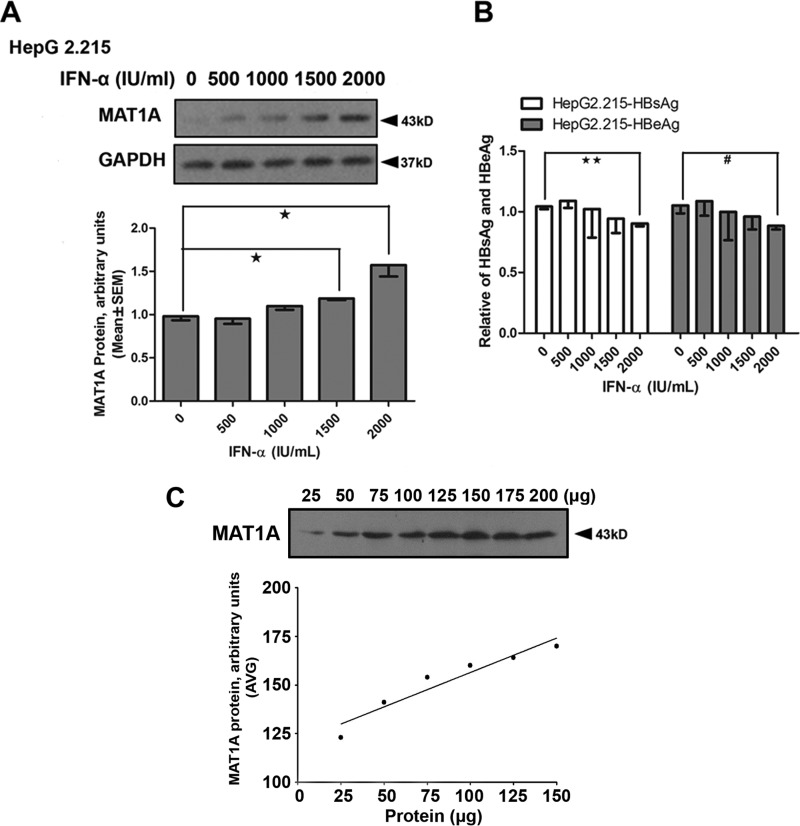
As mentioned above, Dex failed to increase the production of AdoMet in HepG2.2.15, possibly because Dex enhanced the replication of HBV. It was suggested in our previous study that HBV replication can suppress AdoMet production (22). We speculated that the antiviral drug could restore HBV-suppressed MAT1A expression through an antiviral pathway. Therefore, we used IFN-α as an antiviral drug to inhibit viral replication in this study, and we investigated the effects of Dex, AdoMet and IFN-α on the expression of MAT1A, HBsAg, and HBeAg in HepG2.2.15 (Fig. 6). The results showed that IFN-α combined with AdoMet could reduce the expression of HBsAg and HBeAg, and induce expression of MAT1A (Fig. 6, A and D). The expression of MAT1A was induced and the expression of HBsAg and HBeAg was repressed when IFN-α was combined with Dex (Fig. 6, B and E). In addition, the expression of MAT1A was significantly induced when Dex and AdoMet were combined with IFN-α (Fig. 6C), and the antiviral effect was enhanced in HepG2.2.15 (Fig. 6F). Interestingly, IFN-α could suppress the expression of HBsAg and HBeAg at a concentration of 2000 IU/ml, and IFN-α could also induce the expression of MAT1A in a concentration-dependent manner (Fig. 7). As shown in Fig. 7A, the protein levels of MAT1A were significantly increased after the treatment with IFN-α at 1500 units/ml (1.19 ± 0.03 versus 0.98 ± 0.08, p = 0.014) and 2000 units/ml (1.57 ± 0.23 versus 0.98 ± 0.08, p = 0.013) compared with that after the treatment with IFN-α at 0 units/ml. Interestingly, we observed that IFN-α could not affect the protein expression of MAT1A (Fig. 7), but the combination treatment of IFN-α, AdoMet, and Dex significantly increased the protein expression of MAT1A (Fig. 6) when the concentration of IFN-α was 1000 IU/ml. These findings indicated that the induced expression of MAT1A by IFN-α might be due to the suppression of HBV DNA replication. These results suggested that IFN-α might restore HBV-suppressed MAT1A expression through an antiviral pathway, and Dex-induced increase of AdoMet production might enhance the antiviral effect of IFN-α on HBV.
FIGURE 6.
