Abstract
Cockayne syndrome (CS) is a devastating neurodevelopmental disorder, with growth abnormalities, progeriod features, and sun sensitivity. CS is typically considered to be a DNA repair disorder, since cells from CS patients have a defect in transcription-coupled nucleotide excision repair (TC-NER). However, cells from UV-sensitive syndrome patients also lack TC-NER, but these patients do not suffer from the neurologic and other abnormalities that CS patients do. Also, the neurologic abnormalities that affect CS patients (CS neurologic disease) are qualitatively different from those seen in NER-deficient XP patients. Therefore, the TC-NER defect explains the sun sensitive phenotype common to both CS and UVsS, but cannot explain CS neurologic disease. However, as CS neurologic disease is of much greater clinical significance than the sun sensitivity, there is a pressing need to understand its molecular basis. While there is evidence for defective repair of oxidative DNA damage and mitochondrial abnormalities in CS cells, here I propose that the defects in transcription by both RNA polymerases I and II that have been documented in CS cells provide a better explanation for many of the severe growth and neurodevelopmental defects in CS patients than defective DNA repair. The implications of these ideas for interpreting results from mouse models of CS, and for the development of treatments and therapies for CS patients are discussed.
1. Introduction
Patients with the genetic disease xeroderma pigmentosum (XP) are highly sensitive to sunlight exposure, and have a greater than 10,000X increased risk of cancer on sun exposed areas of the body [1]. Based upon the earlier discovery of nucleotide excision repair [2], the landmark observation by Cleaver [3] that patients with XP are unable to carry out nucleotide excision repair (NER) was the first description of a DNA repair disease. The subsequent discovery that cells from patients with Cockayne syndrome (CS) have a defect in a sub-pathway of NER, referred to as transcription-coupled nucleotide excision repair (TC-NER) [4, 5] led to the identification of CS as a DNA repair disease as well. CS is a rare neurodevelopmental disorder with progeriod features, and sun sensitivity, and the NER defect common to both diseases provided a convincing explanation for the shared sun sensitive phenotype.
The discovery of the TC-NER defect in CS cells brought this extremely rare disease to the attention of world-class scientists working on DNA repair and transcription, resulting in several ground breaking scientific publications, and much progress into understanding the mechanistic basis of TC-NER [6, 7]. In addition, knowledge of the DNA repair defect was essential to the cloning of the genes responsible for CS [8, 9]. However, the association between CS and TC-NER deficiency has had a negative impact as well. Numerous publications on the mechanistic basis of TC-NER, utilizing cells from CS patients, reinforced the association between CS and TC-NER deficiency. Furthermore, the shared NER deficiency in both XP and CS fostered the idea that all of the clinical features of CS, including the neurologic disease, are the result of the TC-NER defect (e.g. [10]). The obvious problem is that while both XP and CS patients have neurologic abnormalities, the nature of the neurologic abnormalities in CS are fundamentally different than those in XP [11, 12].
In my view, much of the confusion in the literature is the result of looking at CS through the lens of the well-known TC-NER deficiency. In fact, cells from CS patients have multiple abnormalities in addition to defective TC-NER. In this work, I will take the opposite approach, starting with a description of the clinical features of CS with a particular emphasis on CS neurologic disease. I then consider which of the various molecular abnormalities that have been described in CS cells, including but not limited to the TC-NER defect, provides the best explanation for the different pathologies of CS neurologic disease. Specifically, I will propose an updated version of the original transcription syndrome hypothesis of CS [13–16] expanded to include an important role for defective transcription by both RNA polymerase I (RNAPI) and RNA polymerase II (RNAPII) in CS , and argue that this expanded transcription hypothesis provides a better explanation for many aspects of CS neurologic disease than defective DNA repair. I will also consider the implications of the expanded transcription syndrome hypothesis for interpreting recent findings from mouse models of CS. Finally, I will consider the translational implications of different hypotheses for the pathophysiology of CS. As such, this work is not intended to be a comprehensive review of CS, but a different way of looking at the mechanistic basis of CS neurologic disease. Since neurologic disease is of far greater clinical significance to CS patients than sun sensitivity, understanding its mechanistic basis has important implications for the development of treatments and therapies which are urgently needed.
2. Cockayne syndrome (CS) and CS neurologic disease
CS is a rare autosomal recessive disease, characterized by severe growth failure, neurologic disease, developmental abnormalities, degeneration of multiple organ systems including the eye and ear, cataracts, and, in most (but not all) patients, sun sensitivity [17–20]. CS can result from mutations in either of two genes; ERCC6 (CSB) or ERCC8 (CSA). In addition, a subset of patients with mutations in the XPB, XPD, and XPG genes have the somatic features of CS, as well as the increased risk of skin cancer on sun exposed areas of the body that is characteristic of XP [19]. Therefore, any explanation for a CS phenotype must be able to explain how some mutations in any of five genes (CSA, CSB, XPB, XPD, XPG) can result in the CS phenotype.
Different types of CS are referred to in the literature [17, 21, 22]. Patients with the most severe form, Type II, show prenatal growth retardation, and are therefore affected at birth. These patients typically do not survive past 6–7 years of age. Patients with the more common presentation of CS, Type I, are normal at birth, but begin to show growth and developmental abnormalities by 1–2 years of age. Other patients, referred to as Type 3, show later age of onset symptoms, and a slower disease course, and some patients survive into their 40s. The distinction between these types is not absolute and is becoming less distinct as additional patients are identified.
2.1 Cockayne syndrome neurologic disease
The neurologic defects observed in CS patients, collectively referred to as CS neurologic disease, are quite complex and multifaceted. Some of the major pathologic features of CS neurologic disease are described below, based upon multiple publications [20, 23–27]. Readers should consult the original references for more detailed information.
2.1.1 Brain growth defects, and atrophy
One of the most striking features of the brain of CS patients at the time of death is the extremely small size. As stated by Weidenheim et al [20] this likely reflects a combination of both brain growth defects and profound brain atrophy. In severe CS cases, the cerebellum is already small at the time of first evaluation, indicative of early cerebellar hypoplasia [25]. Atrophy of the white matter is typically more severe than in the gray matter.
2.1.2. Myelin defects
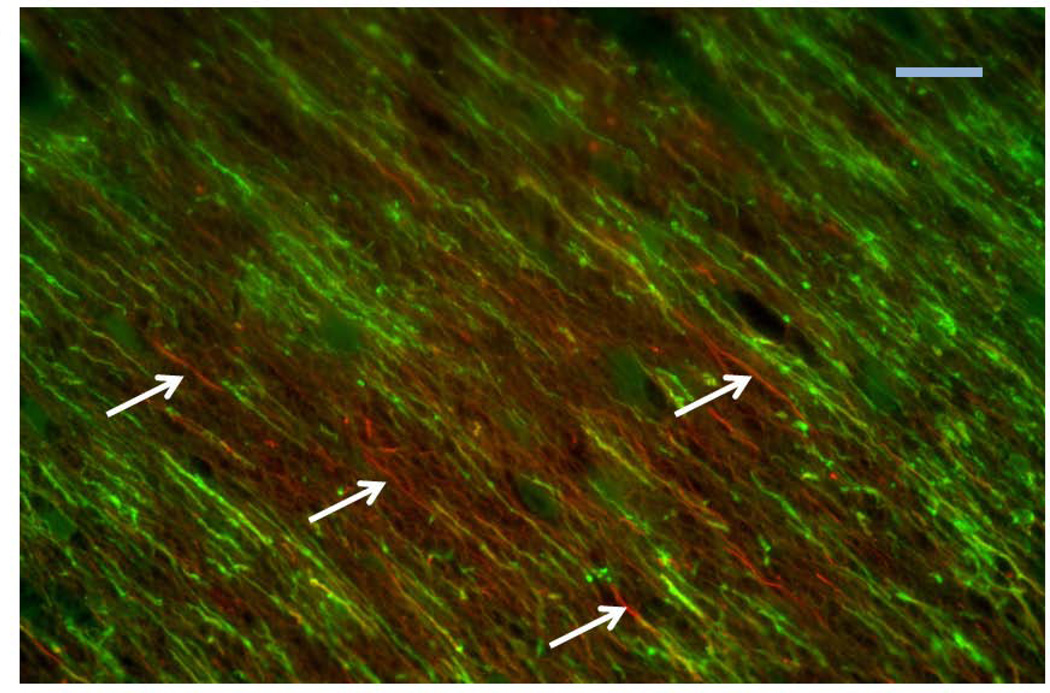
There are two distinct types of white matter (myelin) abnormalities in CS. One is a defect in brain myelination during development, referred to as hypomyelination or dysmyleination. Based on brain imaging and MR spectroscopy, Koob et al [25] concluded that CS is primarily a hypomyelinating disease. In addition to this global hypomyelination, a patchy loss of myelination, referred to as tigroid demyelination, has been described. This pathology has been most well documented in post mortem studies of CS patients, although abnormalities consistent with tigroid demyelination can be observed with brain imaging (see [25] and references therein). Figure 1 is an immunofluoresence image taken of the brain of a CS patient, which illustrates this patchy demyelination at the cellular level.
Fig. 1.
Photomicrograph of a section through the white matter of the brain of a patient with CS, stained with antibodies against neurofilament (red), a marker for axons, and proteolipid protein (green), a marker of myelin. Colors refer to the online version of the manuscript. Arrows point to some segments of axons lacking myelination, illustrating the patchy demyelination. Scale bar = 10 microns.
2.1.3. Calcification
A striking and unusual feature of CS neurologic disease is the presence of calcium and other mineral deposits within the brain. In vivo, brain calcification can be readily detected by brain imaging, and is most commonly seen in the basal ganglia and cerebellum. In severe CS cases, additional calcification is seen below the depths of sulci in the cerebral cortex, and in association with the large leptomenigeal blood vessels [25].
Microscopic analysis of the brain of CS patients postmortem also demonstrates different types of calcification. One type is small crystals (concretions) of calcium found in the brain tissue which are easily seen with the light microscope. In addition, calcium and/or iron can be detected in and around brain vascular elements.
2.1.4. Brain vasculopathy
An important aspect of CS neurologic disease that is frequently overlooked is brain vasculopathy, affecting both large and small blood vessels [20, 26]. The remains of obliterated blood vessels, referred to as string vessels, can be found in the brains of CS patients by microscopic investigation, and represent evidence of brain microvasculopathy [23, 26]. Rapin et al [23] stated that string vessels appeared more frequent in areas of the brain lacking white matter, suggesting a relationship between the two pathologies. However, additional quantitative data would be necessary to further evaluate this possibility.
2.1.5 Other Cellular abnormalities
Finally several authors have noted the existence of unusual and bizarre cell types in the brains of CS patients. These include dystrophic and degenerating cerebellar Purkinje neurons, some containing so-called axonal torpedoes, multinucleated astrocytes and neurons, and ferruginated (mineral-containing) degenerating neurons [27]. Determining which of these cellular phenotypes are the result of the disease progression, versus which are clues to the underlying pathophysiology, is an important question for future research.
In summary, CS neurologic disease is complex and heterogeneous set of pathologies, including both developmental defects and degenerative processes, and perhaps other pathogenic mechanisms as well. We now turn to a consideration of the possible mechanistic explanations for the different features of CS neurologic disease, beginning with the DNA repair defect.
3. Transcription-coupled NER and its relevance to CS
Before assessing the relevance of defective TC-NER for CS neurologic disease, it is necessary to provide some basic information about NER and TC-NER. Since several comprehensive and detailed reviews of various aspects of NER have been recently published in a special issue of DNA Repair [28] here I will give only a basic description of the processes, and refer interested readers to these reviews.
3.1 NER and TC-NER, a brief overview
NER is the DNA repair mechanism responsible for removing UV-light induced lesions as well other types of DNA damage that cause significant distortion of the helical structure of DNA. The major type of UV-induced lesions are cyclobutane pyrimidine dimers (CPDs), with a smaller fraction of 6–4 photoproducts. The classical approach of studying NER in the laboratory is to expose cells in culture to UV light, then follow the repair of the UV-induced lesions over time. This can be done either by measuring DNA repair synthesis with radioactive tracers, or by isolation of cellular DNA followed by Southern blotting (for review see [29]). In the Southern blotting method, isolated DNA is first treated with enzymes that cleave at the site of DNA lesions prior to electrophoresis, such that the repair of DNA lesions is detected as the disappearance of enzyme-sensitive sites. In parallel with these cellular studies, biochemical experiments carried out in several laboratories [30–32] have provided a fairly detailed picture of the mechanistic basis of NER.
