Abstract
Background:
Peucedanum praeruptorum L., a traditional Chinese medicine known as Qian-hu, is commonly used for dispelling wind-heat and expectorant and loss of energy. However, due to similar morphological characters and high market demand, there are many substitutes and adulterants of P. praeruptorum. DNA barcoding is an approach to identify species based on sequences from a short, standardized DNA region.
Objective:
To authenticate P. praeruptorum from its substitutes and adulterants.
Materials and Methods:
The differential identification of P. praeruptorum and 13 regional substitutes and 23 adulterants was investigated by means of DNA sequence analysis of internal transcribed spacer (ITS) region of the nuclear ribosomal DNA (nrDNA), a bootstrap neighbor-joining (NJ) tree according to Kimura's 2-parameter method was also calculated.
Results:
The data showed that P. praeruptorum, its substitutes and adulterants could be easily distinguished at the DNA level, while almost all species were well resolved, and successfully identified on the NJ tree.
Conclusion:
The ITS sequence can be used for the identification of P. praeruptorum and to distinguish it from common substitutes and adulterants.
Keywords: Apiaceae, DNA barcoding, identification, nrDNA ITS, Peucedanumpraeruptorum L.
INTRODUCTION
The family Apiaceae (Umbelliferae) includes some of the world's most important medicinal and poisonous plants, such as angelica, bupleurum, anise (aniseed), celeriac, water hemlock and fool's parsley.[1] Among which, Peucedani Radix, derived from the roots of Peucedanum praeruptorum L., is a well-known traditional Chinese medicine called Qian-hu (Bai-hua Qian-hu), together with Peucedani decursivi Radix derived from Angelica decursiva L., which has been separately recorded as Zi-hua Qian-hu in the current Pharmacopoeia of the People's Republic of China.[2] P. praeruptorum was recorded as medicinal plant as early as the Liang Dynasty in Ming-yi-bie-lu (Apendant Records of Famous Physicians) and was also included in the ancient encyclopedia, China Compendium of Materia Medica of Ming Dynasty.[3] Phytochemical studies have shown that the active ingredients of P.praeruptorum include volatile oils and coumarins such as praeruptorin A and praeruptorin B,[4,5,6] which can deal with anemopyretic cold, cough with abundance of phlegm, impeded chest as well as removing nebula for improving eyesight. However, due to morphological similarities, high market demands and regional factors, 41 species in 16 genera of the Apiaceae are often misused or used as substitutes for P. praeruptorum.[3] Available criteria and methods to authenticate P. praeruptorum, are mainly based on its morphological characters and analysis of chemical compounds[7,8,9,10], which are susceptible to intrinsic and extrinsic factors such as the time of harvest, availability of experts and processing methods etc.[11,12] Therefore, a reliable method to discriminate P. praeruptorum precisely from its substitutes and adulterants is needed.
DNA barcoding, an approach to identify species based on sequences from a short, standardized DNA region, opens up a unique avenue for the identification of organisms.[13,14] Among the several candidate DNA barcodes, the internal transcribed spacer (ITS) of the nuclear ribosomal DNA (nrDNA) has been recently proposed for incorporation into the core barcode, and has the potential to be used as a standard DNA barcode to identify medicinal plants and their close relatives.[15,16] Furthermore, ITS is the best and most popular marker for lower level phylogenetic analysis of Apiaceae and has demonstrated excellent reliability for species resolution.[17,18,19,20,21] In this study, the ITS regions of P. praeruptorum and its substitutes and adulterants were sequenced and compared to explore the possibility of using them to differentiate between them. The classification tree constructed by their sequences is also discussed.