Effect of the combination of IFN-α, AdoMet (SAMe), and Dex on expression of MAT1A, HBsAg, and HBeAg in HepG2.2.15 cells. A–C, MAT1A protein levels were detected in HepG2.2.15 cells after treatment with AdoMet combined with IFN-α, Dex combined with IFN-α, or AdoMet and Dex combined with IFN-α. The inset shows representative immunoblots of MAT1A with different treatments. D–F, HBsAg and HBeAg were determined by ELISA after treatment with AdoMet combined with IFN-α, Dex combined with IFN-α, or AdoMet and Dex combined with IFN-α in HepG2.2.15 cells. **, p < 0.01, and ***, p < 0.001; #, p < 0.05, and ##, p < 0.01. Shown is a representative result from three independent experiments.
FIGURE 7.
Effect of IFN-α on expression of MAT1A, HBsAg, and HBeAg in HepG2.2.15 cells. A, MAT1A protein levels were detected in HepG2.2.15 cells after treatment with IFN-α. The inset shows representative immunoblots of MAT1A with different treatments. B, HBsAg and HBeAg were determined by ELISA after treatment with IFN-α in HepG2.2.15 cells. C, dilution curve of the total protein shows linear MAT1A protein levels from 25 to 150 μg of protein. *, p < 0.05, and **, p < 0.01; #, p < 0.05. Shown is a representative result from three independent experiments.
Dex-induced Increase of AdoMet Production Restored STAT1 Methylation Rather than Phosphorylation
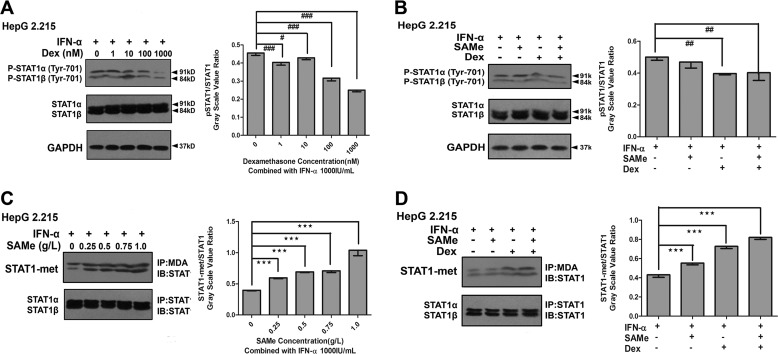
Recent evidence suggests that HBV has evolved strategies to block the nuclear translocation of STAT1 to limit IFN-α-induced cellular antiviral responses (18). Because of the crucial role of STAT1 phosphorylation in IFN-α signaling, we investigated whether Dex and AdoMet could potentially influence the phosphorylation of STAT1 responding to IFN-α in HepG2.2.15. We pretreated HepG2.2.15 cells with different doses of Dex, followed by treatment with IFN-α, and we then detected the phosphorylated STAT1 by immunoblot analysis using a specific anti-phospho-STAT1 antibody. The results showed that Dex repressed the phosphorylation of STAT1 responding to IFN-α in a concentration-dependent manner (Fig. 8A). As shown in Fig. 8B, the phosphorylation of STAT1 was decreased by 20.80% (0.40 ± 0.01 versus 0.50 ± 0.02, p = 0.004) after the treatment with IFN-α and Dex compared with that after the treatment with IFN-α alone. The phosphorylation of STAT1 was lower (0.40 ± 0.05 versus 0.50 ± 0.02, p = 0.006) after the treatment with IFN-α, AdoMet, and Dex than that after the treatment with IFN-α alone. However, AdoMet did not enhance the suppression by Dex of the phosphorylation of STAT1 responding to IFN-α (Fig. 8B).
FIGURE 8.
