Abstract
We report a genome-wide association study (GWAS) and admixture analysis of glaucoma in 12 008 African-American and Hispanic women (age 50–79 years) from the Women's Health Initiative (WHI). Although GWAS of glaucoma have been conducted on several populations, this is the first to look at glaucoma in individuals of African-American and Hispanic race/ethnicity. Prevalent and incident glaucoma was determined by self-report from study questionnaires administered at baseline (1993–1998) and annually through 2005. For African Americans, there was a total of 658 prevalent cases, 1062 incident cases and 6067 individuals who never progressed to glaucoma. For our replication cohort, we used the WHI Hispanics, including 153 prevalent cases, 336 incident cases and 2685 non-cases. We found an association of African ancestry with glaucoma incidence in African Americans (hazards ratio 1.62, 95% CI 1.023–2.56, P = 0.038) and in Hispanics (hazards ratio 3.21, 95% CI 1.32–7.80, P = 0.011). Although we found that no previously identified glaucoma SNPs replicated in either the WHI African Americans or Hispanics, a risk score combining all previously reported hits was significant in African-American prevalent cases (P = 0.0046), and was in the expected direction in the incident cases, as well as in the Hispanic incident cases. Additionally, after imputing to 1000 Genomes, two less common independent SNPs were suggestive in African Americans, but had too low of an allele frequency in Hispanics to test for replication. These results suggest the possibility of a distinct genetic architecture underlying glaucoma in individuals of African ancestry.
INTRODUCTION
Glaucoma is a heritable and irreversible degenerative optic neuropathy that is the second-leading cause of blindness worldwide (1). The majority of glaucoma cases are caused by open angle glaucoma (OAG), although angle closure glaucoma (ACG) is also common in some racial/ethnic groups. The primary risk factor for open angle glaucoma is intraocular pressure. Heritability estimates for the intraocular pressure endophenotype range from 0.29 to 0.36 (2–4).
The prevalence of glaucoma (OAG and ACG combined) varies by race/ethnicity/ancestry with, for example, individuals from Europe having an estimated prevalence of 2.2%, China 2.7%, Japan 3.7%, Latin America 3.4% and Africa 4.3% (1). Of these, the proportion of individuals due to OAG in Europe is 88.6%, China 52.6%, Japan 89%, Latin America 94.3% and Africa 96.2%, with the remainder due to ACG. Individuals of African ancestry have the highest prevalence of glaucoma, although individuals of white race/ethnicity have a steeper incline in recent years (5). This racial disparity in terms of glaucoma cases suggests that further study of African Americans may provide valuable insights into this disease. In addition, there is a sex disparity in prevalence, as women comprise 59% of all glaucoma cases (1).
Genome-wide association studies (GWAS) of glaucoma have been conducted on individuals of white race/ethnicity (6–10), individuals of Chinese race/ethnicity (11) and individuals of Japanese race/ethnicity (12–16), but none to our knowledge on African Americans or Hispanics. Here, we present a GWAS of self-reported glaucoma prevalence and incidence in the Women's Health Initiative (WHI) SNP Health Association Resource (WHI SHARe), which includes 7787 African-American and 3174 Hispanic women.
RESULTS
Demographic characteristics of the cohort are given in Table 1. The baseline prevalence of glaucoma in African Americans was 8.4% (95% CI 7.8–9.1%) for women of an average baseline age of 61.6 (SD 7.04), and in Hispanics was 4.8% (95% CI 4.1 to 5.6%) for women of an average baseline age of 60.2 (SD 6.68). Results of multivariable regression analyses are provided in Table 2. As expected, age significantly increases the risk for glaucoma. Body mass index (BMI) is positively associated with risk (except in prevalent African Americans), and is statistically significant for African-American incident cases. As has been reported previously (17,18), diabetes is also positively associated with risk of glaucoma in both prevalent and incident cases, and statistically significant for all but the Hispanic incident cases. Systolic blood pressure (SBP), diastolic blood pressure (DBP) and hypertension were generally not associated with glaucoma. Global (genome-wide) African ancestry was associated with an increased hazard of developing glaucoma in the African-American individuals, even though we were controlling for education and income (which showed no significant association) along with the other covariates (hazards ratio 1.62, 95% CI 1.023–2.56, P = 0.038). African ancestry is also associated with an even greater increase in risk in the Hispanic individuals, even though we adjusted for income, education, nationality and the other covariates (hazards ratio 3.21, 95% CI 1.32–7.80, P = 0.011), although the confidence interval overlaps with that of African Americans. Global African ancestry was not formally significant in either the African-American or Hispanic prevalent cases, though the direction was the same as that of the incident cases. Among the Hispanics, Native American ancestry was positively associated with glaucoma risk in analysis of both prevalent and incident cases, but did not reach statistical significance.
Table 1.
Demographic and clinical factors of women with and without glaucoma in the Women's Health Initiative (WHI) SNP Health and Association Resource (SHARe) cohort
| African American |
Hispanic |
|||||
|---|---|---|---|---|---|---|
| Prevalent cases | Incident cases | Non-cases | Prevalent cases | Incident cases | Non-cases | |
| N | 658 | 1062 | 6067 | 153 | 336 | 2685 |
| BMI | 30.77 (0.25) | 31.49 (0.19) | 31.23 (0.08) | 29.58 (0.48) | 29.19 (0.28) | 29.07 (0.11) |
| SBP | 132.85 (0.6) | 131.71 (0.45) | 130.79 (0.19) | 128.86 (1.18) | 126.99 (0.73) | 124.75 (0.27) |
| DBP | 75.55 (0.31) | 76.16 (0.25) | 76.77 (0.1) | 74.18 (0.67) | 73.98 (0.4) | 73.75 (0.14) |
| Hypertension | 398 [61.5%] | 591 [56.6%] | 3269 [54.6%] | 64 [41.8%] | 113 [33.7%] | 754 [28.2%] |
| Diabetes | 146 [22.2%] | 187 [17.6%] | 762 [12.6%] | 28 [18.3%] | 31 [9.2%] | 203 [7.5%] |
| Income < $35,000 | 367 [59.4%] | 577 [57.1%] | 2832 [48.9%] | 80 [60.6%] | 185 [58.9%] | 1359 [54.3%] |
| High School or less | 180 [27.6%] | 274 [26.2%] | 1386 [23.1%] | 64 [43.2%] | 151 [45.5%] | 983 [37.2%] |
| Age | 65.06 (0.28) | 62.15 (0.22) | 61.07 (0.09) | 63.74 (0.58) | 61.21 (0.35) | 59.89 (0.13) |
| Puerto Rican nationality | – | – | – | 16 [12.6%] | 51 [16.5%] | 273 [11.6%] |
| Mexican American nationality | – | – | – | 56 [44.1%] | 144 [46.5%] | 1146 [48.6%] |
| Cuban nationality | – | – | – | 8 [6.3%] | 34 [11%] | 184 [7.8%] |
| Other Spanish nationality | – | – | – | 38 [29.9%] | 66 [21.3%] | 639 [27.1%] |
Counts [% of non-missing] are given for dichotomous variables, and mean (SE) for continuous variables.
Table 2.