MATERIALS AND METHODS
Plant material
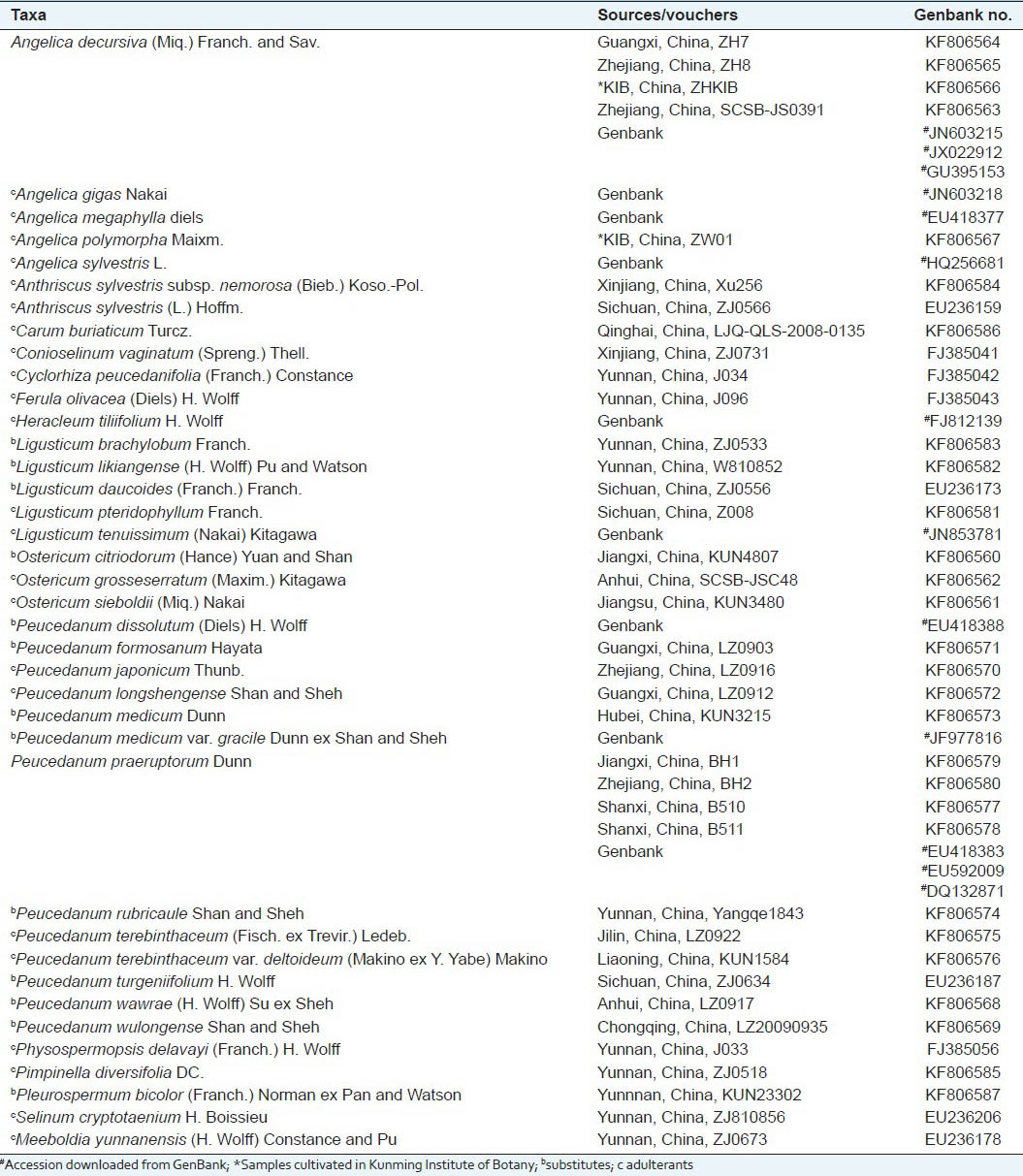
Samples for analysis were obtained from collections of natural populations by the authors or from specimens deposited in Kunming Institute of Botany, Chinese Academy of Sciences (KUN). All accessions were identified using published keys by the first author [Table 1], and corresponding vouchers were deposited at KUN (Kunming Institute of Botany, Chinese Academy of Sciences). For P. praeruptorum and A. decursiva, seven accessions collected from different locations or downloaded from Genbank, which were examined for possible infraspecific molecular variation. In total, 50 accessions representing 38 species/variants covering P. praeruptorum and most of its substitutes and adulterants were included.
Table 1.