Effect of combination treatment with Dex and AdoMet (SAMe) on IFN-α-dependent STAT1 phosphorylation and methylation in HepG2.2.15 cells. A, cells were pretreated with different concentrations (0–1000 nm) of Dex for 16 h, followed by treatment with IFN-α (1000 IU/ml) for 8 h. B, cells were pretreated with or without Dex (100 nm) and/or AdoMet (SAMe) (0.75 g/liter) for 16 h, followed by treatment with IFN-α (1000 IU/ml) for 8 h. The inset shows the ratio of pSTAT1/STAT1 with different treatments. STAT1 methylation (immunoprecipitation (IP) with antibody to methyl- and dimethylarginine (MDA), Western blot with anti-STAT1 antibody) was detected by co-IP analysis. STAT1 protein was used as a loading control. C, cells were pretreated with different concentrations (0–1 g/liter) of AdoMet for 16 h, followed by treatment with IFN-α (1000 IU/ml) for 8 h. D, cells were pretreated with or without Dex (100 nm) or/and AdoMet (0.75 g/liter) for 16 h, followed by treatment with IFN-α (1000 IU/ml) for 8 h. The inset shows the ratio of STAT1-met/STAT1 with different treatments. ***, p < 0.001; #, p < 0.05; ##, p < 0.01, and ###, p < 0.001. Shown is a representative result from three independent experiments. IB, immunoblot.
Furthermore, methylation is functionally essential for STAT1, as unmethylated STAT1 can be bound and inactivated by a protein inhibitor of activated STAT1 (PIAS1) (25, 26). We investigated whether AdoMet and Dex could influence the methylation of STAT1 responding to IFN-α in HepG2.2.15. To test whether the combination of AdoMet and IFN-α can improve the methylation of STAT1, we pretreated HepG2.2.15 cells with AdoMet, followed by treatment with IFN-α. As shown in Fig. 8C, the methylation of STAT1 was effectively induced by AdoMet in a concentration-dependent manner. As shown in Fig. 8D, STAT1 methylation was significantly increased by 1.28-fold (0.55 ± 0.02 versus 0.43 ± 0.02, p < 0.001) after combination treatment with IFN-α and AdoMet compared with that after treatment with IFN-α alone. STAT1 methylation was increased by 1.70-fold (0.73 ± 0.02 versus 0.43 ± 0.02, p < 0.001) after treatment with IFN-α and Dex compared with that after treatment with IFN-α alone. STAT1 methylation was increased by 1.91-fold (0.82 ± 0.02 versus 0.43 ± 0.02, p < 0.001) after treatment with IFN-α, AdoMet, and Dex compared with that after treatment with IFN-α alone. These results showed that the combination treatment of AdoMet and Dex significantly induced the methylation of STAT1 responding to IFN-α and the Dex-induced increase of AdoMet production restored STAT1 methylation rather than phosphorylation.
GC-induced Increase of AdoMet Production Altered Arginine Methylation of STAT1 by the Protein-arginine Methyltransferase (PRMT1)
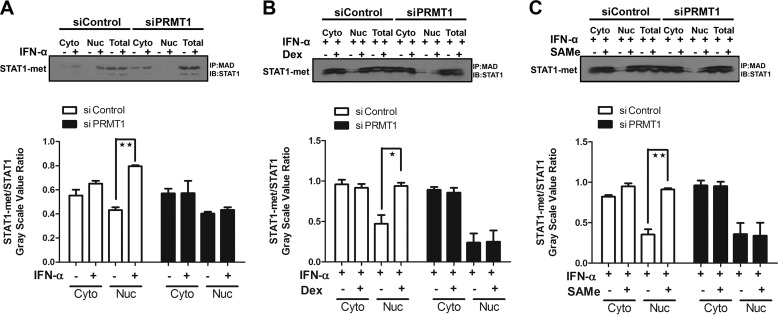
Arginine methylation of STAT1 is an additional post-translational modification regulating transcription factor function, and alteration of arginine methylation might be responsible for the lack of interferon responsiveness observed in hepatoma cells. To demonstrate the mechanistic insight into the effect of GCs on IFN action, we knocked down PRMT1 with siRNA (5′-CGUCAAAGCCAACAAGUUA-3′). The results showed that methylation of STAT1 was induced by IFN-α, but IFN-α failed to promote the methylation of STAT1 when PRMT1 was knocked down with siPRMT1 (Fig. 9A). As shown in Fig. 9, B and C, similar results were observed after treatment with IFN-α and Dex, as well as IFN-α and AdoMet. These results indicated the effect of GCs on the antiviral response of IFN-α action through altering arginine methylation status of STAT1, which was catalyzed by PRMT1.
FIGURE 9.