Multivariable regression on glaucoma prevalence/incidence
| African American |
Hispanic |
|||
|---|---|---|---|---|
| Prevalent | Incident | Prevalent | Incident | |
| BMI | 0.9927 (0.978,1.0072) | 1.013 (1.003,1.024) | 1.013 (0.975,1.051) | 1.0051 (0.982,1.029) |
| Age | 1.071 (1.057,1.086) | – | 1.067 (1.033,1.10) | – |
| SBP | 0.99905 (0.9919,1.0062) | 0.9926 (0.987, 0.998) | 1.0059 (0.986,1.026) | 0.997 (0.986,1.0083) |
| DBP | 0.9952 (0.982,1.0081) | 1.012 (1.0022,1.022) | 1.0054 (0.971,1.041) | 1.0087 (0.989,1.029) |
| Diabetes | 1.62 (1.3,2.019) | 1.42 (1.19,1.68) | 2.22 (1.25,3.93) | 1.0029 (0.63,1.59) |
| Hypertension | 1.093 (0.906,1.32) | 0.903 (0.79,1.035) | 1.27 (0.80,2.0021) | 0.984 (0.74,1.30) |
| Income < $35,000 | 1.12 (0.926,1.35) | 1.066 (0.931,1.22) | 0.962 (0.63,1.48) | 0.93 (0.72,1.20) |
| High School or less | 1.018 (0.83,1.25) | 1.049 (0.901,1.22) | 0.82 (0.52,1.29) | 1.31 (1.011,1.69) |
| Puerto Rican | – | – | 0.85 (0.43,1.68) | 1.46 (0.9935,2.16) |
| Mexican American | – | – | 0.70 (0.44,1.11) | 1.18 (0.87,1.60) |
| Cuban | – | – | 0.64 (0.26,1.62) | 1.47 (0.925,2.33) |
| African ancestry | 1.26 (0.68, 2.33) | 1.62 (1.023,2.56) | 1.89 (0.34,10.43) | 3.21 (1.32,7.80) |
| Native American ancestry | – | – | 1.97 (0.59,6.57) | 1.18 (0.54,2.56) |
Odds ratios and 95% CI are from logistic regression (prevalent cases) and hazard rates and 95% CI are from Cox proportional hazards regression (incident cases).
We first tested 18 SNPs that were previously shown to be associated with glaucoma (6–16) obtained from the National Human Genome Research Institute GWAS catalog (19). None of these SNPs replicated in the WHI-SHARe cohorts, as shown in Table 3, even though the majority of the SNPs imputed extremely well (all but one have estimated r2 > 0.95). However, of the 10 SNPs in six chromosome locations that were previously found to be associated with OAG, most showed association in the same direction in both the analysis of prevalent and incident cases (African-American incident 8/10; African-American prevalent 9/10; Hispanic incident 7/10; Hispanic prevalent 8/10).
Table 3.
P-values at SNPs associated with Glaucoma in previous studies. Tests of individual SNPs
| SNP | Chr | Pos | Gene | A1 | A2 | African-American incident |
African-American prevalent |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 | Case A1F | Non-case A1F | HR/OR (95% CI) | P | Case A1F | Non-case A1F | HR/OR (95% CI) | P | ||||||||
| rs4656461 OAG | 1 | 165 687 205 | TMCO1 | G | A | 0.986 | 0.212 | 0.211 | 1.012 (0.91,1.12) | 0.82 | 0.237 | 0.212 | 1.17 (1.021,1.34) | 0.024 | ||
| rs7588567 OAG | 2 | 134 363 032 | NCKAP5 | C | T | 0.517 | 0.396 | 0.401 | 0.95 (0.84,1.073) | 0.41 | 0.419 | 0.4 | 1.14 (0.97,1.34) | 0.12 | ||
| rs4236601 OAG | 7 | 116 162 729 | CAV2- CAV1 | A | G | 0.988 | 0.353 | 0.362 | 0.97 (0.89,1.066) | 0.57 | 0.376 | 0.361 | 1.081 (0.96,1.22) | 0.2 | ||
| rs284489 OAG,NR | 8 | 105 958 020 | LRP12- ZFPM2 | A | G | 1 | 0.395 | 0.386 | 1.055 (0.97,1.15) | 0.23 | 0.398 | 0.388 | 1.059 (0.94,1.19) | 0.34 | ||
| rs1063192 OAG | 9 | 22 003 367 | CDKN2B | A | G | 0.975 | 0.907 | 0.905 | 1.0099 (0.87,1.18) | 0.9 | 0.924 | 0.905 | 1.33 (1.06,1.66) | 0.013 | ||
| rs523096 OAG | 9 | 22 019 129 | CDKN2BAS | A | G | 1 | 0.909 | 0.905 | 1.036 (0.89,1.21) | 0.65 | 0.922 | 0.906 | 1.29 (1.036,1.61) | 0.023 | ||
| rs7865618 OAG | 9 | 22 031 005 | CDKN2BAS | A | G | 1 | 0.91 | 0.907 | 1.018 (0.87,1.19) | 0.82 | 0.926 | 0.908 | 1.33 (1.062,1.66) | 0.013 | ||
| rs2157719 OAG,NR | 9 | 22 033 366 | CDKN2BAS | T | C | 0.999 | 0.91 | 0.907 | 1.017 (0.87,1.19) | 0.83 | 0.926 | 0.908 | 1.33 (1.062,1.66) | 0.013 | ||
| rs4977756 OAG | 9 | 22 068 652 | CDKN2BAS | A | G | 0.981 | 0.656 | 0.648 | 1.033 (0.94,1.13) | 0.48 | 0.664 | 0.649 | 1.079 (0.95,1.22) | 0.23 | ||
| rs10483727 OAG | 14 | 61 072 875 | SIX1- SIX6 | T | C | 1 | 0.88 | 0.858 | 1.18 (1.027,1.35) | 0.019 | 0.859 | 0.861 | 0.98 (0.82,1.16) | 0.81 | ||
| rs3753841 ACG | 1 | 103 379 918 | COL11A1 | G | A | 0.997 | 0.847 | 0.841 | 1.015 (0.9,1.15) | 0.81 | 0.829 | 0.842 | 0.89 (0.76,1.049) | 0.17 | ||
| rs10881542 ACG | 1 | 104 852 738 | FTLP17- CDK4PS | C | T | 0.95 | 0.6 | 0.586 | 1.056 (0.97,1.15) | 0.23 | 0.564 | 0.588 | 0.9 (0.8,1.014) | 0.083 | ||
| rs1015213 ACG | 8 | 52 887 541 | PCMTD- ST18 | T | C | 1 | 0.266 | 0.28 | 0.94 (0.85,1.03) | 0.17 | 0.261 | 0.278 | 0.92 (0.8,1.043) | 0.18 | ||
| rs11024102 ACG | 11 | 17 008 605 | PLEKHA7 | C | T | 0.99 | 0.07 | 0.07 | 1.051 (0.88,1.25) | 0.57 | 0.069 | 0.07 | 0.98 (0.77,1.23) | 0.84 | ||