Of particular relevance for CS was the discovery, using the Southern blotting method, that the rate of NER across the genome varies depending upon the transcriptional state of the DNA [33, 34]. These observations led to the concept of two distinct sub-pathways of NER, referred to as global genome NER (GG-NER) and transcription-coupled-NER (TC-NER). The GG-NER pathway removes DNA lesions throughout the genome, whereas TC-NER is responsible for the more rapid repair of transcription blocking lesions on the transcribed strand of active genes [6]. Cells from CS patients have a specific defect in TC-NER, while the GG-NER pathway is normal in CS cells [4, 5]. The CSA and CSB proteins, encoded by the ERCC8 and ERCC6 genes, respectively, play an essential role in TC-NER, but not in GG-NER [35]. The primary mechanistic difference between the two sub-pathways is in the manner of lesion detection. For GG-NER, the lesion is first detected by the XPC protein, in association with different partners, whereas in TC-NER, stalling of an elongating RNA polymerase at a transcription blocking DNA lesion acts as a signal to initiate and carry out NER [6, 36]. The CSA and CSB proteins are involved in localized the NER machinery to the transcription-blocking DNA lesion [37].
Once the DNA lesion has been identified, the subsequent steps in NER are thought to be essentially the same. The basic steps include: 1) unwinding the DNA to form an approximately 30 nucleotide “bubble” around the lesion, carried out by the XPB and XPD helicases that are components of the transcription factor TFIIH [38, 39]; 2) cleaving the lesion containing DNA strand on both sides of the lesion, carried out by the nucleases XPF-ERCC1 (which cleaves 5’ to the lesion) and XPG (which cleaves 3’ to the lesion) [40]; 3) DNA synthesis to fill the gap resulting from removal of the lesion containing DNA, carried out by one or more DNA polymerases [41]; and 4) sealing the nick resulting from DNA repair synthesis, carried out by a DNA ligase, which completes the repair. Another protein, XPA, plays an essential role in both TC-NER and GG-NER, though the precise nature of its role remains under investigation [42].
3.2. Limitations on the role of TC-NER in the genome and in terminally differentiated cells
As originally defined, TC-NER refers to the more rapid rate of repair of transcription blocking lesions from the transcribed strand of active genes, compared to the rate of repair of the genome overall (GG-NER). In contrast to this quantitative difference in the rate of repair, some schematic diagrams of the NER pathway [43] can give the impression that the genome is divided into two domains, one in which TC-NER acts, and one in which GG-NER acts. To those reading about this field for the first time, these diagrams can give a misleading impression. While TC-NER only acts on DNA lesions located on the transcribed strand of active genes, the GG-NER pathway can function anywhere within the genome, including the transcribed strand of active genes. Whether repair of a blocking lesion on the transcribed strand of an active gene is repaired by TC-NER or GG-NER depends upon whether RNAPII or the XPC protein reaches the lesion first, which in turn will depend upon many factors, including the extent of helical distortion caused by the lesion, how many other lesions are in the cell, and the rate of transcription of the gene. Indeed, on the basis of experiments using fluorescently tagged proteins, which allow estimation of the time that different proteins are engaged in different functions, van den Boom et al concluded that for some very long genes expressed at a low level, TC-NER may be slower than GG-NER [44]. Furthermore, it is also worth noting that while most studies of TC-NER were done on a relatively small subset of genes, there is published evidence that not all genes are subject to TC-NER [45], perhaps due to differences in chromatin structure.
The GG-NER TC-NER distinction applies to dividing cells. For terminally differentiated cells, the compartmentalization of NER throughout the genome is fundamentally different. Work by Nouspikel, Hanawalt, and colleagues has shown that in neuronal cells, as well as other terminally differentiated cells, UV-induced cyclobutane pyrimidine dimer (CPD) lesions are repaired at the same rapid rate on both the transcribed and non-transcribed strands of active genes. In contrast, repair of CPDs in non-transcribed DNA is nearly undetectable [46]. This pattern of repair was originally referred to as differentiation associated repair (DAR), then renamed domain associated repair. Of particular relevance to CS, DAR is not dependent on CSB, and therefore considered a sub pathway of GG-NER [47].
In a landmark technical achievement, Giglia-Mari, Vermuelen, and co-workers from the Hoeijmakers laboratory [48] generated a knock-in mouse expressing GFP-tagged Xpb protein, which allows studies of the mobility of TFIIH in multiple cells including neurons, in vivo and in the normal cellular context. One striking finding from this study was that in contrast to dividing cells in tissue culture, the mobility of TFIIH is greatly reduced in neurons and other terminally differentiated cells, and that this change in mobility occurs in a differentiation-dependent and developmentally regulated manner. This finding itself raises questions about the generality of the TC-NER/GG-NER model in different cell types, as the original model is primarily based on proliferative fibroblasts in cell culture.
3.3 The TC-NER Defect Cannot Explain CS Disease
The original rationale for studying NER in CS cells was the cutaneous sun sensitivity of CS patients, a clinical feature shared with XP patients. As such, the observation that cells from CS specifically lack the ability to carry out TC-NER [4, 5] provided a convincing mechanistic explanation for the sun sensitive phenotype. In addition, Venema et al [4] suggested that a defect in TC-NER could also explain the degenerative phenotypes of CS as well. This hypothesis was based on the earlier work by Robbins and colleagues, who proposed that XP neurologic disease was the result of defective NER of endogenous DNA damage [49]. Given the information available at the time, such a proposal was not unreasonable.
However, since the TC-NER defect in CS was discovered, much progress has been made in our understanding of the mechanisms of TC-NER, the formation and repair of endogenous DNA damage, and the multiple cellular roles of the CS proteins and TFIIH outside of DNA repair. In addition, several other human diseases with defects in NER have been identified, including trichothiodystophy (TTD), and UV-sensitive syndrome (UVsS) (see Table 1). Last but not least, as described above, we also now have a more detailed understanding of the complex neurologic abnormalities in CS neurologic disease, which are fundamentally different from the neurodegeneration observed in some XP patients (XP neurologic disease). Based upon these considerations, the concept that defective TC-NER can explain the internal features of CS, including CS neurologic disease, is no longer tenable, for the reasons discussed below.
| Disease |
Some clinical features |
Mutant genes |
|---|---|---|
| Xeroderma Pigmentosum (XP) |
sun sensitivity, 10,000X increase skin cancer, XP neurologic disease (some patients, especially XP-A) |
XPA-G, V |
| Cockayne Syndrome (CS) |
sun sensitivity, no skin cancer, CS neurologic disease, progeriod features, growth defects |
CSA, CSB |
| XP/CS complex | sun sensitivity, increased skin cancer, CS neurologic disease |
XPB, XPD, XPG |
| Trichothiodystrophy (TTD) |
sun sensitivity (some patients), brittle hair, ichthyosis, TTD neurologic disease (neurodevelopmental and myelination abnormalities) |
XPD, TTDA, others? |
| UV-sensitive syndrome (UVsS) |
sun sensitivity, no increased skin cancer, no neurologic disease |
CSA, CSB, UVSSA |
3.3.1. CS is not a mild form of XP
If the clinical phenotypes of XP and CS were to be understood solely through the lens of the TC-NER/GG-NER model [43], there is a clear prediction. XP patients should be the most severely affected, since they lack all NER, including TC-NER, GG-NER, and DAR. In contrast, CS patients should be less affected than XP, since they only lack the TC-NER sub-pathway. However, even a cursory understanding of the clinical phenotype of CS and XP shows that this prediction is not confirmed. CS is not a mild form of XP, it is a fundamentally, qualitatively different disease. In fact, the primary clinical feature that CS and XP have in common is sun sensitivity, but even here there is a major difference. XP patients have a greater than 10,000X increased risk of cancer on sun exposed areas of the body [1], whereas CS patients never get skin cancer.
The difference between CS and XP is most obvious when comparing the neurologic phenotypes. XP neurologic disease is a primary neurodegenerative disease, characterized by the loss of large neurons throughout the brain, spinal cord, and peripheral nervous system. In contrast, as described above, CS neurologic disease is much more heterogeneous, including dysmyelination, demyelination, brain calcification, and vasculopathy, none of which are seen in XP neurologic disease [11, 50].
In my view, part of the confusion in the literature stems from terminology. Venema et al [4] used the term neurological degeneration to refer to both XP and CS. It is certainly true that in both diseases, some patients experience a profound degeneration of the nervous system. However, as indicated above, the nature of the degenerative process, and the brain cell types affected, are quite different in the two diseases. More problematic is the word neurodegeneration, which can technically refer specifically to the degeneration of neurons [51]. While XP is a primary neurodegeneration, the major features of CS neurologic disease including microcephaly, de and dysmyleination, vasculopathy, and calcification, are not neurodegeneration, although some neurons, notably including Purkinje neurons, do degenerate in the brain of CS patients. To avoid confusion, the term XP neurologic disease can be used to describe the neurologic abnormalities in XP patients [52], and the term CS neurologic disease used to describe the combined neurologic abnormalities in CS patients.
It should be emphasized that the issue here is not merely semantic. The use of the term neurodegeneration to refer to both CS and XP confuses the literature, and perpetuates the idea that the both XP and CS neurologic disease result from defective TC-NER (e.g. [10]). Even more importantly, understanding which cell types are affected in the different diseases is essential for the development of rational therapeutics, and using the term neurodegeneration to describe both XP and CS is unhelpful in this regard. For example, therapeutics that could be used to prevent the degeneration of neurons in XP would have to cross the blood-brain barrier and enter neurons. In contrast, therapeutics to target the vasculopathy in CS might target brain vascular endothelial cells, which are on the blood side of the blood-brain barrier.
3.3.2 UV-sensitive syndrome (UVsS): Defective TC-NER leads to sun sensitivity but no neurologic disease
The other key piece of evidence demonstrating that TC-NER cannot explain CS neurologic disease comes from studies of patients with UV-sensitive syndrome (UVsS) [53]. Patients with UVsS are sun sensitive, as are patients with XP and CS. Like CS patients, UVsS patient also do not develop cancer on sun exposed areas of the body. Also in common with CS, cells from UVsS patient lack the capacity for TC-NER. Importantly however, UVsS patients do not have any neurologic defects [53], in contrast to the severe neurodevelopmental abnormalities seen in CS patients. Thus, defective TC-NER can only explain the sun sensitive phenotype common to UVsS and CS, but cannot explain the somatic features of CS, including CS neurologic disease.
UVsS can be caused by mutations in CSA [54], CSB [55], or UVSSA [56–58]. The recently described UVSSA protein plays a role in regulating the turnover and recycling of RNAPII stalled at DNA lesions, which requires ubiquitination and deubiquitination. In cells exposed to UV light, large numbers of RNA polymerases will stall at DNA lesions. Therefore, in the absence of UVSSA, RNAPII cannot resume transcription, and since continued transcription is by definition necessary for TC-NER, the failure to restart transcription provides an explanation for the TC-NER defect in UVsSA cells.
As indicated in Table 1, mutations in CSA and CSB can cause UVsS. The Tanaka laboratory [55] made the surprising observation of a patient having the clinical features of UVsS (sun sensitivity with no evidence of neurologic abnormalities) with an apparently inactivating mutation in CSB. Specifically, the patient is homozygous for a stop codon mutation at amino acid 77, near the N-terminus of the 1493 amino acid CSB protein. The authors considered various explanations for the very mild features of this patient, including the use of a downstream ATG start codon at amino acid 227 of CSB, but could not detect a protein of the size predicted by this mechanism. However, since only cultured fibroblasts were available for study, the possibility that alternative splicing and/or alternative translational initiation takes place in a subset of cells, resulting in an alternate form of CSB that protects against CS neurologic disease, cannot be ruled out. It is also possible that this patient, who was 33 years old at the time the paper was published, will develop neurologic disease later in life. Indeed, Hashimoto et al [59] described another patient who developed some symptoms of CS neurologic disease at the age of 47. The use of the downstream translational initiation codon in CSB could potentially explain the mild symptoms in this patient as well.
Nardo et al [54] reported a UVsS patient with a amino acid substitution (p.trp361cys) in CSA. Whether this patient was homozygous or hemizygous for the mutation could not be determined. Cells from this patient were hypersensitive to killing by UV light, but not to exposure to menadione (a vitamin K analog that generates oxidative stress). These findings indicate that trp361 is important for TC-NER, but not for other functions of CSA which prevent CS neurologic disease. The authors suggest defective repair of oxidative DNA damage as the other function, but other interpretations are possible (see section on Defective repair of oxidative DNA damage in CS and its relationship to CS neurologic disease below).