Arginine methylation of STAT1 was catalyzed by PRMT1. STAT1 methylation (immunoprecipitation (IP) with antibody to methyl- and dimethylarginine (MDA), Western blot with anti-STAT1 antibody) was detected by co-IP analysis. STAT1 protein was used as a loading control. STAT1 methylation levels were detected after HepG2.2.15 cells were transfected with siControl or siPRMT1. A, cells were treated with vehicle or IFN-α (1000 IU/ml) for 24 h. B and C, cells were pretreated with or without Dex (100 nm) or AdoMet (0.75 g/liter) for 16 h, followed by treatment with IFN-α (1000 IU/ml) for 8 h. The inset shows the ratio of STAT1-met/STAT1 with different treatments. *, p < 0.05; **, p < 0.01. Shown is a representative result from three independent experiments. IB, immunoblot; Nuc, nuclear protein; Cyto, cytoplasmic protein.
DISCUSSION
HBV infection is a serious global health problem, with 2 billion people infected worldwide, and 350 million suffering from chronic HBV infection. Currently, treatment with IFN-α is one of the major therapies that have been approved for CHB patients. Conventional use of IFN-α has produced encouraging results, with HBeAg loss rates of 20–50% (27). However, HBV, as a hepatotropic DNA virus, may have a low sensitivity to IFN-induced ISGs and may counteract IFN actions at different levels, including the IFN signal transduction and antiviral functions of ISG products (28). Although the effect of IFN seems indisputable, response rates are unsatisfactory, from a clinical point of view. Pretreatment with GCs is one of the proposed ways to increase the response to IFN-α treatment. The rationale for GC pretreatment therapy stems from an early clinical observation that patients with chronic HBV infection often cleared markers of viral replication following tapering or discontinued GC treatment (7). The exact mechanism underlying the effectiveness of combination regimen has not been fully elucidated.
As a major methyl donor, the availability of AdoMet potentially has profound effects on liver metabolism, and AdoMet synthesis is depressed in chronic liver disease (12). Thus, there has been considerable interest in the utility of AdoMet to ameliorate disease severity (13). Furthermore, hepatocellular injury in cholestasis is frequently associated with glutathione depletion, and thus, AdoMet may help correct this problem (29, 30). These findings suggest that any drug that can increase the steady-state level of AdoMet could offer substantial clinical benefits for restoring liver function. Recently, studies have shown that AdoMet may improve IFN signaling and antiviral effects (31, 32). GCs strongly up-regulate AdoMet synthetase both in vivo and in vitro (14, 15). Therefore, we speculated that the GC-induced increase of AdoMet production enhances IFN signaling in HBV-infected cells. To confirm our speculation, we investigated the effect of GCs and IFN-α on AdoMet production and MAT1A expression in HepG2.2.15 cells. We found that AdoMet homeostasis was disrupted by pharmacologic concentrations of GCs. AdoMet and the ratio of AdoMet/AdoHcy were markedly increased in Dex-treated cells, including normal hepatic L02 cells and HepG2 cells. However, Dex could not induce MAT1A expression, even at a high dose in HepG2.2.15 cells, which may be due to the induction of the expression of HBsAg and HBeAg by promoting the replication of HBV. The expression of HBsAg and HBeAg was repressed with the use of IFN-α at a dose of 2000 IU/ml, which was consistent with previous studies (18–20), and the expression of MAT1A was induced, and AdoMet production was increased in HepG2.2.15 cells. Interestingly, IFN-α can also induce the expression of MAT1A in a concentration-dependent manner, which may be due to IFN-α suppression of HBV DNA replication. These results indicated that GCs could increase antiviral effects by inducing AdoMet production when HBV was effectively suppressed by IFN-α. Furthermore, we observed that HBV suppressed AdoMet production and MAT1A expression induced by Dex. To investigate the mechanism of the transcriptional regulation of the MAT1A gene by Dex, we evaluated the 5′-flanking sequence of the MAT1A gene within 1474 bp upstream of the transcription start site by a transient transfection assay. We found that the GRE in the promoter was an important cis-regulatory element and that the sequence between nt −1474 and −974 of the MAT1A promoter along with two GRE sites (nt −876 to −862 and nt −1022 to −1008) were required for the functional induction of MAT1A expression by Dex. The GR participates in Dex-induced MAT1A expression by being translocated to the nucleus. We observed that GCs facilitated the binding of the GR to the MAT1A promoter in GRE1 (nt −876 to −862) and GRE2 (nt −1022 to −1008). To further verify the role of HBV and GCs in the regulation of MAT1A expression, we studied whether post-transcriptional regulation is involved in HBV-repressed MAT1A mRNA expression induced by GCs. Our results suggested that Dex-induced MAT1A expression was disrupted by HBV, which may be due to HBx recruiting DNMT1 to increase methylation at the putative GRE of the MAT1A promoter. It has been demonstrated that HBx expression increased total DNMT activities by up-regulation of DNMT1, DNMT3A1, and DNMT3A2 and selectively promoted regional hypermethylation of specific tumor suppressor genes leading to regional hypermethylation and global hypomethylation during the formation of HCC (23). HBV inhibited MAT1A expression through CpG2 and CpG3 hypermethylation within the MAT1A promoter. Although CpG3 is not located within the GRE, HBV may affect the methylation status of CpG3 in a direct or indirect manner, which is the neighbor dependence mechanism (33).