| rs3788317 ACG | 22 | 19 889 825 | TXNRD2 | G | T | 0.876 | 0.515 | 0.529 | 0.95 (0.87,1.038) | 0.25 | 0.545 | 0.527 | 1.1 (0.98,1.25) | 0.12 | ||
| rs3213787 NTG,NR | 2 | 45646 824 | SRBD1 | A | G | 1 | 0.983 | 0.985 | 0.88 (0.64,1.22) | 0.45 | 0.986 | 0.984 | 1.049 (0.65,1.7) | 0.84 | ||
| rs735860 NTG,NR | 6 | 53 123 118 | GCM1− ELOVL5 | C | T | 1 | 0.893 | 0.899 | 0.93 (0.81,1.071) | 0.31 | 0.909 | 0.898 | 1.14 (0.93,1.39) | 0.22 | ||
| rs3825942 EXF | 15 | 74 219 582 | LOXL1 | G | A | 0.987 | 0.631 | 0.627 | 1.014 (0.93,1.11) | 0.76 | 0.612 | 0.627 | 0.94 (0.83,1.058) | 0.3 | ||
| SNP | Chr | Pos | Gene | A1 | A2 | Hispanic incident | Hispanic prevalent | Prev OR | Effect | |||||||
| r2 | Case A1F | Non-case A1F | HR/OR (95% CI) | P | Case A1F | Non-case A1F | HR/OR (95% CI) | P | ||||||||
| rs4656461 OAG | 1 | 165 687 205 | TMCO1 | G | A | 0.988 | 0.151 | 0.138 | 1.081 (0.87,1.34) | 0.47 | 0.147 | 0.139 | 1.052 (0.76,1.46) | 0.76 | 1.51 | ++++ |
| rs7588567 OAG | 2 | 134 363 032 | NCKAP5 | C | T | 0.573 | 0.459 | 0.449 | 1.085 (0.89,1.32) | 0.42 | 0.458 | 0.45 | 1.036 (0.77,1.4) | 0.82 | 1.18 | −+++ |
| rs4236601 OAG | 7 | 116 162 729 | CAV2- CAV1 | A | G | 0.996 | 0.244 | 0.249 | 0.96 (0.81,1.14) | 0.66 | 0.242 | 0.249 | 0.96 (0.73,1.25) | 0.74 | 1.27 | −+−− |
| rs284489 OAG,NR | 8 | 105 958 020 | LRP12- ZFPM2 | A | G | 1 | 0.638 | 0.624 | 1.092 (0.93,1.28) | 0.29 | 0.572 | 0.625 | 0.82 (0.64,1.043) | 0.11 | 1.61 | +++− |
| rs1063192 OAG | 9 | 22 003 367 | CDKN2B | A | G | 0.973 | 0.762 | 0.757 | 0.998 (0.83,1.2) | 0.98 | 0.772 | 0.757 | 1.029 (0.77,1.37) | 0.84 | 1.33 | ++−+ |
| rs523096 OAG | 9 | 22 019 129 | CDKN2BAS | A | G | 1 | 0.746 | 0.755 | 0.93 (0.78,1.11) | 0.42 | 0.765 | 0.754 | 1.0081 (0.76,1.33) | 0.95 | 2.13 | ++−+ |
| rs7865618 OAG | 9 | 22 031 005 | CDKN2BAS | A | G | 1 | 0.769 | 0.763 | 1.0035 (0.83,1.21) | 0.97 | 0.778 | 0.764 | 1.025 (0.77,1.36) | 0.87 | 1.79 | ++++ |
| rs2157719 OAG,NR | 9 | 22 033 366 | CDKN2BAS | T | C | 0.998 | 0.769 | 0.763 | 1.0036 (0.83,1.21) | 0.97 | 0.778 | 0.764 | 1.023 (0.77,1.36) | 0.88 | 1.72 | ++++ |
| rs4977756 OAG | 9 | 22 068 652 | CDKN2BAS | A | G | 0.994 | 0.728 | 0.745 | 0.94 (0.79,1.11) | 0.48 | 0.751 | 0.743 | 1.019 (0.78,1.33) | 0.89 | 1.39 | ++−+ |
| rs10483727 OAG | 14 | 61 072 875 | SIX1- SIX6 | T | C | 1 | 0.42 | 0.412 | 1.015 (0.87,1.18) | 0.85 | 0.477 | 0.413 | 1.3 (1.027,1.64) | 0.029 | 1.29 | +−++ |
| rs3753841 ACG | 1 | 103 379 918 | COL11A1 | G | A | 0.999 | 0.375 | 0.346 | 1.069 (0.92,1.25) | 0.4 | 0.359 | 0.349 | 1.028 (0.81,1.3) | 0.82 | 1.2 | +−++ |
| rs10881542 ACG | 1 | 104 852 738 | FTLP17- CDK4PS | C | T | 0.97 | 0.526 | 0.5 | 1.1 (0.94,1.29) | 0.23 | 0.481 | 0.503 | 0.94 (0.73,1.19) | 0.59 | 1.17 | +−+− |
| rs1015213 ACG | 8 | 52 887 541 | PCMTD- ST18 | T | C | 1 | 0.071 | 0.083 | 0.85 (0.63,1.15) | 0.29 | 0.059 | 0.082 | 0.68 (0.42,1.12) | 0.13 | 1.49 | −−−− |
| rs11024102 ACG | 11 | 17 008 605 | PLEKHA7 | C | T | 0.996 | 0.26 | 0.255 | 1.043 (0.88,1.24) | 0.63 | 0.284 | 0.256 | 1.16 (0.9,1.49) | 0.26 | 1.22 | +−++ |
| rs3788317 ACG | 22 | 19 889 825 | TXNRD2 | G | T | 0.87 | 0.744 | 0.732 | 1.11 (0.92,1.33) | 0.29 | 0.775 | 0.734 | 1.3 (0.96,1.75) | 0.09 | 1.21 | −+++ |
| rs3213787 NTG,NR | 2 | 45646 824 | SRBD1 | A | G | 1 | 0.876 | 0.873 | 1.019 (0.81,1.28) | 0.87 | 0.869 | 0.873 | 0.99 (0.7,1.39) | 0.94 | 2.8 | −++− |
| rs735860 NTG,NR | 6 | 53 123 118 | GCM1− ELOVL5 | C | T | 1 | 0.649 | 0.645 | 0.9997 (0.85,1.17) | 1 | 0.641 | 0.645 | 0.96 (0.75,1.21) | 0.71 | 1.69 | −+−− |
| rs3825942 EXF | 15 | 74 219 582 | LOXL1 | G | A | 0.992 | 0.819 | 0.835 | 0.9 (0.74,1.1) | 0.31 | 0.853 | 0.833 | 1.16 (0.84,1.62) | 0.37 | 20.1 | +−−+ |
Chr, chromosome; Pos, base pair position; A1, allele 1, risk allele previously associated; A2, allele 2; A1F, frequency of A1 allele; HR, hazards ratio (incident); OR, odds ratio (prevalent); P, P-value; Prev OR, odds ratio previously reported for SNP; Effect, going across the columns for African-American incident, African-American prevalent, Hispanic incident and Hispanic prevalent, a “+” indicates that it is in the same direction as previously reported and a “−” indicates that it is in the opposite direction; OAG, previously found to be associated with open angle glaucoma; ACG, angle-closed glaucoma; NTG, normal tension glaucoma; EXF, exfoliation glaucoma; NR, previously associated but not replicated SNP. Thus, the first 13 SNPs are the ones we are most confident of being true previous hits.