Taxa, sources, vouchers and GenBank accession numbers for species investigated in this study
DNA extraction, amplification, sequencing, and data analysis
Total genomic DNA was extracted from fresh, silica-gel-dried or herbarium leaf material using the modified hexadecyltrimethylammonium bromide (CTAB) procedure of Doyle and Doyle.[22] Double-stranded DNAs of the complete ITS region (including ITS1, 5.8S and ITS-2) were polymerase chain reaction (PCR)-amplified using primers ITS-4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) and ITS-5 (5′-GGA AGT AAA AGT CGT AAC AAG G-3′).[23] These PCR reactions contained 2.0 μl of 10 × Taq DNA polymerase reaction buffer (TaKaRa Biotechnology Dalian Co., Ltd.), 2.5 mM/L of each dNTP (TaKaRa), 1.5 mM/L of MgCl2, 1.0 μl of 5% dimethyl sulfoxide, 0.2 mM/L of each primer (Shanghai Sangon Biological Engineering Technology and Service Co., Ltd.), 1.5 Units of AmpliTaq DNA polymerase (TaKaRa), 1.5 μl of unquantified genomic template DNA, and sterile water to a final volume of 20 μl. The PCR parameters were as follows: Initial denaturation for 3 min at 94°C, followed by 30 cycles of denaturation (94°, 45 s), annealing (55°C, 1 min) and extension (72°C, 3 min), and a final extension for 7 min at 72°C. Purifying and bidirectional sequencing were completed by Sangon Co., Ltd. DNA sequences for each accession was produced using SeqMan (DNAstar), aligned and corrected manually using the BioEdit (version 7.0.5). The Molecular Evolutionary Genetics Analysis (MEGA 4.0) was used to generate Kimura 2-parameter (K2P) distance matrices and for intraspecific and interspecific sequence similarity. Additionally, a bootstrap Neighbor-Joining (NJ) tree was calculated according to K2P method with bootstrap testing of 1000 replicates. The alignment of the ITS sequences are available upon request.
RESULTS
All samples analyzed were successfully amplified and sequenced with the universal primers “ITS4” and “ITS5”. The final aligned data matrix contained 623 positions, ranging in length from 593bp to 605bp. Of these, 217 sites were parsimony informative, 318 were constant, and 88 were autapomorphic. On average, ITS1 was slightly shorter than ITS2, but it provided parsimony informative sites almost as much as ITS2. The means of guanine-cytosine (GC) content were same in ITS1 and ITS2 sequences [Table 2]. Each of the seven accessions of P. praeruptorum and A. decursiva yielded identical DNA sequences, showing that their ITS sequences are homologous regardless of geographical origin. The sequence divergence among P. praeruptorum and its substitutes varied from 0.00% (P. turgeniifolium H. Wolff) to 21.60% (Pleurospermum bicolor (Franch.) Norman ex Pan and Watson), while divergence values between P. praeruptorum and its adulterants varied from 2.50% (P. japonicum Thunb.) to 24.0% (Physospermopsis delavayi (Franch.) H. Wolff). Across the matrix, the informative and variable sites were widely dispersed, indicating that it is feasible to use sequence alignments to distinguish the P. praeruptorum from its substitutes and adulterants accurately. Based on the NJ tree constructed under K2P method, all accessions are resolved into eight clades with moderate to high support [Figure 1]. Except for P. turgeniifolium, which intermingled with accessions of P. praeruptorum, P. praeruptorum and its substitutes and adulterants could be differentiated successfully.
Table 2.
Sequences characteristics of ITS in this study
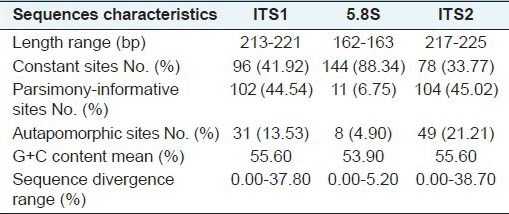
Figure 1.