3.3.3. Why defective TC-NER cannot explain CS Neurologic disease: Qualitative and quantitative differences between UV-induced and endogenous DNA damage
The inability of TC-NER deficiency to explain the internal manifestations of CS is likely explained by two factors: the existence of a DNA damage response in cells acutely exposed to high levels of DNA damage, and the vastly different levels of DNA lesions generated by solar UV light versus endogenous lesion levels. Considering the DNA damage response first, two papers published in 2007 [60, 61] demonstrated that exposing cells to either UV or IR not only generates DNA damage, but also triggers a massive coordinated response involving changes in the phosphorylation of hundreds of cellular proteins. The net result of this DNA damage response is a qualitatively different functional cellular state than existed without DNA damage. In fact, altered protein ubiquitination is also a part of the DNA damage response, an observation which was utilized to identify the UVSSA gene [57]. As such, it is not surprising that the clinical result of mutation in this gene is UVsS, which is only manifest on sun exposed areas of the body that experience high levels of UV radiation.
There are several classes of endogenous DNA lesions that are substrates for NER but not other repair pathways, and can therefore be viewed as endogenous analogues of the UV-induced CPDs. These lesions include the cyclopurine deoxynucleosides (cyPudNs) [62, 63], certain classes of intrastrand crosslinks resulting from reactive oxygen species [64], and DNA lesions resulting from lipid peroxide products such as the malondialdehyde-deoxyguanosine adduct M1G [65, 66]. Regarding endogenous lesion levels, it has now been shown by two different laboratories [67, 68], using state of the art HPLC-tandem mass spectrometry, that cyPudNs are present in untreated mammalian cells and tissues at a level on the order of between 1 adduct in 107 to 108 normal nucleotides (10−7 to 10−8). Recent studies have shown that oxidatively induced crosslinks are detectable in tissue from normal human and animals at a similarly low level 10−8 [69], as are lipid peroxidation induced adducts [70].
In contrast to these low adduct levels, the UV exposure used to study TC-NER in the laboratory (typically 10 J/m2) results in the levels of CPDs on the order of 1 lesion/104 nucleotides. Such high levels are necessary to carry out the Southern blot based TC-NER assay, which requires roughly 1 adduct per 10 kb restriction fragment [33, 34]. Exposing cells to 10 J/m2 of UV is a valid and appropriate for an in vitro model of human sunlight exposure (see [71]), as well as for understanding the DNA damage response as discussed above. However, for understanding CS somatic disease, which affects internal organs not exposed to sunlight, the key point is that endogenous levels of DNA lesions that are substrates for NER are roughly 3 orders of magnitude lower than the levels of UV induced adducts generated during experiments used to detect TC-NER.
The dramatic difference in the level of UV induced lesions under experimental conditions compared to endogenous lesion levels has correspondingly different effects on cellular transcription. As calculated by McKay and colleagues [72], at 10 J/m2, an average size mammalian gene will have about 2.4 lesions or roughly 1 lesion in the transcribed strand. Under such conditions, the importance of rapidly removing transcription-blocking DNA lesions from the transcribed strand of active genes can be easily appreciated. However, for rare endogenous DNA lesions that are exclusively, repaired by NER, the situation is very different. Assuming for the purpose of discussion that transcription blocking lesions repaired by NER are present at 10−7, three orders of magnitude lower levels than 10 J/m2 of UV, then under basal conditions only 1 per 1000 average size genes will have a blocking lesion on the transcribed strand. Since mammalian genes are diploid, the probability that both copies of the same gene in the same given cell would have a blocking lesion on the transcribed strand is therefore 1 per million. In those 1 in a million cells, the effect of CS gene deficiency would only be a delay until the lesion was repaired by GG-NER, because TC-NER affects the rate of NER. Furthermore, in terminally differentiated cells from CS patients, lesions would be rapidly repaired by DAR, which does not require the CSB protein [47].
In summary, both patients with UVsS and CS both lack the capacity to carry out TC-NER, and both diseases are characterized by sun sensitivity. Therefore, defective TC-NER provides a convincing explanation for the sun-sensitive phenotype common to both diseases. TC-NER is important on sun exposed areas of the body, which experience acute high levels of transcription blocking lesions, as well as a DNA damage response. In contrast, the lack of CS neurologic disease in UVsS patients despite the deficient TC-NER, as well as the fundamental differences in between CS neurologic disease and XP neurologic disease, demonstrate that CS neurologic disease cannot be caused by defective TC-NER. The importance of TC-NER under conditions of UV exposure but not under basal conditions is likely explained by of low levels of endogenous DNA lesions that are specifically repaired by NER, the lack of a DNA damage response, and the existence of the DAR mechanism in terminally differentiated cells.
3.4. TC-NER is the only documented type of TCR
Several years ago, the hypothesis that the CS proteins as well as XPG, were involved in transcription coupled repair of non-bulky oxidative lesions by a mechanism distinct from NER (so called TC-BER) was dominant in the DNA repair field and literature. However, as discussed elsewhere [73] the primary data supporting this hypothesis have been retracted. As such, no direct evidence supporting transcription-coupled BER remains in the peer reviewed literature. As stated by Spivak and Hanawalt [6], oxidatively-induced DNA lesions such as thymine glycol and 8-oxoG are rapidly removed by BER, and there is no compelling biochemical evidence of coupling their repair to transcription. Even in the rare case that RNAPII might stall at such a lesion in a specific sequence context before it can be repaired by BER, there is no need to invoke a separate TC-BER mechanism. Using yeast as a model system, Kim and Jinks-Robertson [74] found that abasic sites on the transcribed strand of an active gene can be repaired by TC-NER, involving rad26, the yeast homolog of CSB. Abasic sites can arise from spontaneous hydrolytic depurination, and are also obligatory intermediates in BER. Since TC-NER is triggered by transcription-blocking lesions, this observation is consistent with studies showing that abasic sites are strong blocks to transcription by RNAPII [75, 76]. Given the wide variety of DNA lesions that can be repaired by NER, it is likely that in a similar manner, TC-NER could also serve a backup repair pathway for other non-bulky lesions such as 8-oxo-dG, thymine glycol, or others that might stall RNAPII in certain sequence contexts. The cut and patch mechanism of NER makes it well suited to the removal of structurally different types of transcription-blocking DNA lesions, and in fact in vitro studies have shown that NER can repair these lesions [77]. In contrast, BER is initiated by different DNA glycosylases that are specific for particular DNA lesions. Therefore, a separate TC-BER mechanism would seem to be not only redundant, but mechanistically difficult to conceptualize.
In summary, defective TC-NER explains the cutaneous sun-sensitivity phenotype in CS, but cannot explain the somatic features of CS, including CS neurologic disease. Since TC-NER is the only documented type of TCR, it follows that defective TCR cannot explain the somatic clinical features of CS. This does not mean that cells lacking TC-NER are completely normal, but that if TC-NER deficiency alone does have any cellular consequences other than sun sensitivity, they do not appear to be clinically significant, based on the reports of UVsS patients in the literature.
4. Defective repair of oxidative DNA damage in CS and its relationship to CS neurologic disease
The inability of defective TC-NER to explain the somatic features of CS does not rule out a possible role for defective repair of oxidative DNA damage in the CS phenotype. The concept that CS results from defective repair of oxidative DNA damage is a very popular concept in the literature, and has been reviewed by others [78, 79]. In my view, however, part of the attraction of the oxidative DNA damage hypothesis of CS is based in part on two mutually reinforcing but flawed concepts: CS as a premature aging disease, and the free-radical theory of aging.
4.1 CS is a Progeriod disorder, not premature aging
Like Werner’s syndrome and Hutchinson-Gilford progeria, CS is sometimes referred to as a premature aging disorder, or progeria. However, as discussed by the gerontologist George Martin [80], these diseases are more accurately referred to as progeriod disorders. The suffix “-oid” , meaning “-like”, emphasizes the fact that while some aspects of these diseases resemble features of aging, this does not necessarily mean that the underlying mechanisms are the same as in normal aging. Indeed, the mechanistic basis of normal aging itself is incompletely understood. To be sure, some aspects of CS, including cataracts, the wizened facial appearance, kyphosis, and perhaps brain vasculopathy are progeriod. However, other features are open to multiple interpretations. For example, brain calcification is sometimes observed in the brains of elderly individuals, and considered a non-specific sign of aging [81]. In contrast, the brain calcification in CS occurs in the developing brain, is often bilaterally symmetrical, and can be very severe [25]. As such, calcification in CS neurologic disease is more similar to several childhood neurologic conditions having nothing to do with aging, including birth anoxia/hypoxia [82], viral infection of the brain [83] and Aicardi-Goutieres syndrome [12, 84]. Finally, early microcephaly, cerebellar hypoplasia and hypomyelination seen in CS neurologic disease are clearly developmental abnormalities, and as such are fundamentally different than aging.
4.2 The Free Radical Theory of Aging is Under Revision
One of the most widely cited hypotheses for the mechanism of aging is the free radical theory of aging (FRTA), proposed by Harman over 50 years ago [85]. The essence of the FRTA is that the degenerative aspects of aging could be attributed to the damaging effects of oxygen radicals on cellular macromolecules, including proteins, lipids, and nucleic acids. However, the FRTA is currently undergoing a serious challenge (see [86] and references therein). Recent evidence against the FRTA comes from studies showing that some long-lived species have elevated levels of oxidative stress, in contrast to prediction by the FRTA [87]. Another key point is the realization that reactive oxygen species (ROS) are not simply toxic by-products of cellular metabolism, but important signaling molecules in their own right [88]. When the signaling function of ROS is considered, the interpretation of the documented relationship between ROS generation and aging looks quite different [87]. Of particular relevance to the neurodevelopmental defects in CS, proliferating neural stem cells have recently been found to have high endogenous ROS levels that play an important signaling role in maintaining the capacity for self-renewal [89].
4.3 Interpretational issues with the defective repair of oxidative DNA damage hypothesis of CS
Studies showing that cells from CS patients or mouse models are hypersensitive to killing by ionizing radiation or oxidative stress are sometimes interpreted as evidence of defective repair of oxidative DNA damage [54, 90]. However, such studies are open to multiple interpretations. Hypersensitivity to ionizing radiation or oxidative stress could as well be explained by mechanisms other than defective DNA repair, such as a defective DNA damage response [60], a defective transcriptional response to oxidative stress [91], or simply generalized cellular fragility and hypersensitivity to cell killing as a result of disordered transcription [92] (see “one-hit” neurodegeneration below). Indeed, cells from CS patients are hypersensitive to other stressors in addition to oxidative stress [93, 94] The results of host-cell reactivation studies, in which plasmids encoding transcribed reporter genes are exposed to oxidative DNA damage and transfected into cells, are also difficult to interpret definitively, because they are indirect measures of DNA repair and also depend upon transcription for the experimental end-point (see [95, 96]). It is also notable that in humans, mutations in both CSA and CSB result in CS, but some cell types from Csa and Csb deficient mice are differentially susceptible to killing by oxidative stressors [90]. To the extent that these mice are relevant models for CS in humans, this finding also complicates the oxidative DNA repair defect hypothesis.
In summary, although the hypothesis that defective repair of oxidative DNA damage is involved in the pathogenesis of CS is attractive, in my view it is based upon questionable assumptions, and much of the data that is interpreted to support the hypothesis could be explained by other mechanisms. For these reasons, this hypothesis needs to be critically evaluated for relevance to different aspects of CS.
4.4. Mitochondrial and metabolic abnormalities in CS: A different perspective on the oxidative stress in CS
The discussion above focused on repair of nuclear DNA. However, there is now accumulating evidence that defects in mitochondrial DNA repair, and other mitochondrial defects, may play a role in CS. A fraction of both CSA and CSB proteins can be detected in the mitochondria of human cells, [97, 98], where they are proposed to play a role in base excision repair of mitochondrial DNA. Other recent data implicates CSB in transcription elongation [99], and CSB was recently reported to play a role in autophagy of damaged mitochondria, a finding that has potential translational implications [100]. Also, Pascucci et al [101] carried out a detailed analysis of mitochondrial function and intermediary metabolism in CS cells, and obtained evidence for altered redox balance and oxidative stress in cells from both CS-A and CS-B patients.
Though of increasing interest, the full significance of reported mitochondrial defects for different aspects of CS remains to be established. While some aspects of the CS phenotype overlap with those of diseases resulting from defects in mitochondrial function, others do not [20]. One major complication with the idea that the somatic features of CS are the result of mitochondrial defects is how to explain the CS phenotype in patients with certain mutations in XPB, XPD, and XPG. To date, there is no evidence for these proteins in mitochondria, yet the description of neuropathology in a CS patient with a mutation in XPG [24] is unmistakably that of CS.