Previous studies have demonstrated that nucleocapsid proteins of HBV could be involved in a deficient IFN-α response (34, 35). The primary and most important signaling pathway activated by IFNs is the JAK-STAT pathway. By binding to type I IFN receptors, IFN-α triggers the oligomerization and tyrosine phosphorylation of the receptors followed by the activation of receptor-associated Janus tyrosine kinase (JAK) (36). Recently, studies have suggested that type I IFNs are critical GC targets for regulating STAT1 activity and may account for the overall effectiveness of GCs in inflammation suppression in a clinically relevant time (37). However, type I IFN receptors were expressed to a much higher extent in HepG2.2.15 cells than in HepG2 cells. Therefore, the potential role of STAT1 methylation remains controversial (18). It is thus necessary to further investigate the effect of the GC-induced increase of AdoMet production on the STAT pathway to obtain a more accurate picture. Recent studies have shown that AdoMet can increase the induction of ISGs and the antiviral effects of IFN-α by increasing STAT1 methylation, possibly affecting STAT1-DNA binding (31). Inhibition of STAT1 methylation is involved in the resistance of hepatitis B virus to IFN-α (18). These studies suggest that AdoMet can restore STAT1 methylation and improve IFN-α signaling in vitro. In this study, we found that the combination of AdoMet and Dex significantly induced the methylation of STAT1 responding to IFN-α. Although Dex suppressed STAT1 phosphorylation, the addition of AdoMet had no effect on STAT1 phosphorylation. These results showed that the Dex-induced increase of AdoMet production enhanced the antiviral effect of IFN-α by restoring STAT1 methylation rather than phosphorylation in HBV-infected cells. Furthermore, Mowen et al. (38) have demonstrated that methylation of an arginine in STAT1 is catalyzed by PRMT1, which is a novel requirement for IFNα/β-induced transcription. Alignment of the N termini of the seven mammalian STATs reveals a region of high homology and an invariant arginine at position 31 (Arg-31), which is an efficient substrate for methylation (38). For STAT1 methylation, PRMT1 always uses AdoMet, which is one of the most frequently used enzyme substrates and is recognized as the major methyl donor in all living organisms (39). In this study, the results indicated that the effect of GCs on IFN-α action through altering arginine methylation status of STAT1, which catalyzed by PRMT1.
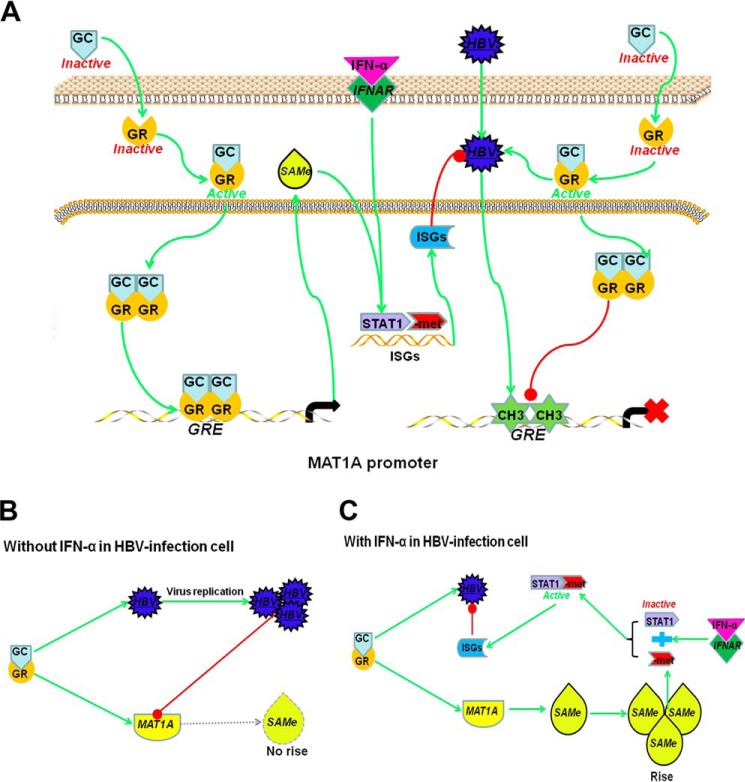
Our data demonstrated that GCs directly regulated the MAT1A expression in vitro by enhancing the binding of the GR to GRE in the MAT1A promoter. GCs can also activate HBV replication by enhancing the binding of the GR to GRE in the HBV genome. HBV infection leads to hypermethylation in the MAT1A promoter by recruiting DNMT1 and disturbs GR binding to GRE in the MAT1A promoter. Thus, GC-induced AdoMet production and MAT1A expression were disrupted by HBV through site-specific hypermethylation at GRE sites within the MAT1A promoter and competitive binding with the GR in vitro. However, when HBV replication was effectively suppressed by IFN-α, GCs induced an increase of AdoMet production through a positive feedback loop, which enhanced the antiviral effect of IFN-α by improving arginine methylation of STAT1, rather than phosphorylation (Fig. 10). These findings suggest that combination therapy of GCs, AdoMet, and IFN-α is possibly useful for patients with CHB.