All ORs/RRs in Table 3 are smaller than the originally reported value. This may be due to winner's curse (inflation of values in the original studies) (20), diminished effects in the race/ethnicity groups in this study or attenuation due to imprecise diagnosis in the current study. To assess the impact of the latter, for each of the 10 SNPs in Table 3 for OAG, we calculated RR-1 (or OR-1) for the original study and then for each of our four study groups (African-American and Hispanic incident and prevalent), and then to the ratio of (RR-1) for each SNP in each of our study groups to the original SNP (RR-1). The mean (median) value of this ratio across 10 SNPs for each of the four study groups is 0.054 (0.027) African-American incident, 0.377 (0.310) African-American prevalent, 0.175 (0.146) African-American incident and prevalent combined, 0.048 (0.005) Hispanic incident, 0.110 (0.040) Hispanic prevalent and 0.070 (0.014) Hispanic incident and prevalent combined. These results suggest somewhat stronger replication evidence in the prevalent cases (who are younger at diagnosis) and African Americans. However, overall, the relative risks are substantially attenuated compared with the originally observed values.
To evaluate the impact of imprecise diagnosis, we employed Formula 2 from Materials and Methods. Formula 2 is more influenced by the specificity (b) of self-report than sensitivity (a). Therefore, we consider the single value of a = 0.9 and vary b from 0.75 to 0.95 and consider two values for the prevalence (u) in non-genotype carriers of 0.10 and 0.20. From Formula 2, the ratio (RRo − 1)/(RRt − 1) ranges from 0.21 at b = 0.75–0.63 at b = 0.95 when u = 0.10 and from 0.34 at b = 0.75–0.77 at b = 0.95 when u = 0.20. These values are generally higher, even at a moderate specificity of 0.75, than those above based on the observed SNP association results, suggesting that diagnostic imprecision may not be the only factor attenuating the observed RRs.
We then formed an SNP risk score, weighting each variant by the previously reported log odds ratio of the OAG-associated SNPs (see Materials and Methods), the score was significant in African-American prevalent cases (P = 0.00086, Table 4), and in the expected positive direction though not significant for two of the three other groups. Of note, all of the SNPs previously found to be associated with glaucoma were originally identified in studies of white, Chinese and Japanese subjects, who have a different LD structure than the African Americans and Hispanics in the WHI. To assess if a different SNP that was highly correlated with the original GWAS finding might actually be the true causal locus, and the GWAS failed replication because of different LD patterns in the current study, local association plots using the LD pattern in the population in which the SNP was originally discovered are shown in Supplementary Material, Figure S2. There is a very mild suggestion of a few potential hits that are in strong LD with the SNP in the discovery panel, but no longer in African Americans or Hispanics (because of the different LD structure in these races/ethnicities); none is particularly notable nor would survive multiple comparison correction.
Table 4.
Association of risk score combining previously replicated OAG SNPs by multivariate logistic (prevalent cases) or Cox proportional hazards (incident cases) regression analysis
| Population | Risk score |
|
|---|---|---|
| HR/OR (95% CI) | P | |
| African-American incident | 1.075 (0.955, 1.209) | 0.23 |
| African-American prevalent | 1.020 (1.008, 1.033) | 0.00086 |
| Hispanic incident | 1.096 (0.907, 1.325) | 0.34 |
| Hispanic prevalent | 0.9997 (0.9867, 1.0128) | 0.96 |
Chr, chromosome; Pos, base pair position; A1, allele 1, risk allele previously associated; A2, allele 2; A1F, frequency of A1 allele; HR, hazards ratio (incident); OR, odds ratio (prevalent); P, P-value; Prev OR, odds ratio previously reported for SNP; Effect, going across the columns for African-American incident, African-American prevalent, Hispanic incident and Hispanic prevalent, a “+” indicates that it is in the same direction as previously reported and a “−” indicates that it is in the opposite direction; OAG, previously found to be associated with open angle glaucoma; ACG, angle-closed glaucoma; NTG, normal tension glaucoma; EXF, exfoliation glaucoma; NR, previously associated but not replicated SNP. Thus, the first 13 SNPs are the ones we are most confident of being true previous hits.
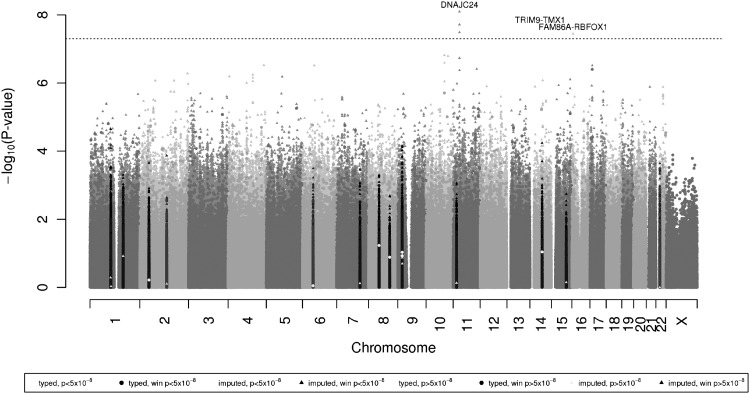
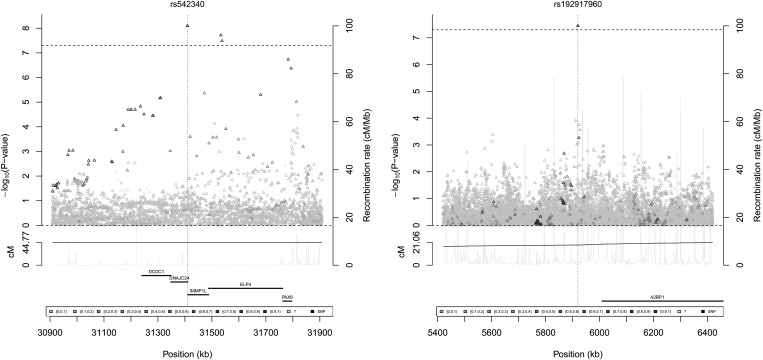
None of the directly genotyped SNPs in the combined incidence and prevalence analysis in African Americans showed genome-wide significant association (defined as the conventional P < 5 × 10−8) with glaucoma. We then examined SNPs imputed from the 1000 Genomes dataset (http://1000genomes.org), shown in a Manhattan plot in Figure 1 and Q–Q plot in Supplementary Material, Figure S1a (individual incident and prevalent in Supplementary Material, Figure S1b and c, respectively). The SNP rs542340 in the DNAJC24 gene on chromosome 11 was the most significant (P = 8.0 × 10−9), and is described in Table 5, along with a local association plot in Figure 2. The frequency of the SNP we observed in the WHI African-American controls (0.018, Table 5) is consistent with those found in 1000 Genomes populations (10/246 YRI carriers, MAF 0.0203; 1/181 AMR, MAF 0.0028; 0/286 ASN; and 0/379 EUR). As expected, this SNP is very rare in the WHI Hispanics and does not have a sufficiently high allele frequency to test for replication. Two other neighboring imputed SNPs in the introns of ELP4, rs555091 and rs506227, both nearly perfectly correlated with rs542340, also showed genome-wide significance; neither was significant after adjusting for rs542340. We examined the recent ENCODE results (21) and queried RegulomeDB (22) to evaluate the regulatory potential score of these SNPs. There was no data found for rs542340, and only very minimal support for the other highly correlated imputed SNPs around it. We identified two additional SNPs, one on chromosome 14 and one on chromosome 16 that were genome-wide significant (Table 5). The minor allele of intergenic SNP rs192917960, on chromosome 16, was also too rare to test for replication in the WHI Hispanics and has no encode data supporting it. The final SNP that was genome-wide significant in African Americans, with 1000 Genomes identifier chr14:51604618:I, failed to replicate in the Hispanics, although the summary odds ratio and the odds ratio for incident cases were in a consistent direction.