5. A New Look at an Old Hypothesis: CS as a RNAPI and RNAPII transcription disease
The identification of the XPD and XPB helicases as components of the transcription factor TFIIH [102, 103], was an important discovery which dramatically impacted our understanding of the relationship between DNA repair, transcription, and human disease. This finding and subsequent work led to the concept that while XP is a DNA repair disorder, CS and trichothiodystrophy (TTD), could be thought of as “transcription syndromes”, i.e. diseases resulting from abnormalities of transcription [13, 16, 104]. TTD neurologic disease is phenotypically somewhat similar to CS, including neurodevelopmental abnormalities and white matter pathology [105]. TTD can result from mutations in XPB, XPD, TTD-A, all encoding proteins that are components of TFIIH [105].
In this context it is important to emphasize that, although much attention has been placed on the helicase activities of XPB and XPD in both RNA polymerase II transcription initiation and NER, TFIIH can also regulate transcription by interaction with other factors [106]. Also, TFIIH contains a kinase activity, which phosphorylates RNAPII as well as other nuclear hormone receptors [107], and is involved in cell-cycle regulation [108]. In addition, work from the Kornberg laboratory showed that, in yeast, TFIIH acts as an ubiquitin ligase in response to DNA damage [109]. Recent studies in mammalian cells indicate an interrelationship between nuclear hormone receptor phosphorylation and ubiquitin-dependent proteolysis [110]. Therefore, the role of TFIIH in transcriptional regulation is more than a basal RNAPII transcription initiation factor (for a recent review see [16]).
The concept that TTD is a transcription disorder has been fairly well accepted [111–113], and studies in mice provided evidence that myelination defects observed in a mouse model of TTD are the result of dysregulation of genes involved in the synthesis of myelin related proteins [114]. However, the discovery of patients with the phenotypic features of CS caused by mutations in XPG provided an early challenge to the transcription syndrome hypothesis for CS, since at the time no direct evidence linking XPG to transcription was available [115]. In the intervening years, multiple publications from different laboratories and using different methodologies have provided additional support for a role for XPG in transcription, and for the hypothesis that CS is a transcription syndrome.
Regarding XPG, the Prakash lab demonstrated that deletion of the yeast XPG homolog, Rad2, resulted in defective transcription [116]. Notably, this paper showed that a Rad2 mutant corresponding to the XPG mutation in a patient with XP-CS also showed a transcription defect. In mammalian cells, a collaborative effort by the laboratories of Egly and Tanaka [117] showed that human XPG stabilizes TFIIH, and thereby regulates its modulation of transcription by nuclear hormone receptors. Since nuclear hormone receptors influence many cellular processes, including lipid metabolism, defective nuclear hormone signaling provides a compelling explanation for many of CS features in XPG-CS patients [118].
Other work from the Egly lab indicates that CSB is a master regulator of a transcriptional program following exposure to cellular stressors, such as UV [119]. This paper provided evidence that CSB and P53 play a reciprocal role in regulating the transcriptional program in response to UV, a concept that has been mechanistically investigated by Fan and colleagues [120, 121]. Also, Velez-Cruz et al [122] recently described a defect in reassembling gene promoters following UV exposure in cells from an XP-D/CS patient, but not in cells from a XP-D patent with pure XP, further supporting the concept of CS as a transcription disorder. Of more physiologic relevance to the internal manifestations of CS, CS cells have a defective response to hypoxia [94]. The interrelationship between CSB, P53, and other transcription factors adds further support to the transcription syndrome hypothesis [123].
5.1 Microarray evidence of transcription defects by RNAPII in CS and the possible role of a transposable element
A particularly important publication in support of the transcription hypothesis of CS was that of Weiner and colleagues [92] who used a microarray approach to investigate the effects of CSB on gene expression. In contrast to many other studies, these authors exclusively focused on the effects of CSB on transcription in cells in the basal state, i.e., not exposed to UV. Their results support a general role for CSB protein in maintaining chromatin structure, and are consistent with the hypothesis that CS results from the deregulation of transcription of a subset of genes, resulting in abnormalities of growth-related, inflammatory, and proapoptotic pathways.
Subsequent work by these authors, as well as the Tanaka laboratory, has focused on a transposable element, referred to as PGBD3, which is found in intron 5 of the human CSB gene [124]. This transposable element is not present in rodents, which could be of relevance for interpreting mouse models of CS. As a result of alternative splicing, a fusion protein containing the N-terminal portion of CSB and the PGBD3 is produced that can regulate both DNA repair and transcription. A recent publication [125] provides evidence that in the absence of CSB, the fusion protein can induce an interferon and innate immune-like transcriptional response, which may have implications for CS neurologic disease and other phenotypes. This paper also discusses the potential role of genetic background and homozygosity on the expression of the CS phenotype.
5.2 Evidence of defective transcription by RNAPI in CS
As originally proposed, the transcription syndrome hypothesis focused on RNAPII, which was generally consistent with a role for CSB in TC-NER of genes transcribed by RNAPII. However, subsequent work from the laboratories of Egly and Grummt showed that both CSB [126] and TFIIH [127], are involved in transcription by RNA polymerase I, which is responsible for the transcription of ribosomal RNA in the nucleolus. Notably, Bradsher et al [126] identified a protein complex containing CSB, TFIIH, RNAPI and XPG, linking XPG to RNAPI transcription. Subsequent work from the Iben laboratory has extended these findings providing evidence that truncated forms of CSB, as found in some CS patients, can interfere with RNAPI transcription [129]. Most recently, this same group found that TFIIH can act as an elongation factor for RNAPI, and that mutations in XPB and XPD that cause CS specifically impact RNAPI transcription [130]. Other recent studies have demonstrated a role of CSB in regulating RNAPI gene expression via chromatin remodeling effects and DNA methylation [131, 132].
The concept that CSB and TFIIH function in RNAPI transcription is also consistent with evidence showing that a substantial fraction of CSB and XPB are localized to the nucleolus in the absence of DNA damage. Importantly, this conclusion does not solely depend upon studies of fixed tissue but is supported by studies in living cells using fluorescently tagged proteins [44, 48, 133]. An important paper by Hoogstraten et al [133] utilized live cell imaging with fluorescent XPB to demonstrate that TFIIH can rapidly switch between transcription by RNAPI, RNAPII, and NER in vivo. Based on the results, the authors presented a model of the residence time of TFIIH engaged in NER, RNAPI, and RNAPII transcription. Under conditions of UV exposure, the model predicted that TFIIH would spend much time engaged with NER. However, as emphasized above, the relevant comparison for understanding the somatic features of CS are cells without UV exposure, where endogenous levels of DNA lesions repaired by NER are very low. In the absence of UV, the kinetic model would predict that both TFIIH and CSB would spend the majority of time functioning in transcription by RNAPI and II. It seems reasonable that in cells of CS patients that are not exposed to sunlight, mutations in CSB and TFIIH would primarily impact the functions that these proteins carry out in the absence of UV, which are transcription by RNAPI and RNAPII.
A characteristic feature of cells from CS patients is defective recovery of RNA synthesis (RRS) following UV irradiation [134]. RRS was generally thought to correspond to TC-NER, until studies by Mullenders and colleagues demonstrated that defective RRS actually represents a defect in transcription initiation after UV [135]. Interestingly, a recent study [136] observed that the RRS after UV is primarily nucleolar RNA synthesis, consistent with transcription by RNA polymerase I.
In summary, there is direct evidence linking CSB and TFIIH (including XPB, XPD, and XPG) to transcription by both RNAPI and II. The missing factor is CSA. However, CSA is part of the ubiquitin ligase complex (the COP9 signalosome) [137], and there are multiple lines of evidence supporting a role for ubiquitination in the control of transcription [138], including transcription by RNAPI [139]. Thus a plausible hypothesis is that CSA functions to regulate transcription of RNAPI and II, perhaps via interaction with CSB and TFIIH. Notably, purified CSB alone can trigger the ubiquitin ligase activity of CSA-containing COP9 signalosome, demonstrating that DNA damage is not required for this process [140]. The current lack of evidence for a role for CSA in transcription may simply reflect the relative lack of attention that has been paid to the function of CSA outside of TC-NER compared to CSB and TFIIH. Perhaps the recent elucidation of the structure of CSA [140] will stimulate a greater research focus into the functions of CSA.
6. An expanded version of the transcription syndrome hypothesis: CS as a RNA polymerase I and II transcription disorder
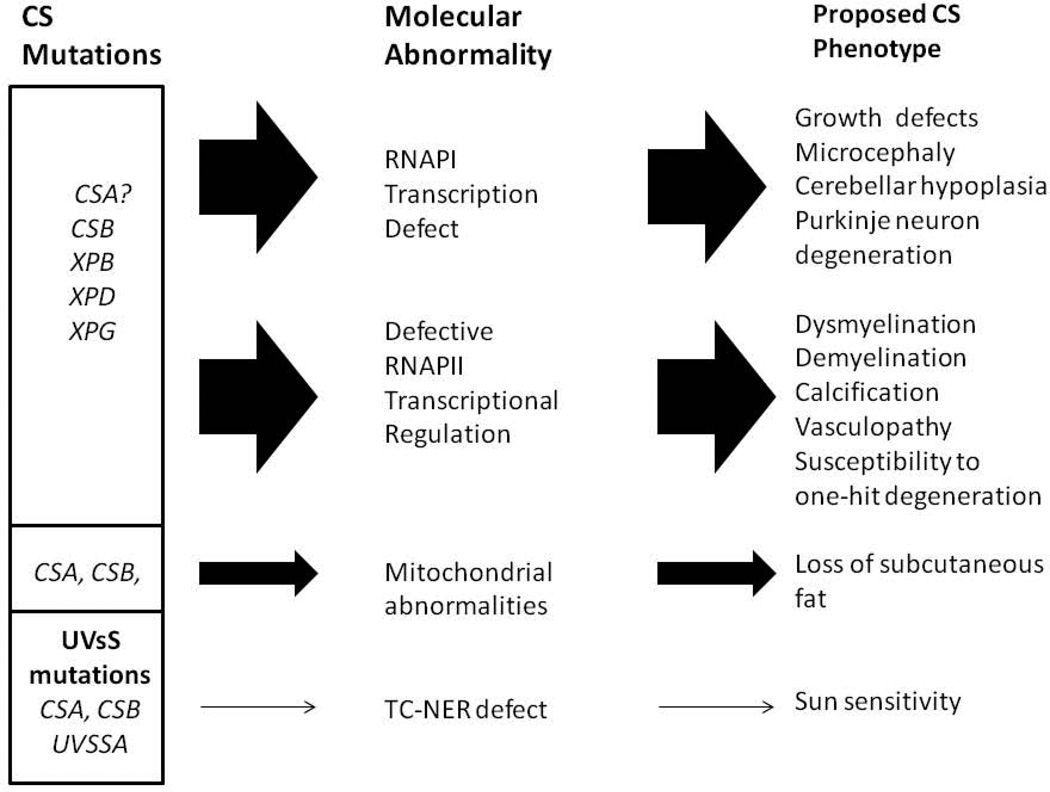
A combination of defective transcription by RNAPI, coupled with abnormalities in the regulation of transcription by RNAPII, provides plausible explanation for many of the somatic features of CS, including growth defects and aspects of CS neurologic disease. Some examples are given below, and a schematic diagram summarizing this hypothesis is given in Fig. 2. In Fig. 2 and the following text, I have separated the discussion of RNAPI and RNAPII for clarity, and to emphasize the potential role of defective RNAPI transcription in different aspect of CS neurologic disease. However, in vivo, the effects of defective RNAPI and RNAPII transcription are not separate but interacting, and together likely influence many aspects of CS disease.
Fig. 2.
An expanded version of the transcription syndrome hypothesis of CS, including defective transcription by both RNAPI and RNAPII. The panel on the right lists different features of CS proposed to result from defects in transcription by RNAPI or II. As discussed in the text, I have separated RNAPI and II for clarity, but in vivo, abnormalities in transcription by both polymerases will likely impact multiple clinical features. Mitochondrial abnormalities could explain the loss of subcutaneous fat, based on [97, 100], but could be involved in other pathologies as well. Multiple mechanisms are likely to be involved in many of the pathologic aspects of CS.
6.1 Growth defects
The ability to synthesize ribosomes is a fundamental cellular process that determines growth and cell division rates [141]. Ribosomes are comprised of protein and ribosomal RNA (rRNA), which is synthesized by RNAPI. In addition to their role in protein synthesis, there is evidence that the rate of ribosome formation determines entry into S-phase [142], similar to a cell cycle checkpoint. Thus a defect in transcription of rRNA genes provides a simple explanation for growth defects observed in CS patients, one of the canonical features of the disease. Interestingly, patients with Werner’s syndrome and Bloom’s syndrome have growth defects, and reduced nucleolar transcription has been described in cells from both diseases [143, 144]. A reduction in RNAPII transcription in permeabilized WS cells has also been reported [145]. In addition to CS, Bloom’s and Werner’s, growth defects are also observed in patients with other “ribosomopathies” as well, further supporting a link between abnormal RNAPI transcription and growth deficiency [144].