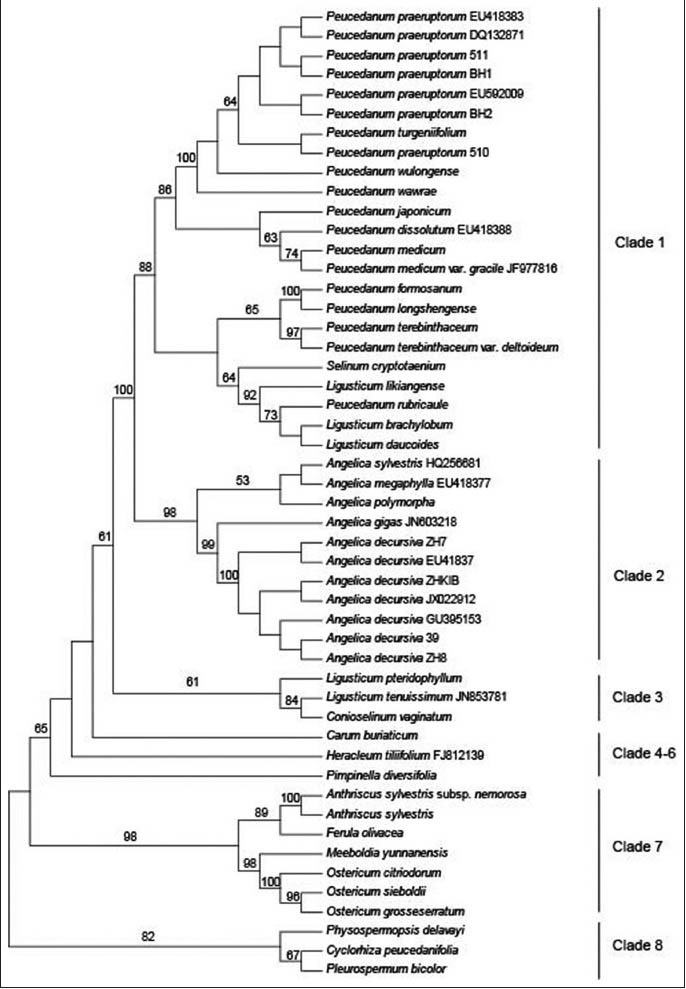
Classification tree of ITS sequences using the NJ method. Branch length was calculated by Kimura's 2-parameters method. Bootstrap (1000 replicates) analysis was performed to estimate the confidence of the topology of the consensus tree
DISCUSSION
According to surveys, there are 11,146 medicinal plant species from 2,309 genera of 383 families in China, representing a rich biodiversity. Accurate and rapid authentication of these plants and their adulterants is difficult to achieve at the scale of international trade in medicinal plants.[15] Furthermore, many commercial products are sold either in dried form or as processed material, rendering their authentication by morphological methods very difficult, if not impossible.[24] However, DNA-based methods, e.g. barcoding can be useful in quickly and efficiently pinpointing adulterated or misidentified raw materials without further need for time- and resource-consuming morphological, physical, and phytochemical examinations.[25,26]
A suitable barcode must exhibit high interspecific but low intraspecific divergence.[27] ITS was initially proposed as a universal DNA barcode for plants because of its high sequence divergence[28], and it also has been successfully used as a genetic marker for molecular authentication and identification of several medicinal plants. Panax ginseng C.A.Mey.[29,30], Dendrobium Species[25,26,31], Euphorbia pekinensis[32], Bupleurum species[33], Chimaphila species[34] and Gentianopsis paludosa (Hook. f.) Ma[24] are all successful examples. Furthermore, Chen et al. tested the discrimination ability of ITS2 (4800 species from 753 distinct genera) and found that it has the potential to be used as a standard DNA barcode to identify medicinal plants and their closely related species.[15] Li et al. proposed that ITS/ITS2 should be incorporated into the core barcode for seed plants after investigating several candidate barcodes.[16] In our barcoding results, a single-region of ITS sequence can distinguish P. praeruptorum from its substitutes and adulterants. This was supported by sequence alignment analyses, which revealed the high sequence variation to be sufficient for species identification.