Figure 1.
Manhattan plot of glaucoma association for African Americans. P-values combine incident and prevalent coefficients using an inverse variance method. The dotted line here marks genome-wide significance at 5 × 10−8. Inflation factor λ = 1.022. In the plot/legend, the color code of the P-values corresponds to the P-value in previously reported studies; in the legend, for example, “genotyped, P < 5 × 10−8” indicates that a previous study had a P-value of <5 × 10−8, i.e. genome-wide significant, and “genotyped, win P < 5 × 10−8” indicates that it is within 0.5 Mb of such an SNP (a 1 Mb window). Triangles are imputed SNPs and circles are genotyped SNPs.
Table 5.
Results for SNPs that reach genome-wide significance (P < 5 × 10−8) in the WHI-SHARe African Americans
| SNP | Chr | Pos | Gene | Location | RDB | A1 | A2 | African-American incident |
African-American prevalent |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 | Case | Non-case | HR/OR | P | Case | Non-case | HR/OR | P | ||||||||
| rs542340 | 11 | 31 409 438 | DNAJC24 | Intron | – | G | A | 0.72 | 0.028 | 0.018 | 1.86 (1.4,2.48) | 2.1 × 10−5 | 0.032 | 0.019 | 2.1 (1.45,3.039) | 8.5 × 10−5 |
| rs555091 | 11 | 31 532 959 | ELP4 | Intron | 5 | C | T | 0.728 | 0.029 | 0.018 | 1.85 (1.39,2.47) | 2.3 × 10−5 | 0.032 | 0.02 | 2.037 (1.4,2.97) | 2e−04 |
| rs506227 | 11 | 31 537 439 | ELP4 | Intron | – | T | C | 0.741 | 0.028 | 0.018 | 1.83 (1.38,2.44) | 3.4 × 10−5 | 0.032 | 0.02 | 2.026 (1.39,2.95) | 0.00023 |
| chr14:51604618:I | 14 | 51 604 618 | TRIM9- TMX1 | Intergenic | – | TG | T | 0.609 | 0.031 | 0.02 | 2.075 (1.54,2.8) | 1.6 × 10−6 | 0.03 | 0.022 | 1.88 (1.23,2.88) | 0.0036 |
| rs192917960 | 16 | 5 919 655 | FAM86A- RBFOX1 | Intron | – | T | C | 0.862 | 0.032 | 0.015 | 2.14 (1.65,2.78) | 1.2 × 10−8 | 0.022 | 0.018 | 1.31 (0.86,2.0049) | 0.21 |
| African American combined | Hispanic incident | Hispanic prevalent | Hispanic combined | |||||||||||||
| SNP | HR/OR | P | r2 | Case | Non-case | HR/OR | P | Case | Non-case | HR/OR | P | HR/OR | P | Effect | ||
| rs542340 | 1.95 (1.55,2.44) | 8.0 × 10−9 | 0.669 | 0.001 | 0.001 | – | – | 0.001 | 0.001 | – | – | – | – | ++(+)??(?) | ||
| rs555091 | 1.92 (1.53,2.41) | 1.9 × 10−8 | 0.678 | 0.001 | 0.001 | – | – | 0.001 | 0.001 | – | – | – | – | ++(+)??(?) | ||
| rs506227 | 1.9 (1.51,2.39) | 3.3 × 10−8 | 0.72 | 0.001 | 0.001 | – | – | 0.001 | 0.001 | – | – | – | – | ++(+)??(?) | ||
| chr14:51604618:I | 2.009 (1.57,2.57) | 2.2 × 10−8 | 0.62 | 0.051 | 0.043 | 1.34 (0.88,2.052) | 0.17 | 0.034 | 0.044 | 0.71 (0.31,1.6) | 0.4 | 1.17 (0.81,1.71) | 0.41 | ++(+)+−(+) | ||
| rs192917960 | 1.87 (1.5,2.33) | 3.6 × 10−8 | 0.742 | 0.004 | 0.004 | – | – | 0.001 | 0.004 | – | – | – | – | ++(+)+−(?) | ||
RDB, Regulome database score.
Figure 2.
Local plot of rs542340 and rs192917960 in glaucoma in African Americans with 1000 Genomes African-American LD structure. Triangles are imputed SNPs and circles are genotyped SNPs.
Though we were inadequately powered to draw firm conclusions, we further ran exploratory GWAS analysis on Hispanic incident and Hispanic prevalent cases (Supplementary Material, Fig. S1d–f), but no SNPs were genome-wide significant (Supplementary Material, Table S1).
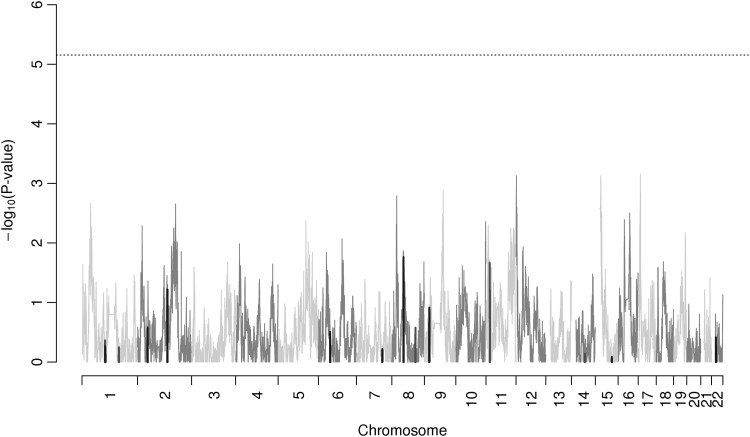
In addition, we performed admixture mapping analysis in the African-American subjects with results shown in Figure 3. We did not find any genome-wide significant regions (threshold of P < 7 × 10−6) under this approach. Supplementary Material, Figure S3 shows the separate incident and prevalent admixture mapping analysis results.
Figure 3.
Admixture results for glaucoma in African Americans. P-values combine incident and prevalent coefficients using an inverse variance method. None of the points reach genome-wide significance of 7 × 10−6. Black lines indicate SNPs that are in within 0.5 Mb of an SNP that was previously found associated with glaucoma with P < 5 × 10−8; dark gray is those previously associated with glaucoma with P > 5 × 10−8.
DISCUSSION
Although it has been shown that African Americans have a higher incidence of glaucoma (1,23), our study is the first to demonstrate that African ancestry, both in African Americans and Hispanics, is associated with an increased risk of glaucoma. Although we tried to control for as many potential confounders as were known, including income, education, nationality, blood pressure and diabetes, it is important to note that these signals may still represent an unmeasured confounding factor, rather than reflecting causal genetic factors.
Although none of the previously identified SNPs associated with glaucoma formally replicated in our cohort, many of them were in the same direction, and a risk score combining them was significant in the prevalent African-American cases. This is likely due in part to sample size and different genetic architecture, including LD patterns and allele frequencies in our populations. Lack of diagnostic precision is also likely a factor, although according to our calculations the attenuation of the observed relative risks is not likely fully explained by poor diagnostic specificity. Winner's curse may also play a role, where the original relative risks were inflated (20). A post hoc analysis of the minimum detectable odds ratio/hazards ratio, correcting for the 13 tests performed here (α = 0.05/13 = 0.0038), for MAFs of 0.05, 0.10 and 0.20, is 1.57, 1.41 and 1.30, respectively, for prevalent African Americans; 2.31, 1.91 and 1.67 for prevalent Hispanics; 1.40, 1.31 and 1.26 for incident African Americans; and 1.74, 1.58 and 1.50 for incident Hispanics (24). These would be slightly larger for markers that are imputed instead of genotyped.