6.2 Microcephaly, brain development, and neurologic disease
The importance of the nucleolus for neuronal function and its role in neurologic disease has been studied for many years, and has recently attracted renewed research interest [146]. In terms of RNAPI transcription specifically, an elegant set of experiments [147] demonstrated the differential importance of RNAPI transcription in proliferating versus postmitotic neurons. These authors used an inducible Cre-Lox strategy to delete the gene encoding TIF-1A, an essential basal transcription factor for RNAPI, in either developing or post mitotic neurons. Strikingly, deletion of TIF-1A in developing neurons resulted in an essentially complete lack of all brain tissue by embryonic day 14. Given this dramatic effect of abolishing RNAPI transcription in developing neurons, it seems reasonable to propose that even a small reduction in RNAPI transcription elongation, as in CS cells, could interfere with the development of the brain, providing a plausible explanation for the microcephaly that is a common feature of CS. Furthermore, since the most abundant cell type in the cerebellum are granule cell neurons, a defect in the proliferation of cerebellar granule neurons resulting from defective RNAPI transcription could also explain the cerebellar hypoplasia observed in severely affected CS type II patients.
In contrast to the effect in developing neurons, deletion of the TIF-1A gene in postmitotic neurons resulted in a slowly progressing neuronal death, taking place over a period of months [147]. These findings illustrate the differential importance of rRNA transcription in post mitotic versus differentiating neural cells, which parallels the pathology seen in the brains of CS patients; i.e. severe defects in brain development, with relatively limited degeneration of post mitotic neurons.
6.3 Purkinje neuron degeneration
One exception to the relative sparing of post mitotic neurons from degeneration in CS are Purkinje neurons of the cerebellum. However, this may be an exception that proves the rule. Purkinje neurons are amongst the largest neurons in the brain, and must synthesize high amounts of ribosomal RNA and protein to maintain the massive dendritic tree that is essential for their function. Consistent with this requirement, the size of the nucleolus in human Purkinje neurons approaches the size of the entire nucleus of adjacent cerebellar granule neurons (see [148]). Human Purkinje neurons have very high levels of toposiomerase 1 in the nucleolus, consistent with a role in rRNA gene expression [148]. In addition, studies in mice expressing a GFP-tagged XPB protein showed that XPB, a core component of TFIIH, is almost exclusively localized in the nucleus of Purkinje neurons in postnatal mice [48], consistent with a role in RNAPI transcription in these cells. Based on all of these considerations, defective transcription by RNAPI is a likely explanation for the small size, dysmorphology, and collapsed dendritic tree that is observed in Purkinje neurons in the brain of patients with CS. Notably, nucleolar stress can also trigger P53 activation [149], and increased P53 staining has been observed in Purkinje neurons and other brain cells in mouse models of CS [150, 151].
6.4 Myelin defects
Other aspects of the neurologic abnormalities observed in the brain of CS patients can be explained by abnormal regulation of gene expression by RNAPII. As noted by Koob et al [25], the hypomyelination observed in the brains of CS patients is reminiscent of what is seen in Pelizaeus-Merzbacher disease (PMD), a rare developmental white matter disorder. Koob et al [25] also pointed out that mutations in the MTC8 gene, encoding a thyroid hormone transporter, result in PMD-like brain phenotype [152]. In view of these considerations, a thyroid hormone transcription co-activator defect is a compelling explanation for the global hypomyelination observed in the brain of CS patients. Consistent with this idea, it has been known for decades that thyroid hormone plays an important role in normal brain development in general, and myelination in particular [153], supporting the hypothesis that defects in thyroid-hormone dependent gene transcription could result in myelination defects.
The other type of the myelination phenotype in CS is tigroid demyelination. On the basis of analogy with PMD, Koob et al [25] suggest that the tigroid demyelination could be secondary to defective myelination during development. Alternatively, localized hypoxia resulting from reduced blood flow due to brain microvasculopathy, could also cause focal oligodendrocyte death, as oligodendrocytes are particularly sensitive to hypoxia [154]. In support of the localized hypoxia hypothesis, Rapin et al [23] noted an apparent relationship between the presence of string vessels and demyelinated areas in the brain of CS patients, although no quantitative data were presented. Here it should also be noted that the effect of hypoxia in the brain of CS patients will be amplified, since as noted above, CS cells show an abnormal transcriptional response to hypoxia [94]. More studies of brain tissue from CS patients will be necessary to determine whether either of these two explanations is correct.
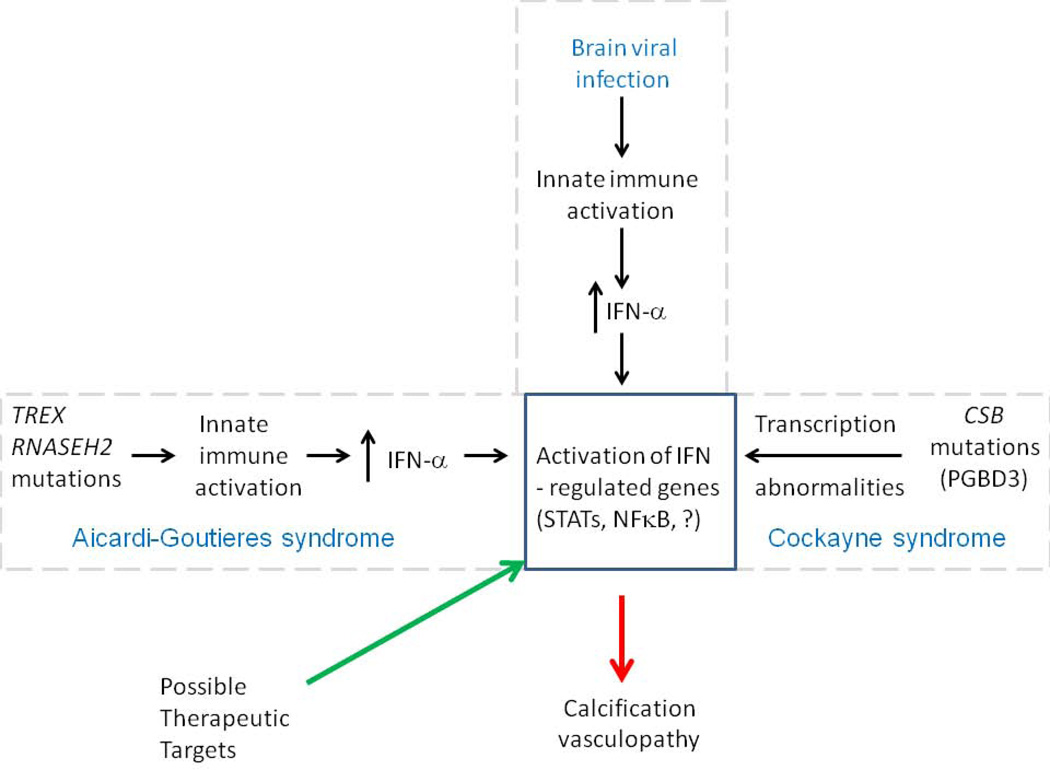
6.5 Neuroinflammatory phenotypes and similarities to Aicardi-Goutieres syndrome
In a previous publication [12] we noted the overlap between the neurologic phenotypes in another rare nucleic acid processing disorder, Aicardi-Goutieres syndrome (AGS) and CS. Figure 3 is an updated version of this concept. AGS results from the inappropriate activation of the innate immune system in the brain, resulting in brain vasculopathy and calcification. A key aspect of the brain pathophysiology in AGS is the secretion of interferon alpha (IFN-α) which acts on other cells in the brain to cause vasculopathy and calcification. Since interferon alpha exerts biologic activity by regulating the expression of a subset of genes, we proposed that if transcriptional deregulation resulting from CSB deficiency mimics the gene expression pattern induced by IFN-α, this could explain the similar brain phenotypes in the two diseases. Notably, our hypothesis [12] does not predict increased IFN-α levels in CS, but rather that CS cells will have transcriptional alterations mimicking those induced by IFN-α. This hypothesis was based upon the findings of Weiner and colleagues [92], which concluded that cells lacking CSB exhibit the gene expression pattern of cells under inflammatory stress. In this context, it is worth noting that the failure to clear damaged mitochondria by autophagy can trigger an innate immune response [155], suggesting a possible mechanistic link between the role of CSB in removing damaged mitochondria [100] and IFN-α regulated gene expression pattern.
Fig. 3.
IFN-α regulated genes as a common pathologic mechanism underlying calcification and vasculopathy in AGS, viral infection of the brain, and CS, as well as a potential therapeutic target for CS. Adapted from [12].
6.6. Brain vasculopathy
The mechanistic basis of the brain vasculopathy in CS neurologic disease is a particularly important question, as it is possible that in addition to tigroid demyelination, other brain phenotypes could be the result of the vascular disease. For example, brain calcification can also be caused by hypoxia, raising the possibility that the brain calcifications are secondary to the vasculopathy. The observation that the pattern of gene expression changes resulting from CSB expression overlaps that of genes regulated by vascular endothelial growth factor [92] could be of relevance understanding brain vasculopathy in CS, but here again much more data are needed.
In summary, an expanded version of the transcription hypothesis of CS, including defects in RNAPI and RNAPII, can potentially explain several features of CS neurologic disease, as well as other aspects of CS. The implications of the expanded transcription syndrome hypothesis for interpreting recent findings from mouse models of CS are considered below.
7. Mouse models of CS neurologic disease and their mechanistic interpretation
Several mouse models of CS have been generated, mostly from the Hoeijmakers laboratory, which have had an important impact on the CS field. The first of these was a mouse with a stop codon mutation in Csb [156]. While fibroblasts from these mice were sensitive to killing by UV light, and showed defects in measures of TC-NER, they had an increased incidence of skin cancer in response to UV, in contrast to human CS patients. These mice were slightly smaller than wild-type controls, and displayed some behavioral abnormalities, but did not show the severe growth retardation and neurologic disease characteristic of human CS patients. Similarly, mice lacking Csa also did not show the somatic or neurologic defects of human CS [157].
7.1. Pathology in single gene knockout mouse models of CS
Jaarsma et al from the Hoeijmakers laboratory [151] carried out a very detailed and careful analysis of the brains from Csa−/−, Csb−/−, and other Cs mouse models. The authors observed an increased frequency of activated microglial cells and astrocytosis in the white matter. Notably, clusters of activated microglia were observed in close association with mature oligodendrocytes. Given that patchy demyelination is characteristic of human CS patients, this phenotype is a potentially important finding. From the mechanistic standpoint, it is notable that activated microglia were already present in 10 week old animals, and the number did not increase with age, as would have been expected from an accumulated DNA damage model. Thus this finding is compatible with a transcription defect. Notably, a subsequent paper by the Cleaver laboratory [158] also described white matter abnormalities in mice lacking both Xpc and Csb, which were interpreted as resulting in part from transcription defects.
7.2 Neuron-specific deletion of Xpa and Csb cause neurodegeneration: A mouse model of XP neurologic disease?
A second important aspect of the paper by Jaarsma et al [151] was the production and analysis of mice lacking both Xpa and Csb in postmitotic forebrain neurons. Mice lacking both Xpa and Csb in all tissues show severe runting, and do not survive past weaning [159], which prevents studies of age-related neurologic disease. To circumvent this problem, the authors produced mice with a neuron-specific deletion of the Xpa gene (flanked by LoxP sites) in Csb−/− mice, by expression of Cre recombinase under the control of the CamKII promoter. This promoter becomes active in postmitotic neurons around the time of birth. These mice were phenotypically normal until around 6 months of age, but then began showing weight loss and neurologic abnormalities, including seizures beginning between 9–12 months of age. All of these neuron-specific Xpa−/− Csb−/− mice died prematurely, between 12–22 months of age. Postmortem analysis revealed progressive neurodegeneration with profound atrophy of the forebrain and hippocampus, accompanied by a massive increase in the volume of forebrain ventricles. The overall appearance of the brains of these animals was strikingly similar to the appearance of the brain in severe cases of human XP neurologic disease [52] (unpublished observations).