Traditionally, because of their similar effects on dispelling wind-heat, expectorant and loss of energy, A. decursiva, together with P. praeruptorum were both regarded as certified products of Qian-hu.[35] However, subsequent chemical analysis revealed that they differ greatly in type of coumarins. Bai-hua qian-hu mainly contains angle-type dihydro-pyran-coumarins, while line-type furan- or pyran- coumarins are the main components of Zi-hua qian-hu[4,5], and they have been separately recorded in the current Pharmacopoeia of the People's Republic of China.[2] Angelica decursiva was previously ascribed to the genus Peucedanum as P. decursivum (Miq.) Maxim. Recent phylogenetic studies considered that A. decursiva/P. decursivum should be classified into Angelica.[36] In our results, seven accessions of A. decursiva formed a well-supported clade distant from P. praeruptorum, with the divergence value of 6.20%. In the background of Chinese herb standardization, it is difficult to specify a unanimous quality standard for Qian-hu from two different resources. Therefore, we consider that it is more reasonable to consider A. decursiva to be a regional substitute for P. praeruptorum.
Through historical textual research and chemical analyses, thirteen species, Ligusticum brachylobum Franch., L. likiangense (H. Wolff) Pu and Watson, L. daucoides (Franch.) Franch., Ostericum citriodorum (Hance) Yuan and Shan, Peucedanum dissolutum (Diels) H. Wolff, P. formosanum Hayata, P. medicum Dunn, P. medicum var. gracile Dunn ex Shan and Sheh, P. rubricaule Shan and Sheh, P. turgeniifolium H. Wolff, P. wawrae (H. Wolff) Su ex Sheh, P. wulongense Shan and Sheh and Pleurospermum bicolor are regarded as regional substitutes for P. praeruptorum.[3] Sequence divergence values between P. praeruptorum and these substitutes, all of which mainly clustered into Clade 1 except for Pleurospermum bicolor and Ostericum citriodorum [Figure 1], ranged from 0.00% to 21.60%. Pleurospermum bicolor is a traditional medical plant used by the Naxi ethnic group as Qian-hu. Despite their great pair-wise distance (21.60%), it can be substituted for P. praeruptorum because of similar active ingredients.[37] Except for P. turgeniifolium, all the other substitutes were well resolved on the NJ tree. Peucedanum turgeniifolium occurs in S Gansu (Jone, Têwo) and northern Sichuan and is important in Sichuan folk medicine. According to Rao et al., it has chemical constituents similar to those of P. praeruptorum and can be regarded as a substitute.[38] In our study, the sequence divergence between them was 0.00% (the divergence values between them for other barcodes such as psbA-trnH, matK and rbcL were also zero, unpublished data). As P. praeruptorum is distributed widely in China, covering Gansu and Sichuan, we consider P. turgeniifolium to be a geographical variety of P. praeruptorum pending further investigation.
Pairwise sequence divergence estimates ranged from 2.50% to 24.0% of nucleotides within P. praeruptorum and its adulterants, the latter of which were scattered throughout the NJ tree (Clade 1–Clade 8) [Figure 1]. All adulterants were well resolved and could be distinguished from P. praeruptorum. Most of the adulterants, such as P. terebinthaceum (Fisch. ex Trevir.) Ledeb., Ligusticum pteridophyllum Franch. and Angelica sylvestris (L.) Hoffm., also used differed medicinally from P. praeruptorum, and so should be used under their original herbal medicine names.
In conclusion, Peucedanum praeruptorum could be identified by analysis of a single DNA barcoding–ITS sequence that provides enough variability for authentication in contrast to the difficulties in using morphological characters. We consider nrDNA ITS sequence analysis to be reliable and convenient for authenticating P. praeruptorum and its substitutes, adulterants and other medicinal species.
ACKNOWLEDGMENTS
The authors thank David E. Boufford from Harvard University Herbaria for language polishing. This work was supported by the National Natural Science Foundation of China (no. 31000098), the Open-access Project of the State Key Laboratory of Systematic and Evolutionary Botany (LSEB) and Open Research Foundation of Yunnan Key Laboratory of Pharmacology for Natural Products (No. 2013G006).
Footnotes
Source of Support: Nill
Conflict of Interest: None declared.