FIGURE 10.
Proposed mechanism/model for the rationale of treatment with a combination regimen of GCs and IFN-α in HBV-infected cell. A, GR is stimulated by GCs and translocates to the nucleus. GCs induce MAT1A expression by enhancing the binding of GR to GREs in the MAT1A promoter, which induces the production of AdoMet (SAMe). GC-induced production of AdoMet, which enhances the antiviral effect of IFN-α. HBV infection leads to hypermethylation in the MAT1A promoter and disturbs GR binding to GRE in the MAT1A promoter. B, in HBV-infected cells not treated with IFN-α, HBV was able to compete with MAT1A for binding to GR at the GRE site. GCs activate HBV replication, which suppresses the expression of MAT1A and production of AdoMet. C, in HBV-infected cells treated with IFN-α, HBV replication was effectively suppressed by IFN-α, GCs induced an increase of AdoMet production through a positive feedback loop, which promoted expression of the ISGs and enhanced the antiviral effect of IFN-α by improving STAT1 methylation rather than phosphorylation.
Acknowledgments
We thank the editors at American Journal Experts for valuable contributions in editing and revising the manuscript. We are grateful to Dr. Ying Zhu and the State Key Laboratory of Virology (College of Life Sciences, Wuhan University) for the generous gift of the pCMV-HBV-1.3 plasmid.
This work was supported by National Natural Science Foundation of China Grants 30872491/C160402, 81372552, and 81172349/H1617.
- HBV
- hepatitis B virus
- CHB
- chronic hepatitis B
- Dex
- dexamethasone
- DNMT
- DNA methyltransferase
- GC
- glucocorticoid
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid-response element
- HBeAg
- hepatitis B e antigen
- HBsAg
- hepatitis B surface antigen
- HBx
- the X protein of hepatitis B virus
- HCC
- hepatocellular carcinoma
- ISG
- interferon-stimulated genes
- AdoHcy
- S-adenosylhomocysteine
- AdoMet
- S-adenosylmethionine
- nt
- nucleotide.