An important limitation of our study is that glaucoma was defined by self-report, though studies have shown substantial agreement between self-report and medical records (24,25). In addition, data from the WHI Sight Examination (WHISE) study had 89.8% sensitivity and 98.0% specificity when comparing self-reported treatment to self-reported diagnosis (26). Using self-report, we find a slightly higher prevalence of the disease than has been found in other populations of similar race/ethnicity (1). We also have no distinction between different subtypes of glaucoma which may have different underlying genetic architectures (or for purely environmental causes, such as steroid-induced glaucoma), although the large majority of the African-American and Hispanic individuals analyzed here are likely to have OAG (1). OAG and ACG have very different clinical manifestations. Two-thirds of OAG cases have elevated ( > 21 mmHg) IOP, whereas others may have normal IOP (normal tension glaucoma), and drainage is inadequate. Secondary mechanisms include developmental anomalies, trauma/infection scarring and plugging of channels by pigment dispersion syndrome or pseudo-exfoliation syndrome. In contrast, ACG is caused by factors that push/pull the iris up into the angle, physically blocking aqueous drainage and elevating IOP (27). In addition, IOP may not distinguish between glaucoma itself and ocular hypertension.
Of note, some of the allele frequencies of previously identified hits are lower in African Americans and Hispanics. In particular, this is true of the SNPs in the CDKN2B/CDKN2BAS region which have much lower allele frequencies in African Americans and Hispanics than in the 1000 Genomes whites (MAF of rs523096 0.43, rs7865618 0.42, rs2157719 0.42, rs4977756 0.40 and rs1063192 0.43). These correlated SNPs are protective against glaucoma, and so the reduced frequency of these alleles may contribute to the higher prevalence of disease in African Americans and Hispanics.
We also found two potential novel SNPs associated with glaucoma in African Americans that require further follow-up. Currently, no such replication cohort exists, to the best of our knowledge. The SNPs did not have RegulomeDB scores (i.e. no evidence of functionality), and both had too low of an allele frequency to be tested in the WHI Hispanics. However, we anticipate that the development of other large cohorts of African-American glaucoma cases may allow for a direct assessment of replication, which will be critical. We also note that the SNP with 1000 Genomes identifier chr14:51604618:I, which was genome-wide significant in African Americans but did not replicate in Hispanics, is in the location of a previous linkage peak at chromosome 14q22 (hg19 position 50 900 001–58 100 000) (28).
In summary, although our study fails to replicate previous findings from different races/ethnicities after correcting for multiple testing, many of the associations are in the same direction. Larger sample sizes are necessary to fully understand the degree to which these associations are replicable in African Americans and Hispanics, and the relative magnitude of genetic effects. We also identified several genome-wide significant SNPs by imputation that should be tested for replication in additional cohorts with African ancestry. Finally, it is the first study to demonstrate an association of glaucoma with African ancestry.
MATERIALS AND METHODS
Participants
The WHI is a US-wide study consisting of an observational cohort and three clinical trials examining common health problems in generally healthy postmenopausal women. Study baseline was from 1993 to 1998 with annual follow-up for 8–12 years and two 5-year extensions (from 2005 to 2010 and 2010 to 2015). Details of the study design and cohort characteristics have been previously described (29,30). The nested WHI-SHARe cohort analyzed here consisted of 8515 individuals of African-American race/ethnicity and 3642 of Hispanic race/ethnicity from both the WHI Observational Study and the Clinical Trials, who had consented to genetic research. All participants provided written informed consent as approved by local Human Subjects Committees. Phenotypes were based on the phs000200.v3.p1 release of the dataset on October 21, 2010.
Genotyping and data quality control
Individuals in the WHI-SHARe cohort were genotyped on the Affymetrix 6.0 array. Genotype quality control criteria included call rate, sex discrepancy and blinded and unblinded duplicates. Furthermore, individuals whose genetic ancestries differed from self-reported races/ethnicities, as well as any relatives (one from each set) were removed. A total of 8153 individuals of African-American race/ethnicity and 3587 individuals of Hispanic race/ethnicity passed all sample and genotype quality control criterion. Details of these quality control procedures have been described previously (31–33).
Glaucoma phenotype definition
Women self-reported prevalent and incident of glaucoma through a series of questionnaires. For the prevalence analysis, at WHI baseline (1993–1998) an initial Medical History form was sent out to WHI participants (Form 30) asking individuals if they had ever had glaucoma. There were 658 African-American women who self-reported having glaucoma at this time, and 7129 women who did not; as well as 153 Hispanic women who self-reported glaucoma at baseline, and 3021 who did not.
For the incidence analysis, we followed up on individuals who did not have glaucoma at baseline. We also excluded individuals who were missing a response to the glaucoma question on the baseline Medical History form, though including them as unaffected at baseline did not change the overall conclusions in the manuscript (results not shown). After baseline, participants self-reported annually (Form 33) until 2005, on the Medical History Update form whether they had first been told they had glaucoma since their last update. This resulted in 1062 African-American and 336 Hispanic individuals with incident glaucoma. A total of 6067 African American and 2685 Hispanic individuals free of self-reported glaucoma at baseline never progressed to glaucoma and were censored at the last time they self-reported their unaffected status (Form 33).
To get some assessment of the accuracy of the self-report medical history information, we used an independent set of individuals from the WHISE substudy of WHI, which obtained information on self-reported glaucoma treatment (drops, medication or surgery) (26). Treating self-reported treatment as the closest thing available to a true doctor's diagnosis, self-reported glaucoma has sensitivity 89.8% and specificity 98.0% (220 self-report glaucoma and glaucoma treatment; 4015 self-report no glaucoma and no treatment; 82 self-report glaucoma but not glaucoma treatment; and 25 self-report treatment but not glaucoma).
Evaluation of demographic and medical characteristics
Study subjects were evaluated for possible glaucoma-associated demographic factors including age, sex, race/ethnicity/nationality, income and education. Medical traits also possibly associated with glaucoma were obtained from the baseline Medical History form and included BMI, SBP, DBP, hypertension and diabetes. These factors were tested for association with glaucoma using a multivariable logistic regression model for prevalent cases and Cox proportional hazards model for incident cases.
Genomic imputation
Imputation was performed using a cosmopolitan reference panel of all of the individuals of all races/ethnicities from the 1000 Genomes March 2012 interim release (http://1000genomes.org). Genotypes were pre-phased with SHAPE-IT v1.ESHG (34), and then imputed using Impute2 v2.2.2 (35). We then discarded all SNPs with imputation r2 < 0.3. To assure sufficient allele counts in tests of association with glaucoma, we used only SNPs with MAF > 0.01 in the analysis of African-American incident cases (15 784 307 SNPs) and MAF > 0.02 in the analysis of African-American prevalent cases (14 803 533 SNPs), MAF > 0.03 in Hispanic incident case analyses (7 791 354 SNPs), and MAF > 0.05 in Hispanic prevalent case analyses (6 450 021 SNPs). These MAF thresholds give an expected cell count of at least 20 in the smallest (case) cell including minor homozygotes plus heterozygotes.