REFERENCES
- 1.Sheh ML, Watson MF. Apiaceae Lindley. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 14. Beijing: Science Press and St.Louis: Missouri Botanical Garden Press; 2005. pp. 1–205. [Google Scholar]
- 2.Chinese Pharmacopoeia Commission. Vol. 1. Beijing: People's Medical Publishing House; 2010. Pharmacopoeia of the People's Republic of China; pp. 501–826. [Google Scholar]
- 3.Rao GX, Liu QX, Dai ZJ, Yang Q, Dai WS. Textual research for the traditional Chinese medicine Radix Peucedani and discussion of its modern varieties. J Yunnan Coll Tradit Chin Med. 1995;18:1–6. [Google Scholar]
- 4.Zhang C, Xiao YQ, Taniguchi M, Baba K. Studies on chemical constituents in roots of Peucedanum praeruptorum. Chin J Chin Mat Med. 2005;30:675–7. [PubMed] [Google Scholar]
- 5.Zhang C, Xiao YQ, Taniguchi M, Baba K. Studies on chemical constituents in roots of Peucedanum praeruptorum. Chin J Chin Mat Med. 2006;16:1333–5. [PubMed] [Google Scholar]
- 6.Xue JC. Review on active ingredient and related pharmacological action of Peucedanum praeruptorum. Strait Pharm J. 2012;24:34–8. [Google Scholar]
- 7.Lan RF, Zheng X, Lin SQ, Chen ZT. Comparison on the Chemical Constituents of P. japonicum Thunb with P. praeruptorum Dunn. Strait Pharma J. 2000;12:45–7. [Google Scholar]
- 8.Liu BM, Xu Q. Analysis of dl-Praeruptorin A, dl-Praeruptorin B and Nodakenetin in Qianhu by GC/MS and GC. Guangxi Sci. 2002;9:294–7. [Google Scholar]
- 9.Zhu GY, Chen GY, Li QY, Shen XL, Fang HX. HPLC/MS/MS method for chemical profiling of Radix Peucedani (Baihua Quianhu) Chin J Nat Med. 2004;2:304–8. [Google Scholar]
- 10.Qi C, Chen ZJ. SDS-PAGE to identify with purple Peucedanum and Peucedanum praeruptorum Dunn and Counterfeit. J Hubei Uni Chin Med. 2012;14:30–2. [Google Scholar]
- 11.Joshi K, Chavan P, Warude D, Patwardhan B. Molecular markers in herbal drug technology. Curr Sci. 2004;87:159–65. [Google Scholar]
- 12.Xue CY, Li DZ, Lu JM, Yang JB, Liu JQ. Molecular authentication of the traditional Tibetan medicinal plant Swertia mussotii. Planta Med. 2006;72:1223–6. doi: 10.1055/s-2006-951695. [DOI] [PubMed] [Google Scholar]
- 13.Hebert PD, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc R Soc Biol Sci Ser B. 2003;270:313–21. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebert PD, Gregory TR. The promise of DNA barcoding for taxonomy. Syst Biol. 2005;54:852–9. doi: 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]
- 15.Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, et al. Validation of the ITS2 Region as a Novel DNA Barcode for identifying Medicinal Plant Species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.China Plant BOL Group. Li DZ, Gao LM, Li HT, Wang H, Ge XJ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Nat Acad Sci U S A. 2011;108:19641–6. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downie SR, Plunkett GM, Watson MF, Spalik K. Tribes and clades within Apiaceae subfamily Apioideae: The contribution of molecular data. Edinb J Bot. 2001;58:301–30. [Google Scholar]
- 18.Winter PJ, Magee AR, Phephu N, Tilney PM, Downie SR, van Wyk BE. A new generic classification for African peucedanoid species (Apiaceae) Taxon. 2008;57:347–64. [Google Scholar]
- 19.Zhou J, Peng H, Downie SR, Liu ZW, Gong X. A molecular phylogeny of Chinese Apiaceae subfamily Apioideae inferred from nuclear ribosomal DNA internal transcribed spacer sequences. Taxon. 2008;57:402–16. [Google Scholar]