REFERENCES
- 1. Liaw Y. F., Chu C. M. (2009) Hepatitis B virus infection. Lancet 373, 582–592 [DOI] [PubMed] [Google Scholar]
- 2. Locarnini S. (2004) Molecular virology of hepatitis B virus. Semin. Liver Dis. Suppl. 1, 24, S3–S10 [DOI] [PubMed] [Google Scholar]
- 3. Wong D. K., Cheung A. M., O'Rourke K., Naylor C. D., Detsky A. S., Heathcote J. (1993) Effect of α-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann. Intern. Med. 119, 312–323 [DOI] [PubMed] [Google Scholar]
- 4. Férir G., Kaptein S., Neyts J., De Clercq E. (2008) Antiviral treatment of chronic hepatitis B virus infections: the past, the present and the future. Rev. Med. Virol. 18, 19–34 [DOI] [PubMed] [Google Scholar]
- 5. Rakela J., Redeker A. G., Weliky B. (1983) Effect of short-term prednisone therapy on aminotransferase levels and hepatitis B virus markers in chronic type B hepatitis. Gastroenterology 84, 956–960 [PubMed] [Google Scholar]
- 6. Hayata T., Nakano Y., Yoshizawa K., Sodeyama T., Kiyosawa K. (1991) Effects of interferon on intrahepatic human leukocyte antigens and lymphocyte subsets in patients with chronic hepatitis B and C. Hepatology 13, 1022–1028 [PubMed] [Google Scholar]
- 7. Mellerup M. T., Krogsgaard K., Mathurin P., Gluud C., Poynard T. (2005) Sequential combination of glucocorticosteroids and α interferon versus α interferon alone for HBeAg-positive chronic hepatitis B. Cochrane Database Syst. Rev. 3, CD000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu S. C., Mato J. M. (2012) S-Adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 92, 1515–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu S. C., Mato J. M. (2005) Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol. 35, 227–234 [DOI] [PubMed] [Google Scholar]
- 10. Avila M. A., Berasain C., Torres L., Martín-Duce A., Corrales F. J., Yang H., Prieto J., Lu S. C., Caballería J., Rodés J., Mato J. M. (2000) Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 33, 907–914 [DOI] [PubMed] [Google Scholar]
- 11. Cai J., Mao Z., Hwang J. J., Lu S. C. (1998) Differential expression of methionine adenosyltransferase genes influences the rate of growth of human hepatocellular carcinoma cells. Cancer Res. 58, 1444–1450 [PubMed] [Google Scholar]
- 12. Cai J., Sun W. M., Hwang J. J., Stain S. C., Lu S. C. (1996) Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology 24, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 13. Anstee Q. M., Day C. P. (2012) S-Adenosylmethionine (SAMe) therapy in liver disease: a review of current evidence and clinical utility. J. Hepatol. 57, 1097–1109 [DOI] [PubMed] [Google Scholar]
- 14. Gil B., Pajares M. A., Mato J. M., Alvarez L. (1997) Glucocorticoid regulation of hepatic S-adenosylmethionine synthetase gene expression. Endocrinology 138, 1251–1258 [DOI] [PubMed] [Google Scholar]
- 15. García-Trevijano E. R., Latasa M. U., Carretero M. V., Berasain C., Mato J. M., Avila M. A. (2000) S-Adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: a new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J. 14, 2511–2518 [DOI] [PubMed] [Google Scholar]
- 16. Mato J. M., Corrales F. J., Lu S. C., Avila M. A. (2002) S-Adenosylmethionine: a control switch that regulates liver function. FASEB J. 16, 15–26 [DOI] [PubMed] [Google Scholar]
- 17. Li J., Ramani K., Sun Z., Zee C., Grant E. G., Yang H., Xia M., Oh P., Ko K., Mato J. M., Lu S. C. (2010) Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenicity in mice. Am. J. Pathol. 176, 2456–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Chen F., Zheng M., Zhu H., Zhao D., Liu W., Liu W., Chen Z. (2010) Inhibition of STAT1 methylation is involved in the resistance of hepatitis B virus to interferon α. Antiviral Res. 85, 463–469 [DOI] [PubMed] [Google Scholar]
- 19. Wang J., Jiang D., Zhang H., Lv S., Rao H., Fei R., Wei L. (2009) Proteome responses to stable hepatitis B virus transfection and following interferon α treatment in human liver cell line HepG2. Proteomics 9, 1672–1682 [DOI] [PubMed] [Google Scholar]
- 20. Sun D., Nassal M. (2006) Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus. J. Hepatol. 45, 636–645 [DOI] [PubMed] [Google Scholar]