7.3 Mechanistic interpretations of neuropathology in mouse models of CS
From a translational standpoint, the findings of Jaarsma et al [151] are very important, as they provide mouse models that can be used to test potential therapeutics in XP and CS neurologic disease. The more complicated and controversial issue concerns the mechanistic interpretation of these findings. The title of the paper [151] refers to the combination of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. As discussed above, the only documented TCR is TC-NER, and Xpa−/− mice already lack all NER. The authors refer to “non-NER TCR”, but it is unclear from the discussion and references provided what specifically is meant by this term. However, Jaarsma et al [151] also state that the question of whether the CS phenotype really results from a repair deficiency remains to be further investigated, and note the possible role of transcription abnormalities as well.
If the important function of CSB is to regulate transcription by RNAPI and II, the progressive degeneration of neurons lacking both Xpa and Csb is readily interpretable by accumulation of transcription-blocking DNA damage on a background of a transcription abnormality. According to this model, the loss of Csb alone would result in a transcription defect, but one that is insufficient to cause neuronal death. However, when NER is disabled in the same neurons by deletion of Xpa, transcription-blocking DNA lesions that would normally be repaired by NER accumulate over time, further interfering with transcription, until the combined transcriptional defects cause neuronal death. And since (with few exceptions) neurons in the mammalian brain cannot be replaced, the result is progressive neurodegeneration and brain atrophy.
7.4 Evidence that accumulated DNA damage does not explain retinal degeneration in CS mice: One-hit neurodegeneration
Another potentially important insight into CS somatic disease comes from studies of retinal degeneration in Csa−/− mice [160]. To understand the implications of these studies, it is useful to briefly review the concept of “one-hit neurodegeneration” [161] which demonstrates how studies of the kinetics of cell death can elucidate pathologic mechanisms.
7.4.1 The “one-hit” model of neurodegeneration
In their influential theoretical paper, Clarke et al [161] began by considering a commonly accepted mechanism of neurodegeneration, in which oxidatively induced damage accumulates over time and eventually causes cell death. They noted that such a mechanism predicts that the probability of cell death increases over time. At early time points, the level of damage will be low, and therefore the probability of cell death is also low. As the damage accumulates, the probability of cell death increases, until late the in the disease, the rate of cell death is rapid as many cells have accumulated high levels of damage. If such a mechanism is operative, a plot of the percentage of starting cell number over time will be sigmoidal in shape. In contrast, if the probability of cell death remains constant over time, the plot will show an exponential decline (see Fig. 1 in [161]).
To test this accumulated damage model, Clarke et al plotted cell number over time for several neurodegenerative diseases, to determine the rate of cell death. Surprisingly, they found that in 17 examples studied, the kinetics of cell death followed an exponential decline. As noted above, such kinetics are not consistent with the accumulated damage model. Instead, this pattern is consistent with an elevated probability of cell death, which remains constant over time.
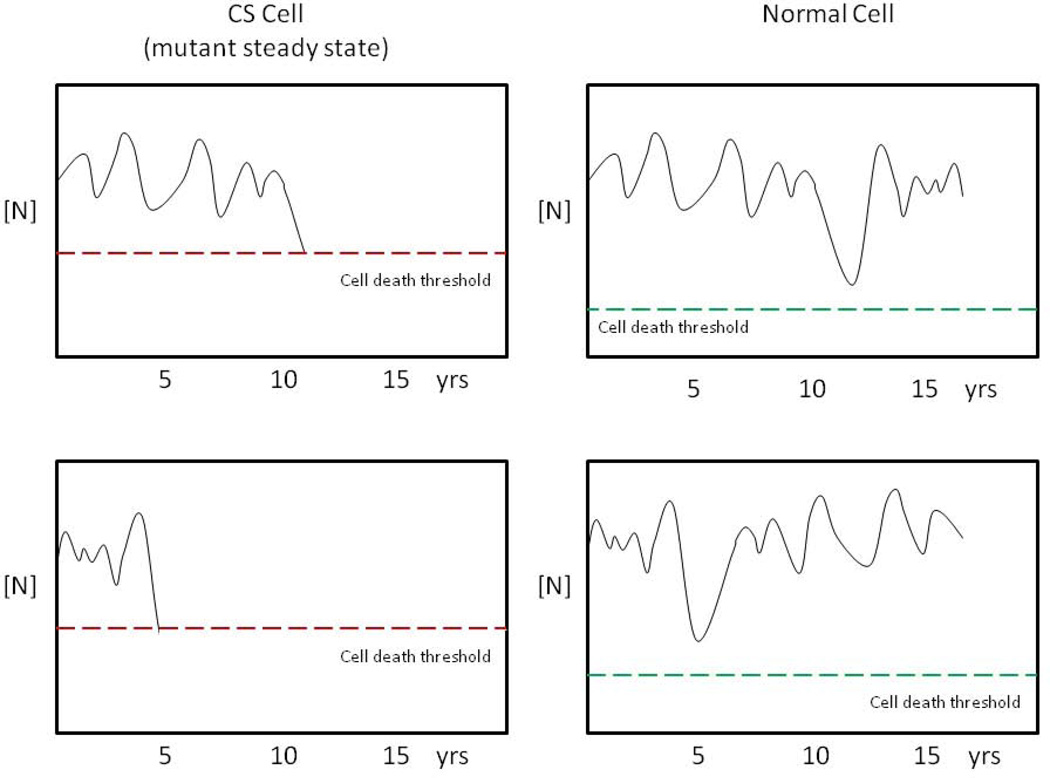
Based on these analyses, Clarke et al proposed the “one-hit neurodegeneration” mechanism. According to this model, vulnerable cells exist in a mutant steady state (MSS), which does not directly result in cell death, but is characterized by a constant, elevated risk of cell death from a single catastrophic rare event, which occurs randomly in time. For example, such a rare event could be a transient large-magnitude fluctuation in the level of some essential nutrient, or toxic metabolite. Importantly, in a normal cell, an event of the same magnitude would be tolerated, and not result in cell death. Figure 4, based upon a subsequent paper [162], is an schematic illustration of this one-hit degeneration mechanism.
Fig. 4.
Schematic representation of the one-hit neurodegeneration hypothesis as applied to CS. The Y axis represents the concentration of a hypothetical critical nutrient, N, (e.g. oxygen) the levels of which fluctuate over time. In CS cells, (left panels) transcription defects lead to a mutant steady state (MSS), characterized by a reduced threshold for cell death (dotted line), which can be triggered by low levels of the nutrient. In the examples shown, transient reductions in [N] result in cell death at around 11 years (top cell) or 5 years (bottom cell). Fluctuations in [N] of same magnitude do not cause cell death in the normal cells (right panels). The same principle could apply to transient elevated levels of a toxic metabolite [162].
7.4.2 Retinal degeneration in Csa mice: Evidence for one-hit degeneration
Returning to CS, Gorgels et al [160] documented evidence of photoreceptor cell loss over time in the retina of Csa−/− mice. As retinal degeneration is also observed in CS patients [20], this is a particularly relevant phenotype. Photoreceptors are terminally differentiated neural cells, and therefore suitable for such an analysis. A careful analysis of the kinetics of photoreceptor loss showed that the data were statistically consistent with a model including a constant probability of cell death over time. The alternative model, explicitly including an increasing rate of cell death as would be predicted by accumulation of oxidative DNA damage, was also tested and was found not to be consistent with the data. Interpreting their results in terms of defective repair of oxidative DNA damage and the free radical theory of aging, the authors proposed that constant probability of retinal cell death over time was the result of elevated steady-state levels of transcription-blocking oxidative DNA damage, due to defective DNA repair. Alternatively however, transcription abnormalities provide an equally plausible explanation for the elevated steady state probability of cell death due to transcription defects, without invoking a form of TCR different from TC-NER.
7.4.3 Implications of one-hit degeneration mechanism for other aspects of CS
The finding that the kinetics of retinal degeneration in Csa−/− mice are consistent with “one-hit” degeneration has potentially broad implications for understanding retinal degeneration as well as other degenerative features of CS. As suggested above, RNAPI and RNAPII transcription abnormalities could explain the elevated susceptibility to cell death in CS. Since different sets of genes are expressed in different cell types, the nature of the transcription abnormalities could vary across different cell types, and therefore the nature of the rare catastrophic events causing cell death could be different in different cells and tissues as well. As one example, hypoxia, perhaps resulting from vasculopathy, could be one such event, leading to the localized death of oligodendrocytes in CS patients.
In summary, many of the recent findings from mouse models of CS neurologic disease can be explained by transcription defects, rather than defects in “non-NER TCR”. The kinetics of cell death during retinal degeneration in Csa−/− mice do not support a causative role for accumulated DNA damage, but are consistent with a transcriptional defect resulting in increased susceptibility to “one-hit” degeneration, which is potentially applicable to other degenerative phenotypes in CS. In this context, it would be of great interest to determine the kinetics of neurodegeneration in mice with neuron-specific deletion of Xpa and Csb described above, as that information could provide evidence for an accumulated DNA damage model of neurodegeneration in XP neurologic disease.
8. Therapeutic Implications
Understanding the mechanistic basis of a human disease is essential for the rational development of treatments and therapies for the patients. At present, the only documented treatment for CS neurologic disease is for tremors and other motor complications that are observed in some CS type I patients [163]. For this reason, I have taken a critical look at multiple mechanisms of CS neurologic disease, because they have quite different implications for additional potential therapies.
As discussed above, the hypothesis that some aspects of CS are the result of defective repair of oxidative DNA damage is common in the literature. While I have questioned the rationale for this idea above, other evidence of elevated oxidative stress and mitochondrial defects in CS cells [100, 101] suggest a potential therapeutic value of antioxidants. However, in view of the signaling role of some ROS, particularly in maintaining the proliferative capacity of neural stem cells [89] as well as recent evidence that some antioxidants can actually cause DNA damage [164], the choice of antioxidant could be critical. To the extent that oxidative stress secondary to mitochondrial abnormalities play a role in the pathophysiology of CS, antioxidants targeted to the mitochondria could be potentially promising in this regard [165].
The possibility that CS is the result of defective transcription regulation is less readily translated into a therapeutic strategy, unless a consistent pattern of transcription abnormality can be identified that can be related to some aspect of the disease, and is “druggable”. While much more work needs to be done on this hypothesis, an example in this regard are the interferon-α regulated genes that are activated in CSB deficient cells [125], and could be involved in some aspects of CS neurologic disease [12] (see Figure 2). If one or more transcription factors could be identified that mediate the abnormal transcriptional response (e.g. [166]), these could be therapeutic targets. However, a limitation here is that because of the involvement of the PGBD3 transposon, which is not present in rodents, currently available mouse models are not suitable for this approach.
The “one-hit” degeneration model also has translational implications. Since the key feature of this model is an increased probability of cell death that remains constant over time, this implies that a cell is as equally susceptible to rescue by a drug or other therapeutic late in the disease as early in the disease [162]. This would not be the case with an accumulated DNA damage mechanism. One major challenge for the “one-hit” hypothesis is determining the nature of the rare catastrophic events that cause cell death, which are likely to vary across different cell types. Since the original evidence for the one-hit hypothesis was obtained in a mouse, mouse models could be potentially valuable in this context.
9. Summary and concluding remarks
CS is a devastating neurodevelopmental disorder, with growth abnormalities, progeriod features, and sun sensitivity. While it was the sun sensitivity that led to the discovery of the TC-NER defect in CS patients, sun sensitivity is a relatively trivial aspect of the clinical picture compared to the neurologic disease. As such, a better understanding of the mechanistic basis of CS neurologic disease is urgently needed, as it essential to the development of rational therapeutic strategies. In this work, I have proposed that while defective TC-NER explains sun sensitivity in CS, a combination of transcription abnormalities affecting RNAPI, and II provides a better explanation for many aspects of CS neurologic disease and other internal features of CS than defective DNA repair. The recent evidence for mitochondrial defects in CS cells is potentially important, but currently difficult to reconcile with the CS phenotypes seen in patients with XPB, XPD, or XPG mutations. Clearly, this will also be a topic for future studies, as will other aspects of the issues I have raised here. There is no doubt that we still have much more to learn about this complex human disease.
In closing, it is important to acknowledge the value and impact of mechanistic studies of TC-NER for CS. While defective TC-NER cannot explain CS neurologic disease, much of our current knowledge of the functions of the CS proteins and TFIIH has come from studies of TC-NER. Moreover, investigations into the mechanisms of TC-NER continue to produce important insights into the mechanistic basis of interactions, communication and signaling between CS proteins, TFIIH, RNAPII, and other proteins (e.g. [167]). Such mechanistic studies will certainly generate additional important insights in the future, which will likely contribute to full understanding of all aspects of CS.