- 20.Zhou J, Gong X, Downie SR, Peng H. Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: Additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol Phylog Evol. 2009;53:56–68. doi: 10.1016/j.ympev.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 21.Feng T, Downie SR, Yu Y, Zhang XM, Chen WW, He XJ, et al. Molecular systematics of Angelica and allied genera (Apiaceae) from the Hengduan Mountains of China based on nrDNA ITS sequences: Phylogenetic affinities and biogeographic implications. J Plant Res. 2009;122:403–14. doi: 10.1007/s10265-009-0238-4. [DOI] [PubMed] [Google Scholar]
- 22.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf issue. Phytochem Bull. 1987;19:11–5. [Google Scholar]
- 23.White TJ, Bruns TD, Lee SB, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols A guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–22. [Google Scholar]
- 24.Xue CY, Li DZ. Use of DNA barcode to identify traditional Tibetan medicinal plant (Gentianaceae) Gentianopsis paludosa. J Syst Evol. 2011;49:267–70. [Google Scholar]
- 25.Lau TW, Shaw PC. Authentication of medicinal Dendrobium species by the internal transcribed spacer of ribosomal DNA. Planta Med. 2001;67:456–60. doi: 10.1055/s-2001-15818. [DOI] [PubMed] [Google Scholar]
- 26.Ding XY, Xu LS, Wang ZT, Zhou KY, Xu H, Wang YQ. Authentication of stems of Dendrobium officinale by rDNA ITS region sequences. Planta Med. 2002;68:191–2. doi: 10.1055/s-2002-20239. [DOI] [PubMed] [Google Scholar]
- 27.Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008;105:2923–8. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005;102:8369–74. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngan F, Shaw P, But P, Wang J. Molecular authentication of Panax species. Phytochemistry. 1999;50:787–91. doi: 10.1016/s0031-9422(98)00606-2. [DOI] [PubMed] [Google Scholar]
- 30.Kim OT, Bang KH, In DS, Lee JW, Kim YC, Shin YS, et al. Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5. 8S rDNA sequences. Plant Biotechnol Rep. 2007;1:163–7. [Google Scholar]
- 31.Xu H, Wang ZT, Ding XY, Zhou KY, Xu LS. Differentiation of Dendrobium species used as “Huangcao Shihu” by rDNA ITS sequence analysis. Planta Med. 2006;72:89–92. doi: 10.1055/s-2005-916228. [DOI] [PubMed] [Google Scholar]
- 32.Xue HG, Zhou SD, He XJ, Yu Y. Molecular authentication of the traditional chinese medicinal plant Euphorbia pekinensis. Planta Med. 2006;73:91–3. doi: 10.1055/s-2006-951769. [DOI] [PubMed] [Google Scholar]
- 33.Yang ZY, Chao Z, Huo K, Xie H, Tian ZP, Pan SL. ITS sequence analysis used for molecular identification of the Bupleurum species from northwestern China. Phytomedicine. 2007;14:416–23. doi: 10.1016/j.phymed.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu ZW, Zhao QR, Zhou J. A test of four candidate barcoding markers for the identification of geographically widespread Chimaphila species (Pyroleae, Ericaceae) Acta Bot Gallica. 2013;160:11–7. [Google Scholar]
- 35.Chinese Pharmacopoeia Commission. Vol. 1. Beijing: People's Medical Publishing House; 2000. Pharmacopoeia of the People's Republic of China. [Google Scholar]
- 36.Xue HJ, Yan MH, Lu CM, Wang NH, Wu GR. Taxonomic study of Angelica from East Asia: Inferences from ITS sequences of nuclear ribosomal DNA. Acta Phytotax Sin. 2007;45:783–95. [Google Scholar]
- 37.Rao GX, Dai WS, Yang Q, Dai YX, Liu QX, San HD. Chemieal constituents of Pleurospermum govanianum (Wall.) Chemieal constituents of Pleurospermum govanianum (Wall.) Benth ex C.B. Clarke var. bicolor Wolff. Chin J Chin Mater Med. 1995;20:740–2. [PubMed] [Google Scholar]
- 38.Rao GX, Sun HD, Lin ZW, Niu FD. Study on the chemical basis of Qianhu. Nat Prod Res Dev. 1993;5:1–17. [Google Scholar]