- 21. Li Y., Xie J., Xu X., Liu L., Wan Y., Liu Y., Zhu C., Zhu Y. (2013) Inducible interleukin 32 (IL-32) exerts extensive antiviral function via selective stimulation of interferon λ1 (IFN-λ1). J. Biol. Chem. 288, 20927–20941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Q., Chen J., Liu L., Zhang J., Wang D., Ma L., He Y., Liu Y., Liu Z., Wu J. (2011) The X protein of hepatitis B virus inhibits apoptosis in hepatoma cells through enhancing the methionine adenosyltransferase 2A gene expression and reducing S-adenosylmethionine production. J. Biol. Chem. 286, 17168–17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park I. Y., Sohn B. H., Yu E., Suh D. J., Chung Y. H., Lee J. H., Surzycki S. J., Lee Y. I. (2007) Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology 132, 1476–1494 [DOI] [PubMed] [Google Scholar]
- 24. Nordeen S. K., Suh B. J., Kühnel B., Hutchison C. A., 3rd (1990) Structural determinants of a glucocorticoid receptor recognition element. Mol. Endocrinol. 4, 1866–1873 [DOI] [PubMed] [Google Scholar]
- 25. Liao J., Fu Y., Shuai K. (2000) Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1-Stat1 interaction. Proc. Natl. Acad. Sci. U.S.A. 97, 5267–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu B., Mink S., Wong K. A., Stein N., Getman C., Dempsey P. W., Wu H., Shuai K. (2004) PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 5, 891–898 [DOI] [PubMed] [Google Scholar]
- 27. Lavanchy D. (2005) Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J. Clin. Virol. 34, S1–S3 [DOI] [PubMed] [Google Scholar]
- 28. Chen J., Wu M., Zhang X., Zhang W., Zhang Z., Chen L., He J., Zheng Y., Chen C., Wang F., Hu Y., Zhou X., Wang C., Xu Y., Lu M., Yuan Z. (2013) Hepatitis B virus polymerase impairs interferon-α-induced STAT activation through inhibition of importin-α5 and protein kinase C-δ. Hepatology 57, 470–482 [DOI] [PubMed] [Google Scholar]
- 29. Frezza M., Surrenti C., Manzillo G., Fiaccadori F., Bortolini M., Di Padova C. (1990) Oral S-adenosylmethionine in the symptomatic treatment of intrahepatic cholestasis. A double-blind, placebo-controlled study. Gastroenterology 99, 211–215 [DOI] [PubMed] [Google Scholar]
- 30. Qin B., Guo S., Zhao Y., Zou S., Zhang Q., Wang Z., Zeng W., Zhang D. (2000) A trial of ademetionine in the treatment of intrahepatic biliary stasis viral hepatitis. Zhonghua Gan Zang Bing Za Zhi 8, 158–160 [PubMed] [Google Scholar]
- 31. Feld J. J., Modi A. A., El-Diwany R., Rotman Y., Thomas E., Ahlenstiel G., Titerence R., Koh C., Cherepanov V., Heller T., Ghany M. G., Park Y., Hoofnagle J. H., Liang T. J. (2011) S-Adenosyl methionine improves early viral responses and interferon-stimulated gene induction in hepatitis C nonresponders. Gastroenterology 140, 830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Filipowicz M., Bernsmeier C., Terracciano L., Duong F. H., Heim M. H. (2010) S-Adenosyl-methionine and betaine improve early virological response in chronic hepatitis C patients with previous nonresponse. PLoS One 5, e15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonello N., Sampson J., Burn J., Wilson I. J., McGrown G., Margison G. P., Thorncroft M., Crossbie P., Povey A. C., Santibanez-Koref M., Walters K. (2013) Bayesian inference supports a location and neighbour-dependent model of DNA methylation propagation at the MGMT gene promoter in lung tumours. J. Theor. Biol. 336, 87–95 [DOI] [PubMed] [Google Scholar]
- 34. Foster G. R., Goldin R. D., Hay A., McGarvey M. J., Stark G. R., Thomas H. C. (1993) Expression of the terminal protein of hepatitis B virus is associated with failure to respond to interferon therapy. Hepatology 17, 757–762 [PubMed] [Google Scholar]
- 35. Rosmorduc O., Sirma H., Soussan P., Gordien E., Lebon P., Horisberger M., Bréchot C., Kremsdorf D. (1999) Inhibition of interferon-inducible MxA protein expression by hepatitis B virus capsid protein. J. Gen. Virol. 80, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 36. Der S. D., Zhou A., Williams B. R., Silverman R. H. (1998) Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 95, 15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhattacharyya S., Zhao Y., Kay T. W., Muglia L. J. (2011) Glucocorticoids target suppressor of cytokine signaling 1 (SOCS1) and type 1 interferons to regulate Toll-like receptor-induced STAT1 activation. Proc. Natl. Acad. Sci. U.S.A. 108, 9554–9559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mowen K. A., Tang J., Zhu W., Schurter B. T., Shuai K., Herschman H. R., David M. (2001) Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell 104, 731–741 [DOI] [PubMed] [Google Scholar]
- 39. Grillo M. A., Colombatto S. (2005) S-Adenosylmethionine and protein methylation. Amino Acids 28, 357–362 [DOI] [PubMed] [Google Scholar]