Acknowledgements
This paper was inspired by meeting patients with CS, and discussions with their families, at events organized by the Share and Care Cockayne syndrome network. http://cockaynesyndrome.net/main/ I also thank Cheryl Marietta, Dr. Rebecca Laposa, and anonymous referees for suggestions and helpful comments on the manuscript, and Nicole Patten for the photomicrograph used in Fig. 1.
The human tissue used in Fig. 1, was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. (NICHD contract #HHSN275200900011C, Ref. No. NO1-HD-9-0011).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg EC. Nucleotide excision repair of DNA: The very early history. DNA Repair (Amst) 2011;10:668–672. doi: 10.1016/j.dnarep.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 4.Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci U S A. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LH, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 7.Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH. DNA damage response and transcription. DNA Repair (Amst) 2011;10:743–750. doi: 10.1016/j.dnarep.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV, Friedberg EC. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 9.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6 a member of a subfamily of putative helicases is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 10.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 11.Itoh M, Hayashi M, Shioda K, Minagawa M, Isa F, Tamagawa K, Morimatsu Y, Oda M. Neurodegeneration in hereditary nucleotide repair disorders. Brain Dev. 1999;21:326–333. doi: 10.1016/s0387-7604(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 12.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7:834–848. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Gool AJ, van der Horst GT, Citterio E, Hoeijmakers JH. Cockayne syndrome: defective repair of transcription? EMBO J. 1997;16:4155–4162. doi: 10.1093/emboj/16.14.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bootsma D, Weeda G, Vermeulen W, van Vuuren H, Troelstra C, van der Spek P, Hoeijmakers J. Nucleotide excision repair syndromes: molecular basis and clinical symptoms. Philos Trans R Soc Lond B Biol Sci. 1995;347:75–81. doi: 10.1098/rstb.1995.0012. [DOI] [PubMed] [Google Scholar]
- 15.Hoeijmakers JH, Egly JM, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 16.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 17.Laugel V. Cockayne Syndrome. 1993 [Google Scholar]
- 18.Pasquier L, Laugel V, Lazaro L, Dollfus H, Journel H, Edery P, Goldenberg A, Martin D, Heron D, Le Merrer M, Rustin P, Odent S, Munnich A, Sarasin A, Cormier-Daire V. Wide clinical variability among 13 new Cockayne syndrome cases confirmed by biochemical assays. Arch Dis Child. 2006;91:178–182. doi: 10.1136/adc.2005.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weidenheim KM, Dickson DW, Rapin I. Neuropathology of Cockayne syndrome: Evidence for impaired development premature aging, and neurodegeneration. Mech Ageing Dev. 2009;130:619–636. doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 22.Natale V. A comprehensive description of the severity groups in Cockayne syndrome. Am J Med Genet A. 2011;155A:1081–1095. doi: 10.1002/ajmg.a.33933. [DOI] [PubMed] [Google Scholar]
- 23.Rapin I, Weidenheim K, Lindenbaum Y, Rosenbaum P, Merchant SN, Krishna S, Dickson DW. Cockayne syndrome in adults: review with clinical and pathologic study of a new case. J Child Neurol. 2006;21:991–1006. doi: 10.1177/08830738060210110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindenbaum Y, Dickson D, Rosenbaum P, Kraemer K, Robbins I, Rapin I. Xeroderma pigmentosum/cockayne syndrome complex: first neuropathological study and review of eight other cases. Eur J Paediatr Neurol. 2001;5:225–242. doi: 10.1053/ejpn.2001.0523. [DOI] [PubMed] [Google Scholar]
- 25.Koob M, Laugel V, Durand M, Fothergill H, Dalloz C, Sauvanaud F, Dollfus H, Namer IJ, Dietemann JL. Neuroimaging in Cockayne syndrome. AJNR Am J Neuroradiol. 2010;31:1623–1630. doi: 10.3174/ajnr.A2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi M, Miwa-Saito N, Tanuma N, Kubota M. Brain vascular changes in Cockayne syndrome. Neuropathology. 2012;32:113–117. doi: 10.1111/j.1440-1789.2011.01241.x. [DOI] [PubMed] [Google Scholar]
- 27.Brumback RA, Brooks P, Leech R. Cockayne syndrome. In: Harding GJAB, editor. Pathology and Genetics: Developmental Neuropathology. Basel: ISN Neuropathology Press; 2004. pp. 318–323. [Google Scholar]
- 28.Scharer OD. Multistep damage recognition, pathway coordination and connections to transcription, damage signaling, chromatin structure, cancer and aging: current perspectives on the nucleotide excision repair pathway. DNA Repair (Amst) 2011;10:667. doi: 10.1016/j.dnarep.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenburger T. DNA Repair and Mutagenesis. Second. ASM Press; 2006. [Google Scholar]
- 30.Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 31.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 32.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 34.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 35.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 36.Diderich K, Alanazi M, Hoeijmakers JH. Premature aging and cancer in nucleotide excision repair-disorders. DNA Repair (Amst) 2011;10:772–780. doi: 10.1016/j.dnarep.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 38.Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egly JM, Coin F. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 2011;10:714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2011;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann AR. DNA polymerases and repair synthesis in NER in human cells. DNA Repair (Amst) 2011;10:730–733. doi: 10.1016/j.dnarep.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 42.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 43.Hoeijmakers JH. DNA damage aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 44.van den Boom V, Citterio E, Hoogstraten D, Zotter A, Egly JM, van Cappellen WA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J Cell Biol. 2004;166:27–36. doi: 10.1083/jcb.200401056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Z, Hu W, Komissarova E, Pao A, Hung MC, Adair GM, Tang MS. Transcription-coupled DNA repair is genomic context-dependent. J Biol Chem. 2002;277:12777–12783. doi: 10.1074/jbc.M112297200. [DOI] [PubMed] [Google Scholar]
- 46.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26:8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, Fousteri M, Hoeijmakers JH, Vermeulen W. Differentiation driven changes in the dynamic organization of Basal transcription initiation. PLoS Biol. 2009;7:e1000220. doi: 10.1371/journal.pbio.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews AD, Barrett SF, Robbins JH. Xeroderma pigmentosum neurological abnormalities correlate with colony-forming ability after ultraviolet radiation. Proc Natl Acad Sci U S A. 1978;75:1984–1988. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks PJ. The case for 8,5’-cyclopurine-2’-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–1417. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Przedborski S, Vila M, Jackson-Lewis V. Neurodegeneration: What is it and where are we? J Clin Invest. 2003;111:3–10. doi: 10.1172/JCI17522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robbins JH, Brumback RA, Mendiones M, Barrett SF, Carl JR, Cho S, Denckla MB, Ganges MB, Gerber LH, Guthrie RA, et al. Neurological disease in xeroderma pigmentosum. Documentation of a late onset type of the juvenile onset form, Brain. 1991;114(3):1335–1361. doi: 10.1093/brain/114.3.1335. [DOI] [PubMed] [Google Scholar]
- 53.Itoh T, Ono T, Yamaizumi M. A new UV-sensitive syndrome not belonging to any complementation groups of xeroderma pigmentosum or Cockayne syndrome: siblings showing biochemical characteristics of Cockayne syndrome without typical clinical manifestations. Mutat Res. 1994;314:233–248. doi: 10.1016/0921-8777(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 54.Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC, Sarasin A, Stefanini M. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A. 2009;106:6209–6214. doi: 10.1073/pnas.0902113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M, Tanaka K. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T, Yasui A, Tanaka K. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet. 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 57.Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, Marteijn JA. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 58.Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, Takahashi Y, Ohyama K, Ito K, Mishima H, Nomura M, Kinoshita A, Ono S, Takenaka K, Masuyama R, Kudo T, Slor H, Utani A, Tateishi S, Yamashita S, Stefanini M, Lehmann AR, Yoshiura K, Ogi T. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet. 2012;44:586–592. doi: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- 59.Hashimoto S, Suga T, Kudo E, Ihn H, Uchino M, Tateishi S. Adult-onset neurological degeneration in a patient with Cockayne syndrome and a null mutation in the CSB gene. J Invest Dermatol. 2008;128:1597–1599. doi: 10.1038/sj.jid.5701210. [DOI] [PubMed] [Google Scholar]
- 60.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 61.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5’-(S)-cyclo-2’-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 63.Kuraoka I, Robins P, Masutani C, Hanaoka F, Gasparutto D, Cadet J, Wood RD, Lindahl T. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase eta and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J Biol Chem. 2001;276:49283–49288. doi: 10.1074/jbc.M107779200. [DOI] [PubMed] [Google Scholar]
- 64.Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde a product of lipid peroxidation, is mutagenic in human cells. J Biol Chem. 2003;278:31426–31433. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- 66.Cline SD, Riggins JN, Tornaletti S, Marnett LJ, Hanawalt PC. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7275–7280. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaruga P, Xiao Y, Nelson BC, Dizdaroglu M. Measurement of (5’R)- and (5’S)-8,5’-cyclo-2’-deoxyadenosines in DNA in vivo by liquid chromatography/isotope-dilution tandem mass spectrometry. Biochem Biophys Res Commun. 2009;386:656–660. doi: 10.1016/j.bbrc.2009.06.107. [DOI] [PubMed] [Google Scholar]
- 68.Wang J, Yuan B, Guerrero C, Bahde R, Gupta S, Wang Y. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal Chem. 2011;83:2201–2209. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Cao H, You C, Yuan B, Bahder R, Gupta S, Nishigori C, Niedernhofer L, Brooks P, Wang Y. Endogenous Formation and Repair of Oxidatively Induced G[8-5m]T Intrastrand Cross-link Lesion Nucleic Acids Res. 2012 doi: 10.1093/nar/gks357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nair U, Bartsch H, Nair J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radic Biol Med. 2007;43:1109–1120. doi: 10.1016/j.freeradbiomed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 71.Callegari AJ, Kelly TJ. UV irradiation induces a postreplication DNA damage checkpoint. Proc Natl Acad Sci U S A. 2006;103:15877–15882. doi: 10.1073/pnas.0607343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC. Regulation of ultraviolet light-induced gene expression by gene size. Proc Natl Acad Sci U S A. 2004;101:6582–6586. doi: 10.1073/pnas.0308181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Friedberg EC. Retraction notice to “The yeast RAD2, but not RAD1, gene is involved in the transcription-coupled repair of thymine glycols”. [Mutat. Res. 1995;337:169–178. doi: 10.1016/j.dnarep.2006.08.004. [DOI] [PubMed] [Google Scholar]; DNA Repair (Amst) 2006;5:1507. doi: 10.1016/j.dnarep.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 74.Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tornaletti S, Maeda LS, Hanawalt PC. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem Res Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Sheppard TL, Tornaletti S, Maeda LS, Hanawalt PC. Transcriptional inhibition by an oxidized abasic site in DNA. Chem Res Toxicol. 2006;19:234–241. doi: 10.1021/tx050292n. [DOI] [PubMed] [Google Scholar]
- 77.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frosina G. The current evidence for defective repair of oxidatively damaged DNA in Cockayne syndrome. Free Radic Biol Med. 2007;43:165–177. doi: 10.1016/j.freeradbiomed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Berquist BR, Wilson DM., 3rd Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 81.Heron E, Hernigou A, Chatellier G, Fornes P, Emmerich J, Fiessinger JN. Intracerebral calcification in systemic sclerosis. Stroke. 1999;30:2183–2185. doi: 10.1161/01.str.30.10.2183. [DOI] [PubMed] [Google Scholar]
- 82.Midroni G, Willinsky R. Rapid postanoxic calcification of the basal ganglia. Neurology. 1992;42:2144–2146. doi: 10.1212/wnl.42.11.2144. [DOI] [PubMed] [Google Scholar]
- 83.Perlman JM, Argyle C. Lethal cytomegalovirus infection in preterm infants: clinical radiological, and neuropathological findings. Ann Neurol. 1992;31:64–68. doi: 10.1002/ana.410310112. [DOI] [PubMed] [Google Scholar]
- 84.Stephenson JB. Aicardi-Goutieres syndrome (AGS) Eur J Paediatr Neurol. 2008;12:355–358. doi: 10.1016/j.ejpn.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 85.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 86.Martin GM. The biology of aging: 1985–2010 and beyond. FASEB J. 2011;25:3756–3762. doi: 10.1096/fj.11-1102.ufm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hekimi S, Lapointe J, Wen Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 2011;21:569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 89.Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Waard H, de Wit J, Andressoo JO, van Oostrom CT, Riis B, Weimann A, Poulsen HE, van Steeg H, Hoeijmakers JH, van der Horst GT. Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol Cell Biol. 2004;24:7941–7948. doi: 10.1128/MCB.24.18.7941-7948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kyng KJ, May A, Brosh RM, Jr, Cheng WH, Chen C, Becker KG, Bohr VA. The transcriptional response after oxidative stress is defective in Cockayne syndrome group B cells. Oncogene. 2003;22:1135–1149. doi: 10.1038/sj.onc.1206187. [DOI] [PubMed] [Google Scholar]