Population structure and genome-level ancestry analysis
Principal components analysis was performed separately in individuals of African-American race/ethnicity and Hispanic race/ethnicity using Eigenstrat (36). We also used the software Frappe (37) to determine individual ancestry proportions using 656 852 autosomal markers and 475 publicly available samples to represent the ancestral populations (YRI and CEU from HapMap and East Asian and Native Americans from the Human Genome Diversity Project (38)), as has been described (32). The individual ancestry proportions so derived were used as the dependent variables in a logistic regression analysis of prevalent cases and survival analysis of incident cases (as described above for the demographic variables) to test the effect of ancestry on glaucoma risk.
Local ancestry estimation and admixture mapping
For admixture mapping analysis, local ancestry was estimated using the program SABER+ (39), as has been described in a previous analysis of WHI-SHARe data (32). The threshold for genome-wide significance is less than for genome-wide association tests because recent admixing history causes extensive correlation in local ancestry. Previous theoretical analysis and simulations suggest that a threshold of 7 × 10−6 provides the correct type I error (40). Admixture analysis was only performed in individuals of African-American race/ethnicity, and not in those of Hispanic race/ethnicity because of the reduced sample size for the latter (32). The analysis was based on logistic regression for prevalent cases and proportional hazards analysis for incident cases, testing a covariate for local ancestry at each SNP location (instead of for an SNP genotype as in a GWAS analysis, as described below).
Genome-wide association analysis
Association analysis was performed using R (41). A logistic regression model was used to analyze prevalent case data and a Cox proportional hazards model was used to analyze incident case data. Analyses were conducted separately in each race/ethnicity group. Genotypes were analyzed using an additive model for typed markers, and additive dosages for imputed markers, which has been shown to be efficient (42). The first and seventh principal components were used in the analysis with African Americans and the second and fifth principal component were used in the analysis with Hispanics, as they were the only PCs significant in the model. We also included as covariates significant demographic/medical factors described above.
Under certain assumptions, we can combine the results of the incidence and prevalence analyses within each race/ethnicity. Although the individuals in the prevalence and incidence models overlap, the person-time of the individuals does not overlap. If we assume that being free of glaucoma at baseline (the time of prevalence measure) is independent of the time to develop incident glaucoma, conditional on the covariates (which include age), then the logistic regression analysis of prevalent cases and the survival analysis of incident cases are independent under the martingale property of Cox models (43). We can test this assumption to some extent by computing the correlation of the regression coefficients; we saw very low correlation estimates of 0.029 in African Americans and −0.0062 in Hispanics. When this assumption is met, we can combine the coefficients of both results. We use the standard inverse variance method to combine the coefficients, an approach typically employed under a rare-disease assumption, which in our case is only approximate but adequate (in terms of the odds ratio from the analysis of prevalence cases approximating the relative risk).
We used a multiple comparison statistical significance threshold of 5 × 10−8, which we use to declare genome-wide significance. For replication of the 13 SNPs found and replicated in previous studies to be associated with glaucoma, we used Bonferroni-corrected statistical significance threshold of α = 0.05/13 = 0.0038.
The genomic inflation factor (44) was very modest for all GWAS analyses (African-American incident and prevalent λ = 1.022; African-American incident λ = 0.988; African-American prevalent λ = 1.014; Hispanic incident and prevalent λ = 1.025; Hispanic incident λ = 1.036; Hispanic prevalent λ = 0.994).
Test of previously identified hits (combined risk score)
We constructed a risk score for each individual by weighting the additive coding of each OAG-associated SNP by the previously reported log(OR), and then summing them up, excluding variants in high LD with each other (namely the CDKN2B/CDKN2BAS region where we only included the maximally correlated SNP rs7865618, see Supplementary Material, Table S1). We coded the SNP genotypes at each locus so that the predisposing allele had a score of 1 (versus 0 for the protective allele), so that the log(OR) was always positive, and we could assess the consistency of the directionality of the risk score with previous results.
Assessing attenuation of SNP relative risks using self-report diagnosis
Self-reported diagnosis of glaucoma may have reduced sensitivity and specificity with a true diagnosis of glaucoma, and in particular OAG. To assess the potential impact on our SNP association results (i.e. observed relative risks), we develop some formulas. Let RRt denote the true relative risk associated with an SNP variant, and RRo the relative risk observed in our study based on self-report. Let G denote the high-risk genotype(s) and g the low-risk genotype(s). Let X denote the true glaucoma status of an individual ( = 1 if affected and 0 if unaffected) and Y denote the self-report glaucoma status (=1 if affected and 0 if unaffected). Let t = Prob(X = 1|G) and u = Prob(X = 1|g), so that RRt = t/u. Let a = Prob(Y = 1|X = 1) = sensitivity of self-report and b = Prob(Y = 0|X = 0) = specificity of self-report. From these definitions, we derive the following formula:
| (1) |
After some algebra, we obtain
| (2) |
This formula can then be used to assess the attenuation of an SNP relative risk due to an imprecise diagnostic assessment.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported in part by the National Institutes of Health grants R25 CA112355. The Women's Health Initiative (WHI) program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health and the United States Department of Health and Human Services through contracts HHSN268201100046C, HHSN2682011 00001C, HHSN268201100002C, HHSN268201100003C, HHSN2 68201100004C and HHSN271201100004C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the WHI investigators and staff for their dedication and the study participants for making the program possible. A listing of WHI investigators can be found at http://www.whiscience.org/publications/WHI_investigators_shortlist_2010-2015.pdf. We are grateful to an anonymous reviewer for pointing out the overlap of our association finding on chromosome 14 with a prior linkage peak in the same population (African Americans).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Quigley H.A., Broman A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang T.C., Congdon N.G., Wojciechowski R., Muñoz B., Gilbert D., Chen P., Friedman D.S., West S.K. Determinants and heritability of intraocular pressure and cup-to-disc ratio in a defined older population. Ophthalmology. 2005;112:1186–1191. doi: 10.1016/j.ophtha.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein B.E.K., Klein R., Lee K.E. Heritability of risk factors for primary open-angle glaucoma: the Beaver Dam Eye Study. Invest. Ophthalmol. Vis. Sci. 2004;45:59–62. doi: 10.1167/iovs.03-0516. [DOI] [PubMed] [Google Scholar]
- 4.Van Koolwijk L.M.E., Despriet D.D.G., van Duijn C.M., Pardo Cortes L.M., Vingerling J.R., Aulchenko Y.S., Oostra B.A., Klaver C.C.W., Lemij H.G. Genetic contributions to glaucoma: Heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest. Ophthalmol. Vis. Sci. 2007;48:3669–3676. doi: 10.1167/iovs.06-1519. [DOI] [PubMed] [Google Scholar]