- 92.Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci U S A. 2006;103:9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Filippi S, Latini P, Frontini M, Palitti F, Egly JM, Proietti-De-Santis L. CSB protein is (a direct target of HIF-1 and) a critical mediator of the hypoxic response. EMBO J. 2008;27:2545–2556. doi: 10.1038/emboj.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 96.Khobta A, Lingg T, Schulz I, Warken D, Kitsera N, Epe B. Mouse CSB protein is important for gene expression in the presence of a single-strand break in the non-transcribed DNA strand. DNA Repair (Amst) 2010;9:985–993. doi: 10.1016/j.dnarep.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 97.Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, Schaller M, van der Horst GT, van Steeg H, Rocken M, Rapaport D, Krutmann J, Mullenders LH, Berneburg M. Proteins of nucleotide base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat a hallmark of aging. J Exp Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, 3rd, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. Faseb J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berquist BR, Canugovi C, Sykora P, Wilson DM, 3rd, Bohr VA. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012;40:8392–8405. doi: 10.1093/nar/gks565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson Iii DM, Cooper M, Spencer R, de Cabo R, Croteau DL, Bohr VA. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F, Roginskya V, Janjic BM, Van Houten B, Stefanini M, Dogliotti E, D’Errico M. An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell. 2012 doi: 10.1111/j.1474-9726.2012.00815.x. [DOI] [PubMed] [Google Scholar]
- 102.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 103.Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 104.de Boer J, de Wit J, van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 105.Faghri S, Tamura D, Kraemer KH, Digiovanna JJ. Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J Med Genet. 2008;45:609–621. doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, Reinberg D, Levens D. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 107.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 108.Rossignol M, Kolb-Cheynel I, Egly JM. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 1997;16:1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takagi Y, Masuda CA, Chang WH, Komori H, Wang D, Hunter T, Joazeiro CA, Kornberg RD. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol Cell. 2005;18:237–243. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 110.Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 2011;30:468–479. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions. three diseases, Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 112.Bergmann E, Egly JM. Trichothiodystrophy, a transcription syndrome. Trends Genet. 2001;17:279–286. doi: 10.1016/s0168-9525(01)02280-6. [DOI] [PubMed] [Google Scholar]
- 113.Orioli D, Compe E, Nardo T, Mura M, Giraudon C, Botta E, Arrigoni L, Peverali FA, Egly JM, Stefanini M. XPD mutations in trichothiodystrophy hamper collagen VI expression and reveal a role of TFIIH in transcription derepression. Hum Mol Genet. 2013 doi: 10.1093/hmg/dds508. [DOI] [PubMed] [Google Scholar]
- 114.Compe E, Malerba M, Soler L, Marescaux J, Borrelli E, Egly JM. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat Neurosci. 2007;10:1414–1422. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]
- 115.Clarkson SG. The XPG story. Biochimie. 2003;85:1113–1121. doi: 10.1016/j.biochi.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 116.Lee SK, Yu SL, Prakash L, Prakash S. Requirement of yeast RAD2 a homolog of human XPG gene, for efficient RNA polymerase II transcription. implications for Cockayne syndrome. Cell. 2002;109:823–834. doi: 10.1016/s0092-8674(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 117.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 118.Friedberg EC, Wood RD. New insights into the combined Cockayne/xeroderma pigmentosum complex: human XPG protein can function in transcription factor stability. Mol Cell. 2007;26:162–164. doi: 10.1016/j.molcel.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 119.Proietti-De-Santis L, Drane P, Egly JM. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25:1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lake RJ, Basheer A, Fan HY. Reciprocally regulated chromatin association of Cockayne syndrome protein B and p53 protein. J Biol Chem. 2011;286:34951–34958. doi: 10.1074/jbc.M111.252643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Velez-Cruz R, Zadorin AS, Coin F, Egly JM. Sirt1 suppresses RNA synthesis after UV irradiation in combined xeroderma pigmentosum group D/Cockayne syndrome (XP-D/CS) cells. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1213076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Frontini M, Proietti-De-Santis L. Interaction between the Cockayne syndrome B and p53 proteins: implications for aging. Aging (Albany NY) 2012;4:89–97. doi: 10.18632/aging.100439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008;4:e1000031. doi: 10.1371/journal.pgen.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bailey AD, Gray LT, Pavelitz T, Newman JC, Horibata K, Tanaka K, Weiner AM. The conserved Cockayne syndrome B-piggyBac fusion protein (CSB-PGBD3) affects DNA repair and induces both interferon-like and innate antiviral responses in CSB-null cells. DNA Repair (Amst) 2012 doi: 10.1016/j.dnarep.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 127.Iben S, Tschochner H, Bier M, Hoogstraten D, Hozak P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 128.Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 129.Lebedev A, Scharffetter-Kochanek K, Iben S. Truncated Cockayne syndrome B protein represses elongation by RNA polymerase I. J Mol Biol. 2008;382:266–274. doi: 10.1016/j.jmb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 130.Assfalg R, Lebedev A, Gonzalez OG, Schelling A, Koch S, Iben S. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012;40:650–659. doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 132.Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A. 2012;109:8161–8166. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, Vermeulen W, Houtsmuller AB. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol Cell. 2002;10:1163–1174. doi: 10.1016/s1097-2765(02)00709-8. [DOI] [PubMed] [Google Scholar]
- 134.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 135.Rockx DA, Mason R, van Hoffen A, Barton MC, Citterio E, Bregman DB, van Zeeland AA, Vrieling H, Mullenders LH. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc Natl Acad Sci U S A. 2000;97:10503–10508. doi: 10.1073/pnas.180169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hasegawa M, Iwai S, Kuraoka I. A non-isotopic assay uses bromouridine and RNA synthesis to detect DNA damage responses. Mutat Res. 2010;699:62–66. doi: 10.1016/j.mrgentox.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 137.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 138.Hammond-Martel I, Yu H, Affar el B. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012;24:410–421. doi: 10.1016/j.cellsig.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 139.Fatyol K, Grummt I. Proteasomal ATPases are associated with rDNA: the ubiquitin proteasome system plays a direct role in RNA polymerase I transcription. Biochim Biophys Acta. 2008;1779:850–859. doi: 10.1016/j.bbagrm.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 140.Fischer ES, Scrima A, Bohm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, Iwai S, Gut H, Sugasawa K, Thoma NH. The molecular basis of CRL4DDB2/ CSA ubiquitin ligase architecture targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 141.Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 142.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shiratori M, Suzuki T, Itoh C, Goto M, Furuichi Y, Matsumoto T. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene. 2002;21:2447–2454. doi: 10.1038/sj.onc.1205334. [DOI] [PubMed] [Google Scholar]
- 144.Grierson PM, Lillard K, Behbehani GK, Combs KA, Bhattacharyya S, Acharya S, Groden J. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum Mol Genet. 2012;21:1172–1183. doi: 10.1093/hmg/ddr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Balajee AS, Machwe A, May A, Gray MD, Oshima J, Martin GM, Nehlin JO, Brosh R, Orren DK, Bohr VA. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol Biol Cell. 1999;10:2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hetman M, Pietrzak M. Emerging roles of the neuronal nucleolus. Trends Neurosci. 2012;35:305–314. doi: 10.1016/j.tins.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Parlato R, Kreiner G, Erdmann G, Rieker C, Stotz S, Savenkova E, Berger S, Grummt I, Schutz G. Activation of an endogenous suicide response after perturbation of rRNA synthesis leads to neurodegeneration in mice. J Neurosci. 2008;28:12759–12764. doi: 10.1523/JNEUROSCI.2439-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gorodetsky E, Calkins S, Ahn J, Brooks PJ. ATM the Mre11/Rad50/Nbs1 complex, and topoisomerase I are concentrated in the nucleus of Purkinje neurons in the juvenile human brain. DNA Repair (Amst) 2007;6:1698–1707. doi: 10.1016/j.dnarep.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Laposa RR, Huang EJ, Cleaver JE, Increased apoptosis p53 up-regulation. and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc Natl Acad Sci U S A. 2007;104:1389–1394. doi: 10.1073/pnas.0610619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, Rijksen Y, Maas A, van Steeg H, Hoeijmakers JH, van der Horst GT. Age-related neuronal degeneration: complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet. 2011;7:e1002405. doi: 10.1371/journal.pgen.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vaurs-Barriere C, Deville M, Sarret C, Giraud G, Des Portes V, Prats-Vinas JM, De Michele G, Dan B, Brady AF, Boespflug-Tanguy O, Touraine R. Pelizaeus-Merzbacher-Like disease presentation of MCT8 mutated male subjects. Ann Neurol. 2009;65:114–118. doi: 10.1002/ana.21579. [DOI] [PubMed] [Google Scholar]
- 153.Rodriguez-Pena A. Oligodendrocyte development and thyroid hormone. J Neurobiol. 1999;40:497–512. doi: 10.1002/(sici)1097-4695(19990915)40:4<497::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 154.McTigue DM, Tripathi RB. The life death, and replacement of oligodendrocytes in the adult CNS. J Neurochem. 2008;107:1–19. doi: 10.1111/j.1471-4159.2008.05570.x. [DOI] [PubMed] [Google Scholar]
- 155.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.van der Horst GT, van Steeg H, Berg RJ, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR, Bootsma D, Hoeijmakers JH. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 157.van der Horst GT, Meira L, Gorgels TG, de Wit J, Velasco-Miguel S, Richardson JA, Kamp Y, Vreeswijk MP, Smit B, Bootsma D, Hoeijmakers JH, Friedberg EC. UVB radiation-induced cancer predisposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair (Amst) 2002;1:143–157. doi: 10.1016/s1568-7864(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 158.Revet I, Feeney L, Tang AA, Huang EJ, Cleaver JE. Dysmyelination not demyelination causes neurological symptoms in preweaned mice in a murine model of Cockayne syndrome. Proc Natl Acad Sci U S A. 2012;109:4627–4632. doi: 10.1073/pnas.1202621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Murai M, Enokido Y, Inamura N, Yoshino M, Nakatsu Y, van der Horst GT, Hoeijmakers JH, Tanaka K, Hatanaka H. Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc Natl Acad Sci U S A. 2001;98:13379–13384. doi: 10.1073/pnas.231329598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gorgels TG, van der Pluijm I, Brandt RM, Garinis GA, van Steeg H, van den Aardweg G, Jansen GH, Ruijter JM, Bergen AA, van Norren D, Hoeijmakers JH, van der Horst GT. Retinal degeneration and ionizing radiation hypersensitivity in a mouse model for Cockayne syndrome. Mol Cell Biol. 2007;27:1433–1441. doi: 10.1128/MCB.01037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Clarke G, Collins RA, Leavitt BR, Andrews DF, Hayden MR, Lumsden CJ, McInnes RR. A one-hit model of cell death in inherited neuronal degenerations. Nature. 2000;406:195–199. doi: 10.1038/35018098. [DOI] [PubMed] [Google Scholar]
- 162.Clarke G, Lumsden CJ, McInnes RR. Inherited neurodegenerative diseases: the one-hit model of neurodegeneration. Hum Mol Genet. 2001;10:2269–2275. doi: 10.1093/hmg/10.20.2269. [DOI] [PubMed] [Google Scholar]
- 163.Neilan EG, Delgado MR, Donovan MA, Kim SY, Jou RL, Wu BL, Kang PB. Response of motor complications in Cockayne syndrome to carbidopa-levodopa. Arch Neurol. 2008;65:1117–1121. doi: 10.1001/archneur.65.8.1117. [DOI] [PubMed] [Google Scholar]
- 164.Fox JT, Sakamuru S, Huang R, Teneva N, Simmons SO, Xia M, Tice RR, Austin CP, Myung K. High-throughput genotoxicity assay identifies antioxidants as inducers of DNA damage response and cell death. Proc Natl Acad Sci U S A. 2012;109:5423–5428. doi: 10.1073/pnas.1114278109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Frantz MC, Wipf P. Mitochondria as a target in treatment. Environ Mol Mutagen. 2010;51:462–475. doi: 10.1002/em.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gray LT, Fong KK, Pavelitz T, Weiner AM. Tethering of the conserved piggyBac transposase fusion protein CSB-PGBD3 to chromosomal AP-1 proteins regulates expression of nearby genes in humans. PLoS Genet. 2012;8:e1002972. doi: 10.1371/journal.pgen.1002972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM, Svejstrup JQ. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol Cell. 2010;38:637–648. doi: 10.1016/j.molcel.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]