- 5.Rudnicka A.R., Mt-Isa S., Owen C.G., Cook D.G., Ashby D. Variations in primary open-angle glaucoma prevalence by age, gender, and race: a Bayesian meta-analysis. Invest. Ophthalmol. Vis. Sci. 2006;47:4254–4261. doi: 10.1167/iovs.06-0299. [DOI] [PubMed] [Google Scholar]
- 6.Burdon K.P., Macgregor S., Hewitt A.W., Sharma S., Chidlow G., Mills R.A., Danoy P., Casson R., Viswanathan A.C., Liu J.Z., et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 2011;43:574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 7.Gibson J., Griffiths H., De Salvo G., Cole M., Jacob A., MacLeod A., Yang Y., Menon G., Cree A., Ennis S., et al. Genome-wide association study of primary open angle glaucoma risk and quantitative traits. Mol. Vis. 2012;18:1083–1092. [PMC free article] [PubMed] [Google Scholar]
- 8.Thorleifsson G., Magnusson K.P., Sulem P., Walters G.B., Gudbjartsson D.F., Stefansson H., Jonsson T., Jonasdottir A., Jonasdottir A., Stefansdottir G., et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 9.Thorleifsson G., Walters G.B., Hewitt A.W., Masson G., Helgason A., DeWan A., Sigurdsson A., Jonasdottir A., Gudjonsson S.A., Magnusson K.P., et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat. Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiggs J.L., Yaspan B.L., Hauser M.A., Kang J.H., Allingham R.R., Olson L.M., Abdrabou W., Fan B.J., Wang D.Y., Brodeur W., et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vithana E.N., Khor C.-C., Qiao C., Nongpiur M.E., George R., Chen L.-J., Do T., Abu-Amero K., Huang C.K., Low S., et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat. Genet. 2012;44:1142–1146. doi: 10.1038/ng.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meguro A., Inoko H., Ota M., Mizuki N., Bahram S. Genome-wide association study of normal tension glaucoma: common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology. 2010;117:1331–1338. doi: 10.1016/j.ophtha.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Nakano M., Ikeda Y., Taniguchi T., Yagi T., Fuwa M., Omi N., Tokuda Y., Tanaka M., Yoshii K., Kageyama M., et al. Three susceptible loci associated with primary open-angle glaucoma identified by genome-wide association study in a Japanese population. Proc. Natl. Acad. Sci. USA. 2009;106:12838–12842. doi: 10.1073/pnas.0906397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano M., Ikeda Y., Tokuda Y., Fuwa M., Omi N., Ueno M., Imai K., Adachi H., Kageyama M., Mori K., et al. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS ONE. 2012;7:e33389. doi: 10.1371/journal.pone.0033389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman W., Low S.-K., Takahashi A., Kubo M., Nakamura Y. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum. Mol. Genet. 2012;21:2836–2842. doi: 10.1093/hmg/dds103. [DOI] [PubMed] [Google Scholar]
- 16.Takamoto M., Kaburaki T., Mabuchi A., Araie M., Amano S., Aihara M., Tomidokoro A., Iwase A., Mabuchi F., Kashiwagi K., et al. Common variants on chromosome 9p21 are associated with normal tension glaucoma. PLoS ONE. 2012;7:e40107. doi: 10.1371/journal.pone.0040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chopra V., Varma R., Francis B.A., Wu J., Torres M., Azen S.P. Type 2 diabetes mellitus and the risk of open-angle glaucoma: The Los Angeles Latino Eye Study. Ophthalmology. 2008;115:227–232. doi: 10.1016/j.ophtha.2007.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman-Casey P.A., Talwar N., Nan B., Musch D.C., Stein J.D. The relationship between components of metabolic syndrome and open-angle glaucoma. Ophthalmology. 2011;118:1318–1326. doi: 10.1016/j.ophtha.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraft P. Curses—winner's and otherwise—in genetic epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 21.Consortium T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman D.S., Wolfs R.C.W., O'Colmain B.J., Klein B.E., Taylor H.R., West S., Leske M.C., Mitchell P., Congdon N., Kempen J. Prevalence of open-angle glaucoma among adults in the United States. Arch. Ophthalmol. 2004;122:532–538. doi: 10.1001/archopht.122.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLennan P.A., McGwin G., Searcey K., Owsley C. Medical record validation of self-reported eye diseases and eye care utilization among older adults. Curr. Eye Res. 2013;38:1–8. doi: 10.3109/02713683.2012.733054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise L.A., Rosenberg L., Radin R.G., Mattox C., Yang E.B., Palmer J.R., Seddon J.M. A prospective study of diabetes, lifestyle factors, and glaucoma among African-American women. Ann. Epidemiol. 2011;21:430–439. doi: 10.1016/j.annepidem.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haan M.N., Klein R., Klein B.E., Deng Y., Blythe L.K., Seddon J.M., Musch D.C., Kuller L.H., Hyman L.G., Wallace R.B. Hormone therapy and age-related macular degeneration: The women's health initiative sight exam study. Arch. Ophthalmol. 2006;124:988–992. doi: 10.1001/archopht.124.7.988. [DOI] [PubMed] [Google Scholar]
- 27.Porter R.S. The Merck Manual of Diagnosis and Therapy. 19th ed. 2011. Merck Sharpe and Dohme Corp., a subsidiary of Merck & Co., inc., Whitehouse Station, NJ. [Google Scholar]
- 28.Rotimi C.N., Chen G., Adeyemo A.A., Jones L.S., Agyenim-Boateng K., Eghan B.A., Zhou J., Doumatey A., Lashley K., Huang H., et al. Genomewide scan and fine mapping of quantitative trait loci for intraocular pressure on 5q and 14q in West Africans. Invest. Ophthalmol. Vis. Sci. 2006;47:3262–3267. doi: 10.1167/iovs.05-1537. [DOI] [PubMed] [Google Scholar]
- 29.Anderson G.L., Manson J., Wallace R., Lund B., Hall D., Davis S., Shumaker S., Wang C.-Y., Stein E., Prentice R.L. Implementation of the women's health initiative study design. Ann. Epidemiol. 2003;13:S5–S17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 30.Hays J., Hunt J.R., Hubbell F.A., Anderson G.L., Limacher M., Allen C., Rossouw J.E. The women's health initiative recruitment methods and results. Ann. Epidemiol. 2003;13:S18–S77. doi: 10.1016/s1047-2797(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 31.Carty C.L., Johnson N.A., Hutter C.M., Reiner A.P., Peters U., Tang H., Kooperberg C. Genome-wide association study of body height in African Americans: the Women's Health Initiative SNP Health Association Resource (SHARe) Hum. Mol. Genet. 2012;21:711–720. doi: 10.1093/hmg/ddr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coram M.A., Duan Q., Hoffmann T.J., Thornton T., Knowles J.W., Johnson N.A., Ochs-Balcom H.M., Donlon T.A., Martin L.W., Eaton C.B., et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 2013;92:904–916. doi: 10.1016/j.ajhg.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiner A.P., Beleza S., Franceschini N., Auer P.L., Robinson J.G., Kooperberg C., Peters U., Tang H. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am. J. Hum. Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaneau O., Marchini J., Zagury J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 35.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 37.Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 38.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 39.Tang H., Coram M., Wang P., Zhu X., Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 2006;79:1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H., Siegmund D.O., Johnson N.A., Romieu I., London S.J. Joint testing of genotype and ancestry association in admixed families. Genet. Epidemiol. 2010;34:783–791. doi: 10.1002/gepi.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R Development Core Team (2012) R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria.
- 42.Zheng J., Li Y., Abecasis G.R., Scheet P. A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet. Epidemiol. 2011;35:102–110. doi: 10.1002/gepi.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamin E.J., Rice K.M., Arking D.E., Pfeufer A., van Noord C., Smith A.V., Schnabel R.B., Bis J.C., Boerwinkle E., Sinner M.F., et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat. Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 2004;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.